Abstract
Previous studies have shown that the kinase activation loop (KAL) of the oncogenic fusion protein NPM-ALK regulates its overall tyrosine phosphorylation status and tumorigenicity. Using tandem affinity purification-mass spectrometry, we assessed how the KAL of NPM-ALK regulates the phosphorylation status of its individual tyrosines. Using the lysates of GP293 cells transfected with NPM-ALK, our highly reproducible results showed evidence of phosphorylation in all 3 tyrosines in KAL and 8 tyrosines outside KAL. We created 7 KAL mutants, each of which carried a Tyr-to-Phe mutation of ≥1 of the 3 tyrosines in KAL. A complete loss of the 8 phosphotyrosines outside KAL was found in 3 KAL mutants, and their oncogenicity (assessed by cell viability, colony formation, and the ability to phosphorylate effector proteins) was abrogated. A partial loss of the 8 phosphotyrosines was found in 4 KAL mutants, but their oncogenicity did not show simple correlation with the number of residual phosphotyrosines. Tyr-to-Phe mutations of each of the 8 phosphotyrosines outside KAL did not result in a significant decrease in the oncogenicity. In conclusion, we have provided details of how the KAL in NPM-ALK regulates its tyrosine phosphorylation pattern. Our results challenge some of the current concepts regarding the relationship between the tyrosine phosphorylation and oncogenicity of NPM-ALK.
Introduction
It is well established that inappropriate activation of various tyrosine kinases represents one of the important mechanisms underlying tumorigenesis (1, 2). NPM-ALK, an oncogenic fusion protein found exclusively in a subset of ALK-positive anaplastic large cell lymphoma, is believed to promote tumorigenesis via its constitutively active tyrosine kinase (3). The formation of NPM-ALK is a direct consequence of the t(2,5) chromosomal translocation that places the DNA segment encoding the N-terminal/oligomerization domain (1–117 amino acid residues) of the nucleophosmin (NPM) gene directly upstream of the DNA segment encoding the C-terminal/kinase portion (1058–1620 amino acid residues) of the anaplastic lymphoma kinase (ALK) gene (4). There is mounting evidence that NPM-ALK is central to the pathogenesis of ALK-positive anaplastic large cell lymphoma (5). Based on the current concept, NPM-ALK undergoes auto-phosphorylation of the 3 tyrosine residues located in the kinase activation loop (KAL)3 that bears the insulin receptor kinase subfamily motif (“YxxxYY”)(6, 7). Once the autophosphorylation of the KAL tyrosine residues is achieved, NPM-ALK becomes activated, and subsequent phosphorylation of various tyrosine residues outside of the KAL follows. Some of these phosphorylated tyrosine residues have been shown to serve as the docking sites for a number of effector proteins, many of which are known to be involved in cellular signaling. Via their physical interactions with NPM-ALK, these effector proteins are in turn phosphorylated and activated by NPM-ALK, resulting in the constitutive activation and deregulation of a host of important signaling pathways (3, 8–17). The interaction between NPM-ALK and Shc, an adaptor cellular signaling protein, provides an example to illustrate these concepts. Specifically, it has been demonstrated Shc co-immunoprecipitates with NPM-ALK and site-directed mutagenesis of Tyr567 of NPM-ALK was found to abrogate the Shc/NPM-ALK interaction and reduce the phosphorylation of Shc (1, 18).
While this conceptual model regarding the biology of NPM-ALK is widely accepted, details of some of the aspects are incompletely understood. For instance, the importance and functional roles of the KAL in NPM-ALK have not been fully examined. Because the KAL of NPM-ALK carries the motif of the insulin receptor kinase subfamily (YxxxYY)(6, 7), it has been inferred that this KAL regulates the phosphorylation process in a similar fashion as that found in the other members of the insulin receptor kinase subfamily. In our search of the literature, we found only one study directly addressing the biological importance of the KAL of NPM-ALK. Using site-directed mutagenesis of the three tyrosine residues in the KAL of NPM-ALK (Y338xxxY342Y343), Tartari and et al. (2) assessed the biological importance of the KAL by correlating various mutations in the KAL and the oncogenic potential of these mutants. Their results have provided direct evidence that the KAL of NPM-ALK is biologically important, because mutation at Tyr338 (but not that of Tyr342 or Tyr343) completely abrogated evidence of tyrosine phosphorylation of NPM-ALK and its oncogenic potential (2). Nevertheless, because the tyrosine phosphorylation status of NPM-ALK was assessed by immunoprecipitation and immunoblotting (with an anti-tyrosine antibody), only an estimate of the overall tyrosine phosphorylation status of NPM-ALK could be made. In other words, whether the reduction in the oncogenic potential of the NPM-ALK mutants is due to a specific change in the tyrosine phosphorylation pattern could not be addressed.
In this study, we aimed to perform functional characterization of the KAL of NPM-ALK. Instead of using immunoprecipitation and immunoblotting, an approach used in most of the previous studies of the KAL in the insulin receptor kinase subfamily, we chose to employ tandem affinity purification (TAP) in combination with a newly developed, highly sensitive liquid chromatography-mass spectrometry (LC-MS) protocol (19–21). The major advantage of using this method is that we are able to comprehensively profile the phosphorylation status of individual tyrosine residues on NPM-ALK and evaluate their changes in response to our experimental manipulation of the KAL. TAP, as used in this study, allows us to effectively purify the full-length NPM-ALK using a relatively small amount of total proteins. We aimed to provide new insights into the functional role of the KAL of NPM-ALK that can serve as a proof of principle for future studies of related oncogenic tyrosine kinases.
MATERIALS AND METHODS
Site-directed Mutagenesis and Construction of Gene Vectors
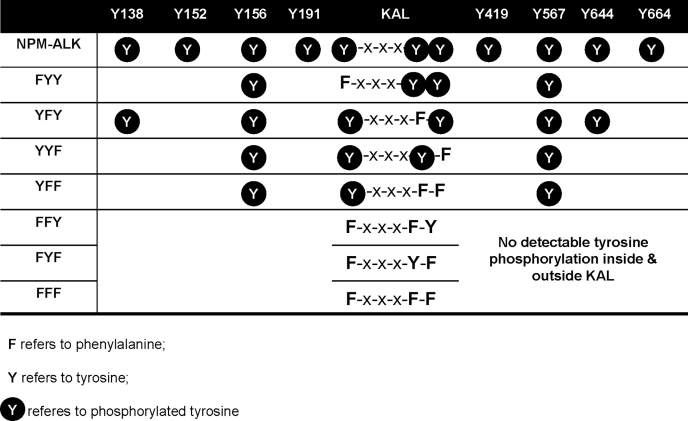
As described previously, our constructed NPM-ALK expression vector can produce full-length human NPM-ALK that is tagged with HB, which consists of an RGS-hexahistidine (H) tag and a bacterially derived biotinylation (B) signal peptide (19). We have also previously found that HB/NPM-ALK expresses functional NPM-ALK at levels similar to those found in ALK-expressing anaplastic large cell lymphoma cell lines (19). Site-directed mutagenesis (Tyr to Phe) was performed using the QuikChange XL kit (Stratagene) according to the manufacturer's instructions. Mutations of ≥1 of the three tyrosine residues present in the KAL (Tyr338, Tyr342, and Tyr343) were introduced using HB/NPM-ALK inserted in a pcDNA3.1(+) backbone. As summarized in Table 1, a total of 7 KAL mutants were generated. Single mutations of individual tyrosine residues outside the KAL of NPM-ALK (“single mutants”) were introduced using the pcDNA3.1(+)/NPM-ALK vector. Primers were designed using the on-line software provided by Stratagene. The coding sequence of NPM-ALK and all of its mutants were confirmed to ensure that no artificial mutations were acquired.
TABLE 1.
Tyrosine phosphorylation patterns for the KAL mutants, as compared to the unaltered NPM-ALK
Cell Culture and Gene Transfection
The GP293 packaging cell line (a modified version of the HEK293 human embryonic kidney cell line, Clontech) was maintained in Dulbecco's modified Eagle's medium (Sigma Aldrich), supplemented with 10% heat-inactivated fetal bovine serum. GP293 cells were transfected with the various NPM-ALK expression vectors using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's suggested protocol. The culture media was supplemented with 4 μm biotin to improve the biotinylation efficiency of HB-tagged proteins.
TAP under Denaturing Conditions
GP293 cells, transfected with HB/NPM-ALK or one of the 7 KAL mutants, were lysed in CellLytic M (Sigma), supplemented with 1 mm phenylmethylsulfonyl fluoride, a phosphatase inhibitor mixture, and a protease inhibitor mixture (Sigma). Lysates were cleared by centrifugation (20,000 × g, 15 min) and the resulting supernatant concentration was determined by the bicinchoninic acid assay following the manufacturer's protocol (Bio-Rad). TAP was performed based on two distinct rounds of purification. For the HB-tagged proteins, purification was performed on Ni2+-Sepharose beads (GE Healthcare), followed by immobilized streptavidin beads as described previously (19) with the addition of 8 m urea to all washing buffers. This modification of our original protocol to denaturing conditions allows for the removal of noncovalently bound binding partners, resulting in increased HB/NPM-ALK purity and the subsequent sequence coverage upon LC-MS analysis.
On-bead Protein Digestion, LC-MS, and Data Analysis
NPM-ALK was digested into peptide fragments suitable for LC-MS by on-bead tryptic or chymotryptic digestions (21–23), with modifications developed by our laboratory (22, 24–26); the different cleavage sites of trypsin and chymotrypsin allow for increased sequence coverage during LC-MS. Data base searches by MASCOT, restricted to Homo sapiens in the Swiss-Prot Database, were performed as described previously (22). Assigned sequence and spectra were imported into a manual data base including the peptide sequences of NPM-ALK and the 7 KAL mutants. An increased molecular mass of 79.96 Da on a tyrosine residue was indicative of phosphorylation. Peptide fragments containing a phosphorylated tyrosine residue, with a matching score above the MASCOT threshold score for identity (95% confident level), were manually examined. The phosphorylation profile for NPM-ALK or 7 KAL mutants was an assembly of three independent experiments (twice using trypsin and one using chymotrypsin). For each experiment, the resulting peptide mix was analyzed with five consecutive runs, with each run carried out using an optimal and maximized sample loading (26); peptide precursor ion exclusion strategy was applied to exclude relatively high abundance peptides identified from the previous runs, thus allowing the identification of relatively lower abundance peptides (27). No further phosphorylated residues were identified after the fourth stage of analysis. The final identified phosphorylated tyrosine residue was only considered as true positive if it was identified from at least two tryptic digestion experiments or one tryptic digestion and one chymotryptic digestion experiment.
Western Blot Analysis
Cell lysates from GP293 cells transfected with HB/NPM-ALK, NPM-ALK, or one of the mutants were subjected to SDS-PAGE and immunoblotting. The following antibodies, all of which were obtained from Cell Signaling Technology (Danver, MA) unless otherwise stated, were used for immunoblotting: phospho-STAT3 (Tyr705), STAT3 (BD Biosciences), phospho-Src (Tyr416), phospho-Src (Tyr527), phospho-Shc (Tyr239/240), Shc, phospho-ZAP70 (Tyr493), ZAP-70, phospho-S6 ribosomal protein (Ser235/236), S6 ribosomal protein, phospholipase Cγ1 (PLCγ1), and phospho-PLCγ1 (Tyr783; AssayDesigns, Ann Arbor, MI). NPM-ALK was detected using either an anti-NPM (LabVision clone NA24, Thermo Fisher Scientific, Fremont, CA) or an anti-ALK (Cell Signaling Technology) monoclonal antibody. For the detection of the KAL mutants, an anti-ALK antibody reactive with an epitope carboxyl to the kinase domain was used. For the detection of the 8 single mutants of NPM-ALK, an anti-NPM monoclonal antibody was used, since none of 8 tyrosine residues were located in the NPM portion of NPM-ALK.
Cell Viability and Colony Formation Assays
GP293 cells transfected with NPM-ALK, one of the 7 KAL mutants or one of the 8 single mutants were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum for 24 h. Transfected GP293 cells (at a final concentration of 0.5 × 104 cells/100 μl) were seeded in each well of a 96-well plate and cultured for 48 h, and the cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay (Promega, Madison, WI) following the manufacturer's protocol (28). For the colony formation assay, transfected GP293 cells (20,000 cells) were seeded in each well of a 6-well plate with soft agarose. The soft agarose consisted of two layers, both of which were prepared from a stock 1.2% agarose (Bio-Rad) that was dissolved in distilled water and autoclaved. For the bottom layer, Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum was added to the stock agarose to achieve a 0.6% agarose concentration. For the top layer, cell suspension was mixed with the stock agarose to achieve a final agarose concentration of 0.35%. Cells in the agar were fed with 200 μl of Dulbecco's modified Eagle's medium/10% fetal bovine serum every 3 days. Colonies were stained and visualized with 0.05% crystal violet after 4 weeks of culture.
RESULTS
Identification of 11 Tyrosine Residues Involved in Phosphorylation
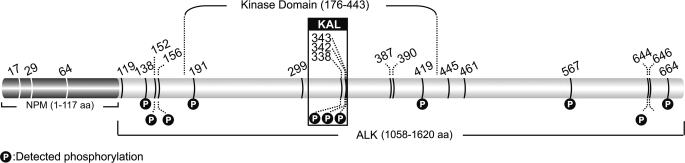
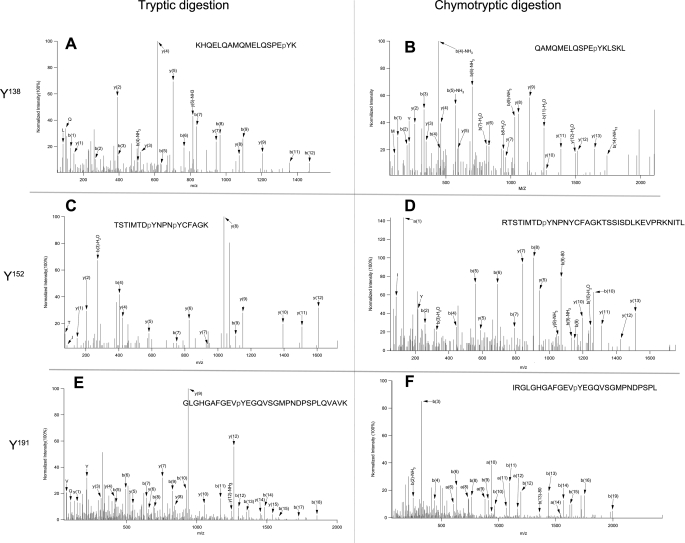
Within NPM-ALK, there are a total of 21 tyrosine residues (Fig. 1). Using two different digestion enzymes (trypsin and chymotrypsin), we were able to assess 95% of the NPM-ALK amino acid sequence using a 95% confident level (Fig. 2). Unfortunately, the first tyrosine residue located in the NPM portion of NPM-ALK (Tyr17) fell into the 5% of the amino acid sequence for which the confident level was <95%. Of the remaining 20 tyrosine residues present on NPM-ALK, we found evidence of phosphorylation in 11, and their locations are summarized in Fig. 1. The tandem mass spectrometry (MS/MS) spectra of these 11 tyrosine residues are illustrated in Fig. 3 and supplemental Fig. 1. All of these 11 tyrosine residues were found in the ALK portion; all of the 3 tyrosine residues (Tyr338, Tyr342, and Tyr343) present in the KAL were found to be phosphorylated, and 8 tyrosine residues found outside the KAL were also phosphorylated (including Tyr138, Tyr152, Tyr156, Tyr191, Tyr419, Tyr567, Tyr644, and Tyr664). Evidence of phosphorylation of 2 additional tyrosine residues (Tyr461 and Tyr646) was found, but only in one of the three independent experiments; in view of their low confident spectra, we excluded these 2 tyrosine sites from our phosphorylation list.
FIGURE 1.
Schematic illustration of the 21 tyrosine residues on NPM-ALK. The locations of the 21 tyrosine residues, with reference to the kinase domain and the YxxxYY motif of the kinase activation loop, are provided. Specific tyrosine residues (n = 11) showing evidence of phosphorylation, and their equivalent positions in the full-length ALK (in parentheses), are summarized as follows: Tyr138 (Tyr1078), Tyr152 (Tyr1092), Tyr156 (Tyr1096), Tyr191 (Tyr1131), Tyr338 (Tyr1278), Tyr342 (Tyr1282), Tyr343 (Tyr1283), Tyr419 (Tyr1359), Tyr567 (Tyr1507), Tyr644 (Tyr1584), and Tyr664 (Tyr1604). aa, amino acid(s).
FIGURE 2.
The sequence coverage of NPM-ALK obtained by LC-MS using peptide precursor ion exclusion strategy. 95% of the NPM-ALK amino acid sequence was covered; sequences obtained by using trypsin are underlined with solid lines, whereas sequences obtained by using chymotrypsin are underlined with dashed lines.
FIGURE 3.
Representative electrospray ionization tandem mass spectrometry spectra of phosphopeptides. Electrospray ionization tandem mass spectrometry spectra of three representative identified phosphotyrosine residues (pTyr138 (A and B), pTyr152 (C and D), and pTyr191 (E and F)) from tryptic (A, C, and E) and chymotryptic digested peptides (B, D, and F). The remaining spectra are available in supplemental Fig. 1.
The Significance of the 3 Tyrosine Residues in the KAL
We then characterized how the 3 tyrosine residues in the KAL regulate the phosphorylation process of NPM-ALK. Using site-directed mutagenesis of one or more of these 3 tyrosine residues in KAL, we created a total of 7 mutants, as summarized in Table 1. Using our TAP/LC-MS method and GP293 cells transfected with NPM-ALK (or one of the KAL mutants), the tyrosine phosphorylation profiles of the expressed NPM-ALK proteins were comprehensively assessed.
Within the KAL
Results are summarized in Table 1. A Tyr-to-Phe mutation of any 1 of the 3 tyrosine residues in the KAL did not affect the phosphorylation of the remaining 2 tyrosine residues. While the phosphorylation of Tyr338 was preserved when both Tyr342 and Tyr343 were mutated (i.e. double mutant YFF), no evidence of phosphorylation was detectable in Tyr342 or Tyr343 in the other 2 double mutants, FYF or FFY, respectively. These data suggests that Tyr338 is likely the first tyrosine residue to be phosphorylated in the KAL.
Outside of the KAL
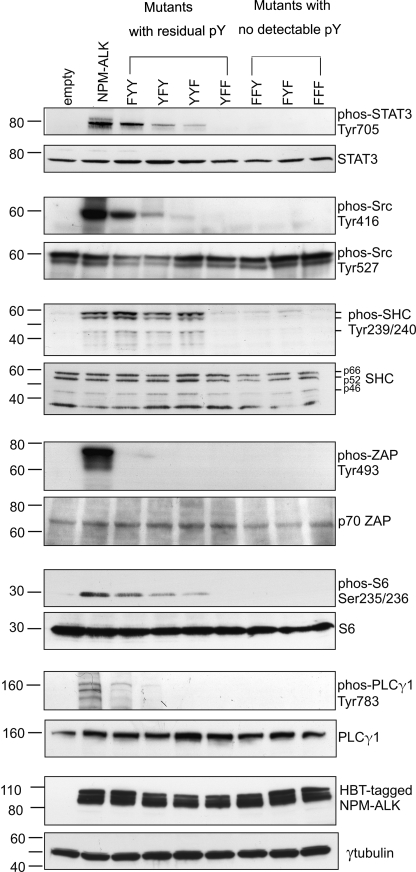
As described above, we found evidence of phosphorylation of 8 tyrosine residues outside the KAL. The phosphorylation of these 8 tyrosine residues was profoundly affected by any mutation of the 3 KAL tyrosine residues, as summarized in Table 1. A simultaneous Tyr-to-Phe mutation of all 3 tyrosine residues (i.e. triple mutant FFF), as well as mutation of Tyr338 in combination with Tyr342 or Tyr343 in the KAL (i.e. FFY, FYF) resulted in a complete loss of phosphorylation of all tyrosine residues, inside or outside the KAL. In the other 4 KAL mutants, abrogation of the phosphorylation outside the KAL was found only in a subset of these 8 tyrosine residues. Specifically, mutants with substitution of Tyr338 (FYY) or Tyr343 (YYF), as well as the double mutant YFF, had two residual phosphotyrosine residues outside the KAL. The most preserved phosphorylation pattern was seen with YFY, which retained 4 phosphotyrosine residues outside the KAL. Interestingly, the loss of phosphorylation due to mutations in the KAL was not a random process; of the 8 tyrosine residues outside the KAL, Tyr156 and Tyr567 were the least affected, whereas Tyr152, Tyr191, Tyr419, and Tyr664 were consistently affected with any mutation of the KAL. We also noted that evidence of tyrosine phosphorylation was found exclusively in the originally identified 8 phosphorylatable sites in these 7 KAL mutants.
The Relationship between the NPM-ALK Phosphorylation Profile and Cell Viability
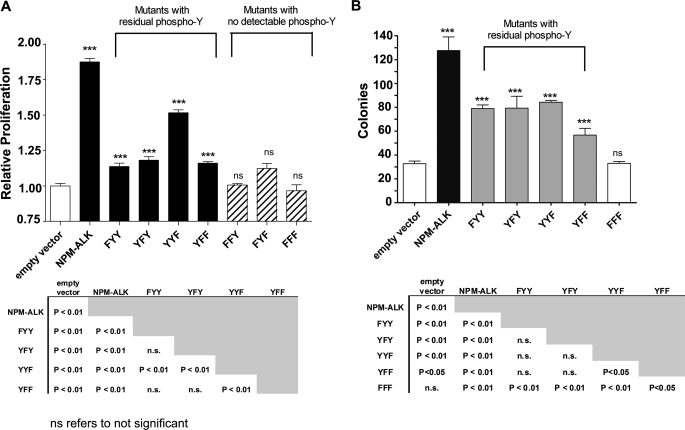
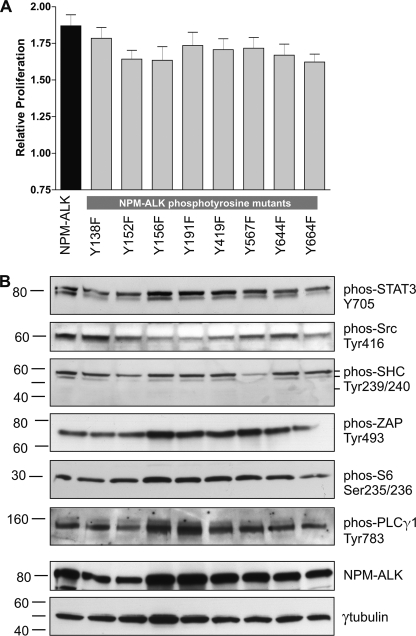
Using our panel of KAL mutants, we then asked whether the KAL of NPM-ALK regulates the oncogenic potential of NPM-ALK by virtue of controlling the number of phosphotyrosine residues. To test this hypothesis, we first assessed the cell viability of GP293 cells transfected with one of the seven KAL mutants using the MTS assay. As shown in Fig. 4, the 3 KAL mutants that showed no detectable tyrosine phosphorylation (i.e. FFF, FFY, and FYF) had no significant difference in the cell viability when compared with cells transfected with an empty vector (i.e. negative control). Cells transfected with the other 4 KAL mutants, all of which had reduced numbers of phosphotyrosine residues outside the KAL, showed a significant increase in cell viability over the negative control (p < 0.01), but the effects were significantly less than cells transfected with unaltered NPM-ALK (p < 0.01). In the 4 KAL mutants with residual phosphotyrosine residues, there was no simple correlation between the number of phosphotyrosine residues and the conferred increase in cell viability. Specifically, cells transfected with the YYF mutant, which had 2 phosphotyrosine residues outside the KAL (i.e. Tyr156 and Tyr567) displayed a significantly higher number of viable cells compared with those transfected with the YFY mutant, which carried 4 phosphotyrosine residues outside the KAL (i.e. Tyr138, Tyr156, Tyr567, and Tyr644) (p < 0.01). Moreover, the number of viable cells in those transfected with YFY was not significantly higher than those transfected with either FYY or YFF, both of which had only 2 phosphotyrosine residues outside the KAL (i.e. Tyr156 and Tyr567). To further evaluate the relationship between the phosphorylation profile of various KAL mutants and their tumorigenicity, colony formation assay was also performed, and our results were essentially aligned with those of the MTS assay (Fig. 4B).
FIGURE 4.
Characterization of the NPM-ALK KAL mutants-associated tumorigenicity. A, cell viability of GP293 cells transfected with HB/NPM-ALK or one of the seven HB-tagged KAL mutants, measured at 48 h post-transfection using MTS assay, is shown. Triplicate experiments were performed. These results were normalized against GP293 cells transfected with the empty HB vector. B, colony formation of GP293 cells transfected with HB/NPM-ALK or one of the three KAL single tyrosine mutant or YFF mutant or FFF mutant is shown. Triplicate experiments were performed. Statistical analysis was performed using one-way analysis of variance. ***, significant difference; ns, not significant.
The Relationship between the NPM-ALK Phosphorylation Profile and the Ability to Phosphorylate Its Downstream Targets
To further assess the oncogenic potential of the seven KAL mutants, we measured their ability to phosphorylate a panel of downstream effector proteins, including STAT3, Src, Shc, ZAP, S6, and PLCγ1, all of which are known to be phosphorylated by NPM-ALK. As shown in Fig. 5, unaltered NPM-ALK phosphorylated all of these 6 substrates strongly. Consistent with our finding that FFF, FFY and FYF do not mediate phosphorylation, all three mutants failed to induce any detectable phosphorylation of the 6 substrates. The 4 KAL mutants that retained residual phosphorylated tyrosine residues were less efficient in phosphorylating these targets. Similar to the MTS and colony formation data, the number of phosphotyrosine residues in these 4 mutants showed no simple correlation with the ability to phosphorylate the 6 substrates. Specifically, the FYY mutant (with 2 residual phosphotyrosine residues outside the KAL) mediated the highest level of substrate phosphorylation among the 4 mutants. In contrast, YFF, which also had the same 2 phosphotyrosine residues outside the KAL as FYY, had no detectable phosphorylation of these 6 substrates. YFY mutant, which carried 4 phosphotyrosine residues outside the KAL, mediated substantially weaker phosphorylation in all but 1 substrate.
FIGURE 5.
The effect of NPM-ALK KAL mutants on the tyrosine phosphorylation of NPM-ALK substrates. GP293 cells were transfected with HB/NPM-ALK or one of the seven HB-tagged KAL mutants. Cell lysates were harvested 24 h after gene transfection and analyzed by immunoblot to detect tyrosine phosphorylation of a number of NPM-ALK substrates. pY, phosphotyrosine; phos, phospho.
The Effects of a Single Tyr-to-Phe Mutation of Tyrosine Residues Outside the KAL
Because we observed that mutations of the KAL resulted in dramatic decreases in the phosphorylation of the 6 effector proteins, we investigated the possibility that 1 of these 8 tyrosine residues may serve as their docking sites, and a Tyr-to-Phe mutation of these sites is expected to be sufficient to abrogate the phosphorylation of these substrates. Thus, we transfected GP293 cells with various single mutants, each of which carried mutation of one of the 8 tyrosine residues outside the KAL. As illustrated in Fig. 6, with the exception of Shc and Src, the phosphorylation of these substrates was not appreciably reduced in these 8 mutants. Shc phosphorylation was dramatically decreased upon mutation of Tyr567, and this finding is consistent with that reported in the literature (18). The phosphorylation of Src was also decreased, but this decrease was found in 4 different mutants, including Tyr156, Tyr191, Tyr419, and Tyr567. We performed MTS assay; no significant difference was found when we compared each of these 8 single mutants with unaltered NPM-ALK (Fig. 6). Similarly, no significant difference using the colony formation was found between each of these single mutants and unaltered NPM-ALK (data not shown).
FIGURE 6.
The effect of single mutations of various tyrosine residues on NPM-ALK. GP293 cells were transfected with pcDNA3.1(+)/NPM-ALK, or one of eight NPM-ALK single mutants. A, MTS-based cell viability assay was performed 48 h post-transfection. Triplicate experiments were performed. Statistical analysis was performed using one-way analysis of variance, and no significant difference was found. Error bar represents S.E. B, cell lysates prepared at 24 h post-transfection were analyzed by Western blot to assess changes in the tyrosine phosphorylation of various NPM-ALK substrates. γ-Tubulin served as the loading control. phos, phospho.
DISCUSSION
To fully characterize the KAL of NPM-ALK, which is believed to be crucial for the activation of this oncogenic fusion protein, we employed a highly sensitive TAP/LC-MS method. As compared with immunoprecipitation and immunoblotting (with an anti-tyrosine antibody), a method used most commonly to study KAL in other members of the insulin receptor kinase subfamily, TAP/LC-MS is superior because this method allows us to comprehensively profile the phosphorylation status of individual tyrosine residues of NPM-ALK. Moreover, the HB tag was specifically selected to be used in this study because it can endure harsh denaturing washing condition (i.e. 8 m urea or 2% SDS can be added to the washing buffer) (21). Thus, we were able to effectively wash away NPM-ALK-binding partners and substantially reduce the background. This aspect is particularly important to this study, as one of the major objectives was to assess the tyrosine phosphorylation status of NPM-ALK itself, and the phosphopeptides derived from NPM-ALK would have been easily outnumbered by those derived from NPM-ALK-binding partners, if washing was not sufficiently harsh. With this powerful tool, we were able to map the tyrosine phosphorylation sites of NPM-ALK, and more importantly, assessed their changes upon our experimental manipulation of the KAL.
Our TAP-LC/MS protocol has proven to produce highly reproducible results, as we achieved consistent conclusions in three independent experiments. In addition, the spectra for all of the phosphorylated tyrosine residues are of high quality, as illustrated in Fig. 3 and supplemental Fig. 1. More importantly, the validity of our data is supported by the fact that our identified 11 phosphorylable tyrosine residues of NPM-ALK largely overlap with those identified in a very recent study (29). Specifically, using proteomic-based approaches, Boccalatte et al. (29) profiled ALK-associated tyrosine phospho-peptides using a panel of ALK-expressing lymphoma cell lines, and they identified 11 phosphorylated tyrosine residues on ALK, with 10 of these 11 tyrosine residues overlapping with our results. The exceptions are Tyr419 and Tyr646, with the former found only in our study and the latter found only in the study by Boccalatte et al. (29). These discrepancies may be related to the differences in the cell lines employed and/or proteomic-based methods. Of note, we found evidence of phosphorylation for Tyr646 in one of three experimental runs, but this tyrosine residue was excluded from our final list because our inclusion criteria requires high consistency (as described in “Materials and Methods”) and a high confident spectrum, both of which lack Tyr646.
Our results led us to conclude that Tyr338 is the first tyrosine residue within the KAL to be phosphorylated, because the phosphorylation of Tyr338 was not dependent on that of either Tyr342 or Tyr343. This conclusion is consistent with that of a previous study employing short synthetic ALK peptides and Edman sequencing (7). Interestingly, in two members of the insulin receptor kinase subfamily, namely the insulin receptor and the insulin-like growth factor 1 receptor, it is the second tyrosine residue in the YxxxYY motif that is initially phosphorylated (30). It is unclear why this order of tyrosine phosphorylation within the KAL is different among the insulin receptor kinase subfamily members, but it may be related to the differences in their amino acid sequence between the first and second tyrosine residues (7). Nevertheless, some of our conclusions regarding the KAL of NPM-ALK are contradictory to the very same study. While it was suggested that Tyr338 is the only tyrosine residue in the KAL that is important in regulating the autophosphorylation of NPM-ALK (7), our results strongly suggest that both Tyr342 and Tyr343 contribute to the phosphorylation and oncogenic potential of NPM-ALK. Specifically, YFY and YYF, as well as the double mutant YFF, were found to have a dramatic decrease in the number of phosphotyrosine residues outside the KAL and a significant decrease in the cell growth-promoting effects. Our study results also clearly showed evidence of phosphorylation of Tyr342 and Tyr343 in the FYY mutant. We believe that these discrepancies are likely due to differences in the methodology, namely the use of synthetic short ALK peptides and Edman sequencing versus TAP-LC/MS. Of note, our conclusions correlate well with the observations that all 3 tyrosine residues within the KAL are phosphorylated independently in the other members of the insulin receptor kinase subfamily (6, 31).
One of our major objectives is to characterize the biological importance of the KAL in mediating phosphorylation of various tyrosine residues in NPM-ALK and how this phosphorylation process is linked to oncogenic potential. Because the tyrosine phosphorylation status of the KAL of the insulin receptor kinase subfamily has been shown to determine whether the kinase is in an inactive or active conformation (6), it has been inferred that the KAL of NPM-ALK is also important in mediating the oncogenic effects of NPM-ALK. As discussed, only one previous study directly addresses this question for NPM-ALK (2). Consistent with the results published by Tartari et al. (2), our data supports that phosphorylation of all 3 tryosine residues in the KAL of NPM-ALK is important, because mutations of ≥1 of these 3 tyrosine sites substantially reduced or completely abrogated its phosphorylation and oncogenic potential. For the first time, with the use of TAP/LC-MS, we were able to reveal details of how the KAL of NPM-ALK regulates the phosphorylation status of individual tyrosine residues on NPM-ALK. Specifically, our study has provided direct proof that the number of phosphorylated tyrosine residues decreased dramatically with mutation of the 3 tyrosine residues in the KAL. Interestingly, we found that the 8 tyrosine residues outside the KAL have different thresholds for losing its phosphorylation status, with Tyr156 and Tyr567 being the best to retain phosphorylation whereas Tyr152, Tyr191, Tyr419, Tyr664 having the lowest threshold level for losing the phosphorylation status. Based on these results, it is tempting to speculate that Tyr156 and Tyr567 carry more biological importance than Tyr152, Tyr191, Tyr419, Tyr664, because the 3 KAL mutants that retain phosphorylation of Tyr156 and Tyr567 only (i.e. FYY, YYF and YFF) produced a high number of viable cells as compared with the negative control. The interesting observation is that none of the KAL mutants show evidence of tyrosine phosphorylation in those residues that are not involved in phosphorylation in the unaltered NPM-ALK. The mechanism underlying their resistance to tyrosine phosphorylation requires further studies.
The lack of significant decrease in oncogenicity in association with mutations of each of the 8 tyrosine residues outside the KAL is relatively surprisingly, because it is widely believed that these phosphorylated tyrosine residues serve as the “docking sites” for the downstream effectors, which mediate the oncogenic effects of NPM-ALK. With the exception of Shc and Src, we could not identify any detectable decrease in the phosphorylation of the remaining 4 substrates, even though their phosphorylation was completely abrogated in many of the KAL mutants. Our finding that mutation of Tyr567 substantially decreased the phosphorylation of Shc is entirely consistent with that described in a previous report (1). However, we did not identify a detectable decrease in the phosphorylation of PLCγ in the Tyr664 mutant, as previously suggested (1, 18). While the phosphorylation of Src was decreased, this decrease was found in 4 different mutants (including Tyr156, Tyr191, Tyr419, and Tyr567), and thus, no specific docking site for Src was identified. Our results argue against the concept that the KAL regulates the oncogenic potential of NPM-ALK simply by controlling the number of phosphotyrosine residues outside the KAL, or by providing the docking sites for the interaction with NPM-ALK effector proteins. Our observation that the number of residual phosphotyrosine residues in the 4 KAL mutants does not simply correlate with the oncogenic potential further supports this notion.
In conclusion, using our TAP/LC-MS method, we have generated a comprehensive profile of the tyrosine phosphorylation status of NPM-ALK. Using this technique, we assessed the biological importance of KAL, and we believe that our data has provided insights into the functional importance of the KAL. Our study has provided evidence that the biological importance of the KAL of NPM-ALK lies beyond the regulation of its tyrosine phosphorylation pattern. Further studies are needed to further characterize the mechanism by which the KAL of NPM-ALK regulates the oncogenic effects of this protein. Importantly, our study provides the proof-of-principle that TAP/LC-MS can be used effectively to characterize the phosphorylation process of various oncogenic tyrosine kinases.
Supplementary Material
Acknowledgments
We thank Dr. Peter Kaiser for providing the HB-vector and Dr. Stephen W. Morris for providing the NPM-ALK cDNA plasmid.
This work was supported by the Canadian Institutes of Health Research and the Alberta Cancer Research Institute (to R. L.). This work was also supported by the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs Program (to L. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- KAL
- kinase activation loop
- TAP
- tandem affinity purification
- LC-MS
- liquid chromatography-mass spectrometry
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt.
REFERENCES
- 1.Fujimoto J., Shiota M., Iwahara T., Seki N., Satoh H., Mori S., Yamamoto T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4181–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tartari C. J., Gunby R. H., Coluccia A. M., Sottocornola R., Cimbro B., Scapozza L., Donella-Deana A., Pinna L. A., Gambacorti-Passerini C. (2008) J. Biol. Chem. 283, 3743–3750 [DOI] [PubMed] [Google Scholar]
- 3.Amin H. M., Lai R. (2007) Blood 110, 2259–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris S. W., Kirstein M. N., Valentine M. B., Dittmer K. G., Shapiro D. N., Saltman D. L., Look A. T. (1994) Science 263, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 5.Delsol G., Ralfkiaer E., Stein H., Wright D., Jaffe E. S. (2001) in Pathology and Genetics Tumors of Haematopoietic and Lymphoid Tissues: World Health Organization Classification of Tumors (Jaffe E. S., Harris N. L., Stem H., Vardiman J. eds.), pp. 230–235, IARC Press, Lyon, France [Google Scholar]
- 6.Zhang B., Tavare J. M., Ellis L., Roth R. A. (1991) J. Biol. Chem. 266, 990–996 [PubMed] [Google Scholar]
- 7.Donella-Deana A., Marin O., Cesaro L., Gunby R. H., Ferrarese A., Coluccia A. M., Tartari C. J., Mologni L., Scapozza L., Gambacorti-Passerini C., Pinna L. A. (2005) Biochemistry 44, 8533–8542 [DOI] [PubMed] [Google Scholar]
- 8.Piva R., Chiarle R., Manazza A. D., Taulli R., Simmons W., Ambrogio C., D'Escamard V., Pellegrino E., Ponzetto C., Palestro G., Inghirami G. (2006) Blood 107, 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan W., Albom M. S., Lu L., Quail M. R., Becknell N. C., Weinberg L. R., Reddy D. R., Holskin B. P., Angeles T. S., Underiner T. L., Meyer S. L., Hudkins R. L., Dorsey B. D., Ator M. A., Ruggeri B. A., Cheng M. (2006) Blood 107, 1617–1623 [DOI] [PubMed] [Google Scholar]
- 10.Han Y., Amin H. M., Franko B., Frantz C., Shi X., Lai R. (2006) Blood 108, 2796–2803 [DOI] [PubMed] [Google Scholar]
- 11.Slupianek A., Nieborowska-Skorska M., Hoser G., Morrione A., Majewski M., Xue L., Morris S. W., Wasik M. A., Skorski T. (2001) Cancer Res. 61, 2194–2199 [PubMed] [Google Scholar]
- 12.Slupianek A., Skorski T. (2004) Exp. Hematol. 32, 1265–1271 [DOI] [PubMed] [Google Scholar]
- 13.Turner S. D., Yeung D., Hadfield K., Cook S. J., Alexander D. R. (2007) Cell Signal 19, 740–747 [DOI] [PubMed] [Google Scholar]
- 14.Marzec M., Kasprzycka M., Liu X., El-Salem M., Halasa K., Raghunath P. N., Bucki R., Wlodarski P., Wasik M. A. (2007) Oncogene 26, 5606–5614 [DOI] [PubMed] [Google Scholar]
- 15.Marzec M., Kasprzycka M., Liu X., Raghunath P. N., Wlodarski P., Wasik M. A. (2007) Oncogene 26, 813–821 [DOI] [PubMed] [Google Scholar]
- 16.Leventaki V., Drakos E., Medeiros L. J., Lim M. S., Elenitoba-Johnson K. S., Claret F. X., Rassidakis G. Z. (2007) Blood 110, 1621–1630 [DOI] [PubMed] [Google Scholar]
- 17.Falini B., Nicoletti I., Bolli N., Martelli M. P., Liso A., Gorello P., Mandelli F., Mecucci C., Martelli M. F. (2007) Haematologica 92, 519–532 [DOI] [PubMed] [Google Scholar]
- 18.Bai R. Y., Dieter P., Peschel C., Morris S. W., Duyster J. (1998) Mol. Cell Biol. 18, 6951–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu F., Wang P., Young L. C., Lai R., Li L. (2009) Am. J. Pathol. 174, 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser P., Meierhofer D., Wang X., Huang L. (2008) Methods Mol. Biol. 439, 309–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) Mol. Cell Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 22.Wang N., Mackenzie L., De Souza A. G., Zhong H., Goss G., Li L. (2007) J. Proteome Res. 6, 263–272 [DOI] [PubMed] [Google Scholar]
- 23.Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 24.Doucette A., Craft D., Li L. (2000) Anal. Chem. 72, 3355–3362 [DOI] [PubMed] [Google Scholar]
- 25.Doucette A., Craft D., Li L. (2003) J. Am. Soc. Mass Spectrom. 14, 203–214 [DOI] [PubMed] [Google Scholar]
- 26.Wang P., Lo A., Young J. B., Song J. H., Lai R., Kneteman N. M., Hao C., Li L. (2009) J. Proteome Res. 8, 3403–3414 [DOI] [PubMed] [Google Scholar]
- 27.Wang N., Li L. (2008) Anal. Chem. 80, 4696–4710 [DOI] [PubMed] [Google Scholar]
- 28.Bard J. D., Gelebart P., Anand M., Amin H. M., Lai R. (2008) Leukemia 22, 1595–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boccalatte F. E., Voena C., Riganti C., Bosia A., D'Amico L., Riera L., Cheng M., Ruggeri B., Jensen O. N., Goss V. L., Lee K., Nardone J., Rush J., Polakiewicz R. D., Comb M. J., Chiarle R., Inghirami G. (2009) Blood 113, 2776–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard S. R., Wei L., Ellis L., Hendrickson W. A. (1994) Nature 372, 746–754 [DOI] [PubMed] [Google Scholar]
- 31.Levine B. A., Clack B., Ellis L. (1991) J. Biol. Chem. 266, 3565–3570 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.