Abstract
The actin-binding protein caldesmon (CaD) reversibly inhibits smooth muscle contraction. In non-muscle cells, a shorter CaD isoform co-exists with microfilaments in the stress fibers at the quiescent state, but the phosphorylated CaD is found at the leading edge of migrating cells where dynamic actin filament remodeling occurs. We have studied the effect of a C-terminal fragment of CaD (H32K) on the kinetics of the in vitro actin polymerization by monitoring the fluorescence of pyrene-labeled actin. Addition of H32K or its phosphorylated form either attenuated or accelerated the pyrene emission enhancement, depending on whether it was added at the early or the late phase of actin polymerization. However, the CaD fragment had no effect on the yield of sedimentable actin, nor did it affect the actin ATPase activity. Our findings can be explained by a model in which nascent actin filaments undergo a maturation process that involves at least two intermediate conformational states. If present at early stages of actin polymerization, CaD stabilizes one of the intermediate states and blocks the subsequent filament maturation. Addition of CaD at a later phase accelerates F-actin formation. The fact that CaD is capable of inhibiting actin filament maturation provides a novel function for CaD and suggests an active role in the dynamic reorganization of the actin cytoskeleton.
Introduction
Caldesmon (CaD)3 is an F-actin-binding protein found in nearly all vertebrate cells. Its multiple actin-binding sites have been shown to bundle actin filaments (1), promote actin nucleation (2), and stabilize actin strands like a staple (3). CaD also interacts with other actin-binding proteins including tropomyosin (4, 5) and cortactin (6). There are two CaD isoforms derived from a single gene (7): the heavy form (h-CaD) is specifically expressed in smooth muscle cells (8–10), and the light form (l-CaD) is ubiquitously expressed (11, 12). In smooth muscle cells the elongated h-CaD binds with its two end domains simultaneously to myosin and actin (13), and regulates the actomyosin interaction in a Ca2+/calmodulin (CaM)- and phosphorylation-dependent manner (14). Significantly, during smooth muscle relaxation h-CaD remains bound to the contractile apparatus, and the actin filaments are only moderately disassembled. The situation is quite different in non-muscle cells, although l-CaD and h-CaD share the same functional domains. In quiescent non-muscle cells l-CaD stabilizes the actin stress fibers in the cytoplasm (15). Upon activation, the cytosolic stress fibers are largely disassembled, whereas l-CaD is phosphorylated and moves to cell peripheries where the actin cytoskeleton is actively reorganized (6, 15). The precise function of phosphorylated l-CaD in the actin dynamics remains unclear.
The dynamic rearrangement of the actin cytoskeleton is a highly regulated process that involves a large number of actin-binding proteins (16, 17). They play an important role in controlling vital cellular activities (18) and contribute to the elastic nature of soft tissues including smooth muscles (19–22). On one hand, the actin cytoskeleton is stable enough to maintain static cellular structures and provide support for motor proteins to generate contractile force. On the other hand, it drives cell shape change during cell movement via rapid cycles of disassembly and reassembly. The mechanisms that regulate the stability and the dynamics of the actin cytoskeleton are under intense investigation.
In this study, we tested the effects of CaD on the kinetics of salt-induced actin polymerization in vitro by using a C-terminal fragment (H32K) of CaD that contains the actin-binding sites and by monitoring the fluorescence increase of the actin-bound pyrene probe. We found that both unphosphorylated and phosphorylated H32K exert different effects on polymerizing actin depending on the timing of addition. This observation is consistent with a model in which actin filaments undergo a maturation process. If present at early stages of actin polymerization, CaD stabilizes an intermediate state and blocks the subsequent filament maturation. In contrast, addition of CaD at a later phase of actin polymerization accelerates the formation of F-actin. That CaD is capable of arresting an intermediate state of actin filaments provides a novel function for CaD and suggests an active role in the dynamic reorganization of the actin cytoskeleton.
EXPERIMENTAL PROCEDURES
Actin and Pyrene Labeling
Actin was extracted and purified from acetone-dried rabbit skeletal muscle (23). The labeling was done by adding pyrene-maleimide (6 mg/ml in dimethyl formamide; from Molecular Probes) into F-actin at a 7.5:1 ratio. The solution was incubated in the dark at 4 °C for 24 h. After dialysis with F-buffer (2 mm Hepes, pH 7.5, 0.2 mm CaCl2, 50 mm NaCl, 2 mm MgCl2, and 0.4 mm ATP), F-actin was collected by centrifugation, and then was de-polymerized by dialysis with G-buffer (2 mm Hepes, pH 7.5, 0.2 mm CaCl2, 0.5 mm dithiothreitol, and 0.4 mm ATP). In some experiments the ATP in both G- and F-buffer was replaced by ADP (Sigma). The insoluble pyrene and F-actin were removed by centrifugation, followed by size-exclusion chromatography. Pyrene-labeled actin was then mixed with unlabeled actin at different ratios for the polymerization assays.
CaD and H32K
Recombinant human full-length h-CaD was prepared from insect cells as described previously (24). The recombinant His6-tagged C-terminal fragment (residues from Leu604 to Val793) of human CaD (H32K) was expressed in Escherichia coli, purified by Ni2+-column, and further purified by using a CaM-Sepharose affinity column (3, 25). To minimize disulfide-based dimerization, H32K was first reduced by 50 mm dithiothreitol and kept in a buffer containing 1.5 mm dithiothreitol. Phosphorylation of H32K: H32K was phosphorylated by ERK2 (New England Biolabs) for 24 h at room temperature. The extent of phosphorylation was checked by MALDI-TOF mass spectrometry (Applied Biosciences, Inc.) after proteolysis with Staphylococcus aureus V8 protease as described previously (25). Both the H32K- and ERK-phosphorylated H32K (pH32K) were dialyzed against F-buffer prior to their use in the actin polymerization assay.
Actin Polymerization Assay
G-actin (mixture of pyrene-labeled and unlabeled actin at a ratio ranging from 1:2.8 to 1:14.4; total concentration ∼60 μm) was first incubated in G-buffer at room temperature; polymerization was initiated by diluting the mixture into F-buffer with or without H32K (or pH32K) so that the final concentrations were 1.5 μm for actin and 0.5 μm for H32K (or pH32K). In another type of experiment, H32K (or pH32K) was added at certain time points after the initiation of actin polymerization. Actin polymerization was monitored by the enhancement of pyrene emission (λex = 365 nm; λem = 407 nm).
Phosphate Release Assay
Continuous monitoring of inorganic phosphate generation during actin polymerization was done by measuring the absorption of the product of a coupled purine nucleoside phosphorylase-catalyzed reaction at 363 nm according to the method of Webb (26) using the EnzCheck® Phosphate Assay kit (E-6646, from Molecular Probes).
Ultracentrifugation
Polymerized actin was collected by centrifugation at 85,000 rpm for 30 min at 4 °C. The pellet was washed twice with F-buffer and dissolved in F-buffer. Both the supernatant and pellet were then analyzed by 10% SDS-PAGE.
Simulation of the Kinetics of Actin Polymerization
The program COPASI (Complex Pathway Simulator) version 4.4.26 (27) was used to simulate the kinetics of actin polymerization.
RESULTS
To probe the effect of CaD on the actin filament assembly, we have followed salt-induced in vitro actin polymerization using pyrene-labeled actin and a recombinant C-terminal fragment (H32K) of CaD that comprises its major actin binding sites. This fragment has been shown previously to inhibit the actomyosin interaction in a Ca2+/CaM-dependent manner like the full-length protein (28). H32K is readily phosphorylated by the extracellular signal-regulated kinases (ERKs). Phosphorylated H32K (pH32K) behaves like phosphorylated full-length CaD, exhibiting diminished inhibition of the actin-activated ATPase activity of myosin (25). This C-terminal fragment is therefore regarded as a good analog of CaD for functional studies. Moreover, because H32K corresponds to a peptide segment common to both h- and l-CaD, results obtained with this fragment are applicable to both isoforms.
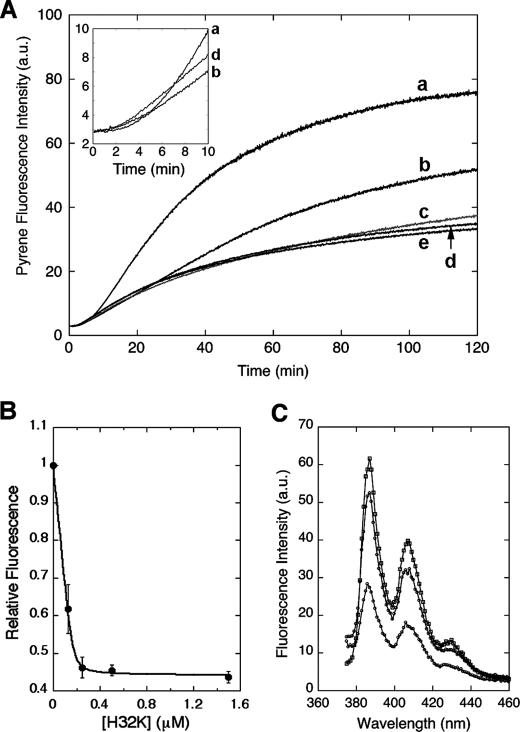
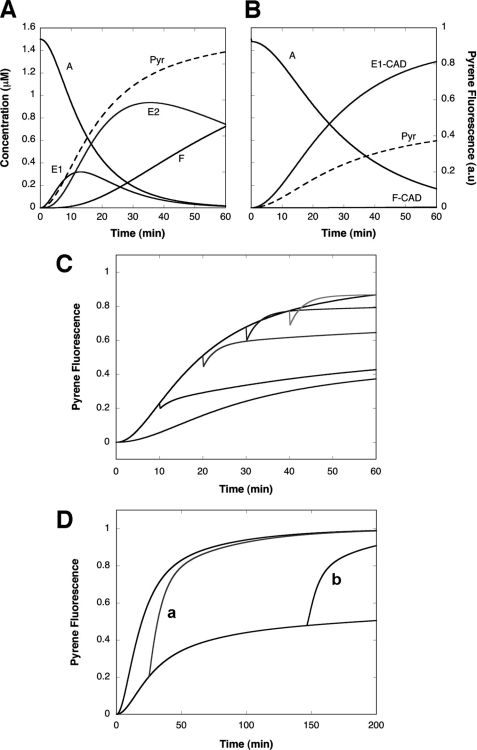
In the presence of H32K the increase in pyrene fluorescence intensity normally associated with actin polymerization was greatly reduced (Fig. 1A). The effect was concentration-dependent; polymerization with increasing amounts of H32K showed that the maximum inhibition (60% suppression of fluorescence intensity) occurred at a molar ratio of H32K/actin = 1:3. This is consistent with the elongated molecular shape of H32K that can cover at least two actin protomers (3). However, the titration of the pyrene-actin fluorescence enhancement yielded a stoichiometry of 0.114 or 1 H32K per 9 actin protomers (Fig. 1B), indicating that the effect the CaD fragment exerts on the filament extends to neighboring regions beyond the bound actin subunits. The apparent binding constant was at least two orders of magnitude higher than the reported value of CaD binding to actin (29). Such a discrepancy suggests that the properties of actin filaments polymerized with and without CaD are quite different (see below). The fluorescence effect was long-lasting, as the pyrene-actin emission intensity of the sample with the CaD fragment remained low after overnight incubation (Fig. 1C). Notably, when H32K was present the fluorescence began to rise sooner than it did for actin alone (Fig. 1A, inset), in agreement with the previous reports that CaD facilitates nucleation of actin polymerization (2, 30). However, this early rise of fluorescence intensity gave way to the subsequent attenuation of the fluorescence signal as compared with that of actin alone. Thus, H32K affects not only the kinetics of actin polymerization, but also the nature of the final products. Like H32K, the ERK-phosphorylated form (pH32K) yielded a similar decrease in the pyrene emission enhancement (see bottom trace in Fig. 2A).
FIGURE 1.
Effect of CaD fragment on the pyrene-actin fluorescence when included at the beginning of the actin polymerization. A, pyrene fluorescence enhancement (in arbitrary units; a.u.) upon actin polymerization was attenuated with increasing amounts of H32K included in the F-buffer (traces labeled a–e from top to bottom represent experiments with 0, 0.125, 0.25, 0.5, and 1.5 μm H32K, respectively; actin concentration, 1.5 μm). Inset, an enlarged rendition of selected traces with 0 μm (trace a), 0.125 μm (trace b), and 0.5 μm (trace d) of H32K showing the pyrene fluorescence increase was accelerated by H32K during the initial 10 min. B, relative intensity of pyrene fluorescence (to that of actin alone, averaged over the period of t = 25 to 120 min) from A is plotted as a function of H32K concentration. The data points are fitted with an equilibrium binding isotherm. The curve represents the best fit with the equation: F = Fo + ΔFν, where Fo is the initial fluorescence, ΔF is the H32K-dependent suppression in the pyrene fluorescence enhancement, and ν is one of the roots of the quadratic equation: nAν2 − (Kd + nA + C)ν + C = 0; n is the stoichiometry defined as the number of H32K molecules bound to one actin subunit, A is the total actin concentration (1.5 μm), Kd is the equilibrium dissociation constant, and C is the total concentration of H32K. The best-fit parameters are: Kd = 3.7 nm and n = 0.114 or ∼1/9. C, emission spectra (excited at 365 nm) of pyrene-actin alone (1.5 μm, top), with H32K (0.5 μm) added at the beginning (bottom) or at 120 min after initiation (middle) of polymerization. All samples were incubated overnight to ensure completion of polymerization.
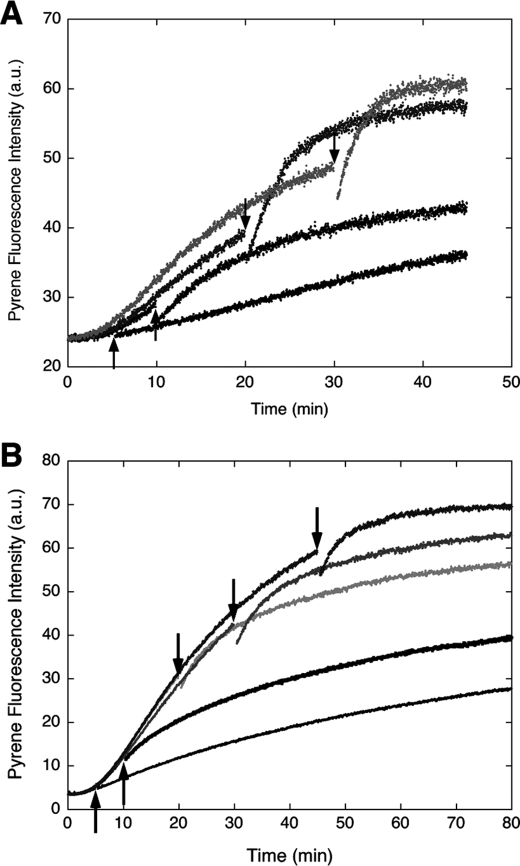
FIGURE 2.
Effect of ERK-phosphorylated H32K (pH32K, A) or unphosphorylated H32K (B) on the pyrene-actin emission change when added after initiation of polymerization. The traces represent experiments with pH32K or H32K added at different time points indicated by arrows. Concentrations of proteins: actin, 3 μm with 13% pyrene labeling; pH32K, 1.0 μm in A and actin, 1.5 μm with 20% pyrene labeling; H32K, 0.5 μm in B.
A very different effect was observed when H32K was added at time points after the polymerization started. Instead of attenuating the pyrene fluorescence enhancement, both H32K and pH32K accelerated the fluorescence increase following an initial drop in the signal (Fig. 2, A and B). The increased pyrene emission was attained by pH32K at a steeper slope than by H32K. Addition of buffer alone did not result in any fluorescence change, which ruled out fragmentation of actin filaments because of agitation as the cause of the faster fluorescence change. Moreover, the CaD effect on the pyrene emission change was different depending on the time of addition with respect to the time course of actin polymerization. An apparent inhibition of actin polymerization was seen when either H32K or pH32K was added 5 min after G-actin was mixed with the F-buffer, whereas addition of the CaD fragment at later time points (t = 10, 20, and 30 min) produced less inhibition but more acceleration. The later the CaD fragment was added, the faster the fluorescence increased. This behavior is not unique to the CaD fragment; full-length h-CaD exhibited the same dual effect as did H32K and pH32K (data not shown).
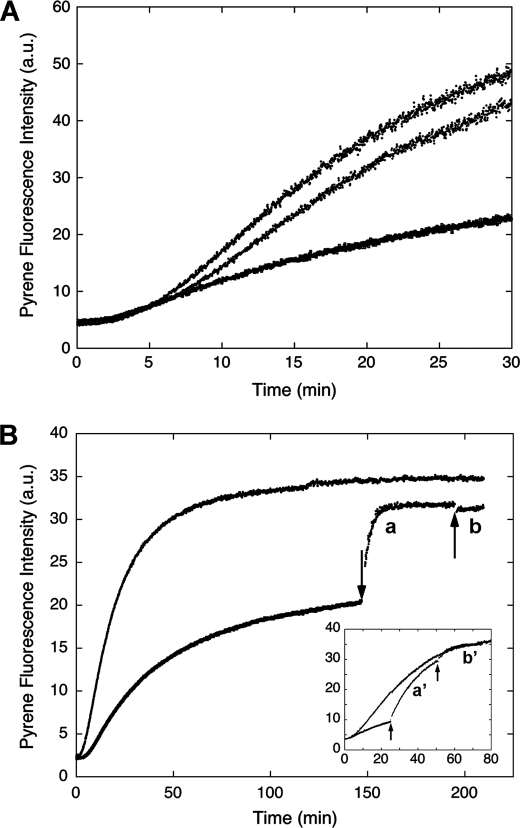
Because Ca2+/CaM prevents CaD (or its C-terminal fragment) from binding to actin (8), we have made use of this property to test the reversibility of the H32K effect. Inclusion of Ca2+/CaM in the polymerization solution restored the fluorescence enhancement (see Fig. 3A). When CaM was added to H32K-suppressed F-actin (at t = 25 min; lower trace labeled a′ in Fig. 3B, inset) in the presence of Ca2+, the pyrene-actin emission increased. The rate of this fluorescence rise, however, was faster than that for actin alone at the corresponding level of fluorescence intensity, suggesting that it represents a different event than simply filament elongation. The fluorescence change fit first-order kinetics and was faster than that when CaM was added at earlier time points (see the two traces labeled a and a′ in Fig. 3B and inset, noting that the buffer contained 0.2 mm CaCl2). The limiting value of the apparent rate constant of the transition was 0.3 min−1 when CaM was added after the polymerization reached completion, which, interestingly, is similar to the reported rate of Pi release (31). Then at a later time point when EGTA was added to remove CaM from H32K, so that H32K was now free to re-bind F-actin, we again observed an acceleration of the pyrene fluorescence increase; no such increase was observed if EGTA was added after polymerization reached the steady state (see the two traces labeled b and b′ in Fig. 3B and inset). The small drop of fluorescence intensity corresponded to the change of pyrene-actin upon binding of H32K to matured actin filaments (Fig. 1C). The difference in the effects of H32K on the fluorescence signal and the dependence of such effects on the time of addition with respect to the progression of actin polymerization are striking. It appears that multiple forms of actin exist in solution during polymerization, each interacting with CaD differently.
FIGURE 3.
Effect of CaM on the pyrene-actin fluorescence change in the presence of H32K. A, when added at the beginning of actin polymerization, inclusion of Ca2+/CaM (middle trace) neutralizes the effect of H32K (bottom trace) on the pyrene-actin fluorescence. Actin alone is represented by the top trace. Concentrations: actin, 1.5 μm; H32K, 0.5 μm; CaM, 3 μm. B, reversible binding of H32K during actin polymerization in ATP-containing F-buffer. H32K (0.5 μm) was added at the beginning of actin (1.5 μm) polymerization as described (bottom trace). At t = 150 min (indicated by arrow) when polymerization approached steady state, CaM (3 μm) was added to displace the actin-bound H32K (trace a); the fluorescence rise fit first-order kinetics with a rate constant of 0.288 ± 0.004 min−1. At t = 195 min EGTA (2 mm) was added to chelate Ca2+ (trace b). The top trace is for actin alone. Inset, earlier addition of CaM (at t = 25 min) resulted in a slower transition (the apparent first-order rate constant was 0.0544 ± 0.0004 min−1), as actin continued to polymerize; when EGTA was added (at t = 50 min), H32K was able to re-bind the post-transitional actin filament causing further acceleration of polymerization. The same protein concentrations and trace labeling (i.e. a′ corresponds to a; b′ corresponds to b) apply to the inset. All fluorescence readings were corrected for dilution effects.
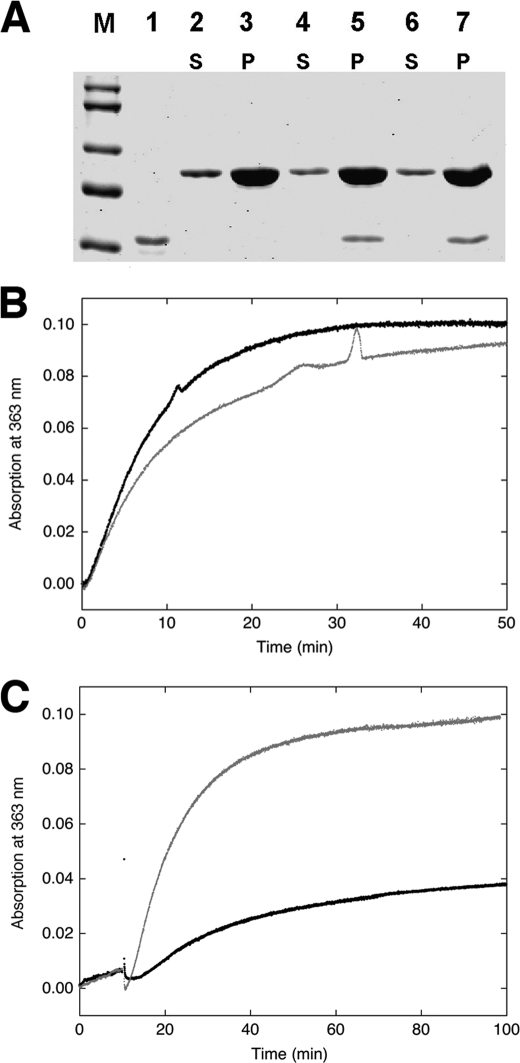
The increase in pyrene fluorescence has been widely used as a measure of actin polymerization. Accordingly, a significantly attenuated fluorescence signal observed in the presence of H32K might indicate a smaller fraction of G-actin being polymerized. On the other hand, the faster kinetics of the fluorescence enhancement upon sequestration of H32K with Ca2+/CaM (see above) argues against simple inhibition of polymerization. To discern whether and how CaD affects actin polymerization, we have subjected the samples of actin polymerized with and without H32K to ultracentrifugation followed by SDS-PAGE analysis. To our surprise, all three samples (actin alone, actin with H32K added at t = 0 and at t = 18 min after the initiation of polymerization) gave the same amount of actin in the pellet, and H32K was found exclusively in the pellet fractions regardless of the time of its addition (Fig. 4A). Thus, actin was fully polymerized under all three conditions.
FIGURE 4.
Effect of CaD on actin polymerization. A, SDS-PAGE of H32K co-sedimented with actin polymerized for 45 min without (lanes 2 and 3) or with H32K added either at the beginning (lanes 4 and 5) or 18 min after the initiation (lanes 6 and 7) of polymerization. M, molecular markers; lane 1, H32K alone. S and P are supernatant (1/20 of total sample loaded) and pellet (1/2 of total sample loaded) fractions, respectively. Protein concentrations are the same as in Fig. 1. B, polymerization monitored by phosphate release of actin alone (black trace) and actin with H32K added at the beginning of reaction (gray trace). Generation of Pi was monitored continuously at room temperature (see “Experimental Procedures”). The baseline, which was obtained for each set of experiments under identical conditions except in the absence of actin, was subtracted from the experimental reading. Protein concentrations used: 12 μm actin and 2 μm H32K. C, comparison between polymerization of actin alone (12 μm) in the presence of F-buffer containing ATP (gray trace) or ADP (black trace), monitored by phosphate release. The residual activity for the ADP sample may have resulted from contamination of ATP.
These results led us to conclude the following: (i) The increase in pyrene fluorescence does not reflect actin polymerization per se; instead, it reports a specific conformational state of polymeric actin. A similar conclusion has been reached by others (32). The observed different fluorescence states may therefore correspond to multiple forms of actin filaments when interacting with CaD. (ii) CaD does not inhibit actin polymerization, but rather, it inhibits a transition of the filament structure that leads to an increase in the pyrene fluorescence. (iii) This transition appears to be irreversible, because the addition of H32K at a late phase of actin polymerization accelerates rather than suppresses the pyrene fluorescence increase.
The key question is which step in the actin polymerization process is inhibited by H32K. According to the generally accepted model, actin polymerization occurs through the addition of ATP-bound G-actin to either end of existing filaments or to transiently formed actin trimers that serve as polymerization nuclei. This is followed by the hydrolysis of ATP and the release of inorganic phosphate (Pi) from the newly incorporated actin monomers (33). Carlier et al. (34) have shown that whereas the fluorescence of pyrene-actin increases initially by about 5-fold upon polymerization, ATP hydrolysis also contributes to the overall fluorescence enhancement. The fluorescence of pyrenyl ATP-F-actin was estimated to be about half of pyrenyl ADP-F-actin, a conclusion supported by a recent simulation study (32). To test the possibility that H32K might inhibit the ATP hydrolysis, we have measured the rate of Pi release during actin polymerization in the presence and absence of H32K. We found that similar amounts of phosphate were generated at comparable rates during actin polymerization irrespective of whether H32K was present or not (Fig. 4B). Actin polymerization was also carried out in a buffer containing mainly (∼95%) ADP rather than ATP. In this case only a small amount of Pi was released as expected (Fig. 4C), but the inhibition of the pyrene fluorescence by H32K was similar as in the presence of ATP (data not shown). These observations led us to the conclusion that H32K-dependent attenuation of pyrene fluorescence must be associated with the blockage of a specific structural transition in the polymeric actin. Because neither ATP hydrolysis nor Pi release is inhibited by H32K, such an additional step in the actin polymerization must occur after the ATP hydrolysis and Pi dissociation. The pyrene probe on the actin subunits is affected by this transition and exhibits the observed fluorescence change.
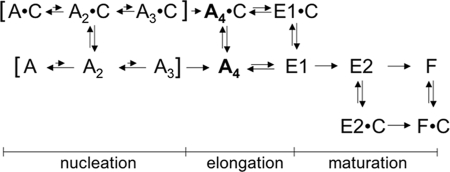
To explain the effects of H32K on the actin polymerization we have simulated numerically the kinetics of actin polymerization and the fluorescence change of pyrene-actin. Our goal was to design a minimum kinetic scheme that would explain three distinct features of the observed CaD effect: 1) accelerated actin nucleation; 2) inhibition of the pyrene-actin fluorescence increase without inhibition of actin polymerization; 3) acceleration of the fluorescence increase when CaD is added at a later phase of polymerization. These effects cannot be explained simultaneously within the framework of the classic nucleation-elongation scheme of actin polymerization. Additional states must be incorporated to satisfy all three boundary conditions. The simplest kinetic mechanism that we have obtained is shown in Scheme 1 where A, C, E, and F represent, respectively, monomeric actin, CaD (or H32K), nascent or young actin filament, and matured actin filament. The distributions of the main actin species during polymerization in the absence (Fig. 5A) and presence of CaD fragment (Fig. 5B) are calculated. Table 1 compiles the reactions and rate constants used in the simulation. The fluorescence signal of the pyrene probe (Pyr, dashed lines) is calculated using the following relative fluorescence coefficients: A, 1; E1, 5.1; E2, 7.9; F, 10; E1-CaD, 4.7; E2-CaD, 6.0; and F-CaD, 9.0. The value for F, the fluorescence increase associated with G- to F-actin transition is an average of published values (53, 54). Other coefficients are determined empirically by testing a range of values to simulate experimentally observed fluorescence changes.
SCHEME 1.
FIGURE 5.
Simulation of the time course of actin polymerization calculated with the program COPASI (27). A and B, time-dependent changes of the concentration of all species (solid curves; left ordinate) and pyrene fluorescence intensity (dashed curve; right ordinate) during actin polymerization in the absence (A) and presence (B) of CaD. Shown here is the distribution of actin in various forms (monomeric actin, A; the young actin filaments, E1 and E2; the mature actin filaments, F; or the CaD-bound young filaments, E1-CaD, and the CaD-bound mature filaments, F-CaD) during polymerization. The starting concentration of G-actin is set to 1.5 μm. All other species are present in negligible concentrations and are not shown. For simplicity the stoichiometry of actin-CaD binding is assumed to be 1:1 with respect to the actin monomer for all polymeric forms of actin. The fluorescence signal (Pyr) is calculated using the following assigned relative fluorescence coefficients: A, 1; E1, 5.1; E2, 7.9; F, 10; and for the CaD-bound species: E1-CaD, 4.7; E2-CaD, 6.0; and F-CaD, 9.0. C, simulated time course of fluorescence changes of pyrene-actin polymerized in the absence and presence of CaD. The kinetic scheme and assumptions for the modeling are described in the text and Table 1. The fluorescence signal is calculated using the same relative fluorescence coefficients as in A and B. Top black trace: actin alone; bottom trace: actin polymerized in the presence of CaD; other traces correspond to (from bottom up) CaD added after 10, 20, 30, and 40 min of actin polymerization, respectively. D, simulated fluorescence changes of pyrene-actin alone (top trace) and in the presence of H32K (bottom trace) with Ca2+/CaM added either at early (trace a) or late (trace b) stage of polymerization (see Fig. 3B and inset). The same set of binding parameters and fluorescence coefficients were used here as in A, B, and C, with additional parameters for the CaM-CaD interaction (Table 1).
TABLE 1.
Species, reactions, and rate constants used in the simulation
A1, A2, A3, and A4 represent the actin monomer, dimer, trimer, and tetramer, respectively. E1, E2, and F represent actin monomers incorporated into the various forms of filamentous actin, i.e. the intermediate “young” forms (E1, E2) and the final “mature” F–actin form (F; see Scheme 1). The units for the rate constants are s−1 for monomolecular reactions and ml·nmol−1·s−1 for bimolecular reactions. The actin tetramer A4 has a dual role in the simulation. It represents an intermediate species during the filament formation; it also represents the number of filaments. For this reason, its formation in reactions 3 and 10 is set to be irreversible. A4 participates in filament elongation (reaction 4 and as a complex with CaD in reaction 11) as a catalyst; i.e. although it is required for the reactions to proceed, its concentration does not change during these reactions. This simplification makes the rates of filament elongation dependent on the filament count [A4]. The simulation was performed with the program COPASI (Complex Pathway Simulator) in deterministic mode (27). Whenever possible, we used the rate constants similar to those published in the literature (16, 32). For the reactions that were not previously considered, we tested a range of values for each constant that yields the most reasonable approximation of the experiment. It should be pointed out that the obtained solutions are neither unique nor complete. In particular, we ignored the fact that one molecule of CaD affects the fluorescence, and hence the structure of several actin monomers in both the intermediate and final forms of the actin filament. Because of these simplifications, the presented model and the underlying simulation should be considered as a starting point for further refinement.
| Reaction | Rate constants |
||
|---|---|---|---|
| Forward | Reverse | ||
| Process | |||
| 1. Nucleation step 1 | A + A = A2 | 10 | 1 × 106 |
| 2. Nucleation step 2 | A2 + A = A3 | 10 | 20000 |
| 3. Filament formation | A3 + A -> A4 | 10 | 0 |
| 4. Elongation | A4 + A -> A4 + E1 | 14 | 0 |
| 5. Maturation 1 | E1 -> E2 | 0.003 | 0 |
| 6. Maturation 2 | E2 -> F | 0.0003 | 0 |
| Reactions involving CaD | |||
| 7. G-actin binding | A + CaD = A-CaD | 10 | 1000 |
| 8. Nucleation step 1 | A-CaD + A = A2-CaD | 10 | 1 × 105 |
| 9. Nucleation step 2 | A2-CaD + A = A3-CaD | 10 | 20000 |
| 10. Filament formation | A3-CaD + A -> A4-CaD | 10 | 0 |
| 11. Elongation | A4-CaD + A -> A4-CaD + E1 | 3.5 | 0 |
| 12. CaD binding to filament | A4 + CaD = A4-CaD | 10 | 0.1 |
| 13. CaD binding to E1 | E1 + CaD = E1-CaD | 10 | 0.01 |
| 14. CaD binding to E2 | E2 + CaD = E2-CaD | 10 | 5 |
| 15. CaD binding to F | F + CaD = F-CaD | 10 | 5 |
| 16. Maturation 2 | E2-CaD -> F-CaD | 0.01 | 0 |
| 17. CaD binding to CaM | CaD + CaM = CaD-CaM | 10 | 0.002 |
Besides the nucleation and elongation steps, this scheme includes a “filament maturation” process involving two irreversible transitions that precede the formation of the final F-actin form. We found that to replicate the “drop-rise” fluorescence changes, it was necessary to assume two intermediate species of filamentous actin (E1 and E2), each having distinct CaD binding and pyrene fluorescence properties. CaD binding to the E1 form of actin prevents the progression of filament maturation by “arresting” the filament at a low fluorescent state. In contrast, CaD binding to the E2 form accelerates the transition to the final F-actin-CaD complex that exhibits enhanced pyrene fluorescence. In addition, the scheme includes a weak interaction between the monomeric actin and CaD, which facilitates the nucleation steps, consistent with the early observation of Dabrowska and co-workers (2). By testing a range of values for the rate constants and fluorescence coefficients (Table 1), we were not only able to generate a series of fluorescence traces (Fig. 5C) that resemble the observed ones (see Fig. 2A), but also the fluorescence change upon addition of Ca2+/CaM (Fig. 5D, see Fig. 3B).
DISCUSSION
The major finding of this study is that CaD exerts multiple effects on the salt-induced polymerization of actin. The fluorescence increase of the actin-bound pyrene probe associated with the G- to F-actin transition is inhibited or accelerated by CaD depending on when it is added relative to the initiation of polymerization. The actual amounts of actin filaments and the total released inorganic phosphate, however, are not affected. This observation is best interpreted with the assumption that actin undergoes an obligatory structural transition after the initial filament formation and before attaining the final, matured F-actin state. We propose that CaD binds to and stabilizes both the intermediate and the final states of the actin filament (Scheme I). Binding of CaD to an early intermediate form of filamentous actin arrests the filament at a “young” stage that exhibits lower pyrene-actin fluorescence. However, once the filament formation proceeds beyond a critical step CaD binding accelerates the transition to the final F-actin state.
Our findings differ from an earlier report by Yamakita et al. (35), who tested the effect of CaD in a pyrene fluorescence-based actin polymerization assay, but did not observe any significant change. In our hands the full-length h-CaD behaved similarly to the C-terminal fragment. One plausible reason for the difference might be different phosphorylation levels of the CaD samples. This however seems unlikely, because (i) both unphosphorylated and ERK-phosphorylated CaD suppressed pyrene fluorescence enhancement in our hands (see Fig. 2); and (ii) the fact that CaD inhibited Arp2/3-induced actin polymerization in the earlier work indicated their CaD is indeed not phosphorylated. Instead, the difference could have stemmed from the CaD preparation itself. CaD is sensitive to proteolytic degradation (28) and also tends to denature upon heat treatment (24), which was used in the earlier work. We have found that aged CaD stocks lost potency of actin binding, and therefore exhibited only mild effects on the pyrene emission. Whether this is sufficient to explain the differences between our results and those of Yamakita et al. (35) requires further investigation.
Significantly, the observed CaD effects on the fluorescence of polymerizing pyrene-actin (Fig. 2A) could be simulated by numerical integration of a simple kinetic scheme (Scheme 1). The simulation not only successfully emulated both the inhibiting and accelerating effects of CaD on the pyrene-actin fluorescence, but also duplicated the observed CaM effect on the pyr-actin fluorescence (the two traces labeled a and b, Fig. 5D). A novel aspect of this scheme as compared with the classic nucleation-elongation mechanism is the inclusion of two intermediate states of filamentous actin, E1 and E2, which precede the final F-actin state. The two irreversible transitions E1 → E2 → F represent the filament maturation process. Inclusion of these steps was found necessary to generate the two opposing effects of CaD on pyrene-actin fluorescence. Without the second intermediate state E2, only the inhibition of fluorescence, but not the acceleration effect, could be simulated. The fact that similar CaD effects were observed in both ADP and ATP solutions, and that the amount of Pi generated was not affected by CaD (Fig. 4B), indicate that the maturation steps occur either after or independently of the ATP hydrolysis and the release of Pi. Thus the intermediate states in Scheme 1 are not directly related to the species in the conventional actin polymerization scheme, which is usually linked to the ATP hydrolysis. In this context it is important to point out that ATP hydrolysis is not required for actin polymerization, because actin containing ADP (36), or non-hydrolyzable ATP analogs (37), or even without nucleotides (38) can polymerize. The proposed “maturation” process is independent of ATP hydrolysis, and therefore represents a general property of polymerizing actin. One possible reason why this step was not considered previously might be because it occurs at a rate similar to that of Pi release (see above). By interacting with the intermediate states CaD blocks the transition, thus revealing this otherwise hidden process. Indeed, the affinity of CaD for the pre-transitional filament was found to be much higher than that for the final, conventional F-actin (Fig. 1B), strongly indicating two distinct types of filaments. According to our kinetic scheme, the 2-fold increase in pyrene-actin that has previously been associated with the conversion of F-actin·ATP to F-actin·ADP (32, 34) may correspond to the conformational change during the maturation process.
Whereas inclusion of the intermediate states E1, E2 accounts well for our experimental data, one has to question whether these states occur during normal actin polymerization, or perhaps they only result from CaD interference. There have been reports of irregular filament structure (the so-called “ragged” filaments) that occur during early phases of actin polymerization (39, 40). These “young” actin filaments were found to be less stable (41), and to form branches more readily (42). They were also found under a broad range of polymerization conditions including different salts, divalent cations, and nucleotides (43). The irregularity in both filament morphology and sensitivity to cross-linking with a bifunctional crosslinker N,N′-(1,4-phenylene)dimaleimide was found to dissipate as a function of time. It has been proposed that the initially irregular filament structure results from the incorporation of actin dimers having an incorrect (antiparallel) orientation (44). All these experiments were done in the absence of CaD. Thus, it appears that there is a natural occurring structural transition in actin during polymerization that takes place on a time scale consistent with our proposed filament maturation process (Scheme 1).
It has been suggested that the young state of actin filaments involves a “tilted” conformation of actin subunits (40), which is promoted either by an internal disulfide bond or by chemical modification of Cys-374, e.g. with tetramethylrhodamine (45). The young actin filaments are eventually converted to F-actin through some structural adjustments. Such a structural change may involve a twist of subdomains of actin from the G-actin configuration to the more flat F-actin configuration, as recently described by Oda et al. (46). It might also involve a change of the relative orientation of the monomers in the filament (40). Whereas supporting evidence for the concept of filament maturation during actin polymerization is growing, the nature of the intermediate conformational state(s) remains elusive, because they only exist transiently. Our finding that CaD arrests actin filaments at such an intermediate form provides an opportunity to examine this otherwise transient state. Searches of further structural evidence for these intermediates are currently underway. However, it should be pointed out that our interpretation for the observed fluorescence effect does not depend on such evidence, because the structural perturbation could be subtle, and may or may not be detectable with low resolution imaging.
The finding that H32K can interact with multiple forms of actin filaments is of particular interest. The C-terminal region of CaD exhibits little tertiary folding (47, 48) and is highly susceptible to proteolytic digestion (28); it easily collapses under rotary shadowing (49), and makes multiple contacts when docked on actin (50). All these properties suggest that H32K is intrinsically unstructured, thus sufficiently flexible to adapt to different topography of actin surface, permitting CaD to play a unique role in the cell with respect to the dynamic properties of the actin cytoskeleton.
As shown previously (40, 42), ADF/cofilin favors the young state of actin filaments. Arp2/3 also prefers to bind newly polymerized actin to form branches (42). Indeed, subtle structural modifications were found in the mother filament at the branch junction of the Arp2/3-actin complex (51). The “ragged” intermediate state of polymerized actin may provide such structural features and facilitate docking of Arp2/3. Binding of CaD to young actin filaments stabilizes this preferred structure and might further promote Arp2/3-mediated branching. Such a mechanism, if confirmed, would help to resolve the conundrum of the apparent low affinity between Arp2/3 and F-actin (52). Furthermore, binding of CaD affects the structure of neighboring actin subunits (Fig. 1B). This cooperative effect might allow CaD to promote Arp2/3 binding despite that the two proteins compete with each other for mature filaments (35). Finally, although unphosphorylated and ERK-phosphorylated H32K exhibited similar effects on actin polymerization, they have very different cellular distributions. In resting cells CaD is unphosphorylated and stays with the stress fibers in the cytosol, whereas upon stimulation, CaD becomes phosphorylated and translocates to the cell peripheries such as the leading edge and focal adhesions (6, 15). The different localization permits CaD and phospho-CaD to assume different functions. In the cytoplasm, unphosphorylated CaD stabilizes the matured actin cytoskeleton, whereas at the cell periphery, where the actin cytoskeleton undergoes dynamic remodeling, phosphorylated CaD inhibits filament maturation, thus promoting Arp2/3-mediated actin branching. It appears that the phosphorylation-dependent trafficking of CaD might enable it to contribute to the regulation of actin dynamics both spatially and temporally. Considering the importance of actin cytoskeleton in cell functions and the ubiquitous presence of CaD in eukaryotic cells, such a model could have profound implications.
Acknowledgments
We thank Dr. Sam Lehrer for helpful discussions and Dr. Hanna Strzelecka-Golaszewska for communicating results to us prior to publication and for many useful comments.
This work was supported, in whole or in part, by National Institutes of Health Grants HL91162 (to Z. G.) and HL92252 (to C. L. A. W.).
- CaD
- caldesmon
- ERK
- extracellular signal-regulated kinase
- CaM
- calmodulin
- H32K
- a C-terminal fragment of CaD
- pH32K
- ERK-phosphorylated H32K
- Pi
- inorganic phosphate.
REFERENCES
- 1.Mornet D., Harricane M. C., Audemard E. (1988) Biochem. Biophys. Res. Commun. 155, 808–815 [DOI] [PubMed] [Google Scholar]
- 2.Galazkiewicz B., Mossakowska M., Osińska H., Dabrowska R. (1985) FEBS Lett. 184, 144–149 [DOI] [PubMed] [Google Scholar]
- 3.Foster D. B., Huang R., Hatch V., Craig R., Graceffa P., Lehman W., Wang C. L. (2004) J. Biol. Chem. 279, 53387–53394 [DOI] [PubMed] [Google Scholar]
- 4.Smith C. W., Pritchard K., Marston S. B. (1987) J. Biol. Chem. 262, 116–122 [PubMed] [Google Scholar]
- 5.Graceffa P. (1987) FEBS Lett. 218, 139–142 [DOI] [PubMed] [Google Scholar]
- 6.Huang R., Cao G. J., Guo H., Kordowska J., Albert Wang C. L. (2006) Arch. Biochem. Biophys. 456, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphrey M. B., Herrera-Sosa H., Gonzalez G., Lee R., Bryan J. (1992) Gene 112, 197–204 [DOI] [PubMed] [Google Scholar]
- 8.Sobue K., Muramoto Y., Fujita M., Kakiuchi S. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 5652–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehman W., Denault D., Marston S. (1992) J. Muscle Res. Cell Motil. 13, 582–585 [DOI] [PubMed] [Google Scholar]
- 10.Wang C. L. (2001) Cell Biochem. Biophys. 35, 275–288 [DOI] [PubMed] [Google Scholar]
- 11.Bretscher A., Lynch W. (1985) J. Cell Biol. 100, 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumura F., Yamashiro S. (1993) Curr. Opin. Cell Biol. 5, 70–76 [DOI] [PubMed] [Google Scholar]
- 13.Wang Z., Jiang H., Yang Z. Q., Chacko S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11899–11904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marston S. B., Huber P. A. J. (eds) (1996) Caldesmon, Academic Press, Inc., San Diego, CA [Google Scholar]
- 15.Kordowska J., Hetrick T., Adam L. P., Wang C. L. (2006) Exp. Cell Res. 312, 95–110 [DOI] [PubMed] [Google Scholar]
- 16.Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 17.Winder S. J. (2003) Curr. Opin. Cell Biol. 15, 14–22 [DOI] [PubMed] [Google Scholar]
- 18.Discher D. E., Janmey P., Wang Y. L. (2005) Science 310, 1139–1143 [DOI] [PubMed] [Google Scholar]
- 19.Gunst S. J., Fredberg J. J. (2003) J. Appl. Physiol. 95, 413–425 [DOI] [PubMed] [Google Scholar]
- 20.Seow C. Y. (2005) Nature 435, 1172–1173 [DOI] [PubMed] [Google Scholar]
- 21.Bursac P., Lenormand G., Fabry B., Oliver M., Weitz D. A., Viasnoff V., Butler J. P., Fredberg J. J. (2005) Nat. Mater. 4, 557–561 [DOI] [PubMed] [Google Scholar]
- 22.Gerthoffer W. T. (2007) Circ. Res. 100, 607–621 [DOI] [PubMed] [Google Scholar]
- 23.Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 24.Zhuang S., Mabuchi K., Wang C. L. (1996) J. Biol. Chem. 271, 30242–30248 [DOI] [PubMed] [Google Scholar]
- 25.Huang R., Li L., Guo H., Wang C. L. (2003) Biochemistry 42, 2513–2523 [DOI] [PubMed] [Google Scholar]
- 26.Webb M. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoops S., Sahle S., Gauges R., Lee C., Pahle J., Simus N., Singhal M., Xu L., Mendes P., Kummer U. (2006) Bioinformatics 22, 3067–3074 [DOI] [PubMed] [Google Scholar]
- 28.Wang C. L., Wang L. W., Xu S. A., Lu R. C., Saavedra-Alanis V., Bryan J. (1991) J. Biol. Chem. 266, 9166–9172 [PubMed] [Google Scholar]
- 29.Velaz L., Hemric M. E., Benson C. E., Chalovich J. M. (1989) J. Biol. Chem. 264, 9602–9610 [PubMed] [Google Scholar]
- 30.Crosbie R. H., Miller C., Chalovich J. M., Rubenstein P. A., Reisler E. (1994) Biochemistry 33, 3210–3216 [DOI] [PubMed] [Google Scholar]
- 31.Carlier M. F., Pantaloni D. (1986) Biochemistry 25, 7789–7792 [DOI] [PubMed] [Google Scholar]
- 32.Brooks F. J., Carlsson A. E. (2008) Biophys. J. 95, 1050–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korn E. D., Carlier M. F., Pantaloni D. (1987) Science 238, 638–644 [DOI] [PubMed] [Google Scholar]
- 34.Carlier M. F., Pantaloni D., Korn E. D. (1984) J. Biol. Chem. 259, 9983–9986 [PubMed] [Google Scholar]
- 35.Yamakita Y., Oosawa F., Yamashiro S., Matsumura F. (2003) J. Biol. Chem. 278, 17937–17944 [DOI] [PubMed] [Google Scholar]
- 36.Hayashi T., Rosenbluth R. (1962) Biochem. Biophys. Res. Commun. 8, 20–23 [DOI] [PubMed] [Google Scholar]
- 37.Cooke R., Murdoch L. (1973) Biochemistry 12, 3927–3932 [DOI] [PubMed] [Google Scholar]
- 38.De La Cruz E. M., Mandinova A., Steinmetz M. O., Stoffler D., Aebi U., Pollard T. D. (2000) J. Mol. Biol. 295, 517–526 [DOI] [PubMed] [Google Scholar]
- 39.Millonig R., Salvo H., Aebi U. (1988) J. Cell Biol. 106, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orlova A., Shvetsov A., Galkin V. E., Kudryashov D. S., Rubenstein P. A., Egelman E. H., Reisler E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17664–17668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kueh H. Y., Brieher W. M., Mitchison T. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16531–16536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ichetovkin I., Grant W., Condeelis J. (2002) Current Biology 12, 79–84 [DOI] [PubMed] [Google Scholar]
- 43.Galińska-Rakoczy A., Wawro B., Strzelecka-Golaszewska H. (2009) J. Mol. Biol. 387, 869–882 [DOI] [PubMed] [Google Scholar]
- 44.Steinmetz M. O., Goldie K. N., Aebi U. (1997) J. Cell Biol. 138, 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudryashov D. S., Phillips M., Reisler E. (2004) Biophys J 87, 1136–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oda T., Iwasa M., Aihara T., Maéda Y., Narita A. (2009) Nature 457, 441–445 [DOI] [PubMed] [Google Scholar]
- 47.Levine B. A., Moir A. J., Audemard E., Mornet D., Patchell V. B., Perry S. V. (1990) Eur. J. Biochem. 193, 687–696 [DOI] [PubMed] [Google Scholar]
- 48.Wang C. L., Chalovich J. M., Graceffa P., Lu R. C., Mabuchi K., Stafford W. F. (1991) J. Biol. Chem. 266, 13958–13963 [PMC free article] [PubMed] [Google Scholar]
- 49.Mabuchi K., Wang C. L. (1991) J. Muscle Res. Cell Motil. 12, 145–151 [DOI] [PubMed] [Google Scholar]
- 50.Gao Y., Patchell V. B., Huber P. A., Copeland O., El-Mezgueldi M., Fattoum A., Calas B., Thorsted P. B., Marston S. B., Levine B. A. (1999) Biochemistry 38, 15459–15469 [DOI] [PubMed] [Google Scholar]
- 51.Rouiller I., Xu X. P., Amann K. J., Egile C., Nickell S., Nicastro D., Li R., Pollard T. D., Volkmann N., Hanein D. (2008) J. Cell Biol. 180, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beltzner C. C., Pollard T. D. (2008) J. Biol. Chem. 283, 7135–7144 [DOI] [PubMed] [Google Scholar]
- 53.Kouyama T., Mihashi K. (1981) Eur. J. Biochem. 114, 33–38 [PubMed] [Google Scholar]
- 54.Cooper J. A., Walker S. B., Pollard T. D. (1983) J. Muscle Res. Cell Motil. 4, 253–262 [DOI] [PubMed] [Google Scholar]