Abstract
Doxorubicin (DOX) is a potent anti-tumor drug known to cause heart failure. The transcription factor GATA4 antagonizes DOX-induced cardiotoxicity. However, the protective mechanism remains obscure. Autophagy is the primary cellular pathway for lysosomal degradation of long-lived proteins and organelles, and its activation could be either protective or detrimental depending on specific pathophysiological conditions. Here we investigated the ability of GATA4 to inhibit autophagy as a potential mechanism underlying its protection against DOX toxicity in cultured neonatal rat cardiomyocytes. DOX markedly increased autophagic flux in cardiomyocytes as indicated by the difference in protein levels of LC3-II (microtubule-associated protein light chain 3 form 2) or numbers of autophagic vacuoles in the absence and presence of the lysosomal inhibitor bafilomycin A1. DOX-induced cardiomyocyte death determined by multiple assays was aggravated by a drug or genetic approach that activates autophagy, but it was attenuated by manipulations that inhibit autophagy, suggesting that autophagy contributes to DOX cardiotoxicity. DOX treatment depleted GATA4 protein levels, which predisposed cardiomyocytes to DOX toxicity. Indeed, GATA4 gene silencing triggered autophagy that rendered DOX more toxic, whereas GATA4 overexpression inhibited DOX-induced autophagy, reducing cardiomyocyte death. Mechanistically, GATA4 up-regulated gene expression of the survival factor Bcl2 and suppressed DOX-induced activation of autophagy-related genes, which may likely be responsible for the anti-apoptotic and anti-autophagic effects of GATA4. Together, these findings suggest that activation of autophagy mediates DOX cardiotoxicity, and preservation of GATA4 attenuates DOX cardiotoxicity by inhibiting autophagy through modulation of the expression of Bcl2 and autophagy-related genes.
Keywords: Cell/Apoptosis, Gene/Regulation, Toxins/Drugs/Xenobiotics/Drug Action, Transcription/Target genes, GATA4, autophagy, cardiomyocyte, doxorubicin
Introduction
Doxorubicin (DOX)4 is a very effective anti-cancer drug with cardiotoxicity that culminates in congestive heart failure (1–3). The prevailing mechanism for DOX cardiotoxicity is oxidative stress that has been supported by the ability of several antioxidants to reduce DOX cardiotoxicity in animal studies (4–8). Unfortunately, clinical trials have failed to reproduce these results in humans (9), suggesting that mechanisms other than oxidative stress might also contribute to DOX-induced heart failure (9–13). In support of this notion, DOX has been shown to cause cardiac mitochondrial injury by both oxidative and non-oxidative mechanisms (14).
Autophagy is the major cellular pathway for degrading and recycling long-lived proteins and organelles that are sequestered in double-membrane vesicles termed autophagosomes. After fusing with the lysosome to form autolysosome, the inner membrane and the contents are degraded and recycled. More than 30 autophagy-related (ATG) genes encode proteins essential for autophagy induction and the generation, maturation, and recycling of autophagosomes or autophagic vacuoles (15, 16). Autophagy has long been recognized to provide a survival pathway that allows the cell to maintain energy homeostasis under starvation conditions (15). Indeed, autophagy-derived energy is required for the survival of neonatal mice deprived of milk right after birth (17). Also, autophagy functions as a cytoplasmic quality control mechanism to remove protein aggregates and damaged organelles. In this respect autophagy has been shown to play an extremely important role in cardiac homeostasis as the inactivation of autophagy gene ATG5 in adults results in myocardial dysfunction (18), and heterozygous disruption of Beclin 1 (ATG6) accelerates heart failure and mortality in desmin-related cardiomyopathic mice (19). In addition, autophagy activation during ischemia stage appears to be cardioprotective (20, 21). These observations demonstrate a beneficial role of autophagy in these specific contexts. However, increased autophagic activity could also be detrimental to the heart under certain pathological conditions (22). For example, diphtheria toxin induces autophagy and triggers heart failure in mice (23). Most intriguingly, the same Beclin 1 knock-out mice that show accelerated heart failure in desmin-related cardiomyopathy (19) display reduced cardiac damage under ischemia-reperfusion or pressure overload condition (20, 24). Together, these results clearly demonstrate that autophagy is a double-edged sword that could be either protective or detrimental depending on the nature of the stimuli and the levels of autophagy induced (20, 25). Because of the dichotomous nature of autophagy in cardiomyocyte survival and death, it is very clear that the functional significance of autophagy under different cardiac conditions has to be individually determined.
The characteristic features of DOX-induced cardiomyopathy are the loss of myofibrils and the vacuolization of cardiac myocytes (2). This is associated with dramatically reduced levels of various long-lived and short-lived proteins including the transcription factor GATA4 (26–28) and structural proteins titin (29) and myosin heavy chain (28). These observations suggest that DOX may activate cellular degradation pathways. Indeed, DOX is able to increase the cardiac activities of calpain (29) and the ubiquitin proteasome system (30). DOX also causes accumulation of autophagic vacuoles (31) in the heart. Importantly, 3-methyadenine (3-MA), an autophagy inhibitor, can reduce DOX-induced cardiomyocyte death (32). This result suggests that autophagy induction may contribute to DOX cardiotoxicity, although it remains unknown if the protective effects of 3-MA are mediated exclusively through autophagy inhibition as the specificity of 3-MA is questionable (33).
Previous studies have shed light on both positive and negative regulatory mechanisms of autophagy (16, 34–36). Forming a complex with several partners, including Beclin 1/Atg6, UVRAG, Ambra1, and Bif-1, the class III phosphatidylinositol 3-kinase is required for autophagy induction. Conversely, the class I phosphatidylinositol 3-kinase-Akt/PKB-mTOR pathway suppresses autophagy by inactivating Atg1 protein kinase. The initiation of autophagy is also negatively controlled by the phosphoinositide 3-phosphatase jumpy (37). The class III phosphatidylinositol 3-kinase inhibitor 3-MA and the mTOR (mammalian target of rapamycin) inhibitor rapamycin have been widely used to inhibit and activate autophagy, respectively. As an energy sensor, the AMP-activated protein kinase stimulates autophagy likely through inhibiting mTOR signaling. Interestingly, the antiapoptotic protein Bcl2 can inhibit autophagy through binding and sequestering Beclin 1, leading to the notion that the interaction between Bcl-2 and Beclin 1 may function as a rheostat that maintains autophagy at levels that are compatible with cell survival rather than cell death (38).
The transcription factor GATA4 regulates the expression of various cardiac genes ranging from contractile proteins to peptide hormones and transcription factors (39, 40). GATA4 plays an important role in cardiac adaptive responses including cardiac hypertrophy and survival (40–44). Importantly, genetic deletion of GATA4 in the heart induces apoptosis and impairs cardiac function (44), underscoring the requirement of GATA4 in maintaining cardiac homeostasis. Previous studies demonstrate that DOX depletes GATA4 transcript and protein levels in cardiomyocytes (26, 27). Preservation of GATA4 levels by the α1-adrenergic agonist phenylephrine or overexpression of GATA4 by adenovirus-mediated gene transfer prevents DOX-induced cardiomyocyte death (26, 27, 45). The protective effect of GATA4 against DOX cardiotoxicity is mediated at least in part by its ability to up-regulate the gene expression of Bcl2 (46), a survival factor that inhibits both apoptosis and autophagy. However, it is largely unknown if GATA4 is able to regulate autophagic activity as one of the mechanisms by which GATA4 attenuates DOX cardiotoxicity.
In the present study we examined the roles of autophagy and GATA4 in DOX cardiotoxicity. Our results demonstrate that DOX dramatically increases autophagic flux in cardiomyocytes, which contributes to DOX cardiotoxicity. Increased GATA4 expression inhibits DOX-induced autophagy and reduces cardiomyocyte death, whereas GATA4 gene silencing triggers autophagy and renders DOX more toxic, suggesting that the cardioprotective effect of GATA4 can be explained partly by its ability to inhibit DOX-induced autophagic activity through modulating the expression of Bcl2 and autophagy-related genes.
EXPERIMENTAL PROCEDURES
Cardiomyocyte Cultures
Neonatal rat ventricular cardiomyocytes were isolated from 0–2-day-old Harlan Sprague-Dawley rat neonates using trypsin and DNase. The crude trypsin was used to dissociate the heart tissues, and DNase was used to prevent cell clumping by digesting sticky DNA released from lysed cells. After digestion of tissue, cells were preplated for 1 h to remove non-myocytes and then plated on gelatinized cell culture dishes and cultured overnight in Dulbecco's modified Eagle's medium (DMEM) with 15% bovine serum and penicillin/streptomycin (100 units/ml). The following day the cells were cultured in DMEM medium with 1% bovine serum unless otherwise indicated. All media contain penicillin and streptomycin (100 units/ml) as well as 100 μm 5-bromo-2′-deoxyuridine (BrdUrd, Sigma). BrdUrd was used to inhibit the growth of contaminating nonmyocytes including fibroblasts.
Drug Treatments
DOX and 3-MA were purchased from Sigma. Rapamycin (Rap) and bafilomycin A1 (BFA) were obtained from LC laboratories. DOX was dissolved in saline and used at 1 μm to induce autophagy and cardiomyocyte death. Rap is a well characterized inhibitor of mTOR complex 1 that can induce autophagy. Rap was dissolved in ethanol and used at 100 nm. 3-MA is a class III phosphatidylinositol 3-kinase inhibitor that inhibits autophagy initiation and was used at 2.5 mm. This dose of 3-MA effectively inhibits autophagy without causing cell injury and is much smaller than 10 mm, which was used in most cell culture studies and may have a number of nonspecific effects (33). BFA is a potent and specific inhibitor of vacuolar proton ATPase that prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes. BFA (50 nm in dimethyl sulfoxide, DMSO) was added to culture dishes 6 h before harvesting cells for measuring autophagic flux.
Western Blot Analysis
Protein extracts from cultured cardiomyocytes were prepared as described previously (47). Protein samples were subjected to polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (GE Healthcare), and blocked in 5% milk for 1 h. We purchased primary antibodies from several vendors including Abgent (SQSTM1/p62), Research Diagnostics (GAPDH), Santa Cruz (GATA4, Bcl2, and Beclin 1), and Cell Signaling, MA (LC3, cleaved caspase 3, poly(ADP-ribose) polymerase (PARP), Akt, phospho-Akt-Ser-473, phospho-Akt-Thr-308, FoxO1, phospho-FoxO1/3a, mTOR, phospho-mTOR-Ser-2448, p70 S6 kinase, phospho-p70 S6 kinase-Thr-389, 4E-BP1, and phospho-4E-BP1-Thr-37/46). Primary antibodies were incubated overnight at 4 °C in 2.5% milk. A horseradish peroxidase-conjugated secondary antibody was incubated for 1 h at room temperature in 2.5% milk and processed for chemiluminescent detection using an ECL Advanced Western blotting kit (GE Healthcare). Protein abundance on Western blots was quantified by densitometry with the Quantity One program from Bio-Rad.
Immunoprecipitation (IP)
Cardiomyocytes were lysed and sonicated in IP buffer: 40 mm Tris-HCl (pH 7.4), 137 mm NaCl, 25 mm sodium β-glycerophosphate, 2 mm sodium pyrophosphate, 2 mm EDTA, 1 mm sodium vanadate, 10% glycerol containing 1% Triton X-100, and Protease Inhibitor Mixture (Sigma). Homogenates were cleared by centrifugation for 5 min at 15,000 × g at 4 °C. For immunoprecipitation, supernatants containing 100 μg of proteins were incubated with anti-Bcl2 monoclonal antibody (Santa Cruz) for 16 h followed by incubation with Protein A/G-agarose beads (Santa Cruz) for 1 h. Beads were washed with IP buffer three times. Precipitants were subjected to immunoblotting using anti-Beclin 1 antibody.
Construction and Utilization of Replication-deficient Adenoviruses
Adenovirus expressing β-galactosidase (Adβgal) or GATA4 (AdG4) has been described previously (41). Adenovirus harboring Bcl2 (AdBcl2) was purchased from Vector Biolabs (Philadelphia, PA). Adenovirus encoding GFP-LC3 was kindly provided by Dr. Tolkovsky (48). Adenovirus expressing short hairpin RNA targeted at mouse and rat GATA4 (AdshG4) was created as described (46). Control short hairpin RNA (shRNA) adenovirus (AdshCon) was generated by cloning an small interfering RNA (siRNA) sequence not having any predicted sequence matches to coding regions in mouse or rat genomes (46). Adenovirus encoding Beclin 1 (AdBCN) was constructed using AdEasy Adenoviral Vector System (Stratagene). The FLAG-tagged human Beclin 1 cDNA was provided by Dr. Yuan (49) and cloned into Pshutle vector, which then underwent homologous recombination with adenoviral genome lacking E1 and E3 sequences. The recombinant was packaged and amplified in AD-293 cells. We used the BLOCK-iT Adenoviral RNAi Expression System (Invitrogen) to construct the adenoviral vector that harbors the shRNA targeting rat Beclin 1 mRNA (AdshBCN). Briefly, we cloned double-stranded oligonucleotide duplexes encoding rat-specific shBCN1 into RNAi entry vector (pENTR/U6). The shBCN1 duplexes contain oligos CACCGCTCAGTACCAGCGAGAATATCGAAATATTCTCGCTGGTACTGAGC and AAAAGCTCAGTACCAGCGAGAATATTTCGATATTCTCGCTGGTACTGAGC (the underline indicates sequences directed against Beclin 1). LR clonase was then used to transfer the shBCN1 expressing cassette to the promoterless Gateway destination cloning vector pAd/BLOCK-iT-DEST. The pAd/BLOCK-iT-DEST vector containing the shBCN1 expression cassette was transfected into 293A cells to produce the replication-incompetent adenovirus (AdshBCN).
Unless otherwise indicated, cardiomyocytes were infected with each adenovirus at a multiplicity of infection of 100 plaque-forming units for 2 h and then cultured in DMEM media containing 1% bovine serum for an additional 24 h in the case of AdG4, AdBCN, AdBcl2, AdGFP-LC3, or Adβgal and 48 h in the case of AdshG4, AdshBCN, or AdshCon before treatment with DOX and other drugs.
Gene Silencing with siRNA
Two different pre-designed siRNAs targeting rat Bcl2 mRNA (1 and 2) and a Silencer Negative Control siRNA #1 (AM4635) were obtained from Ambion (Austin, TX). siRNA transfection was performed using Lipofectamine RNAiMAX according to the manufacturer's instructions (Invitrogen). Briefly, 0.7 × 106 cardiomyocytes were transfected in 3 ml of serum-free and antibiotics-free DMEM containing 500 μl of Opti-MEM (Invitrogen), 6 μl of Lipofectamine RNAiMAX, and 50 nmol/liter concentrations of each siRNA. The media were replaced 24 h later with fresh serum-free DMEM. After another 36 h, the cells were harvested, and gene silencing efficiency was evaluated.
Cell Death and Survival Assays
Cardiomyocyte death was measured by staining with propidium iodide (PI, 1 μg/ml, Roche Applied Science) that was added directly to the culture medium. PI enters dead cells through disrupted membranes to bind to DNA. Myocytes were photographed under both phase contrast and fluorescent conditions. The PI-positive cells (red) were expressed as a percentage of the total number of cells (200 to 250 counted under phase contrast). Cell survival was determined with a thiazolyl blue tetrazolium bromide (MTT) assay as described (50). Briefly, cardiomyocytes were seeded at a density of 10,000 cells/well in 96-well plates. After all treatments, 20 μl of MTT (M5655 Sigma, 5 mg/ml in phosphate-buffered saline) was added to each well. The cells were incubated for 3 h. At this time the media were removed from each well, and 100 μl of DMSO was added and incubated for 5 min. The absorbance, which is directly proportional to the number of living cells in the culture, was measured at 550 nm using a Thermomax microplate reader from Molecular Devices. A blank with DMSO alone was taken and subtracted from all values. The number of viable cells was calculated with a standard curve derived from a suite of wells with given numbers of cells.
Apoptosis Assays
DOX-induced apoptosis was determined by DNA laddering assay and the cleavage of caspase 3 and PARP. For DNA laddering assay, the cells were harvested, and DNA was extracted. A semiquantitative PCR-based DNA laddering kit from Maxim Biotech, Inc. (San Francisco, CA) was used to assess the degree of apoptosis. Cleaved caspase 3 and PARP were determined by Western blot analysis.
Semiquantitative Reverse Transcriptase-PCR
The reverse transcriptase-PCR was carried out using the TaqMan reverse transcription reagents (Applied Biosystems, Foster city, CA). Briefly, 1 μg of total RNA isolated with TRIZOL reagent (Invitrogen) from cardiomyocytes was used for first-strand cDNA synthesis in a 20-μl reaction volume containing 50 pmol of oligo(dT)16 as the primer. Then 2 μl of the reverse transcription reaction was used for PCR amplification in a 50-μl volume. Aliquots of PCR reaction were taken at different cycles for agarose gel analysis to determine the linear range of amplification. The gene-specific primers for GATA4, Bcl2, ATG7, Beclin 1, or GAPDH were derived from mouse mRNA sequences, but they are 100% homologous to rat sequences, some of which were described previously (46). The primer sequences for ATG5 and ATG12 are designed based on rat mRNA sequences. The primer sequences are as follows: ATG5 forward (5′-CCCTCCAGAAGAAAATGGAT) and reverse (5′-ATAGCTCAGATGCTCGCTCA); ATG12 forward (5′-TTCGGTTGCAGTTTCGCC) and reverse (5′-CCATGCCTGTGATTTGCAGTA); ATG7 forward (5′-TGTCAGCCTGGCATTTGATAA) and reverse (5′-TCACTCATGTCCCAGATCTCA); Beclin 1 forward (5′-GACAAATCTAAGGAGTTGCCG) and reverse (5′-AGAACTGTGAGGACACCCA).
Statistical Analysis
Quantitative data were presented as the means ± S.E. Differences between experimental groups were evaluated for statistical significance using one-way or two-way analysis of variance (ANOVA) followed by the Tukey post-test using Prism software (GraphPad). Student's t tests for paired data were also used in some cases as indicated. p values < 0.05 were considered statistically significant.
RESULTS
DOX Induced Autophagy in Cardiomyocytes
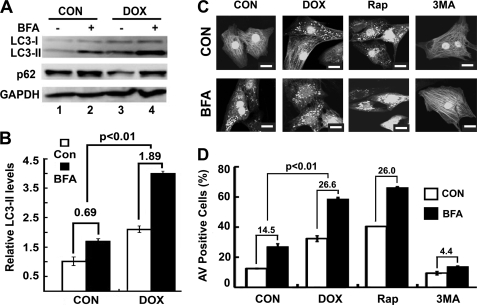
DOX-induced cardiotoxicity is associated with the depletion of a variety of long- and short-lived proteins (26–29, 51, 52), indicating an activation of protein degradation systems including the ubiquitin proteasome system (30) and autophagy (31). When autophagy is induced, microtubule-associated protein light chain 3 (LC3), encoded by autophagy-related gene ATG8, is processed from LC3-I (18 kDa) to LC3-II (16 kDa) and incorporated into autophagic vacuoles (AVs) (53). The levels of LC3-II are proportional to the number of accumulated AVs. To determine whether DOX could induce autophagy in neonatal rat ventricular cardiomyocytes, we cultured the cells in DMEM with 1% bovine serum, treated the cells with DOX (1 μm) for 18 h, and then measured LC3-II levels. Indeed, Western blot analysis showed an increased formation of LC3-II after DOX treatment, suggesting autophagy activation (Fig. 1A, compare lanes 1 and 3). Because the steady state levels of LC3-II or the number of AVs are the net effect of both AV formation and degradation, a more accurate measurement of autophagy activity is autophagic flux. It reflects the number of AVs that are delivered to and degraded in the lysosome and can be measured by the difference in numbers of AVs or levels of LC3-II protein in the absence and presence of lysosomal inhibitors (54). To determine whether DOX-induced accumulation of LC3-II is caused by enhanced autophagic flux rather than an impaired degradation, we treated cells with DOX in the absence or presence of the lysosomal inhibitor BFA. As shown in Fig. 1A, steady state levels of LC3-II were increased in DOX-treated cells compared with those in untreated cells (lane 3 versus lane 1). Further treatment with BFA led to an even larger increase of LC3-II in DOX-treated cells than in control cells (lane 4 versus lane 2), indicating that DOX accelerated autophagic flux in cardiomyocytes, as quantified by the difference of LC3-II in the absence and presence of BFA (Fig. 1B, control 0.69 ± 0.10 versus DOX 1.89 ± 0.02, p < 0.01, n = 3). We also determined p62 protein levels to assess autophagic flux. p62/SQSTM1 is a polyubiquitin-binding protein that is degraded by autophagy, and its protein levels are inversely related to autophagy activity (18, 54, 55). As shown in Fig. 1A, DOX decreased p62 levels. However, BFA treatment led to an accumulation of p62 due to the blockage of lysosomal degradation, further supporting the conclusion that DOX increased autophagic activity.
FIGURE 1.
DOX induced autophagy in cardiomyocytes. Neonatal rat cardiomyocytes were cultured in DMEM with 1% bovine serum and treated with DOX (1 μm) for 18 h. A, Western blots show protein levels of LC3-II and p62 in the absence and presence of the lysosome inhibitor BFA (50 nm). B, DOX increased autophagic flux (DOX 1.89 ± 0.02 versus control (CON) 0.69 ± 0.10), which is defined by the difference in LC3-II protein levels in the absence and presence of BFA. Data are expressed as the mean ± S.E. LC3-II levels were analyzed by 2-way ANOVA, and autophagic flux was analyzed by paired Student's t tests (n = 4). C, DOX induced accumulation of AVs. Cardiomyocytes were infected with AdGFP-LC3 and treated with DOX or saline in the absence or presence of BFA. Upon DOX treatment, GFP-LC3 formed punctate structures or dots, indicative of AVs. The green fluorescent images were converted to black/white for clarity. The scale bar is 20 μm. D, DOX increased autophagic flux as calculated by AV-positive cells with and without BFA (DOX 26.6 ± 1.9 versus CON 14.5 ± 0.4). The autophagic flux is the difference in the number of AV positive cells in the absence and presence of BFA. The AV positive cells were expressed as the percentage of cells that have >40 GFP-LC3 dots over the number of total GFP-expressing cells examined. Roughly 200 cells/6-cm plate were counted. Data were expressed as the mean ± S.E., and autophagic flux was analyzed by paired Student's t tests (n = 3). Rap and 3-MA were used as known autophagy inducer and inhibitor, respectively.
In addition, DOX-induced autophagic flux was also determined using the GFP-LC3 fusion protein, an autophagy reporter widely used to visualize AV formation upon autophagy induction (33, 56). We infected cardiomyocytes with an adenovirus expressing GFP-LC3 and treated them with DOX or saline in the absence or presence of BFA. Images of GFP-LC3 were captured at 600× using an FV1000 Olympus confocal microscope. In saline-treated control cardiomyocytes, GFP-LC3 showed a diffuse staining pattern. However, upon DOX treatment, GFP-LC3 displayed numerous punctate structures or dots in the cytoplasm that are indicative of AV formation (Fig. 1C). BFA treatment not only induced accumulation of AVs in control cells but also caused a further increase in the number of AVs in DOX-treated cells (Fig. 1C), suggesting enhanced autophagic flux by DOX. We counted cells with more than 40 GFP-LC3 dots, an arbitrary cut-off number that defines a cell as positive for increased AVs. At base line, there were 12.5 ± 0.21% of AV-positive cells, which was increased to 27 ± 1.9% by BFA, resulting in an autophagic flux of 14.5. By contrast, DOX caused 32.3 ± 1.9% of AV-positive cells, which was further elevated to 58.9 ± 0.9% by BFA, giving rise to a flux of 26.6, almost doubled as compared with base-line flux (Fig. 1D). Together, these results clearly demonstrated that DOX increased AVs by promoting its formation but not by blocking its degradation. As a positive control, Rap increased GFP-LC3 dots, which was further elevated by BFA, whereas 3-MA, a class III phosphatidylinositol 3-kinase inhibitor, inhibited AV accumulation with or without BFA treatment (Fig. 1D).
Enhanced Autophagy Contributed to DOX-induced Cardiomyocyte Death
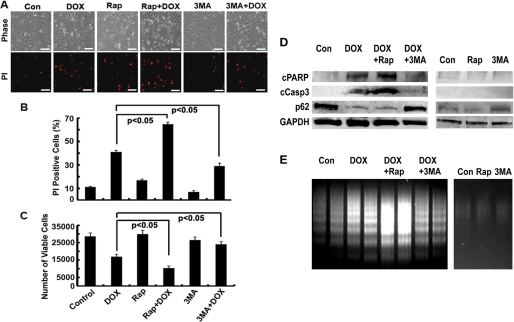
Autophagy could be either protective or detrimental depending on several factors, especially the nature and intensity of the stimuli (20, 25). To determine whether DOX-induced autophagy is an adaptive survival response or a part of mechanism that mediates DOX cardiotoxicity, we manipulated autophagic activity using either pharmacologic or genetic approaches. We then evaluated DOX-induced cardiomyocyte death (Fig. 2) by PI staining, which estimates the number of dead cells regardless of the cause of death, and an MTT assay, which measures the number of cells that survive DOX toxicity. Apoptosis was determined by DNA laddering and cleaved PARP and caspase 3. As expected, DOX treatment (1 μm for 18 h) led to increased PI-positive cells (Fig. 2, A and B), decreased viable cells (Fig. 2C), increased cleavage of PARP and caspase 3 (Fig. 2D), and enhanced DNA laddering (Fig. 2E), demonstrating the sufficiency of DOX to induce both necrosis and apoptosis in cardiomyocytes. However, DOX-induced cell death was markedly exacerbated by Rap (100 nm), an autophagy inducer, and attenuated by 3-MA (2.5 mm), an autophagy inhibitor. Rap or 3-MA was added at the same time with DOX to the culture dish. Neither Rap nor 3-MA alone induced cell death at the dose used despite the ability of either to enhance and inhibit DOX-induced autophagy, respectively, as shown by p62 protein levels (Fig. 2D). These results suggest that activation of autophagy is detrimental contributing to DOX cardiotoxicity.
FIGURE 2.
Rap enhanced DOX toxicity, whereas 3-MA attenuated it. Cardiomyocyte were treated with DOX and Rap (100 nm) or 3-MA (2.5 mm) for 18 h, and cardiomyocyte death was determined by PI staining (A and B), MTT assay (C), cleavage of PARP (cPARP) and caspase 3 (cCasp3, D), and DNA laddering (E). Data in B and C were expressed as mean ± S.E. and analyzed by one-way ANOVA (n = 3). The scale bars in A represent 200 μm.
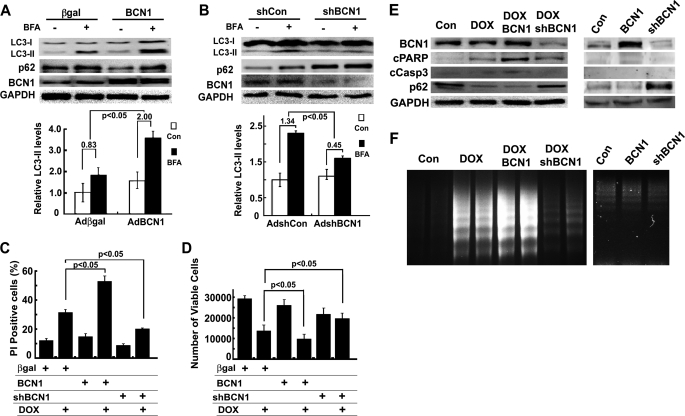
To eliminate the potential nonspecific effects of Rap and 3-MA, we used adenoviral vectors to deliver a FLAG-tagged human Beclin 1 cDNA (AdBCN) or a shRNA against rat Beclin 1 mRNA (AdshBCN) to genetically achieve gain- or loss-of-function of autophagy in cardiomyocytes. Adenovirus expressing β-galactosidase or a scrambled short hairpin RNA (shCon) was used as the control. Infection of cardiomyocytes with AdBCN or AdshBCN led to increased (Fig. 3A) or diminished (Fig. 3B) protein levels of Beclin 1, which was associated with enhanced or inhibited autophagic flux, respectively, as indicated by the difference in protein levels of LC3-II and p62 in the absence and presence of BFA (β-galactosidase 0.83 ± 0.30 versus BCN 2.00 ± 0.34, p < 0.05, n = 6; shCon 1.34 ± 0.23 versus shBCN 0.45 ± 0.33, p < 0.05, n = 3). Autophagic flux was also determined in cardiomyocytes co-infected with AdGFP-LC3 and one of the four adenoviruses expressing β-galactosidase, BCN, shCon, or shBCN. AdBCN increased the number of GFP-LC3 dots at base line, which was further elevated by BFA treatment. By contrast, AdshBCN reduced the number of GFP-LC3 dots regardless of BFA treatment (data not shown), consistent with autophagic flux measured with endogenous LC3-II. Remarkably, although AdBCN or AdshBCN alone did not affect cell viability at base line, DOX-induced cardiomyocyte death was notably aggravated by AdBCN-directed overexpression of Beclin 1 and attenuated by AdshBCN-mediated Beclin 1 gene knockdown as shown by PI-positive cells (Fig. 3C), MTT assay (Fig. 3D), cleavage of PARP and caspase 3 (Fig. 3E), and DNA laddering (Fig. 3F). These results lend further support to the conclusion that DOX-induced autophagy is detrimental and contributes to DOX-induced cardiomyocyte death.
FIGURE 3.
Increased Beclin 1 (BCN1) aggravated DOX toxicity, but knockdown BCN1 reduced it. Cardiomyocytes were infected with AdBCN or AdshBCN for 24 or 48 h and then treated with DOX (1 μm) for another 18 h. Increased Beclin 1 expression accelerated autophagic flux (A, BCN1 2.0 ± 0.34 versus β-galactosidase 0.83 ± 0.30), whereas knocking down Beclin 1 reduced it (B, shBCN1 0.45 ± 0.33 versus shCon 1.34 ± 0.23), as indicated by the difference in protein levels of LC3-II in the absence and presence of BFA. Densitometry data were the mean ± S.E. (n = 6 for BCN1 and 3 for shBCN1) and were analyzed with two-way ANOVA followed by paired Student's t tests. Beclin 1 overexpression exacerbated DOX-induced cell death, and knocking down Beclin 1 reduced it, as determined by PI staining (C), MTT assay (D), cleavage of PARP (cPARP) and caspase 3 (cCasp3, E), and DNA laddering (F). Data in C and D were expressed as the mean ± S.E. and were analyzed by one-way ANOVA (n = 6 for BCN and 3 for shBCN1).
DOX-induced Autophagy Was Associated with Depleted Protein Levels of GATA4
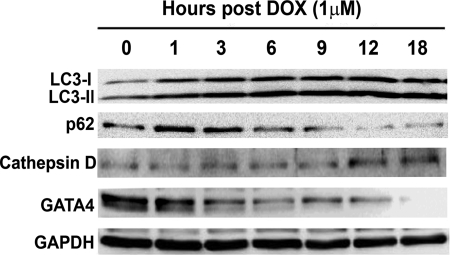
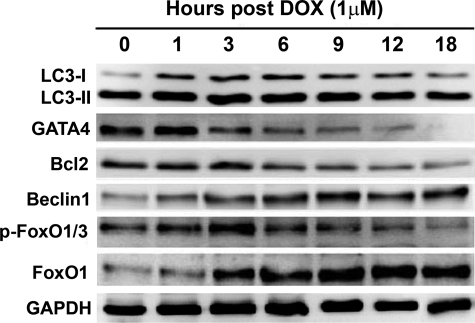
Previous studies demonstrated that DOX depletes GATA4 transcript and protein levels in cultured cardiomyocytes (26, 27). Preservation or overexpression of GATA4 prevents DOX-induced cardiomyocyte death (26, 27, 45), suggesting GATA4 depletion as a critical determinant of DOX cardiotoxicity. We determined if GATA4 depletion and autophagy induction, two important events both contributing to DOX cardiotoxicity, were temporally linked to each other. Cardiomyocytes were treated with DOX for different time periods to assess the time course of autophagy activation (Fig. 4). The LC3-II was increased as early as 1–3 h after DOX treatment and remained elevated throughout the 18-h treatment duration. Conversely, p62 was reduced, albeit starting at a later time point, suggesting that DOX quickly induced autophagy, and its activity was sustained for a long period of time. Interestingly, the protein level of cathepsin D, a lysosomal protease, was not increased until 12 h after DOX treatment, suggesting that the increase in lysosome enzyme follows autophagy induction to contribute to enhanced lysosomal degradation. Importantly, GATA4 protein levels began to decrease as early as 1–3 h and were completely depleted by 18 h post-DOX treatment, demonstrating an intimate temporal relationship between diminished GATA4 and autophagy induction. However, it is unknown whether these two events were functionally associated or they just happened to exist in parallel without any cross-talk.
FIGURE 4.
DOX-induced autophagy was associated with depleted protein levels of GATA4. Cardiomyocytes were treated with DOX for different time periods and then harvested for Western blot analysis. The increase in LC3-II levels was temporally linked with reduced p62 and diminished GATA4 protein levels.
Overexpression of GATA4 Inhibited DOX-induced Autophagy and Cardiomyocyte Death
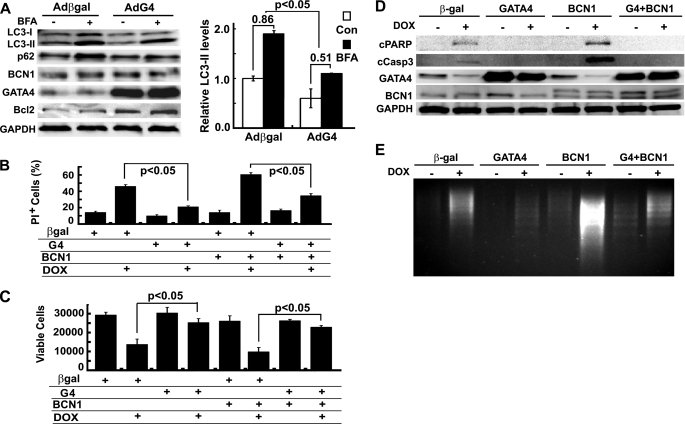
Depletion of GATA4 is a distinct feature of DOX cardiotoxicity, and overexpression of GATA4 can protect cardiomyocytes against DOX-induced cell death (26, 27, 45). However, the underlying mechanisms that mediate the protective effects of GATA4 are not well understood. Given that Bcl2, a downstream effector of GATA4 (46), is able to inhibit autophagy and that GATA4 depletion and autophagy induction are intimately linked, we tested the possibility that GATA4 may affect autophagic activity to attenuate DOX cardiotoxicity. Cardiomyocytes were infected with adenovirus expressing GATA4 or β-galactosidase for 24 h and then cultured in serum-free media overnight to induce base-line autophagy. As shown in Fig. 5A, overexpression of GATA4 slightly reduced LC3-II levels at base line but markedly attenuated BFA-induced accumulation of LC3-II. Specifically, autophagic flux, measured by the difference of LC3-II in the absence and presence of BFA, was substantially reduced in AdG4-infected cells relative to Adβgal-infected cells (β-galactosidase 0.86 ± 0.12 versus GATA4 0.51 ± 0.14, p < 0.05, n = 3). This was supported by a similar change pattern in p62 protein levels. Furthermore, GATA4-inhibited autophagic flux was determined by the difference in GFP-LC3 dots in the presence and absence of BFA. Cardiomyocytes were co-infected with AdGFP-LC3 and Adβgal or AdG4 and treated with BFA and DOX separately or in combination. Similar to endogenous LC3-II levels, the number of GFP-LC3 dots in Adβgal-infected cells was substantially reduced in AdG4-infected cells either with or without BFA or DOX treatment (data not shown), validating the capacity of GATA4 to inhibit base-line or DOX-induced autophagy.
FIGURE 5.
Overexpression of GATA4 inhibited DOX-induced autophagy and cardiomyocyte death. Cardiomyocytes were infected with Adβgal or AdG4 and then treated with DOX for 18 h. A, Western blots show LC3-II and p62 levels (left), and a bar graph demonstrates autophagic flux index (β-galactosidase (β-gal) 0.86 ± 0.12 versus G4 0.51 ± 0.14) as indicated by the difference in protein levels of LC3-II in the absence and presence of BFA (right). Densitometry data were the mean ± S.E. (n = 3) and were analyzed with two-way ANOVA followed by paired Student's t tests. Cardiomyocyte death was determined by PI staining (B), MTT assay (C), cleavage of PARP (cPARP) and caspase 3 (cCasp3, D), and DNA laddering (E). Data in B and C were expressed as the mean ± S.E. and analyzed by one-way ANOVA (n = 3). Con, control.
To determine the ability of GATA4 to inhibit DOX-induced cell death, cardiomyocytes were infected individually with Adβgal, AdG4, and AdBCN1 or co-infected with AdG4 and AdBCN1. Overexpression of GATA4 dramatically attenuated DOX-induced cardiomyocyte death as shown by decreased PI-positive cells (Fig. 5B), increased living cells (Fig. 5C), reduced cleavage of PARP and caspase 3 (Fig. 5D), and attenuated DNA laddering (Fig. 5E). Furthermore, overexpression of FLAG-tagged human Beclin 1 exacerbated DOX-induced cell death, and this effect was almost completely blocked by GATA4, supporting the notion that the protective effect of GATA4 is mediated by its capacity to inhibit autophagy.
Knocking Down GATA4 with shRNA Triggered Autophagy and Exacerbated DOX-induced Cardiomyocyte Death
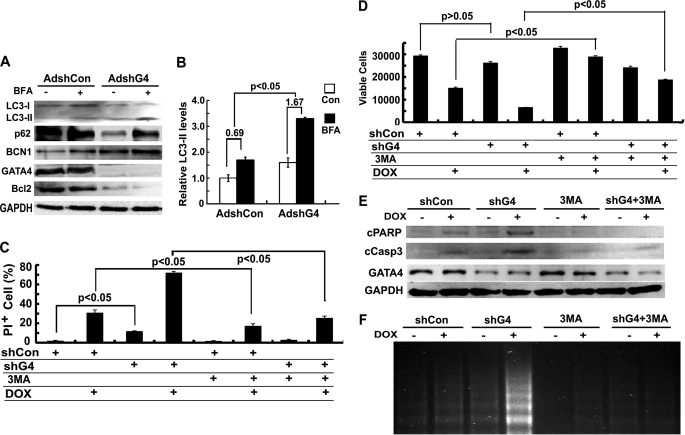
DOX depleted GATA4 protein levels (Fig. 4), which contributed to DOX cardiotoxicity, as preservation of GATA4 not only inhibited autophagy but also blocked DOX-induced myocyte death (Fig. 5). However, it was still unclear whether GATA4 depletion contributed to DOX-induced autophagy or whether it was just an incidental bystander that had nothing to do with autophagy induction. To address if GATA4 depletion per se can induce autophagy in cardiomyocytes, we knocked down GATA4 levels using an adenovirus that encodes a shRNA targeting GATA4 (shG4). Cardiomyocytes were infected with AdshG4 or AdshCon and cultured in DMEM with 1% bovine serum for 48 h. As shown in Fig. 6A, shG4 led to a nearly complete depletion of GATA4 protein levels that was accompanied by a roughly 90% reduction in Bcl2 and 2-fold increase in Beclin 1 levels. Remarkably, shG4-mediated GATA4 depletion induced significant levels of autophagy as indicated by increased LC3-II accumulation and enhanced p62 degradation. This was further validated by increased autophagic flux when cells were treated with BFA, as expressed by the difference of LC3-II in the absence and presence of BFA (Fig. 6B, shCon 0.69 ± 0.26 versus shG4 1.67 ± 0.19, p < 0.05, n = 3). Autophagic flux was also determined in AdGFP-LC3-infected cells. The results showed that shG4 increased the number of GFP-LC3 dots at base line, which was further elevated by BFA treatment (data not shown).
FIGURE 6.
Knocking down GATA4 triggered autophagy and exacerbated DOX-induced cardiomyocyte death. Cardiomyocytes were infected with AdshG4 or AdshCon for 48 h and then treated with DOX for 18 h. A, Western blots show LC3-II and p62 protein levels. B, shown is quantification of autophagic flux index (shCon 0.69 ± 0.26 versus shG4 1.67 ± 0.19) as determined by the difference in protein levels of LC3-II in the absence and presence of BFA. Densitometry data are the mean ± S.E. (n = 3) and were analyzed with two-way ANOVA followed by paired Student's t tests. Cardiomyocyte death was determined by PI staining (C), MTT assay (D), cleavage of PARP (cPARP) and caspase 3 (cCasp3, E), and DNA laddering (F). Data in C and D are expressed as the mean ± S.E. and were analyzed by one-way ANOVA (n = 3). Con, control.
Besides autophagy induction, the functional significance of GATA4 knockdown was evaluated by cell death and survival assays. We found that shG4 slightly but significantly increased cell death, as indicated by PI-positive cells (Fig. 6C), and cleaved PARP and caspase 3 (Fig. 6E) and DNA laddering (Fig. 6F) despite that MTT assay revealed only a trend toward reduced viable cells (Fig. 6D). Nonetheless, shG4 dramatically aggravated DOX-induced cardiomyocyte death as shown by all the parameters measured (Figs. 6, C–F). The results suggested that GATA4 knockdown triggered autophagy and sensitized cardiomyocytes to DOX toxic effects. Not surprisingly, inhibition of autophagy by 3-MA significantly attenuated the cell death effects induced by DOX and shG4 either alone or in combination (Figs. 6, C–F), suggesting that autophagy induction contributed significantly to GATA4 gene silencing and/or DOX-induced cardiomyocyte death.
GATA4 Regulated the Expression of Bcl2 and Autophagy-related Genes
Overexpression of GATA4 inhibited both base-line and DOX-induced autophagy (Fig. 5), whereas GATA4 gene silencing triggered autophagy exacerbating DOX cardiotoxicity (Fig. 6), demonstrating GATA4 as a negative regulator of autophagy. However, it remained unclear how GATA4 achieved its inhibitory effect on autophagy. GATA4 is a positive transcription regulator of many cardiac genes including Bcl2 (46), a factor with both anti-apoptotic and anti-autophagic activities. Indeed, overexpression of GATA4 up-regulated Bcl2 protein levels (Fig. 5A), whereas GATA4 knockdown depleted Bcl2 protein contents (Fig. 6A), confirming that GATA4 is both sufficient and necessary for Bcl2 gene expression. Thus, it is reasonable to assume that GATA4 regulates autophagic activity at least partially through its downstream effector Bcl2. If this is true, DOX-induced GATA4 depletion should lead to diminished Bcl2 levels. As shown in Fig. 7, DOX-induced GATA4 depletion was indeed followed by a reduction in Bcl2 protein levels. Despite that the diminished Bcl2 may not be responsible for initiating autophagy upon exposure to DOX, as the reduction in Bcl2 lagged behind the increase in LC3-II, it may still exacerbate DOX-induced myocyte death.
FIGURE 7.
DOX-induced elevation of LC3-II was temporally associated with depleted levels of GATA4 and Bcl2 but increased levels of Beclin 1 and FoxO1. Cardiomyocytes were treated with DOX for different time periods and then harvested for Western blot analysis.
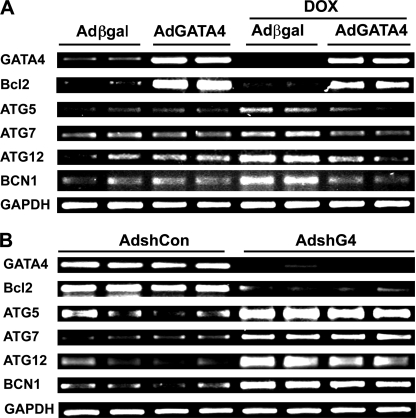
We also examined if DOX-induced autophagy correlated with changes in the expression of autophagy-related genes and if GATA4 could regulate these genes to inhibit DOX-induced autophagy. Not surprisingly, the protein levels of Beclin 1 were increased as early as 1 h after DOX treatment (Fig. 7), which correlated with increased LC3-II levels and were inversely related to GATA4 protein contents throughout the 18-h treatment period. The result suggested that GATA4 may negatively regulate the expression of ATG genes, including Beclin 1, to inhibit autophagy. Therefore, we determined if GATA4 can affect mRNA expression of several ATG genes encoding proteins essential for autophagy induction and the generation and maturation of autophagosomes. As shown by semiquantitative reverse transcriptase-PCR in Fig. 8A, although overexpression of GATA4 was unable to affect mRNA expression of ATG5, -7, -12, and Beclin 1 at base line, it abolished DOX-induced expression of these ATG genes, suggesting that GATA4 may serve as a brake to prevent abnormal activation of these ATG genes under stress conditions. If this is true, releasing the brake would allow excessive expression of these genes. Indeed, knocking down GATA4 with shRNA led to dramatically increased mRNA levels of ATG5, -7, -12 and Beclin 1(Fig. 8B), reinforcing a brake role for GATA4 in maintaining the expression of ATG genes within homeostatic levels. These results suggest that DOX-induced GATA4 depletion disrupted the normal brake function of GATA4, permitting an unchecked up-regulation of ATG genes and, thus, induction of autophagy.
FIGURE 8.
GATA4 regulated the expression of Bcl2 and autophagy-related genes. A, cardiomyocytes were infected with Adβgal or AdG4 and then treated with DOX for 18 h. B, cardiomyocytes were infected with AdshG4 or AdshCon for 48 h. The mRNA expression of Bcl2 and several autophagy-related genes under both conditions were determined by semi-quantitative reverse transcriptase-PCR.
Bcl2 Mediated the Anti-autophagic Effect of GATA4
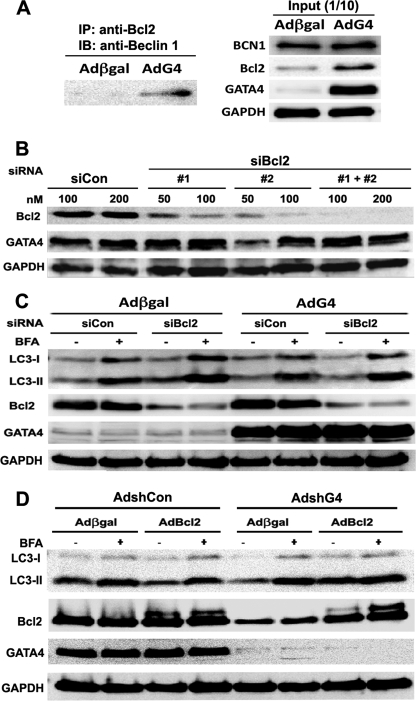
GATA4 is sufficient and necessary for Bcl2 gene expression, suggesting Bcl2 as a potential downstream effector of GATA4 that mediates the anti-autophagic effect of GATA4. The ability of Bcl2 to inhibit autophagy is mediated partly by its interaction with Beclin 1. Therefore, we performed IP to test if GATA4 overexpression can enhance the interaction between Bcl2 and Beclin 1. As shown in Fig. 9A, co-IP with Bcl2 antibodies indeed pulled down more Beclin 1 in AdG4-infected cardiomyocytes than in Adβgal-infected cells, confirming that GATA4 increases Beclin 1 sequestration by Bcl2, likely due to increased availability of Bcl2. To further test the role of Bcl2 in GATA4-induced autophagy inhibition, we determined if diminished Bcl2 levels could attenuate or abolish the ability of GATA4 to inhibit autophagy. We used two different siRNAs against rat mRNA sequence (siBcl2) to knock down Bcl2 in cardiomyocytes. As shown in Fig. 9B, both siRNAs were able to knock down Bcl2 protein levels in a dose-dependent manner, with siBcl2 #2 having a higher efficiency. Cardiomyocyte were infected with Adβgal or AdG4 and then followed by siRNA transfection with a Silencer Negative Control siRNA (siCon) or a mixture of the two siBcl2 (#1 and #2, each used at 50 nm). The ability of GATA4 to inhibit autophagy was determined 48 h later. Expectedly, adenovirus-mediated overexpression of GATA4 up-regulated Bcl2 protein levels and inhibited autophagic flux, as shown by the difference in LC3-II levels in the absence and presence of BFA (Fig. 9C). However, knocking down Bcl2 with siRNA markedly attenuated the inhibitory effect of GATA4 on autophagy, indicating that Bcl2 indeed partly mediates the anti-autophagic effects of GATA4. In addition, to determine whether GATA4 depletion-induced autophagy could be attributed to diminished Bcl2 levels, we knocked down GATA4 for 48 h with shG4 and then overexpressed Bcl2 in cardiomyocytes. As shown in Fig. 9D, GATA4 gene silencing by AdshG4 reduced Bcl2 levels and concurrently triggered autophagy as indicated by LC3-II contents with and without BFA. However, this effect was reversed by late Bcl2 overexpression with AdBcl2, supporting a role for reduced Bcl2 in GATA4 knockdown-induced autophagy. Together, these results suggest that the anti-autophagic effect of GATA4 is mediated at least in part by its downstream effector Bcl2.
FIGURE 9.
Bcl2 mediated the anti-autophagic effect of GATA4. A, increased GATA4 enhanced the interaction between Bcl2 and Beclin 1. Cardiomyocytes were infected with Adβgal or AdG4, and protein samples were prepared 24 h later. 100 μg of protein was used for co-IP with Bcl2 antibodies (left panel), and 10 μg of protein was used as the input control (right panel). IB, immunoblotting. B, two different siRNAs against rat Bcl2 mRNA sequence (siBcl2) were transfected into cardiomyocytes. Both siRNAs were able to knock down Bcl2 protein levels in a dose-dependent manner, with siBcl2 #2 having a higher efficiency. C, cardiomyocytes were infected with Adβgal or AdG4 and then followed by siRNA transfection with a Silencer Negative Control siRNA (siCon) or a mixture of the two siBcl2 (#1 and #2 each at 50 nm). Knocking down Bcl2 markedly attenuated the ability of GATA4 to inhibit autophagy as shown by the difference in protein levels of LC3-II in the absence and presence of BFA. D, cardiomyocytes were infected with AdshG4 or AdshCon for 48 h and then infected with AdBcl2 for another 24 h. Western blot analysis was performed showing that GATA4 depletion-induced autophagy was reversed by Bcl2 overexpression.
DISCUSSION
Oxidative stress due to unopposed ROS generation has been thought to be the major mechanism responsible for DOX cardiotoxicity. Nevertheless, clinical trials have shown very limited efficacy of antioxidant therapy in patients (9–13), suggesting that further studies are needed to identify additional mechanisms that may contribute to DOX cardiotoxicity. DOX depletes the transcription factor GATA4 in cardiomyocytes (26, 27), and restoration of GATA4 levels prevent DOX-induced cardiomyocyte death (26, 27, 45). However, it remains unclear how GATA4 could provide protection against DOX cardiotoxicity. In the present study we demonstrate that the cardioprotective effect of GATA4 is mediated at least partly by its ability to inhibit DOX-induced autophagy through modulating the expression of Bcl2 and autophagy-related genes.
Autophagy has long been depicted as a survival pathway that allows the cell to maintain energy production under various starvation and stress conditions (15). However, autophagy has also been shown to contribute to cell death in other contexts, suggesting autophagy as a double-edged sword that could be either protective or detrimental, depending on the particular cell type, the subcellular environment, the nature and intensity of the stimulus, and the levels of autophagy induced (20, 25, 57). Thus, the functional significance of autophagy induction has to be determined individually within the specific context under study. Added to the complexity, autophagy is often associated with apoptosis, a form of programmed cell death, making it more difficult to determine the role of autophagy in cell survival and death. The interaction of autophagy and apoptosis has been characterized into three different types (57); both autophagy and apoptosis can act as partners to coordinately induce cell death, autophagy acts as an antagonist to block apoptotic cell death by promoting cell survival, or autophagy acts as an enabler of apoptosis that permits apoptosis to occur without leading to death in itself. In the present study we show that DOX dramatically increased autophagic flux in cardiomyocytes (Fig. 1), which was associated with elevated cell death, as measured by PI staining, the MTT assay, which report cell death regardless of the cause, DNA laddering, and cleavage of caspase 3 and PARP, which indicate apoptosis. To tease out the role of autophagy in DOX-induced cardiomyocyte death, we manipulated autophagic activity using both pharmacologic and genetic approaches. We revealed that inhibition of autophagy by 3-MA or Beclin 1 knockdown resulted in significant attenuation of cell death (Figs. 2 and 3). Conversely, activation of autophagy by rapamycin or Beclin 1 expression exacerbated DOX-induced cardiomyocyte death including apoptosis. These results suggest that DOX-induced autophagy does not promote cardiomyocyte survival. Instead, it is intimately linked with apoptosis and acts as a partner to promote cardiomyocyte death. Although it remains to be determined how autophagy interacts with apoptosis to coordinately induce cell death in the context of DOX treatment, our data suggest that inhibition of autophagy constitutes a viable strategy for reducing DOX-induced cardiomyocyte death, thereby alleviating DOX cardiotoxicity.
Induction of autophagy is tightly controlled by many positive and negative regulators (15, 16). Akt and mTOR, which are activated by insulin, several growth factors, and many metabolic signals, are well known to inhibit autophagy. We examined if these two pathways were involved in autophagy induction in response to DOX treatment. Unfortunately, the activities of Akt and mTOR were enhanced by DOX, as indicated by the increased phosphorylation of Akt and mTOR as well as their downstream effectors FoxO1, p70S6K, and 4EBP (data not shown), suggesting that Akt and mTOR signaling pathways are unlikely responsible for DOX-induced autophagy. By contrast, DOX-induced autophagy was correlated with depleted protein levels of GATA4 (Figs. 4 and 7), a transcription factor essential for cardiomyocyte growth and survival (44). Although not known previously to regulate autophagy, GATA4 is both sufficient and necessary for the gene expression of Bcl2, a survival factor with anti-autophagy activity (38) that was also depleted in response to DOX treatment (Fig. 7). It is, thus, possible that GATA4 depletion may play a role in DOX-induced autophagy. Indeed, overexpression of GATA4 inhibited DOX-induced autophagic flux and reduced cardiomyocyte death (Fig. 5), whereas GATA4 gene silencing triggered autophagy and exacerbated DOX toxicity (Fig. 6). These results confirm GATA4 as a negative regulator of autophagy and support the notion that DOX-induced depletion of GATA4 is critical for DOX to induce autophagy and cardiomyocyte death.
We believe that the ability of GATA4 to inhibit autophagy is mediated at least partly by its downstream effector Bcl2. This was supported by the observations that the inhibition of autophagy by GATA4 was attenuated by knocking down Bcl2 with siRNA and that GATA4 gene silencing triggered autophagy, which was blocked by Bcl2 overexpression (Fig. 9). The anti-autophagic effect of Bcl2 is reported to be achieved through its interaction with Beclin 1 (Atg6), a protein essential for autophagy initiation. Beclin 1 is part of a protein complex and serves as a platform, recruiting multiple autophagy activators or repressors including the class III phosphatidylinositol 3-kinase, Bcl2, ICP34.5, UVRAG, Ambra1, and Bif-1 (36). Of note, through binding and sequestering Beclin 1, Bcl2 has been suggested to act as a rheostat that turns autophagy on or off when required (41). Recent data showed that the interaction between Bcl2 and Beclin 1 is inhibited by mitogen-activated protein kinase c-Jun N-terminal kinase 1 that phosphorylates Bcl2 and stimulates autophagy (58), shedding some light on the mechanism that regulates the interplay between Bcl2 and Beclin 1 and further supporting a pivotal role of Bcl2 in controlling autophagy. However, after DOX treatment, the reduction in Bcl2 levels lagged behind the increase in LC3-II (Fig. 7A), suggesting that the diminished Bcl2 may contribute to increased autophagic activity but may not be responsible for initiating autophagy upon exposure to DOX. Therefore, GATA4 may have other downstream effectors that mediate its anti-autophagic activity. Indeed, GATA4 modulates the expression of several ATG genes including Beclin 1 (Fig. 8), which may provide another mechanism by which GATA4 antagonizes DOX-induced autophagy.
Although overexpression of GATA4 did not have any effect on ATG genes at base line, it repressed DOX-induced expression of ATG genes. Importantly, GATA4 gene silencing resulted in a dramatic up-regulation of ATG5, ATG7, ATG12, and Beclin 1 (Fig. 8B), strongly suggesting GATA4 as a negative regulator of ATG genes. Nevertheless, how GATA4 could suppress DOX-induced expression of ATG genes remains unclear. Using MatInspector Professional, we examined the promoter regions of ATG5, -7, and -12 and Beclin 1 genes and identified a few putative GATA motifs. However, these sites are not conserved between human and rodents, suggesting that GATA4 may not directly bind to the regulatory sequences of ATG genes to repress their expression. Interestingly, GATA4 has been shown to inhibit the expression of tryptophan oxygenase gene in fetal hepatocytes by directly binding to the TATA box and interfering with the formation of the transcription initiation complex (59). It is, thus, possible that GATA4 may repress ATG genes through a similar mechanism without the need to bind to a GATA motif. It is also possible that the inhibitory effects of GATA4 on ATG genes are achieved through interaction with another positive or negative regulatory factor that directly binds to the promoters of the ATG genes. In this respect, Yutzey and co-workers (60) have recently shown that the transcription factor FoxO1 or FoxO3 up-regulates the expression of ATG12 gene through directly binding to FoxO consensus motif on the promoter, which contributes to autophagy activation in cardiomyocytes. Coincidently, DOX-induced GATA4 depletion was accompanied by increased FoxO1 protein expression (Fig. 7), raising the possibility that GATA4 may repress ATG genes through transcriptional inhibition of FoxO gene expression or direct interaction with FoxO proteins. In addition, it is possible that GATA4 regulates ATG genes through p53, a transcription factor able to trigger autophagy (61), and implicated in DOX-induced cardiomyocyte death (62). A potential interaction between GATA4 and p53 has been suggested in murine astrocytes (63), and the direct interaction and mutual antagonism between GATA1 and p53 has been confirmed in erythrocytes (64). These possibilities regarding the mechanisms whereby GATA4 inhibits autophagy remain largely speculative and need to be tested experimentally in future studies.
In summary, our study has added GATA4 to the growing list of molecules that negatively impact autophagic activity. Under basal conditions, by modulating the expression of Bcl2 and ATG genes, a normal amount of GATA4 protein serves as a brake to help maintain autophagic activity within a physiological range that favors cell survival but without losing cytoplasmic quality control. By contrast, in response to DOX treatment, GATA4 protein is depleted, and thus, the brake is released, which results in reduced Bcl2 levels and concurrently permits an up-regulation of ATG genes, leading to excessive autophagy activation that contributes to cardiomyocyte death. Thus, therapeutic strategy that blocks GATA4 depletion and/or enhances GATA4-Bcl2 signaling will presumably be able to prevent unchecked activation of autophagy thereby reducing DOX cardiotoxicity.
Acknowledgments
We thank Andy Cypher for preparing cultured NRVC and Stephanie Busch, Troy Lackey, Tricia Krueger, and Yuan Huang for constructing and purifying several adenoviruses used in this work. We are grateful to Dr. Aviva Tolkovsky at the University of Cambridge for providing the adenovirus expressing GFP-LC3 and Dr. Junying Yuan at Harvard University for providing the FLAG-tagged human Beclin 1 cDNA.
This work was supported, in whole or in part, by National Institutes of Health Grant 2P20RR017662-06A1 (Project 3; National Center for Research Resources (to Q. L.)).
- DOX
- doxorubicin
- mTOR
- mammalian target of rapamycin
- DMEM
- Dulbecco's modified Eagle's medium
- 3-MA
- 3-methyladenine
- Rap
- rapamycin
- BFA
- bafilomycin A1
- MTT
- thiazolyl blue tetrazolium bromide
- PARP
- poly(ADP-ribose) polymerase
- IP
- immunoprecipitation
- siRNA
- small interfering RNA
- PI
- propidium iodide
- ANOVA
- analysis of variance
- LC3
- light chain 3
- shRNA
- short hairpin RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- AV
- autophagic vacuole
- GFP
- green fluorescent protein.
REFERENCES
- 1.Swain S. M., Whaley F. S., Ewer M. S. (2003) Cancer 97, 2869–2879 [DOI] [PubMed] [Google Scholar]
- 2.Singal P. K., Iliskovic N. (1998) N. Engl. J. Med. 339, 900–905 [DOI] [PubMed] [Google Scholar]
- 3.Minotti G., Menna P., Salvatorelli E., Cairo G., Gianni L. (2004) Pharmacol. Rev. 56, 185–229 [DOI] [PubMed] [Google Scholar]
- 4.Kumar D., Kirshenbaum L. A., Li T., Danelisen I., Singal P. K. (2001) Antioxid. Redox Signal. 3, 135–145 [DOI] [PubMed] [Google Scholar]
- 5.Yen H. C., Oberley T. D., Vichitbandha S., Ho Y. S., St Clair D. K. (1996) J. Clin. Invest. 98, 1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang Y. J., Chen Y., Epstein P. N. (1996) J. Biol. Chem. 271, 12610–12616 [DOI] [PubMed] [Google Scholar]
- 7.Sun X., Zhou Z., Kang Y. J. (2001) Cancer Res. 61, 3382–3387 [PubMed] [Google Scholar]
- 8.Siveski-Iliskovic N., Hill M., Chow D. A., Singal P. K. (1995) Circulation 91, 10–15 [DOI] [PubMed] [Google Scholar]
- 9.Ladas E. J., Jacobson J. S., Kennedy D. D., Teel K., Fleischauer A., Kelly K. M. (2004) J. Clin. Oncol. 22, 517–528 [DOI] [PubMed] [Google Scholar]
- 10.Dorr R. T. (1996) Semin. Oncol. 23, 23–34 [PubMed] [Google Scholar]
- 11.Legha S. S., Wang Y. M., Mackay B., Ewer M., Hortobagyi G. N., Benjamin R. S., Ali M. K. (1982) Ann. N. Y. Acad. Sci. 393, 411–418 [DOI] [PubMed] [Google Scholar]
- 12.Myers C., Bonow R., Palmeri S., Jenkins J., Corden B., Locker G., Doroshow J., Epstein S. (1983) Semin. Oncol. 10, 53–55 [PubMed] [Google Scholar]
- 13.Olson R. D., Mushlin P. S. (1990) FASEB J. 4, 3076–3086 [PubMed] [Google Scholar]
- 14.Marcillat O., Zhang Y., Davies K. J. (1989) Biochem. J. 259, 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N. (2005) Cell Death Differ. 12, 1535–1541 [DOI] [PubMed] [Google Scholar]
- 16.Klionsky D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 17.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 18.Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., Nishida K., Hori M., Mizushima N., Otsu K. (2007) Nat. Med. 13, 619–624 [DOI] [PubMed] [Google Scholar]
- 19.Tannous P., Zhu H., Johnstone J. L., Shelton J. M., Rajasekaran N. S., Benjamin I. J., Nguyen L., Gerard R. D., Levine B., Rothermel B. A., Hill J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9745–9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsui Y., Takagi H., Qu X., Abdellatif M., Sakoda H., Asano T., Levine B., Sadoshima J. (2007) Circ. Res. 100, 914–922 [DOI] [PubMed] [Google Scholar]
- 21.Yan L., Vatner D. E., Kim S. J., Ge H., Masurekar M., Massover W. H., Yang G., Matsui Y., Sadoshima J., Vatner S. F. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13807–13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine B., Yuan J. (2005) J. Clin. Invest. 115, 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akazawa H., Komazaki S., Shimomura H., Terasaki F., Zou Y., Takano H., Nagai T., Komuro I. (2004) J. Biol. Chem. 279, 41095–41103 [DOI] [PubMed] [Google Scholar]
- 24.Zhu H., Tannous P., Johnstone J. L., Kong Y., Shelton J. M., Richardson J. A., Le V., Levine B., Rothermel B. A., Hill J. A. (2007) J. Clin. Invest. 117, 1782–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang C., Avery L. (2008) Autophagy 4, 82–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y., Ma A. G., Kitta K., Fitch S. N., Ikeda T., Ihara Y., Simon A. R., Evans T., Suzuki Y. J. (2003) Mol. Pharmacol. 63, 368–377 [DOI] [PubMed] [Google Scholar]
- 27.Aries A., Paradis P., Lefebvre C., Schwartz R. J., Nemer M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L., Takemura G., Li Y., Miyata S., Esaki M., Okada H., Kanamori H., Khai N. C., Maruyama R., Ogino A., Minatoguchi S., Fujiwara T., Fujiwara H. (2006) Circulation 113, 535–543 [DOI] [PubMed] [Google Scholar]
- 29.Lim C. C., Zuppinger C., Guo X., Kuster G. M., Helmes M., Eppenberger H. M., Suter T. M., Liao R., Sawyer D. B. (2004) J. Biol. Chem. 279, 8290–8299 [DOI] [PubMed] [Google Scholar]
- 30.Kumarapeli A. R., Horak K. M., Glasford J. W., Li J., Chen Q., Liu J., Zheng H., Wang X. (2005) FASEB J. 19, 2051–2053 [DOI] [PubMed] [Google Scholar]
- 31.Semenov D. E., Lushnikova E. L., Nepomnyashchikh L. M. (2001) Bull. Exp. Biol. Med. 131, 505–510 [DOI] [PubMed] [Google Scholar]
- 32.Lu L., Wu W., Yan J., Li X., Yu H., Yu X. (2009) Int. J. Cardiol. 134, 82–90 [DOI] [PubMed] [Google Scholar]
- 33.Mizushima N. (2004) Int. J. Biochem. Cell Biol. 36, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 34.Codogno P., Meijer A. J. (2005) Cell Death Differ. 12, 1509–1518 [DOI] [PubMed] [Google Scholar]
- 35.Meijer A. J., Codogno P. (2006) Mol. Aspects Med. 27, 411–425 [DOI] [PubMed] [Google Scholar]
- 36.Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. (2008) Biochimie 90, 313–323 [DOI] [PubMed] [Google Scholar]
- 37.Vergne I., Roberts E., Elmaoued R. A., Tosch V., Delgado M. A., Proikas-Cezanne T., Laporte J., Deretic V. (2009) EMBO J. 28, 2244–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 39.Molkentin J. D. (2000) J. Biol. Chem. 275, 38949–38952 [DOI] [PubMed] [Google Scholar]
- 40.Liang Q., Molkentin J. D. (2002) J. Mol. Cell. Cardiol. 34, 611–616 [DOI] [PubMed] [Google Scholar]
- 41.Liang Q., De Windt L. J., Witt S. A., Kimball T. R., Markham B. E., Molkentin J. D. (2001) J. Biol. Chem. 276, 30245–30253 [DOI] [PubMed] [Google Scholar]
- 42.Liang Q., Wiese R. J., Bueno O. F., Dai Y. S., Markham B. E., Molkentin J. D. (2001) Mol. Cell. Biol. 21, 7460–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charron F., Tsimiklis G., Arcand M., Robitaille L., Liang Q., Molkentin J. D., Meloche S., Nemer M. (2001) Genes Dev. 15, 2702–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oka T., Maillet M., Watt A. J., Schwartz R. J., Aronow B. J., Duncan S. A., Molkentin J. D. (2006) Circ. Res. 98, 837–845 [DOI] [PubMed] [Google Scholar]
- 45.Kitta K., Day R. M., Kim Y., Torregroza I., Evans T., Suzuki Y. J. (2003) J. Biol. Chem. 278, 4705–4712 [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi S., Lackey T., Huang Y., Bisping E., Pu W. T., Boxer L. M., Liang Q. (2006) FASEB J. 20, 800–802 [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi S., Mao K., Zheng H., Wang X., Patterson C., O'Connell T. D., Liang Q. (2007) J. Biol. Chem. 282, 21945–21952 [DOI] [PubMed] [Google Scholar]
- 48.Bampton E. T., Goemans C. G., Niranjan D., Mizushima N., Tolkovsky A. M. (2005) Autophagy 1, 23–36 [DOI] [PubMed] [Google Scholar]
- 49.Yuan J. (2008) Autophagy 4, 249–250 [DOI] [PubMed] [Google Scholar]
- 50.Kuzman J. A., Gerdes A. M., Kobayashi S., Liang Q. (2005) J. Mol. Cell. Cardiol. 39, 841–844 [DOI] [PubMed] [Google Scholar]
- 51.Olson R. D., Gambliel H. A., Vestal R. E., Shadle S. E., Charlier H. A., Jr., Cusack B. J. (2005) Cardiovasc. Toxicol. 5, 269–283 [DOI] [PubMed] [Google Scholar]
- 52.Oliveira P. J., Wallace K. B. (2006) Toxicology 220, 160–168 [DOI] [PubMed] [Google Scholar]
- 53.Tanida I., Ueno T., Kominami E. (2004) Int. J. Biochem. Cell Biol. 36, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizushima N., Yoshimori T. (2007) Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 55.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 56.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisenberg-Lerner A., Bialik S., Simon H. U., Kimchi A. (2009) Cell Death Differ. 16, 966–975 [DOI] [PubMed] [Google Scholar]
- 58.Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. (2008) Mol. Cell 30, 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaneoka H., Miyake K., Iijima S. (2008) Cytotechnology 57, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sengupta A., Molkentin J. D., Yutzey K. E. (2009) J. Biol. Chem. 284, 28319–28331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P. R., Gasco M., Garrone O., Crook T., Ryan K. M. (2006) Cell 126, 121–134 [DOI] [PubMed] [Google Scholar]
- 62.Liu J., Mao W., Ding B., Liang C. S. (2008) Am. J. Physiol. Heart Circ. Physiol. 295, H1956–H1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agnihotri S., Wolf A., Picard D., Hawkins C., Guha A. (2009) Oncogene 28, 3033–3046 [DOI] [PubMed] [Google Scholar]
- 64.Trainor C. D., Mas C., Archambault P., Di Lello P., Omichinski J. G. (2009) Blood 114, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]