Abstract
Neurons in the geniculate ganglion, like those in other sensory ganglia, are dependent on neurotrophins for survival. Most geniculate ganglion neurons innervate taste buds in two regions of the tongue and two regions of the palate; the rest are cutaneous nerves to the skin of the ear. We investigated the expression of four neurotrophins, nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin 3 (NT-3), and NT-4, and five neurotrophin receptors, trkA, trkB, trkC, p75, and truncated trkB (Trn-B) in single sensory neurons of the adult rat geniculate ganglion associated with the five innervation fields. For fungiform papillae, a glass pipette containing biotinylated dextran was placed over the target papilla and the tracer was iontophoresed into the target papilla. For the other target fields, Fluoro-Gold was microinjected. After 3 days, geniculate ganglia were harvested, sectioned, and treated histochemically (for biotinylated dextran) or immunohistochemically (for Fluoro-Gold) to reveal the neurons containing the tracer. Single labeled neurons were harvested from the slides and subjected to RNA amplification and RT-PCR to reveal the neurotrophin or neurotrophin receptor genes that were expressed. Neurons projecting from the geniculate ganglion to each of the five target fields had a unique expression profile of neurotrophin and neurotrophic receptor genes. Several individual neurons expressed more than one neurotrophin receptor or more than one neurotrophin gene. Although BDNF is significantly expressed in taste buds, its primary high affinity receptor, trkB, was not prominently expressed in the neurons. The results are consistent with the interpretation that at least some, perhaps most, of the trophic influence on the sensory neurons is derived from the neuronal somata, and the trophic effect is paracrine or autocrine, rather than target derived. The BDNF in the taste bud may also act in a paracrine or autocrine manner on the trkB expressed in taste buds, as shown by others.
Keywords: neurotrophins, NGF, BDNF, NT-3, NT-4, neurotrophin receptors, trk, taste bud, chorda tympani, greater superficial petrosal nerve
The rat geniculate ganglion contains approximately 1,500–1,700 sensory neurons (Miller et al., 1978). Three sensory nerve bundles originate from the ganglion to innervate five innervation fields as follows: (1) the chorda tympani (CT), which divides into two branches when it reaches the tongue (one innervating taste buds in the fungiform papillae and one innervating taste buds in anterior foliate papillae); (2) the greater superficial petrosal nerve (GSP), which divides into two branches when it reaches the palate (one innervating taste buds in the incisive papilla behind the incisor teeth and one innervating the taste buds in the soft palate including the “Geschmack-streifen” [taste stripes]); and (3) the posterior auricular nerve, a cutaneous nerve innervating the skin of the external ear. The neurons in the geniculate ganglion, like those in other sensory ganglia, are dependent on neurotrophins for their survival during development and in adulthood (e.g., for review see Chao, 1992; Schechterson and Bothwell, 1992; Barbacid, 1994; Davies, 1994, 1996, 1997; Bothwell, 1995; Lewin and Barde, 1996; Levi-Montalcini and Angeletti, 1997; Bibel and Barde, 2000; Huang and Reichardt, 2003).
The neurotrophins of interest in this study, nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), and NT-4, are four structurally related secreted factors vital for survival and differentiation of many types of neurons in both the peripheral and central nervous system. There are two classes of receptors for this family of neurotrophins: the low-affinity receptor (p75) that binds all four neurotrophins, and the three high-affinity receptors (trks), all of which are receptor tyrosine kinases. NGF binds to trkA, BDNF and NT-4 bind to trkB and to a truncated isoform designated here as Trn-B (see below), and NT-3 binds to trkC and weakly to trkB, Trn-B, and trkA. We report on the expression of genes coding for neurotrophins and their receptors in neurons innervating the five innervation fields of the geniculate ganglion.
Ever since the seminal work of Nosrat and colleagues (Nosrat and Olson, 1995; Nosrat et al., 1997; Ringstedt et al., 1999), the importance of neurotrophins (particularly BDNF) in development and maintenance of taste buds and the geniculate has been appreciated, although the precise nature of action is not well understood. We examine the expression of BDNF and the other neurotrophins, as well as the neurotrophin receptors including a truncated version of trkB (Middlemas et al., 1991). There are two truncated isoforms of trkB. We have examined the one thought to be expressed more commonly, also known as trkB.T2, but designated herein as Trn-B. Trn-B protein has the ligand-binding part of the receptor but lacks the tyrosine kinase moiety; consequently it can bind the ligand but because the tyrosine kinase moiety is missing it is thought that no signal is transduced. BDNF or NT-4 thus could be bound by Trn-B, which could serve as a response modulator of the typical tyrosine kinase type of response because no signal is transduced (Biffo et al., 1995; Eide et al., 1996; Alderson et al., 2000; Luikart et al., 2003). There is evidence, however, that some kind of signaling can occur (probably by a different mechanism) when BDNF binds to Trn-B (Baxter et al., 1997).
Our results suggest that geniculate ganglion neurons may use neurotrophins released within the ganglion itself, as opposed to the innervation target as their trophic support for survival. Further in the taste buds themselves, autocrine/paracrine reception of trophic factors may be important.
MATERIALS AND METHODS
Injection
All experiments were carried out according to procedures approved by the University of Virginia Animal Care and Use Committee and National Institutes of Health Guidelines. The procedure used here for labeling neurons that innervate fungiform taste buds has been described previously in detail (Krimm and Hill, 1998). Female Sprague-Dawley rats, 40–50 days of age, from Harlan Sprague-Dawley (Indianapolis, IN) were anesthetized with methohexital sodium (Brevital; 50 mg/kg, intra-peritoneally). After anesthetization, the dorsal anterior half of the tongue was exposed from the mouth by gently pulling on the ventral tongue. The tongue was then stabilized by pressing the ventral surface to a glass slide covered with putty. Using a micromanipulator and surgical microscope, a glass pipette (approximately 150-μm diameter) containing biotinylated dextran (3 kDa, 10% wt/vol in distilled water; Molecular Probes) was placed over the target papilla, creating an electrical seal without penetrating the epithelium. A small wire (0.3-mm diameter) was inserted into the ventral tongue as the reference electrode. By applying a square, anodal pulse, the dextran was iontophoresed into the target papilla (2.0 μA positive current; 4 sec on/4 sec off for 5 min). At least five fungiform papillae were labeled for each rat (n = 3).
For all other receptive fields (foliate papillae, incisive papilla, soft palate, and ear), Fluoro-Gold (2% in distilled water; Fluorochrome, Englewood, NJ) was injected into the receptive field with a Hamilton syringe aided by the use of a surgical microscope. The volume used (15–40 μl) depended on the receptive field injected. In all cases, multiple injections were done to label the majority of the field.
Histological Procedures
Rats were sacrificed with a lethal dose of sodium pentobarbital 3 days after labeling, and were perfused intracardially with Krebs solution, followed by 8% paraformaldehyde (pH 7.2). Geniculate ganglia were removed and placed in 30% sucrose overnight. Serial 8-μm sections of the geniculate ganglia were cut with a cryostat and thaw-mounted on gelatin-coated glass slides.
Immunohistochemistry
For ganglia in which fungiform papillae were labeled with biotinylated dextran, sections were reacted with a standard avidin-biotin-peroxidase (ABC) diaminobenzidine reaction. For ganglia containing Fluoro-Gold, sections were blocked with 10% normal goat serum (30 min), followed by a primary antibody directed against Fluoro-Gold (1:2,000; Chemicon, Temecula, CA; for 1 hr), and then visualized through the use of a secondary antibody (biotinylated goat anti-rabbit; 1:250; Jackson Immunochemicals; for 30 min) and ABC diaminobenzidine reactions.
RNA Amplification
Details of the method are found in Eberwine and Crino (1997), and the modifications used in this study are found in Farbman et al., (2004). Briefly, a 1-cm2 reaction well of rubber cement was constructed around the sections on the slide and the sections were hybridized overnight to an oligo(dT)24 primer containing a 5′ extension encoding a T7 RNA polymerase promoter sequence in 50% formamide and 5× saline sodium citrate (SSC). The next morning the slides were rinsed with 2× SSC and cDNA synthesis was carried out directly on the slide with 0.5 U/μl AMV reverse transcriptase in 50 μl cDNA synthesis buffer for 90 min at 37°C. Sections were washed for 2 hr in 0.5× SSC. After this procedure, individual labeled cells were dissected from the sections using an inverted microscope (with a Sony video camera and a monitor) and a micromanipulator armed with glass micropipettes with an external tip diameter of ~1–2 μm (see Fig. 1). Cells were placed in 1.5-ml Eppendorf tubes containing 20 μl DEPC-treated H2O, one cell to a tube. Before and after images for most cells was saved on a computer disc.
Fig. 1.
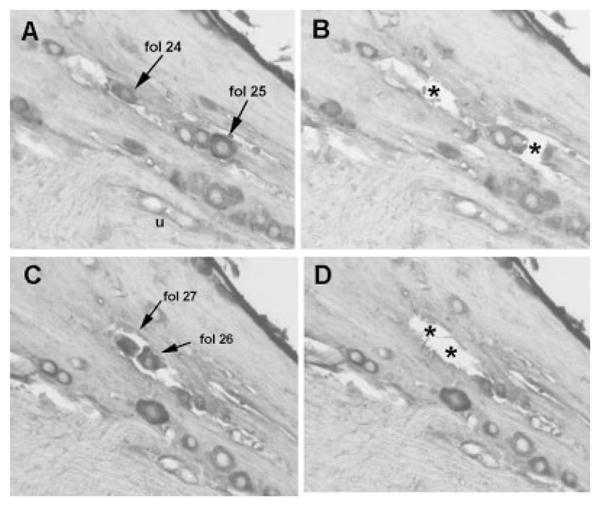
Paired images show sections of geniculate ganglia from animals that were injected in the region of the anterior foliate papillae. Two positively stained neurons (A) have been removed (B) and the spaces occupied by the neurons have been marked by asterisks. The letter “u” in A is positioned beneath two unstained neurons. The two stained neurons indicated in C were removed and the spaces they occupied marked by asterisks in D.
Tubes were heated to 95°C to separate RNA/DNA complexes, quick-cooled, and centrifuged. To make double-stranded cDNA, the following were added to the tube: 10 μl H2O; 4 μl 10× second strand buffer; 4 μl 2.5 mM 4dNTP mix; 1 μl 1U/μl T4 DNA polymerase; and 1 μl Klenow fragment (1U/μl). The tube was incubated for 2 hr at 14°C. This step comprises hairpin loop second-strand synthesis. To degrade single-stranded cDNA, we added 319 μl H2O, 1 μl of S1 nuclease, and 40 μl S1 nuclease buffer, and incubated for 5 min at 37°C. DNA was extracted with 400 μl phenol: chloroform (1:1), vortexed, and tubes were centrifuged for 1 min. Supernatant was transferred to a new sterile tube, 1 ml 100% ethanol was added, and the tube was placed at −80°C for 2 hr to overnight. Tubes were then microcentrifuged for 15 min at 4°C at maximum speed, ethanol was removed, and pellets in the tubes were allowed to air dry. Each pellet was suspended in 20 μl DEPC-treated H2O.
To create blunt ends on cDNA with Klenow fragment, to the suspended DNA we added 2.5 μl Klenow fragment buffer, 0.5 μl of 10 mM of each dNTP, and 0.5 μl Klenow fragment (1U/μl), and incubated for 15 min at 37°C. DNA was extracted by phenol/chloroform, the supernatant was transferred to a new tube, and 2.5 μl of 5 M NaCl and 75 μl ethanol was added. Tubes were placed at −80°C for 2 hr to overnight, then centrifuged for 15 min at 4°C at 13,000 rpm, ethanol was removed, and the tubes were allowed to air dry. The pellet was suspended in 20 μl DEPC-treated H2O.
To remove excess dNTPs, the contents of each tube were drop-dialyzed by placing 10 μl on a 0.025 μm Millipore filter disk floating on 50 ml of RNase-free H2O at room temperature for 4 hr to overnight. After drop-dialysis, RNA amplification was done by placing the dialyzed drop containing double-stranded DNA in a tube and adding 11.5 μl H2O, 2 μl RNA amplification buffer, 2 μl 100 mM dithiothreitol (DTT), 2 μl of 2.5 mM 4NTP mix, 0.5 μl of RNasin (0.1 U/μl), and 1 μl of T7 RNA polymerase (2,000 U/μl). Incubation was carried out for 4 hr at 37°C.
RNA was extracted with phenol/chloroform and sodium acetate. The supernatant was transferred to a new tube and 100% ethanol was added. The tube was placed at −80°C for 2 hr to overnight, and then centrifuged for 15 min at 4°C at maximum speed. The ethanol was then removed and tubes allowed to air dry. The pellet was suspended in 20 μl DEPC-treated H2O.
The amplified RNA (aRNA) was then reverse transcribed using random hexanucleotide primers. First the aRNA was denatured by heating for 3 min at 75°C. The following were added to the tube: 10 ng random hexanucleotide primers; 3 μl 100 mM DTT; 3 μl 2.5 mM 4dNTP mix; 0.5 μl RNasin; 2 μl AMV reverse transcriptase (20U/μl); and cDNA synthesis buffer. This mixture was incubated for 1 hr at 37°C, extracted with phenol/chloroform, and precipitated with ethanol as above. The product was used as a template to synthesize the second DNA strand, as above. Double-stranded DNA was extracted with phenol/chloroform, precipitated with ethanol as above, and the pellet was air dried and dissolved in 20 μl H2O.
PCR
Before PCR was carried out, 1 μl of each sample was mixed with 1 μl of ethidium bromide (2.5 μg/ml), placed on a piece of Parafilm, and exposed to ultraviolet light. A bright spot indicated the presence of DNA in the sample. Only positive samples were carried to the next step.
PCR reactions were carried out on each specimen essentially as described in Cho and Farbman (1999) in a MJ Research PTC-100 Thermal Cycler. Each reaction tube contained 2.5 units of AmpliTaq DNA polymerase, 2 μl of the DNA derived as described above, 50 pmol each of forward and reverse PCR primers, and 2 nmol of dNTPs, all in a 1× PCR buffer mixture. Primers and accession numbers are listed in Table I (Cho and Farbman, 1999). Every primer set used the same basic PCR program as follows, but with primer-specific annealing temperatures (Table I): 94°C for 9 min to dissociate DNA strands, then 94°C for 1 min, ~60°C for 2 min, and 72°C for 40 sec. This was repeated 35 more times but the last cycle at 72°C was extended to 4 min. The reaction was then stopped at 4°C to add more DNA polymerase and both primers, The reaction was then run for another 36 cycles. PCR products were analyzed electrophoretically on a 2% agarose gel consisting of 100 ml of 0.5× Tris-HCl/boric acid/EDTA (TBE) buffer at pH 8.3, 2 g of agarose, and 0.2 mg of ethidium bromide. Samples were run at 100 V for 1 hr in a running buffer consisting of 0.5× TBE. The gels were photographed under ultraviolet light.
TABLE I.
Primers Used in This Study★
| Gene | Accession number | Forward (F) and reverse (R) primer sequences (53 to 33) | Bases | Product size (bp) | Annealing temp (°C) |
|---|---|---|---|---|---|
| TrkA | M85214 |
F-agg gcc aca tca tgg aga ac R-cgt gca gac tcc aaa gaa gc |
1,530–1,549 1,791–1,810 |
281 | 61 |
| TrkB | M55291 |
F-tct cca cca cat ctc caa c R-cac aga cac cgt aga act tga c |
2,101–2,119 2,462–2,483 |
383 | 59.5 |
| Truncated TrkB | M55292 |
F-ctg ttg cct atc cca gga ag R-gag agg cac aat cca atg ag |
1,949–1,968 2,159–2,178 |
230 | 57 |
| TrkC | L03813 |
F-tcc atc aat act cat cag acc R-caa act caa tgc aat gtt cc |
799–819 1,002–1,021 |
223 | 58 |
| p75 | X05137 |
F-tgc agt gtg cag atg tgc cta tgg c R-agg aat gag ctt gtc ggt ggt gcc g |
428–452 851–875 |
448 | 55C |
| NGF | M36589 |
F-gcg agg tga aca tta aca ac R-tta cag gct gag gta ggg ag |
771–790 1,067–1,086 |
316 | 60 |
| BDNF | M61178 |
F-tct acg aga cca agt gta atc R-cat aaa tcc act atc ttc c |
676–696 870–888 |
213 | 56 |
| NT3 | M33968 |
F-gca acc ctt aca gta tat aag R-act gaa tgc caa ata ctg g |
985–1,005 1,116–1,134 |
150 | 55 |
| NT4 | S69323 |
F-ctg act gtg aac tga aat aac c R-tgc tag gca acc aga aac |
933–954 1,228–1,245 |
313 | 57 |
NGF, nerve growth factor; BDNF, brain-derived neurotrophic factor; NT, neurotrophin.
Positive controls for each primer pair were taken from embryonic Day 15 (E15) rat brain. Total RNA was extracted from these brains and reverse transcribed. Negative controls were not exposed to reverse transcriptase.
Subcloning and Sequencing
After the gel was run in some cases the band was cut out with a scalpel, the DNA extracted, and subcloned, and prepared for sequencing to verify its identity.. In each case the sequence was 98–100% identical to the sequence in the NCBI gene bank. Consequently, we followed this procedure for only 15–20% of the bands and were able to verify that each PCR band we tested contained the expected product.
The slice of agarose containing the band was extracted using the QIAEX II procedure (Qiagen, Valencia, CA). The extracted DNA was subcloned into Escherichia coli using the TOPO TA cloning kit (Invitrogen, Baltimore, MD). DNA was isolated and purified from the culture medium using the Perfect Prep DNA extraction procedure (Eppendorf), and sent to the Northwestern University Biotechnology Facility for sequencing. The sequences of the PCR products were then compared to published sequences by doing a BLAST search on the NCBI web site.
RESULTS
Of 265 adult rat geniculate ganglion neurons from the five injected innervation fields examined, 226 (85%) exhibited a signal for at least one mRNA of a neurotrophin or a trk. Neurons from each innervation field had a different profile of trk receptor gene expression and of neurotrophin gene expression. None of the 226 neurons gave a signal for the p75 gene. In many neurons, signals were detectable for the expression of more than one neurotrophin gene (38%) or more than one neurotrophin receptor (19%). Figure 2 is a photograph of a sample agarose gel showing bands for NT-3 and NT-4 from the same 13 neurons isolated from ganglion sections of rats injected with Fluoro-Gold in the soft palate.
Fig. 2.
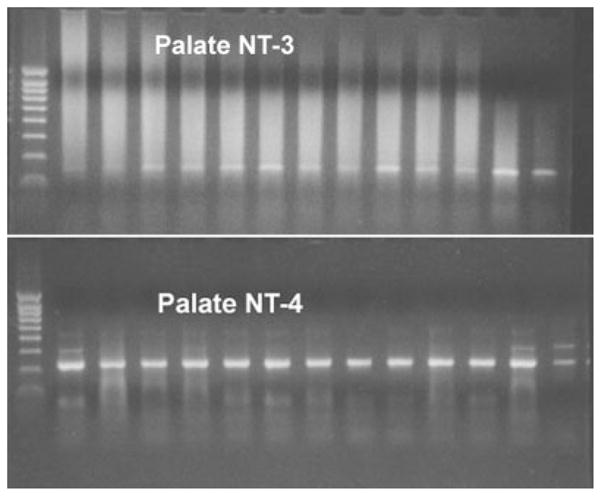
Representative PCR gels showing results from 13 ganglion neurons from specimen injected with a tracer into the soft palate. Top: Bands at 150 base pairs (bp) indicate expression of the amplimer for NT-3. Bottom: Bands at 313 bp indicate expression of the amplimer for NT-4. Left lane in both panels is the DNA ladder. Some neurons in the NT-4 panel exhibited a faint band at 400 bp.
Chorda Tympani Nerve
In terms of neurotrophin receptor expression, the two branches of the CT nerve were different from one another. In 91% of neurons innervating fungiform papillae, trkA message was detected, but messages for trkB, Trn-B, or trkC were not (Table II). It is interesting that this was the only nerve branch in which multiple receptor mRNAs were not expressed, although several of these neurons expressed more than one neurotrophin mRNA (Table III). In the other branch of the CT, trkA message was detected in only 35% of the neurons, but 38% expressed Trn-B and 11% expressed trkC.
TABLE II.
Number of Neurons in Each Nerve Branch That Are Positive for Indicated Neurotrophin or Neurotrophin Receptor★
| Chorda tympani |
Greater superficial petrosal |
Posterior auricular |
|||
|---|---|---|---|---|---|
| Gene | Fungiform | Foliate | Incisive pap | Soft palate | Ear |
| TrkA | 43 | 13 | 6 | 4 | 47 |
| TrkB | 0 | 0 | 7 | 11 | 2 |
| Trn-B | 0 | 14 | 0 | 36 | 15 |
| TrkC | 0 | 4 | 28 | 23 | 21 |
| p75 | 0 | 0 | 0 | 0 | 0 |
| NGF | 18 | 0 | 10 | 0 | 12 |
| BDNF | 5 | 0 | 3 | 0 | 2 |
| NT3 | 16 | 13 | 16 | 32 | 32 |
| NT4 | 16 | 25 | 7 | 41 | 34 |
| n (209) | 47 | 37 | 42 | 44 | 56 |
Value of n may be greater than the sum because some neurons expressed more than one receptor or neurotrophin (see Table III). NGF, nerve growth factor; BDNF, brain-derived neurotrophic factor; NT, neurotrophin.
TABLE III.
Occurrence of Single or Multiple Receptors (Top) or Single or Multiple Neurotrophins (Bottom)★
| Chorda tympani |
Greater superficial petrosal |
Posterior auricular |
|||
|---|---|---|---|---|---|
| NT or NT receptor | Fungiform | Foliate | Incisive pap | Soft palate | Ear |
| NT receptor | |||||
| TrkA | 43 | 9 | 1 | 0 | 23 |
| TrkB | 0 | 0 | 0 | 4 | 0 |
| TrkC | 0 | 1 | 17 | 2 | 1 |
| Trn-B | 0 | 11 | 0 | 9 | 3 |
| TrkA 3 B | 0 | 0 | 0 | 0 | 0 |
| TrkA 3 C | 0 | 2 | 4 | 0 | 12 |
| TrkB 3 C | 0 | 0 | 6 | 1 | 0 |
| TrkB 3 Trn-B | 0 | 0 | 0 | 5 | 0 |
| TrkA 3 B 3 C | 0 | 0 | 1 | 0 | 1 |
| TrkA 3 C3 Trn-B | 0 | 0 | 0 | 1 | 7 |
| TrkA 3 Trn-B | 0 | 2 | 0 | 2 | 4 |
| TrkB 3 C 3 Trn-B | 0 | 0 | 0 | 0 | 1 |
| TrkC 3 Trn-B | 0 | 1 | 0 | 18 | 0 |
| TrkA 3 B 3 C 3 Trn-B | 0 | 0 | 0 | 1 | 0 |
| No receptor | 4 | 11 | 13 | 1 | 4 |
| Sum | 47 | 37 | 42 | 44 | 56 |
| NT | |||||
| NGF | 3 | 0 | 8 | 0 | 2 |
| BDNF | 1 | 0 | 1 | 0 | 0 |
| NT3 | 1 | 3 | 13 | 1 | 13 |
| NT4 | 2 | 15 | 3 | 10 | 10 |
| NGF 3 BDNF | 1 | 0 | 0 | 0 | 1 |
| NGF 3 NT3 | 2 | 0 | 0 | 0 | 0 |
| NGF 3 NT4 | 1 | 0 | 1 | 0 | 3 |
| NT3 3 NT4 | 2 | 10 | 1 | 31 | 15 |
| NGF 3 NT3 3 NT4 | 9 | 0 | 0 | 0 | 4 |
| NGF 3 BDNF 3 NT3 3 NT4 | 1 | 0 | 0 | 0 | 1 |
| NGF 3 BDNF 3 NT4 | 1 | 0 | 1 | 0 | 0 |
| BDNF 3 NT3 | 0 | 0 | 1 | 0 | 0 |
| BDNF 3 NT3 3 NT4 | 1 | 0 | 0 | 0 | 0 |
| No neurotrophin | 22 | 9 | 13 | 2 | 7 |
| Sum | 47 | 37 | 42 | 44 | 56 |
NGF, nerve growth factor; BDNF, brain-derived neurotrophic factor; NT, neurotrophin.
Neither the nerves in the fungiform papilla branch nor those in the foliate papilla branch expressed full-length message for trkB, although in total 14 neurons expressed Trn-B either alone (11/14) or in combination with another trk message (Table III). This was somewhat surprising because we expected to see trkB, an expectation based on the presence of BDNF message and protein in taste buds, the target of these nerves (Nosrat and Olson, 1995; Ganchrow et al., 2003a,b; Yee et al., 2003). We discuss below possible reasons for this.
In other nerve branches, we found cells that expressed message for more than one receptor: foliate, 14%; incisive papilla, 26%; soft palate, 64%; and ear skin, 45%. In an earlier study on neurotrophin receptors in developing geniculate ganglia, we saw that 53% of 212 neurons at all ages expressed mRNAs for multiple receptors (Farbman et al., 2004).
The two branches of the CT differed also in their expression of the neurotrophin genes themselves. Approximately one-third of the fungiform papilla-associated neurons expressed genes for each of the three neurotrophins NGF, NT-3, and NT-4, whereas only about 10% expressed the gene for BDNF. Of 37 foliate papilla-associated neurons, however, about two-thirds expressed the gene for NT-4, 13 of 37 (35%) expressed the gene for NT-3 (Table II) and, of these, 10 (27%) expressed both NT-3 and NT-4 (Table III). The amplimers for NGF and BDNF were not detected.
TrkA is the high-affinity receptor for NGF. It was interesting to note that 14 of 18 neurons that expressed NGF (78%) also expressed trkA, thus opening the possibility that these cells had the potential to be autocrine. TrkA also responds weakly to NT-3. Of 16 neurons expressing the gene for NT-3, 14 (88%) also expressed trkA (Table IV). Again the potential exists here for autocrine stimulation. Finally, 12 of these neurons (75%) expressed both NGF and NT-3.
TABLE IV.
Number of Neurons Coexpressing a Neurotrophic Receptor and Receptor Ligand★
| Chorda tympani |
Greater superficial petrosal |
Posterior auricular |
|||
|---|---|---|---|---|---|
| Coexpressed NT | Fungiform | Foliate | Incisive pap | Soft palate | Ear |
| TrkA 3 NGF (NT3) | 14 (14) | 2 (4) | 3 | 0 (1) | 11 (24) |
| TrkB 3 BDNF, NT4 (NT3) | 0 | 0 | 2 | 10 (5) | 0 |
| TrkC 3 NT3 | 0 | 3 | 10 | 21 | 12 |
| Trn-B 3 BDNF, NT4 (NT3) | 0 | 5 (6)a | 0 | 36 (27)a | 12 (10)a |
Number of neurons with the indicated receptor plus neurotrophin (NT3) are shown in parentheses. NGF, nerve growth factor; BDNF, brain-derived neurotrophic factor.
Many of these neurons expressed both NT3 and NT4 (see Table III).
Greater Superficial Petrosal Nerve
The two branches of the GSP nerve exhibited differences. The main difference in receptors was the presence of Trn-B gene in 82% of soft palate neurons, whereas this gene was not detected in any of 42 neurons associated with the incisive papilla (Table II). TrkC was expressed prominently in both GSP branches, 52% in the soft palate and 67% in the incisive papilla branch (Table II). TrkA on the other hand was expressed in relatively few GSP neurons.
The two branches also differed in expression of the genes for neurotrophins, with 73% of neurons in the soft palate exhibiting NT-3, compared to only 38% in the incisive papilla. Almost all neurons in the soft palate, 42 of 44 (95%), gave a signal for NT-4 whereas only 17% in the incisive papilla branch expressed the gene for this neurotrophin. It should also be noted that a common combination in this group of neurons was the expression of both NT-3 and NT-4 (70%; see Fig. 2).
Posterior Auricular Nerve
Finally, the neurons projecting to the skin of the ear exhibited a pattern of neurotrophin and receptor expression different from the others. Among the receptors, 84% expressed trkA, approximately the same percentage as that expressing trkA in the branch of the CT innervating fungiform papillae, but unlike the latter, 38% of neurons in the posterior auricular expressed trkC (Table II) and 12 of 21 (57%) expressed both TrkA and TrkC (Table III).
Of 56 neurons, 32 expressed NT-3 (57%), 34 expressed NT-4 (61%). Twenty of these expressed both (36%), some with other neurotrophins as well.
DISCUSSION
The data indicate that each nerve branch emanating from the geniculate ganglion has a unique profile with respect to its expression of neurotrophin and neurotrophic receptor genes. It is also clear that individual neurons may express more than one neurotrophin receptor and more than one neurotrophin.
In earlier work on development of geniculate ganglion neurons, we showed that about half the neurons express mRNA for more than one neurotrophin receptor (Farbman et al., 2004; cf. Moshnyakov et al., 1996). In addition, we found that Trn-B was expressed prominently in many neurons, sometimes in combination with full-length trkB or with other trks (Armanini et al., 1995). As indicated above, our interest in Trn-B is based on its putative role as a modulator of the trkB response (Armanini et al., 1995; Biffo et al., 1995; Eide et al., 1996; Alderson et al., 2000; Luikart et al., 2003).
We also found in our earlier study that neurotrophin receptor gene expression in the neurons seems to change during development. Early in rat development (E12–13), neurons express trkB and respond in vitro to BDNF and NT-4 by vigorous neurite outgrowth; later, they lose much of their responsiveness to these neurotrophins (Rochlin et al., 2000).
If one assumes that the expression of a neurotrophin receptor by a neuron implies an important function then one has to determine the origin of the neurotrophin ligand that would bind to the receptor. There are at least five possible nearby sources from which a trophic factor could be derived: (1) the peripheral end organ, which in most of the cases would be a taste bud in one of the terminal fields of the nerve and would contain gustatory epithelial cells adjacent to the nerve terminal; (2) the central nervous system, in this case a region of the nucleus of the solitary tract in the brain stem where the centrally projecting nerve process terminates; (3) the neuron itself, which would permit the possibility of autocrine stimulation; (4) a nearby or adjacent neuron, which would permit the paracrine type of stimulation; or (5) glial or Schwann cells.
Target Organs
The presence of BDNF in some taste bud cells has been documented in fetal, newborn, and adult mammals, rats, mice, and hamsters (Nosrat and Olson, 1995; Ganchrow et al., 2003a,b; Yee et al., 2003). Its function is thought to be important for survival of at least some gustatory sensory neurons because in mutant mice null for the BDNF gene, the number of geniculate ganglion neurons was reduced by 50%. This supports the notion that geniculate ganglion neurons were (at some developmental stage) dependent on BDNF for survival (Ernfors et al., 1994; Liebl et al., 1997). Moreover, taste buds were underdeveloped in BDNF null mutants (Nosrat et al., 1997; Zhang et al., 1997; Cooper and Oakley, 1998; Oakley et al., 1998). Surprisingly, we found only 18 trkB-positive neurons of 86 (21%) we examined in the GSP nerve and only 2 of 56 (4%) in the posterior auricular nerve. Far more neurons expressed the truncated version of the receptor, Trn-B: none in neurons associated with innervation of fungiform papillae or the incisive papilla; 14 of 37 in the foliate (38%); 36 of 44 in the soft palate (82%); and 21 of 56 (38%) in the posterior auricular. We are not aware of any information of neurotrophin or neurotrophin receptor expression in specific neurons in the central target of geniculate ganglion neurons.
Methodologic Advantages and Limitations
The major advantages of the method used here are: (1) the ability to detect very small amounts of message, even from cells stored for years in paraffin sections (Eberwine and Crino, 1997); (2) the ability to validate our results with precision by subcloning and sequencing the PCR product (when each sequence was matched with a BLAST search of the NCBI gene bank we determined that the PCR product was 98–100% identical to the predicted sequence); (3) the innervation field of selected neurons is known, i.e., our approach can presumably distinguish among patterns of neurotrophin or neurotrophin receptor expression associated with innervation fields of the CT versus those of the GSP or the posterior auricular cutaneous nerve; and (4) a substantial number of neurons were sampled (226 in total).
There are disadvantages as well. First, only a small percentage of neurons from each ganglion could be sampled. The failure to detect trkB more widely thus might be a reflection of sampling procedure, particularly if the mRNA of this receptor is found only in some nerve branches arising from geniculate ganglion neurons. In addition to differences in neurotrophin receptor expression, disparities have been reported in biophysical properties and responses to glutamate receptor agonists among geniculate ganglion neurons projecting peripheral axons to different innervation fields (King and Bradley, 2000). In both studies, the absence of evidence for the expression of a particular receptor is not equivalent to evidence for the absence of that receptor. In other words, when our data indicate that the gene for a receptor or a neurotrophin is expressed within a neuron and we can subclone the PCR band and verify the sequence of the PCR product, we are fairly certain that the gene is expressed. If we fail to obtain a signal, however, we cannot rule out the possibility of a technical failure or that the signal is too small and not detectable with our method.
Matsumoto et al. (2001) used in situ hybridization analyses of neurotrophin receptors in adult rat geniculate ganglion neurons and found most cells to be positive to the trkB probe, a few to the trkC probe, and none to the trkA probe. Strictly speaking, their results are not comparable to ours because their probe for trkB, which included nucleotides 632–1048, would have hybridized with both the full-length and truncated versions of this receptor. The most curious difference is in their inability to detect trkA, whereas many neurons in all groups, particularly neurons innervating fungiform papillae and ear skin, express this gene. We confirmed the presence of trkA by sequencing our PCR products.
Nevertheless, we can formulate the following hypothesis to explain the data. Because so many neurons themselves expressed genes for neurotrophins, it is conceivable that these genes are responsible for production of trophic factors in the ganglion itself. If this is so, these neurotrophins could be involved in autocrine or paracrine stimulation (Ernfors et al., 1992). The results of in vitro studies on E12–13 geniculate ganglia showed that the neurons are responsive to BDNF and NT-4 in the early stages of neurite outgrowth, i.e., well before the processes would reach their targets (Rochlin et al., 2000). The most logical source of neurotrophins in these early stages would be other neurons or Schwann cells or extracellular matrix through which the nerve processes are growing in the fetus. Indeed, some of the neuron cell bodies in developing fetuses do produce mRNAs for neurotrophins and could be the source of neurotrophins working by autocrine or paracrine stimulation (Farbman et al., 2004). For example, in the present study if we look at the CT branch to the fungiform papillae, the neurons of which expressed only the gene for trkA, we see that 18 neurons produced NGF, the primary ligand for trkA, and 16 produced NT-3, which can also bind to trkA, albeit with lesser affinity (Table III). Of these, 12 produced both NGF and NT-3. In effect, 18 of 47 neurons (38%) had the potential to be activated in an autocrine manner via trkA and all would have the ability to be paracrine, or a mixture of the two. Similarly, nearly all the neurons from the soft palate expressed either trkB or Trn-B, and nearly all expressed mRNA for NT-4 and/or NT-3. Consequently, it is possible that these neurotrophins could bind weakly to the full-length or truncated trkB (Table IV). Alternatively, 23 neurons (52%) associated with the soft palate branch express mRNA for trkC, so that the NT-3 could be involved in an autocrine or paracrine relationship with this receptor. Similarly, the neurons associated with other innervation fields have the potential for providing trophic support for themselves or their neighbors, without invoking a need for target-derived support from either the periphery or the central nervous system. Because the neurons in the geniculate ganglion do not seem to be clumped according to their innervation fields, there could be cross stimulation by trophic factors irrespective of what is happening in the periphery.
The neurotrophic hypothesis posits that the survival of developing and mature neurons depends on trophic molecules. Survival could involve multiple neurotrophic factors from the same family or from multiple families that might be required simultaneously or sequentially (reviewed by Korsching, 1993; Davies, 1996; Levi-Montalcini and Angeletti, 1997; Bibel and Barde, 2000). The trophic molecules in the nervous system often are derived from the target, and survival of the neuron depends on successful competition by growing nerve terminals for limited amounts of neurotrophins available from target organs. Our data support the possibility, perhaps the likelihood, that autocrine or paracrine trophic support by locally derived neurotrophins from neurons within the sensory ganglion contributes to neuronal survival. The present data support this idea at least for the sensory neurons in the geniculate ganglion, and it may apply to other sensory ganglia (Ernfors et al., 1992) as well as the central nervous system (Lu et al., 1989).
Further, the presence of a neurotrophin in a sensory target organ may not necessarily mean that it is required by the nerve terminals innervating the target. Neurotrophins in so-called target organs may function locally within or near the organ itself, e.g., the taste bud (Farbman, 2003). It has been shown that trkB-like immunoreactivity is present in taste bud cells (Ganchrow et al., 2003a,b). In addition, the use of BDNFlacZ knock-in mice has revealed an interesting effect of BDNF on differentiation of taste buds. In these mice, one allele of the BDNF gene was replaced with the lacZ gene. These mice, then, are hemizygous for BDNF and possibly deficient. The taste buds in these mutant mice are reduced in volume by 31% (Yee et al., 2003). These data support the notion that BDNF/trkB interactions may occur within taste buds and may promote survival or differentiation of the taste epithelium. One cannot rule out the possibility that taste buds may be smaller in these animals, however, because of a reduced number of neurons in the geniculate ganglion resulting from a single BDNF allele being present (Erickson et al., 1996).
In conclusion, the interactions between gustatory nerves and taste buds and among the neurons of the geniculate ganglion may be considerably more complex than was thought previously for the reasons stated.
Acknowledgments
This work was supported by grants from the National Institutes of Health (DC 5R01 04837 to A.I.F., DC 5R01 04846 to S.I.S., and DC R01 00407 to D.L.H.). We thank W. Goodwin Jr. for preparing sections of geniculate ganglia and Ms. M. Bhattacharyya for doing RNA amplifications and PCRs. We also thank Dr. N. Spruston for allowing us free access to his laboratory for fashioning the pipets used for microdissection of the neurons.
Contract grant sponsor: NIH; Contract grant number: DC 5R01 04837, DC 5R01 04846, DC R01 00407.
References
- Alderson RF, Curtis R, Alterman AL, Lindsay RM, DiStefano PS. Truncated TrkB mediates the endocytosis and release of BDNF and neurotrophin-4/5 by rat astrocytes and Schwann cells in vitro. Brain Res. 2000;871:210–222. doi: 10.1016/s0006-8993(00)02428-8. [DOI] [PubMed] [Google Scholar]
- Armanini MP, McMahon SB, Sutherland J, Shelton DL, Philips HS. Truncated and catalytic isoforms of trkB are co-expressed in neurons of rat and mouse CNS. Eur J Neurosci. 1995;7:1403–1409. doi: 10.1111/j.1460-9568.1995.tb01132.x. [DOI] [PubMed] [Google Scholar]
- Barbacid M. The trk family of neurotrophin receptors. J Neurobiol. 1994;25:1386–1403. doi: 10.1002/neu.480251107. [DOI] [PubMed] [Google Scholar]
- Baxter GT, Radeke MJ, Kuo RC, Makrides V, Hinkle B, Hoang R, Medina-Selby A, Coit D, Valenzuela P, Feinstein SC. Signal transduction mediated by the truncated trkB receptor isoforms, trkB.T1 and trkB.T2. J Neurosci. 1997;17:2683–2690. doi: 10.1523/JNEUROSCI.17-08-02683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2929–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Biffo S, Offenhäuser N, Carter GD, Barde YA. Selective binding and internalization by truncated receptors restrict the availability of GDNF during development. Development. 1995;121:2461–2470. doi: 10.1242/dev.121.8.2461. [DOI] [PubMed] [Google Scholar]
- Bothwell M. Functional interactions of neurotrophins and neurotrophin receptors. Annu Rev Neurosci. 1995;18:223–253. doi: 10.1146/annurev.ne.18.030195.001255. [DOI] [PubMed] [Google Scholar]
- Chao MV. Neurotrophin receptors: a window into neuronal differentiation. Neuron. 1992;9:583–593. doi: 10.1016/0896-6273(92)90023-7. [DOI] [PubMed] [Google Scholar]
- Cho TT, Farbman AI. Neurotrophin receptors in the geniculate ganglion. Brain Res Mol Brain Res. 1999;68:1–13. doi: 10.1016/s0169-328x(99)00006-6. [DOI] [PubMed] [Google Scholar]
- Cooper D, Oakley B. Functional redundancy and gustatory development in bdnf null mutant mice. Brain Res Dev Brain Res. 1998;105:79–84. [PubMed] [Google Scholar]
- Davies AM. The role of neurotrophins in the developing nervous system. J Neurobiol. 1994;25:1334–1348. doi: 10.1002/neu.480251103. [DOI] [PubMed] [Google Scholar]
- Davies AM. The neurotrophic hypothesis: where does it stand? Philos Trans R Soc Lond B Biol Sci. 1996;351:389–394. doi: 10.1098/rstb.1996.0033. [DOI] [PubMed] [Google Scholar]
- Davies AM. Neurotrophin switching: where does it stand? Curr Opin Neurobiol. 1997;7:110–118. doi: 10.1016/s0959-4388(97)80128-6. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Crino P. Analysis of mRNA populations from single live and fixed cells of the central nervous system. In: Crawley J, Gerfen C, Rogawski M, Sibley D, Skolnick P, Wray S, McKay R, editors. Current protocols in neuroscience. New York: John Wiley and Sons; 1997. pp. 5.3.1–5.3.5. [DOI] [PubMed] [Google Scholar]
- Eide FF, Vining ER, Eide BL, Zang K, Wang XY, Reichardt LF. Naturally occurring truncated trkB receptors have dominant inhibitory effects on brain-derived neurotrophic factor signaling. J Neurosci. 1996;16:3123–3129. doi: 10.1523/JNEUROSCI.16-10-03123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JT, Conover JC, Borday V, Champagnat J, Barbacid M, Yancopoulos G, Katz DM. Mice lacking brain-derived neurotrophic factor exhibit visceral sensory neuron losses distinct from mice lacking NT4 and display a severe developmental deficit in control of breathing. J Neurosci. 1996;16:5361–5371. doi: 10.1523/JNEUROSCI.16-17-05361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Merlio JP, Persson H. Cells expressing mRNA for neurotrophins and their receptors during embryonic rat development. Eur J Neurosci. 1992;4:1140–1158. doi: 10.1111/j.1460-9568.1992.tb00141.x. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Neurotrophins and taste buds. J Comp Neurol. 2003;459:9–14. doi: 10.1002/cne.10588. [DOI] [PubMed] [Google Scholar]
- Farbman AI, Brann JH, Rozenblat A, Rochlin MW, Weiler E, Bhattacharyya M. Developmental expression of neurotrophin receptor genes in rat geniculate ganglion neurons. J Neurocytol. 2004;33:331–343. doi: 10.1023/B:NEUR.0000044194.71426.ee. [DOI] [PubMed] [Google Scholar]
- Ganchrow D, Ganchrow JR, Verdin-Alcazar M, Whitehead MC. Brain-derived neurotrophic factor-, neurotrophin-3- and tyrosine kinase receptor-like immunoreactivity in lingual taste bud fields of mature hamster. J Comp Neurol. 2003a;455:11–24. doi: 10.1002/cne.2162. [DOI] [PubMed] [Google Scholar]
- Ganchrow D, Ganchrow JR, Verdin-Alcazar M, Whitehead MC. Brain-derived neurotrophic factor-neurotrophin-3- and tyrosine kinase receptor-like immunoreactivity in lingual taste bud fields of mature hamster after sensory denervation. J Comp Neurol. 2003b;455:25–39. doi: 10.1002/cne.2164. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- King MS, Bradley RM. Biophysical properties and responses to glutamate receptor agonists of identified subpopulations of rat geniculate ganglion neurons. Brain Res. 2000;866:237–246. doi: 10.1016/s0006-8993(00)02292-7. [DOI] [PubMed] [Google Scholar]
- Korsching S. The neurotrophic factor concept: a reexamination. J Neurosci. 1993;13:2739–2748. doi: 10.1523/JNEUROSCI.13-07-02739.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm RF, Hill DL. Quantitative relationships between taste bud development and gustatory ganglion cells. Ann N Y Acad Sci. 1998;855:70–75. doi: 10.1111/j.1749-6632.1998.tb10547.x. [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R, Angeletti PU. Nerve growth factor. Physiol Rev. 1997;48:534–569. doi: 10.1152/physrev.1968.48.3.534. [DOI] [PubMed] [Google Scholar]
- Lewin GR, Barde YA. Physiology of the neurotrophins. Annu Rev Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- Liebl DJ, Mbiene JP, Parada LF. NT4/5 mutant mice have deficiency in gustatory papillae and taste bud formation. Dev Biol. 1999;213:378–389. doi: 10.1006/dbio.1999.9385. [DOI] [PubMed] [Google Scholar]
- Lu B, Buck CR, Dreyfus CF, Black IB. Expression of NGF and NGF receptor mRNAs in the developing brain: evidence for local delivery and action of NGF. Exp Neurol. 1989;104:191–199. doi: 10.1016/0014-4886(89)90029-0. [DOI] [PubMed] [Google Scholar]
- Luikart BW, Nef S, Shipman T, Parada LF. In vivo role of truncated TrkB receptors during sensory ganglion neurogenesis. Neuroscience. 2003;117:847–858. doi: 10.1016/s0306-4522(02)00719-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto I, Emori Y, Ninomiya Y, Abe K. A comparative study of three cranial sensory ganglia projecting into the oral cavity: in situ hybridization analyses of neurotrophin receptors and thermosensitive cation channels. Brain Res Mol Brain Res. 2001;93:105–112. doi: 10.1016/s0169-328x(01)00129-2. [DOI] [PubMed] [Google Scholar]
- Middlemas DS, Lindberg RA, Hunter T. TrkB, a neural receptor protein-tyrosine kinase: evidence for a full-length and two truncated receptors. Mol Cell Biol. 1991;11:143–153. doi: 10.1128/mcb.11.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Gomez MM, Lubarsky EH. Distribution of the facial nerve to taste receptors in the rat. Chem Sens Flav. 1978;3:397–411. [Google Scholar]
- Moshnyakov M, Arumäe U, Saarma M. mRNAs for one, two or three members of trk receptor family are expressed in single rat trigeminal ganglion neurons. Brain Res Mol Brain Res. 1996;43:141–148. doi: 10.1016/s0169-328x(96)00168-4. [DOI] [PubMed] [Google Scholar]
- Nosrat CA, Blomlof J, El Shamy WM, Ernfors P, Olson L. Lingual deficits in BDNF and NT3 mutant mice leading to gustatory and somatosensory disturbances, respectively. Development. 1997;124:1333–1342. doi: 10.1242/dev.124.7.1333. [DOI] [PubMed] [Google Scholar]
- Nosrat CA, Olson L. Brain-derived neurotrophic factor mRNA is expressed in the developing taste bud-bearing tongue papillae of rat. J Comp Neurol. 1995;360:698–704. doi: 10.1002/cne.903600413. [DOI] [PubMed] [Google Scholar]
- Oakley B, Brandemihl A, Cooper D, Lau D, Lawton A, Zhang C. The morphogenesis of mouse vallate gustatory epithelium and taste buds requires BDNF-dependent taste neurons. Brain Res Dev Brain Res. 1998;105:85–96. [PubMed] [Google Scholar]
- Ringstedt T, Ibañez CF, Nosrat CA. Role of brain-derived neurotrophic factor in target invasion in the gustatory system. J Neurosci. 1999;19:3507–3518. doi: 10.1523/JNEUROSCI.19-09-03507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin W, O’Connor R, Giger RJ, Verhaagen J, Farbman AI. Comparison of neurotrophin and repellent sensitivities of early embryonic geniculate and trigeminal axons. J Comp Neurol. 2000;422:579–593. [PubMed] [Google Scholar]
- Schechterson LC, Bothwell M. Novel roles for neurotrophins are suggested by BDNF and NT-3 mRNA expression in developing neurons. Neuron. 1992;9:449–463. doi: 10.1016/0896-6273(92)90183-e. [DOI] [PubMed] [Google Scholar]
- Yee C, Jones KR, Finger TE. Brain-derived neurotrophic factor (BDNF) is present in synaptically connected taste cells of the adult mouse. J Comp Neurol. 2003;459:15–24. doi: 10.1002/cne.10589. [DOI] [PubMed] [Google Scholar]
- Zhang CX, Brandemihl A, Lau D, Lawton A, Oakley B. BDNF is required for normal development of taste neurons in vivo. Neuroreport. 1997;8:1013–1017. doi: 10.1097/00001756-199703030-00039. [DOI] [PubMed] [Google Scholar]


