Abstract
TAR DNA-binding protein-43 (TDP-43) has been recently identified as a major component of the ubiquitinated inclusions found in frontotemporal lobar degeneration with ubiquitin-positive inclusions and in amyotrophic lateral sclerosis, diseases that are collectively termed TDP-43 proteinopathies. Several amyotrophic lateral sclerosis-linked mutations of the TDP-43 gene have also been identified; however, the precise molecular mechanisms underlying the neurodegeneration remain unclear. To investigate the biochemical characteristics of TDP-43, we examined truncation, isoforms, and cytoplasmic inclusion (foci) formation using TDP-43-expressing cells. Under apoptosis, caspase-3 generates two 35-kDa (p35f) and 25-kDa (p25f) fragments. However, in caspase-3(−/−) cells, novel caspase-3-independent isoforms of these two variants (p35iso and p25iso) were also detected under normal conditions. With a deletion mutant series, the critical domains for generating both isoforms were determined and applied to in vitro transcription/translation, revealing alternate in-frame translation start sites downstream of the natural initiation codon. Subcellular localization analysis indicated that p35 (p35f and p35iso) expression leads to the formation of stress granules, cellular structures that package mRNA and RNA-binding proteins during cell stress. After applying proteasome inhibitors, aggresomes, which are aggregates of misfolded proteins, were formed in the cytoplasm of cells expressing p35. Collectively, this study demonstrates that the 35-kDa isoforms of TDP-43 assemble in stress granules, suggesting that TDP-43 plays an important role in translation, stability, and metabolism of mRNA. Our findings provide new biological and pathological insight into the development of TDP-43 proteinopathies.
Introduction
Amyotrophic lateral sclerosis (ALS)2 was first reported in 1869 by the French neurologist Jean-Martin Charcot and is one of the most serious neurological diseases. ALS is characterized by progressive degeneration of upper and lower motor neurons, and although the vast majority of ALS cases are sporadic (sALS), almost 10% appear to be familial (fALS). Although mutations in the gene encoding the antioxidant enzyme Cu,Zn-superoxide dismutase-1 (SOD-1) have been detected in 20% of fALS patients (1), the cause of sALS and fALS not associated with SOD-1 remains unclear. Recently, two research groups have identified TDP-43 as a major component of ubiquitinated neuronal cytoplasmic and intranuclear inclusions identified in frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U), as well as in sALS (2, 3). Missense mutations in TDP-43 have been found in autosomal dominant ALS families, suggesting that mutant TDP-43 may be a primary cause of motor neuron degeneration (4–9). Importantly, pathological analysis revealed that abnormal accumulation of TDP-43 does not occur in fALS cases with SOD-1 mutations, suggesting that the pathological process in sALS is distinct from those associated with SOD-1 mutations. Currently, FTLD-U, sALS, and fALS-linked TDP-43 mutations are classified together as TDP-43 proteinopathies (10).
TDP-43 is a ubiquitously expressed nuclear protein that was originally identified as a binding protein of the human immunodeficiency virus, type-1 TAR DNA element (11). Evidence suggests that TDP-43 is involved in the regulation of RNA splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) and survival motor neuron (SMN) (12, 13). Loss of functional TDP-43 is known to affect nuclear membrane stability and induce apoptosis via phosphorylation of the retinoblastoma protein (14).
Limited biochemical evidence has demonstrated the mechanisms underlying the molecular pathogenesis of TDP-43 proteinopathy. Using phosphorylation-specific antibodies, Hasegawa et al. demonstrated that TDP-43 is abnormally phosphorylated in the brain tissue of FTLD-U and ALS patients (15). Zhang et al. focused on proteolytic cleavage and demonstrated that caspase-3 can mediate cleavage of TDP-43 to generate 25- and 35-kDa fragments when progranulin (a candidate gene for familial FTLD-U) is down-regulated (16). They also reported that the 25-kDa C-terminal fragment (CTF) of caspase-cleaved TDP-43 leads to the formation of toxic cytoplasmic inclusions within cells (17). More recently, Igaz et al. identified the cleavage site for the lowest molecular mass TDP-43 CTF (∼22 kDa) in cortical urea extracts of FTLD-U brain and demonstrated that this fragment can recapitulate some of the pathological features of TDP-43 proteinopathy (18). However, little is still known about the biochemical process of the generation of truncated forms of TDP-43 and the characteristics of cytoplasmic inclusion formation of these fragments.
In this study, we first verified the caspase-3-dependent generation of CTFs of TDP-43 protein (p35f and p25f), which were abolished in caspase-3(−/−) mouse embryonic fibroblasts (MEFs). Furthermore, we identified two novel caspase-3-independent isoforms (p35iso and p25iso). Site-specific mutagenesis and in vitro transcription/translation revealed that these isoforms were generated from an alternative translation start site. Our results also show that the p35 forms of TDP-43 (p35f and p35iso) accumulate in two types of cytoplasmic foci, stress granules (SGs) under normal conditions and aggresomes under proteasome inhibition. These findings reveal novel biochemical properties of the TDP-43 protein and suggest the possibility that RNA quality control via the function of SGs is involved in the development of TDP-43 proteinopathy.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Neuro2a mouse neuroblastoma cells, HeLa human carcinoma cells, motor neuron-like hybrid cells, NSC-34 cells, and caspase-3(−/−) MEFs were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum as described previously (19–21). Transfection was performed using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. Stable transfectants were selected in medium containing 0.4 mg/ml G418 (Invitrogen). Proteasome function was inhibited with MG132 (Sigma). Emetine was purchased from Sigma. Z-DEVD-FMK was purchased from Calbiochem.
cDNA
FLAG-tagged human TDP-43 pCMV, a generous gift from Dr. Francisco E. Baralle (International Centre for Genetic Engineering and Biotechnology, Italy), was subcloned into pcDNA3.1/V5-His A. The p35 and p25 cDNAs were generated by PCR using the following primer pairs: p35 (sense (5′-GCTTCATCAGCAGTGAAAGT-3′) and antisense (5′- AAGCTTGTCGTCATCGTCTT-3′)), p25 (sense (5′-GGACGATGGTGTGACTGCAA-3′) and antisense (5′-AAGCTTGTCGTCATCGTCTT-3′)). Deletion mutants, site-directed mutations, non-tagged TDP-43, and 5′-untranslated regions were generated using KOD-Plus-site-directed mutagenesis (Toyobo, Osaka, Japan), according to the manufacturer's instructions. Plasmids containing the genes for GFP-G3BP and RFP-DCP1a (22) were kindly provided by Dr. Paul Anderson (Division of Rheumatology, Immunology and Allergy, Brigham and Women's Hospital and Harvard Medical School).
Antibodies
Mouse monoclonal anti-PABP (10E10), HuR (3A2), and rabbit polyclonal anti-HA (Y-11) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-α tubulin and anti-V5 were from Invitrogen. Mouse monoclonal anti-FLAG M2, M5, polyclonal anti-FLAG, anti-vimentin (clone V9), and anti-V5 agarose affinity gel were from Sigma. Rabbit polyclonal anti-V5 (C464.6) was from Bethyl Labs (Montgomery, TX). Rabbit polyclonal anti-pericentrin was from Abcam (Cambridge, UK). Rabbit polyclonal and monoclonal anti-TDP-43 and anti-G3BP antibodies were from Proteintech (Chicago, IL). Monoclonal p409/410 phospho-specific TDP-43 antibody was from CosmoBio (Tokyo, Japan). Monoclonal 2E2-D3 was from Abnova (Taipei City, Taiwan).
Immunoblotting
Cells were lysed in cold lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 0.25% SDS, 5 mm EDTA, 0.05 μm phenylmethylsulfonyl fluoride, and 1:200 protease inhibitor mixture (Sigma)). Cell lysates were briefly sonicated and then separated via reducing SDS-PAGE on a 4–20% Tris-glycine gradient gel (Invitrogen), or a 12% or 16% Tris-glycine gel, after which proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membrane was then incubated with primary antibodies (1:1000 monoclonal anti-V5, 1:1000 anti-α-tubulin, 1:1000 M5, 1:1000 anti-G3BP, and 1:1000 anti-HuR), followed by horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence reagents (PerkinElmer Life Sciences, Boston, MA). Proteins were then visualized using LAS or Kodak Biomax XAR film (Rochester, NY).
CFTR Exon 9 Skipping Assay
The CFTR minigene splicing assay was conducted as previously described (23–25). HeLa cells grown in 6-well plates were co-transfected with the reporter plasmid pTB-CFTR9 (including a TG13T3 sequence), which was kindly provided by Drs. Francisco E. Baralle and Emanuele Buratti (Dept. of Molecular Pathology, International Centre for Genetic Engineering and Biotechnology, Italy), and wild-type TDP-43, p35, and p25 cDNAs, harvested 48 h post-transfection. Total RNA was extracted by using an RNeasy kit (Qiagen), and cDNA was synthesized from 1 μg of total RNA using the Superscript II system (Invitrogen). Semi-quantitative PCR was carried using primer sets: Bra2, TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA; and a2-3, CAACTTCAAGCTCCTAAGCCACTGC.
Immunofluorescence
HeLa cells cultured on glass coverslips coated with poly-l-lysine were transfected with each TDP-43 expression plasmid. After 48 h, cells were fixed with 4% paraformaldehyde at room temperature for 10 min and then permeabilized in 0.2% Triton X-100 for 5 min. After blocking for nonspecific binding, cells were incubated with polyclonal anti-V5 (1:300), monoclonal anti-V5 (1:500), polyclonal anti-FLAG (1:300), or polyclonal anti-HA (1:300) antibodies along with other antibodies (1:200 anti-PABP, 1:75 anti-vimentin, 1:500 anti-pericentrin, 1:300 anit-G3BP, and 1:300 anti-HuR) diluted in phosphate-buffered saline containing 0.2% Tween 20 and 3% bovine serum albumin. After three washes, cells were incubated with fluorescein isothiocyanate-conjugated anti-rabbit (1:200, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and Alexa 586-conjugated anti-mouse (1:200, Invitrogen) secondary antibodies. Immunofluorescent staining was examined using a confocal microscope (TCS SP5, Leica, Wetzlar, Germany).
In Vitro Transcription/Translation
In vitro transcription/translation was performed using a TnT T7 Quick-coupled Transcription/Translation System (Promega, Madison, WI) in a 25-μl reaction mixture containing 2 μg of plasmid, 20 μl of TnT Quick Master Mix incubated at 30 °C for 90 min. A 2-μl aliquot of each reaction mixture was loaded onto an SDS-polyacrylamide gel and examined by Western blotting as described above.
RESULTS
Characterization of TDP-43 Expression in Cultured Cells
In previous studies, immunoblots of brain tissue from FTLD-U and ALS patients revealed expression of full-length TDP-43 (43 kDa), as well as an increase in several additional smaller fragments (∼25 kDa) (6, 16). Thus, we first performed Western blotting using polyclonal anti-TDP-43 antibody to confirm the presence of full-length TDP-43 and its fragments, including those of 35 kDa and 25 kDa in the human HeLa cell line, as well as those in mouse and rat tissue lysates (supplemental Fig. 1A). For detailed investigation of the biochemical properties of TDP-43 in vitro, we next either transiently or stably overexpressed cDNA encoding human TDP-43 (with an N-terminal FLAG tag and a C-terminal V5-His tag) in various cell lines to analyze expression via immunoblotting with an anti-V5 antibody. Transiently and stably transfected cells expressed full-length TDP-43 (47 kDa, including the FLAG and V5-His tags), and two TDP-43 fragments (37 and 27 kDa, including the V5-His tag) (Fig. 1A), showing results consistent with those observed previously (16). As expected, several smaller fragments (<25 kDa) were also detected on long exposure (Fig. 1A, lower panel). In addition, when the anti-FLAG antibody was used (for N terminus detection) only a faint ∼18-kDa fragment was detected (supplemental Fig. 1B).
FIGURE 1.
TDP-43 expression and generation of its truncated forms in cultured mammalian cells. A, immunoblot analysis of human TDP-43 expression in transiently transfected HeLa cells, and stably transfected Neuro2a, HEK293, and NSC-34 cells. Lysates from cells transiently transfected with FLAG-TDP-43-V5 cDNA were subjected to immunoblotting with an anti-V5 antibody. Full-length and two short forms of TDP-43 (37 and 27 kDa) were detected. B and C, HeLa cells were transfected with wild-type human TDP-43, 409/410AA, or 409/410DD cDNA. The cell lysates were then subjected to immunoblotting using an anti-V5 antibody. Both mutants generated 35- and 25-kDa fragments (B). Cells were labeled with anti-V5 (red) antibody. Nuclear localization of TDP-43 in the nucleus was unchanged by mutation of the serine 409 and 410 phosphorylation sites (C).
A recent study has demonstrated that phosphorylated TDP-43 forms a major component of inclusions, suggesting that abnormal phosphorylation of TDP-43 may be an essential step in the pathogenesis of TDP-43 proteinopathy (15). To test whether phosphorylation of TDP-43 influences generation of its 35- and 25-kDa fragments, we mutated serines 409 and 410 (the most intense phosphorylation sites on immunoblotting and immunohistochemistry of FTLD-U and ALS (15)) to alanine (409/410AA) or to aspartic acid (409/410DD) to mimic dephosphorylation or phosphorylation, respectively. These mutants failed to affect the 35- and 25-kDa fragments (Fig. 1B). Immunocytochemistry and confocal microscopy also revealed normal localization of TDP-43 in the nucleus following mutation of these phosphorylation sites (Fig. 1C). Therefore, we examined additional mutants of three other phosphorylation sites (379A and 403/404AA) and again found no effect on these fragments and nuclear localization (supplemental Fig. 2, A and B). Thus, phosphorylation of TDP-43 is most likely not critical for fragment generation and TDP-43 localization.
Identification of Caspase-independent TDP-43 Isoforms
Zhang et al. have reported the presence of two caspase-3 cleavage consensus sites (DETD (aa 86–89) and DVMD (aa 216–219)) within the amino acid (aa) sequence of TDP-43, and that the generation of the 25- and 35-kDa proteolytic fragments was inhibited by the pan-caspase inhibitor Z-VAD-FMK. These results suggest that the cleavage of TDP-43 in FTLD-U may be mediated by caspase-3 (16). To confirm their findings, we examined proteolytic cleavage in caspase-3(+/−) and (−/−) MEFs treated with staurosporine.
As expected, cleavage of the major 35-kDa TDP-43 fragment (termed p35f) as well as the minor 25-kDa fragment (termed p25f) was observed in caspase-3(+/−) MEFs after induction of apoptosis, whereas enhancement of fragment generation was abolished in caspase-3(−/−) MEFs (Fig. 2A). Surprisingly, caspase-3(−/−), as well as untreated caspase-3(+/−), MEFs can generate some level of these fragments. Moreover, high resolution gel analysis shows that electromobility of this 35-kDa fragment under control conditions is slightly slower than that of caspase-cleaved p35f (Fig. 2B), whereas that of the 25-kDa fragments under control conditions and apoptosis seems to be identical. As shown in Fig. 2C, similar electromobility was also observed for both fragments in HeLa cells treated with staurosporine using well characterized monoclonal anti-TDP-43 antibody, 2E2-D3, which recognizes aa 205–222 residues of human TDP-43 protein (26).
FIGURE 2.
Detection of caspase-independent TDP-43 isoforms. A, caspase(−/−) or (+/−) MEFs were treated for 3 or 6 h with staurosporine (1 μm) or DMSO. Cellular lysates were prepared in lysis buffer and fractionated by SDS-PAGE. Immunoblot with a rabbit polyclonal anti-TDP-43 antibody shows enhancement of proteolytic cleavage in caspase(+/−) MEFs under apoptosis. In contrast, caspase-3(−/−) MEFs showed no increase in fragment generation, but still generated some level of the 35- and 25-kDa fragments. B, on 12% gel, the 35-kDa fragment consisted of two bands, the original fragment (indicated as an asterisk) under normal conditions and the caspase-3-dependent fragment (#) under apoptosis, whereas on 14% gel, the 25-kDa fragment showed the same-sized band under normal conditions and apoptosis. C, HeLa cells were treated for 6 h with staurosporine (1 μm), and then cellular lysates were assessed by Western blotting with monoclonal antibody 2E2-D3. D, caspase-3(−/−) or (+/−) MEFs were transfected with human TDP-43 tagged with V5. The lysates were subjected to immunoblotting with anit-V5 antibody. E, HeLa cells were transfected with wild-type human TDP-43, DETA89, or DVMA219 cDNA. The cell lysates were then subjected to immunoblotting with anti-V5 antibody. Both mutants showed no effect on the generation of the 35- and 25-kDa fragments. The level of the DVMA219 protein was increased for unknown reasons.
To verify these results, we examined caspase-3(−/−) MEFs transfected with TDP-43. Both p35f and p25f were generated in these cells, suggesting fragment generation is independent of caspase-3 (Fig. 2D). Using mutants of asparagine 89 and 219 to alanine (DETA89 and DVMA219) transiently expressed in HeLa cells, we again found no effect on either fragment (Fig. 2E). Collectively, these findings suggest that both fragments are novel caspase-independent TDP-43 isoforms (termed p35iso and p25iso).
Identification of the Critical Domain for Generating p25iso and p35iso
To define the critical domain required for generating p35iso and p25iso, a deletion mutant series of the predicted region was performed based on the size of the isoforms. Mutant constructs were then expressed in HeLa cells to analyze the generated isoforms by immunoblotting (Fig. 3). Among the expressed constructs, those carrying deletions within aa 70–99, 80–89, and 85–89 failed to generate p35iso (Fig. 3, B and C). Furthermore, alanine scanning revealed that generation of p35iso was abolished in the site-directed mutant ADETD, indicating that methionine 85 is critical for generating p35iso (Fig. 3D).
FIGURE 3.
Determination of the critical sites for p35iso and p25iso generation. A, constructs were designed with internal deletions or amino acid substitutions to identify the critical sites for generating TDP-43 fragments. The human TDP-43 amino acid sequence is shown together with the deletion regions. All constructs carried the FLAG tag at the N terminus and the V5 tag at the C terminus. B–D, determination of the critical site for p35iso generation. Protein extracts of cells expressing various deletion constructs were analyzed by immunoblotting with anti-V5 antibody. Among the deletion constructs, Δ80–89 (B) and Δ85–89 (C) did not generate p35iso. D, alanine scanning around the recognition site of cleavage. The candidate recognition site residues (Met85–Asp89) were substituted by alanine, and cleavage was examined by immunoblotting. ADETD abolished p35iso generation, indicating that Met85 is critical for generating this fragment. E–G, determination of the critical site for p25iso generation. Among the serial deletion constructs, Δ160–169 abolished p25iso generation (E). The Δ165–169 (F) and six alanine substitutions (aa 162–167) (G) constructs partially reduced p25iso generation. H, Neuro2a cells were transfected with aa 85–414, 170–414, and full-length cDNAs without tags, and then cell lysates were fractionated by SDS-PAGE with non-transfected HeLa cell lysate and analyzed by immunoblotting with antibody 2E2-D3. The asterisk indicates unknown nonspecific bands that reacted with 2E2-D3. I, alignment of genomic sequences around the third in-frame ATG (underline) among vertebrates. The bold letters show the conserved Kozak consensus sequence. J, Western blot of in vitro transcription and translation products of V5-tagged human TDP-43 cDNA, including 5′-untranslated region, Δ80–89, and Δ160–169 against anti-V5 antibody. HeLa cell-expressed human TDP-43 is presented as a control.
We next examined the events leading to generation of p25iso. The deletion mutant series showed that the Δ160–169 construct was unable to generate p25iso (Fig. 3E), whereas constructs of Δ165–169 and the six alanine substitutions at aa 162–167 were partially able to reduce, but not fully abolish it (Fig. 3, F and G).
To confirm p25iso antigenicity and electromobility in detail, we transfected aa 85–414, 170–414, and full-length cDNAs lacking tags to the mouse neuroblastoma cell line, Neuro2a, and assessed them by Western blotting in parallel with endogenous human TDP-43 protein expression in HeLa cells. As shown in Fig. 3H, both endogenous p25iso and overexpressed residues 170–414 were detected by 2E2-D3 (epitope: 205–222) and showed similar electromobility. Thus, we concluded that the aa 160–169 region is critical for generating p25iso and the N terminus of p25iso appears to be further upstream than the caspase recognition site, DVMD (aa 216–219).
Because the critical site for p35iso (methionine 85) is the third in-frame methionine, with a conserved Kozak consensus sequence (AAAATGG) among vertebrates (Fig. 3I) and there are two methionines (methionine 162 and methionine 167) in the critical region for p25iso (aa 160–169), we examined alternate internal translation initiation sites for generation of these truncated forms by in vitro transcription/translation using human TDP-43 cDNA, including 5′-untranslated region (nucleotides from position −1 to −30). All three products (full-length TDP-43, p35iso, and p25iso) could be translated; however, the Δ80–89 and Δ160–169 constructs could not translate the respective isoforms (Fig. 3J).
These findings confirm that translation of both p35iso and p25iso occurs at alternate translation start sites. Moreover, p25iso comprised several translated fragments, suggesting it may be heterogeneous and generate from several initiation codons (Fig. 1A). We concluded that at least two caspase-3-dependent fragments (p35f and p25f) are produced under apoptosis, and at least two major novel isoforms (p35iso and p25iso) are translated at alternate start codons.
Subcellular Localization of p35 and p25
Biological and pathological findings at the cellular level show that TDP-43, containing a nuclear localization signal (NLS, aa 82–98) on its N terminus, is predominantly localized in the nuclei of cultured cells and intact neurons in the brain tissue. Winton et al. have reported that mutants expressing a defective NLS are redistributed in the cytoplasm, which induces the formation of insoluble aggregates. This finding recapitulates features of the pathological TDP-43 inclusions found in human FTLD-U/ALS cases (27). Based on our results, long (p35) and short (p25) forms of TDP-43 appear to comprise both p35f and p35iso, and p25f and p25iso, respectively, but not the NLS (aa 82–98), suggesting that both forms are redistributed to the cytoplasm. Therefore, we generated p35 (aa 90–414) and p25 (aa 170–414) from expression vectors with N-terminal FLAG and C-terminal V5 tags under the same conditions for the deletion mutant analysis (Fig. 3, B and E) to further examine the biochemical characteristics in transfected cells (Fig. 4A).
FIGURE 4.
Immunofluorescence analysis of TDP-43 expression in HeLa cells. A, HeLa cells transfected with full-length TDP-43, p35, and p25 with the FLAG tag at the N terminus and V5 tag at the C terminus were analyzed by immunoblotting with anti-V5 antibody. A 10-μg aliquot of protein per lane was loaded onto a 4–20% Tris-glycine gradient gel. B, CFTR exon 9 skipping assay of TDP-43 fragments. HeLa cells were co-transfected with the CFTR minigene construct, pTB-CFTR 9 (TG13T3), and full-length, p35, and p25 expression vectors. Two days after transfection, the exon inclusion and exclusion products were examined by RT-PCR. The PCR products were analyzed by electrophoresis in 2% agarose gel. Upper and lower bands correspond to the complete exon 9 inclusion and exclusion, respectively. The middle band corresponds to the cryptic 3′ splice variant. Asterisks are heterodimers with the exon 9 inclusion and exclusion or cryptic 3′ splice variant. Note that neither p35 nor p25 showed exon exclusion activity. C, HeLa cells transfected with full-length TDP-43, p35, and p25 were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and double labeled with anti-FLAG (green) and anti-V5 (red) antibodies. Scale bar = 10 μm.
Because TDP-43 is well characterized as a splicing regulator of CFTR exon 9 (23–25), we first assessed the skipping activity of CFTR exon 9 of p35 and p25, using the CFTR minigene plasmid. Recently, several groups have demonstrated that truncated TDP-43, which is disturbed its nuclear distribution, markedly reduced the exon skipping activity (17, 28). Consistent with these reports, p35 and p25 did not promote exon 9 exclusion, indicating both lost their function as a splicing regulator (Fig. 4B).
We also examined the subcellular localization of p35 and p25 in transfected cells using confocal microscopy (Fig. 4C). Double immunofluorescent staining with anti-FLAG antibody recognizing the N terminus and anti-V5 antibody recognizing the C terminus in HeLa cells expressing full-length TDP-43 showed a typical nuclear localization (Fig. 4C, upper). p35 was localized to both the nucleus and cytoplasm (Fig. 4C, middle), whereas p25 was diffusely distributed throughout the entire cell (Fig. 4C, lower). This finding suggests that the N-terminal region is critical for TDP-43 localization.
Interestingly, p35 formed perinuclear inclusion bodies (foci or IBs) that were recognized by both the N- and C-terminal antibodies (Fig. 4C, middle). Of ∼300 transfected cells in 3 independent experiments, ∼25% of those expressing p35 contained IBs. In contrast, IBs were detected in ∼5% of wild-type cells (mean ± S.D.; anti-FLAG antibody: full-length 3.3 ± 0.3%, p35, 25.0 ± 7.4% (p < 0.001 versus full-length); anti-V5 antibody: full-length 5.2 ± 1.3%, p35, 27.2 ± 5.1% (p < 0.001 versus full-length)) (Fig. 5). These IBs were detected in ∼10% of the motor neuronal cell line NSC-34 transfected with p35, but were rarely identified in full-length TDP-43-expressing cells (supplemental Fig. 3). Similar to the cells expressing full-length TDP-43, ∼5% of HeLa cells and no NSC-34 cells expressing p25 formed IBs. Because expression of p35 without an N-terminal tag (aa 85–414, encoding the sequence for p35iso) resulted in the same IB formation as described above (supplemental Fig. 4), we focused on p35 in the following studies.
FIGURE 5.
Analysis of IB formation in HeLa cells expressing full-length TDP-43 or p35. Transfected cells were treated with or without 10 μm MG132 for 16 h. Approximately 300 transfected cells from 3 independent experiments were counted. The cells expressing p35 demonstrated two types of IB: those found under normal conditions (IBNC) and those found under proteasome inhibition (IBPI). Both anti-FLAG (green) and anti-V5 (red) antibodies detected IBNC, but only anti-V5 antibody labeled IBPI. Scale bar = 10 μm.
Recent studies have demonstrated that short fragments of TDP-43 accumulate in the presence of the proteasome inhibitor MG132, suggesting that TDP-43 is degraded through the ubiquitin-proteasome system (6). Thus, we examined IB formation in cells expressing full-length or p35 treated with MG132. Immunocytochemistry demonstrated that the frequency of cells IB positive for anti-V5 antibody is increased in p35-expressing cells in the presence of proteasome inhibitor, but not in cells expressing full-length TDP-43 (full-length, 4.5 ± 2.3% and p35, 55.5 ± 6.2% (Fig. 5)). In contrast, the frequency of FLAG-positive IBs was not increased in either full-length- or p35-expressing cells (full-length, 3.1 ± 1.0% and p35, 23.8 ± 2.9%) in the presence of protease inhibitor, indicating that p35 could be further cleaved and aggregated in the cytoplasm when proteasome function is inhibited. Thus, p35 appears to accumulate under two conditions: IBs under normal conditions (IBNC) or IBs under proteasome inhibition (IBPI).
Characterization of p35-containing IBs
Winton et al. have reported that overexpression of NLS deletion mutants leads to the sequestering of endogenous TDP-43 in cytoplasmic aggregates, resulting in nuclear depletion of endogenous TDP-43 (27). To confirm whether this also occurs in p35-containing IBs, HeLa cells were transfected with full-length TDP-43 harboring an HA epitope tag and p35 expressing a V5 epitope tag at the C terminus. We found that both full-length HA and p35-V5 were localized to IBNC (Fig. 6), suggesting that the IB recruits full-length TDP-43. Interestingly, IBPI and full-length TDP-43 do not co-localize, suggesting that IBNC and IBPI form via separate pathways.
FIGURE 6.
Co-transfection of full-length TDP-43 and p35. Representative micrographs of HeLa cells transfected with full-length TDP-43 tagged with a HA tag and p35 tagged with a V5 tag, in the absence or presence of proteasome inhibitor. The cells were fixed and immunostained with anti-HA (against full-length TDP-43, green) and anti-V5 (against p35, red) antibodies. Both full-length TDP-43 and p35 assemble in IBNC (arrowhead), but the former is not recruited to IBPI (arrow). Scale bar, 10 μm.
Accumulated TDP-43 found in FTLD-U and ALS brain tissue is hyperphosphorylated and can be easily detected by anti-phosphorylated TDP-43 antibody (15). Therefore, we examined whether p35-containing IBs are also phosphorylated. Immunocytochemical staining with monoclonal antibody (monoclonal antibody pS409/410) directed against phosphoserines 409 and 410 in TDP-43 revealed that IBPI are strongly immunoreactive, whereas IBNC are weakly positive, suggesting both IBs exhibit pathological properties (Fig. 7).
FIGURE 7.
Hyperphosphorylation of p35-containing IBs. HeLa cells at 48 h post-transfection with p35 were treated with or without MG132 for 16 h and immunostained with both polyclonal anti-V5 and monoclonal p409/410 phospho-specific TDP-43 antibodies. Phosphorylated TDP-43 was strongly positive in IBPI, but only weakly positive in IBNC. Scale bar = 10 μm.
Expression of mutant proteins associated with neurodegenerative diseases, including huntingtin, parkin, α-synuclein, and prion, leads to the formation of ubiquitinated aggregates (aggresomes), several features of which have been biochemically characterized (29–35). Aggresomes contain misfolded proteins transported along microtubules that are formed at the microtubule organizing center (MTOC) (29, 31–33). They also contain a deposition of intermediate filaments such as vimentin, which often form a cage-like structure around IBs (29, 31, 32). To determine the similarity between TDP-43 IB and aggresomes, we compared the distribution of TDP-43 IB to that of pericentrin (MTOC marker) and the intermediate filament protein vimentin. As shown in Fig. 8A, IBPI were targeted to and congregated specifically at the MTOC, whereas IBNC were not localized within close proximity of the MTOC. Vimentin formed a ring-like halo surrounding the core of the IBPI (Fig. 8B) (29, 31, 32), but its distribution appeared to be unchanged around IBNC. Taken together, we suggest that IBPI have similar properties to aggresomes, but that formation of IBNC occurs via a different mechanism.
FIGURE 8.
Characterization of TDP-43 IBs. Cells expressing p35 with FLAG at the N terminus and V5 at the C terminus were immunostained with both polyclonal anti-V5 antibody (green) and pericentrin (A), or vimentin antibody (B), in the absence or presence of proteasome inhibitor. IBPI, but not IBNC, aggregated at the MTOC and were surrounded by vimentin. Scale bar = 10 μm.
IBNC Exhibit Characteristics of Cytoplasmic Stress Granules
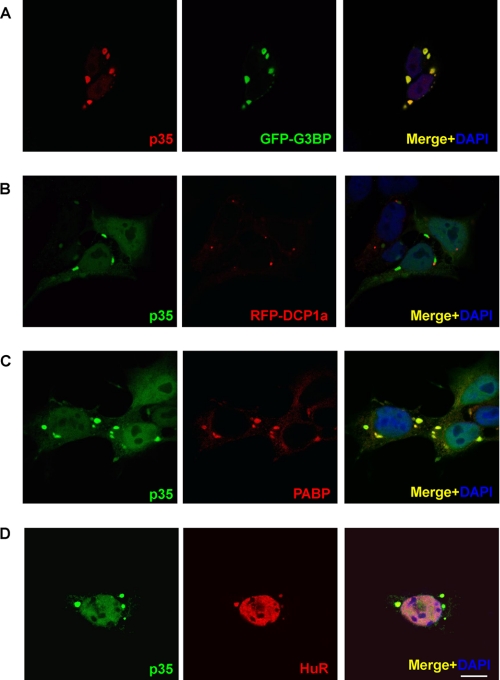
Under certain cellular stresses, housekeeping mRNA- and RNA-binding proteins that are known to regulate translation are assembled in cytoplasmic foci known as SGs that stabilize mRNA and arrest translation to prevent the accumulation of misfolded proteins (22, 36–38). An additional cellular structure known to package mRNA- and RNA-binding proteins associated with RNA decay is the processing body (PB), which contributes to mRNA degradation. Both SG and PB share several protein and mRNA components, and these dynamic relations contribute to compartmentalization in regulating the fate of mRNA (36). Most recently, it has been reported that TDP-43 is up-regulated and immunohistochemically co-localized with the SG marker TIA-1 in axotomized motor neurons (39). We therefore hypothesized that the RNA-binding protein TDP-43 may be recruited to SGs and PBs. To examine this hypothesis, we performed double immunofluorescence staining using fluorescently tagged SG and PB markers (green fluorescent protein (GFP)-G3BP and red fluorescent protein (RFP)-DCP1a, respectively) (22). As shown in Fig. 9, p35 revealed extensive co-localization with GFP-G3BP (Fig. 9A), and partial co-localization with RFP-DCP1a (Fig. 9B). In HeLa cells expressing p35 alone, SG markers, anti-PABP, and HuR were clearly localized to IBNC, suggesting that expression of p35 leads to formation of SGs (Fig. 9, C and D). Although it has been recently reported that the N-terminal tag of TDP-43 artificially affects biochemical characteristics (18), expression of p35 (aa 85–414) without an N-terminal tag similarly formed G3BP-positive IBNC (supplemental Fig. 4). Furthermore, in HeLa cells treated with arsenite, a well established SG inducer (38), endogenous TDP-43 is recruited to SGs (supplemental Fig. 5).
FIGURE 9.
IBNC are associated with SG markers. A and B, cells expressing p35 with V5 at the C terminus were immunostained with both GFP-G3BP (A) or RFP-DCP1a (B) and anti-V5 antibody. p35 and G3BP co-localize to IBNC. C and D, HeLa cells were also transiently transfected with p35 alone. IBNCs were labeled with endogenous SG marker PABP (C) or HuR (D). Scale bar = 10 μm.
Finally, the inhibitor of translational elongation, emetine, stabilizes polysomes and has been shown to suppress SG formation (40). As shown in Fig. 10, treatment with this drug was found to disperse IBNC (% of IBNC-containing cells: 26.3 ± 3.5% without emetine versus 4.3 ± 3.1% with emetine (p < 0.01)) in HeLa cells expressing p35. Taken together, our findings strongly indicate that IBNC are likely to be identical to SGs.
FIGURE 10.
Emetine disassembles IBNC formation. Treatment of 10 μg/ml emetine for 1 h dispersed IBNC in HeLa cells expressing p35. Scale bar = 10 μm.
DISCUSSION
A number of studies have shown that truncation of accumulated neurotoxic proteins associated with neurodegeneration, including the amyloid precursor protein, polyglutamine, and Tau, is critical for neurodegeneration processes (41–46). Truncated forms of TDP-43, whose accumulation is a pathological hallmark of ALS and FTLD-U, have only been recovered from affected central nervous system regions (2, 3, 18). Zhang et al. have suggested that proteolytic cleavage generates the 25- and 35-kDa CTFs present in cultured cells, leading to redistribution of TDP-43 from its nuclear location to the cytoplasm and is associated with cell death through a toxic gain of function (16, 17). Furthermore, Sreedharan et al. have demonstrated that a mutation linked to fALS enhanced fragmentation of TDP-43 in Chinese hamster ovary cells (5), suggesting that truncated TDP-43 is critical in neurodegeneration related to TDP-43 proteinopathies.
Our present study presents several important insights into the proteolytic cleavage and isoforms of TDP-43. Our study identified TDP-43 variant forms: two caspase-3-dependent fragments and two novel isoforms. Under normal conditions, the novel isoforms, p35iso (aa 85–414) and p25iso (aa ∼170–414), are translated from methionine 85 and aa 160–169, respectively, and apoptosis generates two fragments, p35f and p25f, via caspase-3 activation.
The N-terminal amino acids of p25f and p25iso remain unclear. Based on the prediction of caspase-3 cleavage consensus sites (DXXD), p25f should comprise aa 220–414, with a molecular weight of appropriate 19,719, which is obviously smaller than 25 kDa. However, electromobility of overexpressed aa 170–414 (molecular weight: 25,635) of cDNA was compatible to that of the endogenous 25-kDa fragments seen on SDS-PAGE (Fig. 3H). Fig. 2 (B and C) shows no discrepancy in the size between p25f and p25iso on SDS-PAGE. Moreover, well characterized monoclonal anti-TDP-43 antibody, 2E2-D3, could not recognize aa 208–414 and the smaller C-terminal residues of human TDP-43 (26) but could detect both endogenous p25f and p25iso (Fig. 2C), indicating that the N terminus of p25f and p25iso is further upstream than previously reported (16, 18, 28). Therefore, we suggest that DVMD (aa 216–219) may not be the true cleavage site of TDP-43 for generation of p25f and that additional unknown protease(s) activated by caspase-3 may generate p25f under apoptosis. We cannot however exclude the other possibility that p25iso and p25f are identical and caspase-3-dependent signaling suppresses degradation and/or enhances translation of p25iso, resulting in the accumulation of p25iso under staurosporine treatment.
Recently, Igaz et al. have reported two major fragments of ∼22 and 24 kDa that were recognized in immunoprecipitation studies using an anti-TDP-43 antibody and cortical extracts of FTLD-U brain tissue (18). Both this group and Nonaka et al. identified several cleavage sites (aa 208, 219, and 247) for generation of the smaller 22-kDa CTF of TDP-43 by protein sequencing (18, 28). As several smaller fragments (<25 kDa) appear following longer exposure in this study (Fig. 1A), several cleavage sites as well as alternate initial codons appear to exist within aa 160–250 for generating smaller fragments and isoforms.
Another important finding was the characterization of TDP-43-containing IBs that appear in cells expressing p35. Two distinct IBs (IBPI and IBNC) were identified in either the presence or absence of proteasome inhibitors. IBPI demonstrated properties similar to aggresomes, which are formed by the assembly of misfolded proteins at the MTOC, and contained the redistributed intermediate filament protein vimentin in addition to other pathogenic proteins including mutant forms of huntingtin, α-synuclein, parkin, and prion (29–35) (Fig. 8). As IBPI was strongly positive for pS409/410, this inclusion appeared to be suitable for the pathological accumulation of TDP-43 in FTLD-U and ALS brain tissue. Our findings also showed that IBNC contains well established components of SGs, including G3BP, PABP, and HuR (22, 38). SGs, which are induced by stress events, including hypoxia, heat shock, and arsenite, are multimolecular aggregates present in the cytosol and are responsible for the recruitment of ribonucleoprotein and mRNA. SGs are thought to function in the protection of mRNA against harmful stress events and to arrest translation, preventing the accumulation of misfolded proteins (36, 38). We found that p35 forms a part of these SGs and may facilitate the assembly of SGs when overexpressed, similar to other SG-associated proteins: G3BP, TIA-1/TIAR, and SMN (38, 47–49). Although further studies are needed to determine whether p35-induced SGs contribute to RNA stability and regulation of translation, our findings suggest that p35f and p35iso are involved in RNA quality control under cellular stress via the function of SGs.
Fig. 11 outlines our hypothetical model of the relationship between SGs and the variant forms of TDP-43. Full-length TDP-43 is located in the nucleus of the cell and functions as a splicing regulator. p35iso is translated from an alternate initiation codon, and p35f is generated by caspase-3 under apoptosis. As both are localized to the cytoplasm but lack the NLS, they subsequently form aggregates with a structure comparable to intracellular inclusions pathologically observed in the brain tissue of FTLD-U and ALS patients. p35 is also involved in SG formation. Recent evidence suggests that pathological aggregations of toxic proteins, including polyglutamine, and that β-amyloid are not directly linked to pathogenesis and rather might be neuroprotective (50–54), inappropriate SG assembly may be induced by p35, thereby chronically disturbing RNA stability and translation arrest, which is a key process of pathogenesis, compared with pathological aggregations such as neuronal cytoplasmic and skein-like inclusions in TDP- 43 proteinopathy. Interestingly, a recent study has shown that SMN, a protein associated with autosomal recessive motor neuron disease and spinal muscular atrophy, facilitates SG formation, and that loss-of-function of SMN in motor neurons may influence neurodegeneration via the disruption of RNA stability through SG function (49). Moreover, it has been reported that editing of the RNA of GluR2, a subunit of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor, is markedly decreased in the motor neurons of sALS patients (55, 56). Therefore, dysfunction of RNA quality control systems may be a common feature of the pathological processes occurring in the development of motor neuron diseases.
FIGURE 11.
TDP-43 isoforms and formation of SGs or aggresomes. This model suggests that p35iso, which is derived from an alternate initiation codon, localizes to the cytoplasm. Full-length TDP-43 is proteolytically cleaved dependently of caspase-3 to release p35f, which is lacking the NLS and redistributed to the cytoplasm. Under certain stress or pathological conditions, intracellular accumulation of p35iso and p35f leads to the formation of either SGs or aggresomes that may be associated with motor neuron degeneration.
Further investigations are required to determine whether fALS-linked mutant TDP-43 affects the formation and RNA metabolism of SGs. Moreover, although future studies are needed to elucidate the precise molecular pathogenesis of TDP-43 proteinopathies, our study contributes significantly toward a greater understanding for the treatment of the serious neurological diseases ALS and FTLD-U.
Supplementary Material
Acknowledgments
We are grateful to Drs. Francisco E. Baralle and Emanuele Buratti (Dept. of Molecular Pathology, International Centre for Genetic Engineering and Biotechnology, Italy) for providing the FLAG-tagged human TDP-43 plasmid and pTB-CFTR9, to Dr. Paul Anderson (Division of Rheumatology, Immunology and Allergy, Brigham and Women's Hospital and Harvard Medical School) for providing the GFP-G3BP and red RFP-DCP1a plasmids, and to Dr. Richard Flavell (Dept. of Immunobiology, Yale University School of Medicine) for providing the caspase-3(−/−) cells. We also thank Dr. Shin Kwak (Dept. of Neurology, Graduate School of Medicine, University of Tokyo) and Dr. Shoichi Ishiura (Dept. of Life Sciences, Graduate School of Arts and Sciences, University of Tokyo) for their helpful discussions.
This work was supported by Eisai Co., Ltd., the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants 18590955 and 21790850), the ALS Foundation, the Japan ALS Association, and Keio Medical Science Fund Research Grants for Life Science and Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- ALS
- amyotrophic lateral sclerosis
- TDP-43
- TAR DNA-binding protein-43
- FTLD-U
- frontotemporal lobar degeneration with ubiquitin-positive inclusions
- CTF
- C-terminal fragment
- SG
- stress granule
- SOD-1
- Cu,Zn-superoxide dismutase-1
- CFTR
- cystic fibrosis transmembrane conductance regulator
- MEF
- mouse embryonic fibroblast
- SMN
- survival motor neuron
- NLS
- nuclear localization signal
- IBNC
- inclusion body under normal condition
- IBPI
- inclusion body under proteasome inhibition
- MTOC
- microtubule organizing center
- GFP
- green fluorescent protein
- sALS
- sporadic ALS
- fALS
- familial ALS
- CMV
- cytomegalovirus
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- aa
- amino acid(s)
- IB
- inclusion body
- PB
- processing body
- RFP
- red fluorescent protein
- PABP
- poly(A)-binding protein
- HuR
- Hu antigen R
- G3BP
- GAP SH3 domain-binding protein.
REFERENCES
- 1.Shaw C. E., Enayat Z. E., Chioza B. A., Al-Chalabi A., Radunovic A., Powell J. F., Leigh P. N. (1998) Ann. Neurol. 43, 390–394 [DOI] [PubMed] [Google Scholar]
- 2.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 3.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 4.Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., Steinbart E., McCluskey L., Grossman M., Neumann M., Wu I. L., Yang W. S., Kalb R., Galasko D. R., Montine T. J., Trojanowski J. Q., Lee V. M., Schellenberg G. D., Yu C. E. (2008) Lancet Neurol. 7, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford N. J., Zhang Y. J., Baker M., Gass J. M., Finch N. A., Xu Y. F., Stewart H., Kelley B. J., Kuntz K., Crook R. J., Sreedharan J., Vance C., Sorenson E., Lippa C., Bigio E. H., Geschwind D. H., Knopman D. S., Mitsumoto H., Petersen R. C., Cashman N. R., Hutton M., Shaw C. E., Boylan K. B., Boeve B., Graff-Radford N. R., Wszolek Z. K., Caselli R. J., Dickson D. W., Mackenzie I. R., Petrucelli L., Rademakers R. (2008) PLoS Genet 4, e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 8.Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., Hatanpaa K. J., White C. L., 3rd, Bigio E. H., Caselli R., Baker M., Al-Lozi M. T., Morris J. C., Pestronk A., Rademakers R., Goate A. M., Cairns N. J. (2008) Ann. Neurol. 63, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., Okamoto K., Nishizawa M., Takahashi H., Onodera O. (2008) Ann. Neurol. 63, 538–542 [DOI] [PubMed] [Google Scholar]
- 10.Neumann M., Kwong L. K., Sampathu D. M., Trojanowski J. Q., Lee V. M. (2007) Arch. Neurol. 64, 1388–1394 [DOI] [PubMed] [Google Scholar]
- 11.Ou S. H., Wu F., Harrich D., García-Martínez L. F., Gaynor R. B. (1995) J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., Baralle F. E. (2001) EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bose J. K., Wang I. F., Hung L., Tarn W. Y., Shen C. K. (2008) J. Biol. Chem. 283, 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayala Y. M., Misteli T., Baralle F. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., Akiyama H. (2008) Ann. Neurol. 64, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y. J., Xu Y. F., Dickey C. A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., Petrucelli L. (2007) J. Neurosci. 27, 10530–10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igaz L. M., Kwong L. K., Chen-Plotkin A., Winton M. J., Unger T. L., Xu Y., Neumann M., Trojanowski J. Q., Lee V. M. (2009) J. Biol. Chem. 284, 8516–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito D., Suzuki N. (2007) Ann. Neurol. 61, 237–250 [DOI] [PubMed] [Google Scholar]
- 20.Ito D., Walker J. R., Thompson C. S., Moroz I., Lin W., Veselits M. L., Hakim A. M., Fienberg A. A., Thinakaran G. (2004) Mol. Cell. Biol. 24, 9456–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito D., Fujisawa T., Iida H., Suzuki N. (2008) Neurobiol. Dis. 31, 266–277 [DOI] [PubMed] [Google Scholar]
- 22.Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. (2008) Nat. Cell Biol. 10, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayala Y. M., Pagani F., Baralle F. E. (2006) FEBS Lett. 580, 1339–1344 [DOI] [PubMed] [Google Scholar]
- 24.Buratti E., Baralle F. E. (2001) J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 25.Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F. E. (2000) J. Biol. Chem. 275, 21041–21047 [DOI] [PubMed] [Google Scholar]
- 26.Zhang H. X., Tanji K., Mori F., Wakabayashi K. (2008) Neurosci. Lett. 434, 170–174 [DOI] [PubMed] [Google Scholar]
- 27.Winton M. J., Igaz L. M., Wong M. M., Kwong L. K., Trojanowski J. Q., Lee V. M. (2008) J. Biol. Chem. 283, 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. (2009) Hum. Mol. Genet. 18, 3353–3364 [DOI] [PubMed] [Google Scholar]
- 29.Kopito R. R. (2000) Trends Cell Biol. 10, 524–530 [DOI] [PubMed] [Google Scholar]
- 30.García-Mata R., Bebök Z., Sorscher E. J., Sztul E. S. (1999) J. Cell Biol. 146, 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston J. A., Ward C. L., Kopito R. R. (1998) J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Junn E., Lee S. S., Suhr U. T., Mouradian M. M. (2002) J. Biol. Chem. 277, 47870–47877 [DOI] [PubMed] [Google Scholar]
- 33.Tanaka M., Kim Y. M., Lee G., Junn E., Iwatsubo T., Mouradian M. M. (2004) J. Biol. Chem. 279, 4625–4631 [DOI] [PubMed] [Google Scholar]
- 34.Kalchman M. A., Graham R. K., Xia G., Koide H. B., Hodgson J. G., Graham K. C., Goldberg Y. P., Gietz R. D., Pickart C. M., Hayden M. R. (1996) J. Biol. Chem. 271, 19385–19394 [DOI] [PubMed] [Google Scholar]
- 35.Sieradzan K. A., Mechan A. O., Jones L., Wanker E. E., Nukina N., Mann D. M. (1999) Exp. Neurol. 156, 92–99 [DOI] [PubMed] [Google Scholar]
- 36.Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005) J. Cell Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. (2008) Nat. Cell Biol. 10, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 38.Kedersha N., Anderson P. (2007) Methods Enzymol. 431, 61–81 [DOI] [PubMed] [Google Scholar]
- 39.Moisse K., Volkening K., Leystra-Lantz C., Welch I., Hill T., Strong M. J. (2009) Brain Res. 1249, 202–211 [DOI] [PubMed] [Google Scholar]
- 40.Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. (2000) J. Cell Biol. 151, 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sisodia S. S., Price D. L. (1995) FASEB J. 9, 366–370 [DOI] [PubMed] [Google Scholar]
- 42.DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 43.Wellington C. L., Singaraja R., Ellerby L., Savill J., Roy S., Leavitt B., Cattaneo E., Hackam A., Sharp A., Thornberry N., Nicholson D. W., Bredesen D. E., Hayden M. R. (2000) J. Biol. Chem. 275, 19831–19838 [DOI] [PubMed] [Google Scholar]
- 44.Mende-Mueller L. M., Toneff T., Hwang S. R., Chesselet M. F., Hook V. Y. (2001) J. Neurosci. 21, 1830–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arai T., Ikeda K., Akiyama H., Nonaka T., Hasegawa M., Ishiguro K., Iritani S., Tsuchiya K., Iseki E., Yagishita S., Oda T., Mochizuki A. (2004) Ann. Neurol. 55, 72–79 [DOI] [PubMed] [Google Scholar]
- 46.Selkoe D. J., Yamazaki T., Citron M., Podlisny M. B., Koo E. H., Teplow D. B., Haass C. (1996) Ann. N.Y. Acad. Sci. 777, 57–64 [DOI] [PubMed] [Google Scholar]
- 47.Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., Tazi J. (2003) J. Cell Biol. 160, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. (2004) Mol. Biol. Cell 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hua Y., Zhou J. (2004) FEBS Lett. 572, 69–74 [DOI] [PubMed] [Google Scholar]
- 50.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 51.Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. (2003) Hum. Mol. Genet. 12, 749–757 [DOI] [PubMed] [Google Scholar]
- 52.Ito D., Suzuki N. (2009) Brain 132, 8–15 [DOI] [PubMed] [Google Scholar]
- 53.Orr H. T. (2004) Nature 431, 747–748 [DOI] [PubMed] [Google Scholar]
- 54.Selkoe D. J. (2002) Science 298, 789–791 [DOI] [PubMed] [Google Scholar]
- 55.Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S. (2004) Nature 427, 801. [DOI] [PubMed] [Google Scholar]
- 56.Nishimoto Y., Yamashita T., Hideyama T., Tsuji S., Suzuki N., Kwak S. (2008) Neurosci. Res. 61, 201–206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.