Abstract
Fragile X Syndrome results from loss of the Fragile X mental retardation protein (FMRP), an RNA-binding protein regulating a variety of cytoplasmic mRNAs. FMRP regulates mRNA translation and has been suggested to play a role in mRNA localization to dendrites. We report a third cytoplasmic regulatory function for FMRP – control of mRNA stability. We find in mice that FMRP binds, in vivo, the mRNA encoding PSD-95, a key molecule regulating neuronal synaptic signalling and learning. This interaction occurs through the 3′ untranslated region of the PSD–95 mRNA, increasing message stability. Moreover, stabilization is further increased by mGluR activation. While we also find that the PSD–95 mRNA is synaptically localized in vivo, localization occurs independently of FMRP. Through our functional analysis of this FMRP target we provide evidence that misregulation of mRNA stability may contribute to the cognitive impairments in Fragile X Syndrome patients.
Keywords: Fragile X Mental Retardation Protein, PSD-95, G-quartet, U-rich regions, ARE, dendritic mRNA, mRNA stability
Fragile X Syndrome (FXS), caused by a trinucleotide expansion in the X–linked Fragile X mental retardation gene (FMR1), leading to subsequent loss of the FMR protein (FMRP), is the most common cause of X–linked mental retardation. FMRP has multiple RNA–binding motifs and is thought to be involved in mRNA localization and translational regulation in neurons, two processes required for synaptic plasticity (reviewed in1). As the only obvious abnormality in the brains of FXS patients is the presence of longer, immature-appearing spines1 and references therein current models have focused on the possible misregulation of synaptic mRNAs as an underlying cause of FXS mental deficits.
A wide variety of mRNAs have been identified as potential targets of mammalian FMRP both in vitro and in vivo2,3. FMRP binds various mRNA elements1 including a G–rich RNA structure (G–quartet)4-6 and U–rich stretches7. FMRP is also indirectly recruited to some target mRNAs via binding the noncoding RNAs BC1 and BC2008-11. Finally, both mammalian and Drosophila FMRP are present in microRNA complexes12 and references therein and may be recruited to mRNAs bound by miRNAs.
Within the FMRP protein, the RGG box recognizes G–quartet sequences present in some FMRP targets4 while the N-terminus recognizes a bulge within BC RNAs10. Interestingly, while FMRP contains two KH domains, a known RNA–binding motif, no endogenous neuronal targets recognized by this domain have been identified13. Functionally, FMRP acts as a translational repressor of a subset of neuronal mRNAs3 and may be involved in synaptic mRNA localization as FMRP is present in mRNP localization complexes14. A limited number of studies also suggest that FMRP may regulate transcription15-17.
Despite much research, it remains unclear precisely how loss of FMRP leads to alterations in the neuronal mechanisms responsible for cognition. One proposal suggests that alterations in metabotropic glutamate receptor (mGluR) mediated signaling might underlie a number of the cognitive deficits associated with FXS18. Disruption of N–methyl–D–aspartate (NMDA) receptors19, or associated signaling components20-22, also lead to impairments in synaptic plasticity. Interestingly, mGluRs and NMDA receptors coexist in a large scale signaling complex23. PSD–95 (DLG4), a component of the MAGUK family of adaptor proteins that includes SAP102 (DLG3) and PSD–93, binds directly to the NMDA receptor and links other adaptors to mGluRs24. Mice lacking PSD–95 have learning20 and cortical plasticity21 impairments. Similarly, SAP102 mutant mice show learning impairments25 and human Dlg3/SAP102 mutations are implicated in mental retardation26. Interestingly, PSD–95 mutant mice also show dendritic spine abnormalities in the striatum and hippocampus27, one of the key hallmarks in FXS patients and FMRP mutant mice1. A quantitative neuroimaging study also found larger right and left hippocampal volumes in fragile X patients compared with the controls, suggesting an involvement of this region the behavioral and cognitive abnormalities associated with FXS28.
A recent report indicated that FMRP regulates PSD–95 protein levels in response to mGluR signalling29. However, putative FMRP–binding sites were identified by sequence analysis, and direct interactions between the PSD–95 mRNA and FMRP were not tested. While authors concluded that these effects were due to translational regulation of PSD–95 mRNA, the above–mentioned results could not formally distinguish between effects on mRNA export, mRNA stability or translation.
RESULTS
PSD–95 mRNA interacts directly with FMRP
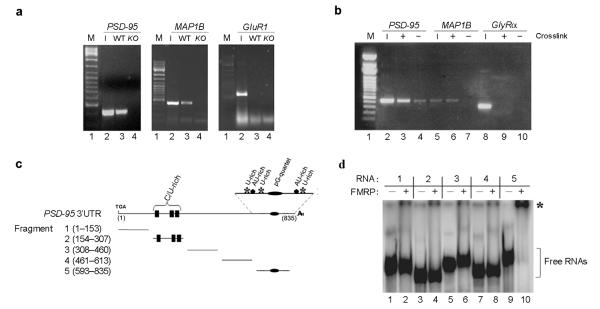
To address whether FMRP directly regulates the PSD–95 mRNA we examined whether PSD–95 mRNA was present in the FMRP complex. We found PSD–95 mRNA in FMRP immunoprecipitates from wildtype mice but not from FMR1 knockout mice (Fig. 1a). A known FMRP–interacting mRNA, MAP1B mRNA8,30,31, was also coprecipitated (Fig. 1a) while a negative control mRNA (GluR1) was not (Fig. 1a). Using reversible crosslinking–immunoprecipitation (CLIP)32 from primary hippocampal neurons (Fig. 1b) we show that FMRP binds directly to the PSD–95 mRNA, as crosslinking only occurs if FMRP and PSD–95 are in close proximity in vivo. MAP1B, but not GlyRα mRNA, was also crosslinked to FMRP (Fig. 1b). Combined, these data indicate that PSD–95 mRNA is part of the FMRP mRNP complex in vivo.
Figure 1. FMRP interacts directly with the 3′UTR of PSD–95 mRNA.
(a) Brain lysates from wildtype (WT) and FMR1 knockout mice (KO) were immunoprecipitated with FMRP antibodies. RT–PCR was performed using oligos for the PSD–95, MAP1B and GluR1 mRNAs. Input (1/5) is reported in lanes 2. Lanes not relevant to this experiment were removed between the marker and lanes 1 and 2. (b) CLIP assay. Hippocampal cell extracts were immunoprecipitated with FMRP antibodies. RT–PCR was performed using oligos for the PSD–95, MAP1B and GlyRα mRNAs. Input (1/5) is reported in lanes 2, 5, 8. (c) PSD–95 3′UTR fragments utilized in EMSA experiments. Potential functional motifs are indicated. (d) 32P radiolabelled fragments (1–5) of the PSD-95 3′UTR were incubated in the presence of FMRP (+, lanes 2, 4, 6, 8, 10). Control reactions were performed in buffer alone (−, lanes 1, 3, 5, 7, 9). RNA:protein complexes were resolved on native polyacrylamide gel. Unbound RNA fragments (]), and RNA:protein complexes (*) are indicated.
To map the FMRP:PSD–95 mRNA interaction we performed direct protein:RNA binding assays. We focused on the 3′ UTR of PSD–95 mRNA because in silico analysis of this region revealed the presence of a putative G–quartet29 and three U–rich stretches33 (Fig. 1c and Supplementary Fig. 1), sequence elements previously shown to recruit FMRP to RNAs4,7. Of the five short RNAs that spanned the entire 3′UTR of the mouse PSD–95 mRNA (Fig. 1c), only fragment 5 contained FMRP–binding ability in electrophoretic mobility shift assays (EMSAs) with purified baculovirus–expressed human FMRP protein (Fig. 1d). This RNA fragment was also bound by mouse brain extracts (data not shown). Lack of FMRP binding to fragments 1–4 (Fig. 1d), to the antisense strand (data not shown) and the ability of excess unlabeled fragment 5 RNA to compete indicated the FMRP:RNA interaction was specific and did not simply reflect general RNA affinity. The protein–binding ability of fragment 5 RNA was also specific as it did not bind other RNA binding proteins (i.e. the microbial transcription and translation modulator NusG or the spliceosomal 15.5KD/hSnu13p protein – data not shown).
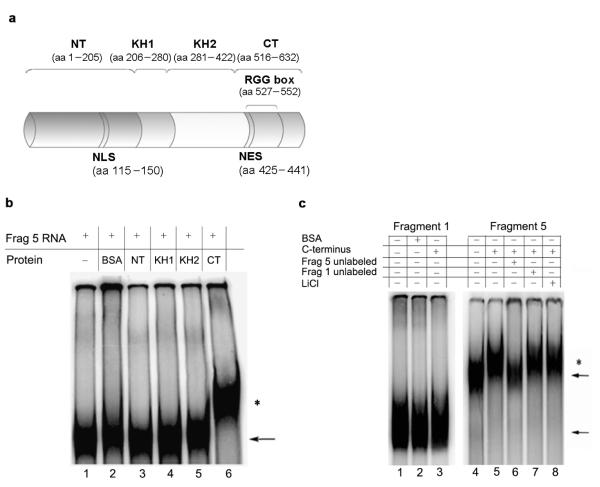
We also investigated which protein domain of FMRP (N–terminus, KH1, KH2, C–terminus)34 was involved in binding to the PSD–95 mRNA (Fig. 2a). We found that only the C–terminus contained PSD–95 mRNA binding ability (Fig. 2b). This domain bound with high affinity as binding was still present under high stringency conditions (50 mM LiCl) (Fig. 2c). Binding was specific as the C–terminus did not interact with fragment 1, and binding to fragment 5 was competed by excess unlabelled fragment 5, but not fragment 1 (Fig. 2c).
Figure 2. The C–terminal domain of FMRP is able to specifically interact with the PSD–95 mRNA 3′UTR.
(a) Schematic representation of FMRP protein and its principal domains: the N terminus (NT), KH1, KH2 and the C terminus (CT) containing the RGG box. The nuclear localization sequence (NLS) and the nuclear export sequence (NES) are also indicated. (b) 32P radiolabeled fragment 5 of the PSD–95 3′UTR was incubated alone (lane 1), in the presence of BSA (lane 2) or in the presence of FMRP domains: N terminus (lane 3), KH1 (lane 4), KH2 (lane 5) and C terminus (lane 6). (c) 32P radiolabeled fragments 1 and 5 of the PSD–95 3′UTR were incubated alone (lane 1 and 4), or in the presence of FMRP C terminus (lane 3 and 5). Fragment 1 was incubated also in the presence of BSA (lane 2). To assess the specificity of interaction between the fragment 5 and the C–terminus, the RNA binding assay was performed in the presence of competitor RNAs (unlabeled fragment 5, lane 6 or fragment 1, lane 7) or in the presence of the chaotropic salt LiCl (lane 8). RNA:protein complexes were resolved on native polyacrylamide gel. Unbound RNA fragments (←) and RNA:protein complexes (*) are indicated.
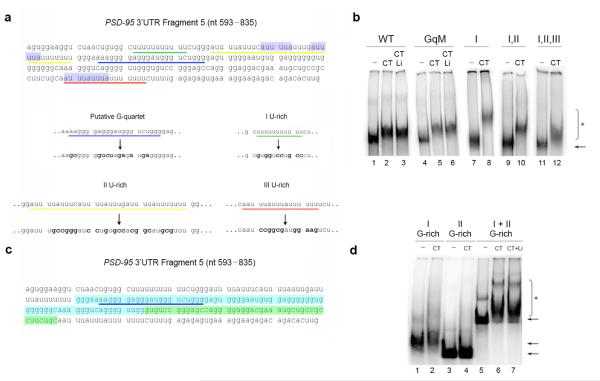
We further mapped the mRNA region within fragment 5 that was responsible for FMRP binding by scrambling the G–rich region to eliminate all similarity to the G–quartet consensus and converting the U–rich regions into mixed sequences (Fig. 3a). High lithium (50 mM), a condition that destabilizes G–quartet structures4,35, did not interfere with FMRP binding to either the wildtype (Fig. 3b) or the mutagenized fragment 5 (Fig. 3b). Interestingly, mutagenesis of all three U–rich regions did not prevent FMRP binding (Fig. 3b). As previous studies suggested that FMRP has high affinity for poly–(rG) in vitro34, we further examined the G–rich region. While the entire G–rich region exhibited binding to FMRP (Fig. 3c and d; I + II G-rich) even in the presence of high lithium salt (Fig. 3d), no binding was detected when we used two short RNA fragments (Fig 3c and d) of that region (Fig. 3c). Our findings argue that FMRP recognizes a structured G–rich sequence within the 3′UTR of the PSD–95 mRNA or a region spanning the two fragments, but that this structure is not forming a G–quartet.
Figure 3. A G–rich region in the PSD–95 3′UTR is responsible for FMRP C–terminus binding.
(a) Sequence of fragment 5 (nucleotide 593–835) and mutagenesis of the putative G–quartet and U–rich regions. The first U–rich region is underlined in green, the second U–rich region in yellow, the third U–rich in red and the putative G–quartet in blue. The blue highlighted regions represent two ARE elements. Bold characters represent introduced substitutions. (b) Wildtype fragment 5 (WT) and the putative G–quartet mutated fragment 5 (GqM) were incubated alone (lanes 1 and 4), with FMRP C-terminus (lanes 2 and 5), or in the presence of LiCl 50 mM (lanes 3 and 6). The first U–rich mutant (I), the first and second U–rich double mutant (I,II) and the triple U–rich mutant (I,II,III) were incubated alone (lanes 7, 9, 11), or with the C–terminus (lanes 8, 10, 12). Unbound RNA fragments (←) and RNA:protein complexes (*) are indicated. (c) The first G–rich region is highlighted in blue while the second G–rich in green. (d) The first G–rich region (nucleotide 666–741) of the fragment 5, the second G–rich region (nucleotide 742–786) or the entire G–rich region (nucleotide 666–786) were incubated alone (lanes 1, 3, 5) or with the C–terminus of FMRP (lanes 2, 4, 6). The C–terminus and the entire G–rich were incubated in the presence of 50 mM of LiCl (lane 7).
PSD–95 polysomal profile is similar in wildtype and FMR1 knockout
FMRP can act as a translational repressor3,31 and references therein and local translation of synaptic mRNAs has been increasingly implicated in neuronal plasticity, learning and memory formation (reviewed in36). Interestingly, a number of localized mRNAs encode synaptic proteins (e.g. Arc, MAP1B, , SAPAP4) that are translationally repressed by FMRP8,16. Thus far, our experiments indicate that FMRP can directly interact with the PSD–95 mRNA, but do not address the functional role of this interaction.
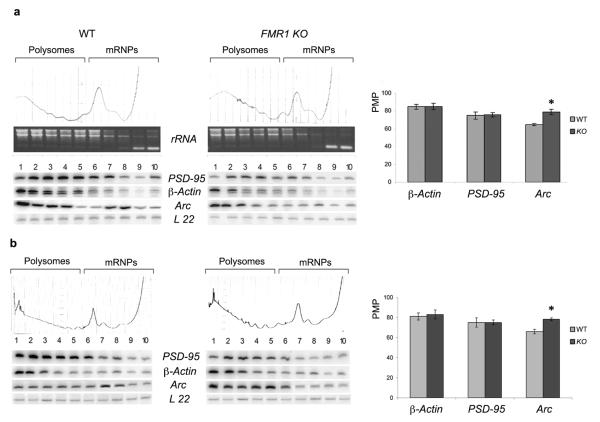
We assessed whether PSD–95 mRNA translation was regulated by FMRP, as was previously proposed29, by performing sucrose gradient fractionation of cytoplasmic (Fig. 4a) and hippocampal (Fig. 4b) brain extracts from wildtype and FMR1 knockout mice. Surprisingly, the percentage of PSD–95 mRNA associated with polysomes did not change in the FMR1 knockout animals compared to wildtype animals in either whole brain or hippocampal extracts. While the profile of the negative control, β-Actin mRNA also remained unchanged, Arc mRNA, which is known to be translationally regulated by FMRP8, showed the expected shift towards a more translationally active polysome pool in FMR1 knockout extracts. We cannot formally rule out the possibility that FMRP changes the translation efficiency of the PSD–95 mRNA without changing the PMP ratio (e.g. by altering miRNA–regulated translation37 and references therein). However, since other FMRP regulated mRNAs do change their PMP ratio (e.g. Arc), the above findings indicate that FMRP does not regulate PSD–95 protein synthesis in a similar manner to other well–studied FMRP targets.
Figure 4. PSD–95 mRNA polysomal profile is similar in wildtype and FMR1 knockout mice.
(a) Cytoplasmic brain extract was fractioned by centrifugation on a 5–70% sucrose gradient. 10 fractions were collected while 254 nm absorbance was recorded. RNA was extracted from each fraction and visualized on a denaturing agarose gel. rRNA 28S, 18S and 5S/tRNA are shown in each fraction. Radioactive RT–PCR analysis of total RNA in each fraction with primers specific for PSD-95, β-Actin, Arc and L22 RNAs was performed. The efficiency of translation is reported as a graphic profile of Percentage Messenger on Polysomes (PMP) which was calculated, after normalization to L22, by comparing the radioactivity of the first 5 fractions containing active polysomes to radioactivity from the entire 10 fractions. The PSD–95, β-Actin and Arc PMP in each fraction of wildtype or FMR1 knockout gradients was normalized for L22 RNA. (b) Same as in (a) using cytoplasmic extracts from the hippocampus. PMP value of three independent experiments with standard error is reported. *, p<0.05 for knockout versus wildtype by Student’s t test.
PSD–95 mRNA is dendritically localized with FMRP in vivo
It has been estimated that hundreds of mRNAs are present in dendrites, but whether the entire population or only a subset are near synapses in currently unknown38. As this list includes mRNAs that are known targets of FMRP (e.g. Arc, ), and as PSD–95 is an integral component of the post-synaptic density, we assessed whether the PSD–95 message was localized in dendrites and, if so, whether this localization was dependent upon FMRP.
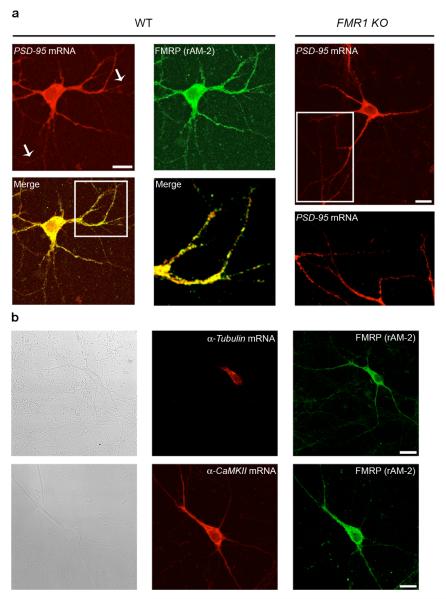
By analyzing the presence of PSD–95 mRNA in synaptoneurosomes from total brain, we found that PSD–95 mRNA showed a remarkable dendrite/soma enrichment ratio (Supplementary Fig. 2), suggesting that the mRNA was localized at synapses. This was further confirmed by in situ hybridization in neuronal cultures (Fig. 5). We found that PSD–95 mRNA localized in both cell bodies and along dendrites of hippocampal (Fig. 5a) and cortical (data not shown) neurons with a typical punctate pattern. Similarly, a recent large–scale screen also suggested putative targeting of the PSD–95 mRNA to both proximal and distal dendrites39. Surprisingly, while PSD–95 mRNA (red) largely colocalized with FMRP (green) throughout the cell and into neurites (Fig. 5a), the PSD–95 mRNA was still localized in dendrites from FMR1 knockout hippocampal (Fig. 5a) and cortical (data not shown) cultures. Control experiments indicated that we could specifically detect dendritic () and cell body (α-Tubulin) mRNAs38 (Fig. 5b) and that the sense probes did not show any specific mRNA staining (Supplementary Fig. 3a). These data further confirm that the PSD–95 message is part of an FMRP mRNP complex, but suggest that FMRP function is not necessary to localize the PSD–95 message.
Figure 5. PSD–95 mRNA is dendritically localized in neuronal cell cultures.
(a). Left panel, in situ hybridization performed using an antisense riboprobe specific for PSD–95 mRNA (red), combined with an immunofluorescence for FMRP (green) on wildtype (WT) hippocampal cultures (DIV 10). Merged image and a merged magnification are also shown (yellow). Right panel, in situ hybridization for PSD–95 in FMR1 knockout hippocampal cultures (top) and magnification (bottow). (b). Upper panels. Bright field. In situ hybridization performed using an antisense riboprobe specific for the cell body-specific α–Tubulin mRNA (red), combined with an immunofluorescence for FMRP (green) on hippocampal cultures (DIV 10). Lower panels. Bright field (left panel). In situ hybridization performed using an antisense riboprobe specific for the dendritically localized α–CaMKII mRNA (red, middle panel), combined with an immunofluorescence for FMRP (green, right panel) on hippocampal cultures (DIV 10).
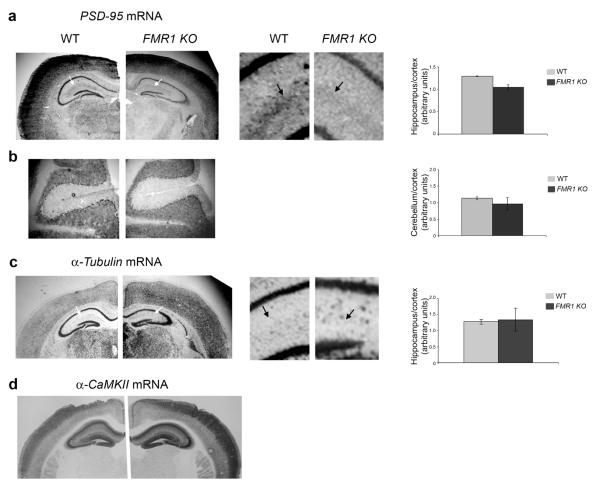
We confirmed that PSD–95 mRNA was dendritically localized using both DIG (data not shown) and radioactive in situ hybridization (Fig. 6) on brain slices. PSD–95 mRNA was present in the hippocampus, cortex (Fig. 6a) and cerebellum (Fig 6b). The unlocalized control mRNA () stained only cell bodies in the hippocampus and dentate gyrus (Fig. 6c and Supplementary Fig. 4). Although PSD–95 mRNA localization was distinct from another localized mRNA (α–CaMKII; Fig. 6d and Supplementary Fig. 4), PSD–95 mRNA was clearly present within hippocampal dendrites of both wildtype and FMR1 knockout mice in a region corresponding to the stratum lacunosum–moleculare (Fig. 6a and Supplementary Fig. 4). A control PSD–95 mRNA sense probe did not show any specific mRNA staining (Supplementary Fig. 3b) Interestingly, quantification of PSD–95 mRNA levels showed a clear, though non significant, reduction in hippocampal mRNA in the FMR1 knockout animals relative to cortical mRNA levels (Fig. 6a). This tendency was not observed when comparing PSD–95 cerebellar to cortical mRNA (Fig. 6b) or hippocampal to cortical mRNA (Fig. 6c) ratios between wildtype and FMR1 knockout mice.
Figure 6. PSD–95 mRNA is dendritically localized in vivo.
Radioactive in situ hybridization on trasversal brain sections from wildtype (WT; left image in each case) and FMR1 knockout mice (KO; right image in each case) for PSD–95 (a-b), β–Tubulin (c) and α–CaMKII (d) mRNAs for PSD–95 mRNA in both wildtype and knockout mice. Arrows point to the stratum lacunosum-moleculare. Right panels show an enlargement of the CA2 areas marked by the white arrows in the smaller image. Black arrows in the enlargement point to the hippocampal region enriched in PSD–95 mRNA. Quantification of PSD–95 mRNA level in hippocampus relative to cortex is reported (average value from 3 sections are reported with standard error). (b) Cerebellar sections are shown. Quantification of PSD–95 mRNA level in cerebellum relative to cortex is reported (average value from 3 sections are reported with standard error). (c) In situ hybridization on brain sections from wildtype and FMR1 knockout mice for α–Tubulin mRNA. A blown–up of the areas marked by the white arrows is shown. Black arrows point the lack of any detectable signal in this area. Quantification of α–Tubulin mRNA level in hippocampus relative to cortex is reported (average value from 3 sections are reported with standard error). d. In situ hybridization on brain sections from wildtype and FMR1 knockout mice for α–CaMKII mRNA.
Combined, these data provide evidence that the PSD–95 mRNA is localized in dendrites in vitro and in vivo. As there is less PSD–95 mRNA in the stratum lacunosum–moleculare in FMR1 knockout mice (Fig 6a), we cannot exclude the possibility that FMRP might be involved in a subtle modulation of PSD–95 mRNA localization. However, as PSD–95 mRNA is clearly present in dendrites in the absence of FMRP (Fig. 5a and 6a), our data suggest that the FMRP is not playing a primary role in PSD–95 mRNA localization.
Impaired PSD-95 mRNA and protein levels in FMR1 knockout
Our results suggest that FMRP is not directly regulating translation (Fig. 4) or transport (Fig. 5 and 6) of PSD–95 mRNA. However, prior reports have hinted that FMRP might also control mRNA abundance via transcriptional regulation15-17. Interestingly, our radioactive in situ hybridization data indicated a possible decrease in PSD–95 mRNA intensity in hippocampal neurons from FMR1 knockout mice (Fig. 6a), suggesting that mRNA abundance may be regulated by FMRP.
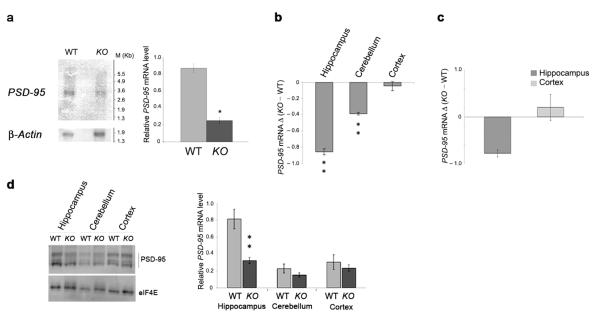
To determine if FMRP controls mRNA abundance, we first examined the total PSD–95 mRNA level in wildtype and FMR1 knockout mice. In total brain, PSD–95 mRNA levels were significantly decreased in FMR1 knockout compared to wildtype mice (Fig. 7a). Interestingly, quantitative RT–PCR analyses performed on the three principal brain areas (hippocampus, cerebellum and cortex) showed that the decrease in PSD–95 mRNA was very pronounced in the hippocampus, less in the cerebellum and not observed in the cortex (Fig. 7b). Quantitative RT–PCR analyses of the PSD–95 mRNA from hippocampal neurons of wildtype and FMR1 knockout mice confirmed this hippocampal–specific decrease in PSD–95 mRNA (Fig. 7c). While there was a subtle trend towards lower levels of PSD–95 mRNA in the hippocampus as detected with radioactive in situ hybridization (Fig. 6), this was not statistically significant and we suggest that these differences may be due to different sensitivities of the techniques. Differential PSD–95 expression was also reflected at the protein level, with a statistically significant decrease occurring in the hippocampus and a non–significant decrease in the cerebellum (Fig. 7d).
Figure 7. PSD–95 mRNA and protein levels are altered in the FMR1 knockout mice.
(a) PSD–95 and β–Actin mRNA levels from total brain were analyzed by Northern blot in wildtype (WT) or FMR1 knockout (KO) mice. The same membrane was hybridized and normalized for β–Actin mRNA (lower panel). PSD–95 mRNA/β–Actin ratio is reported as a histogram with standard error. (b) PSD–95 mRNA levels in three different brain regions were estimated by quantitative RT–PCR from three wildtype and three FMR1 knockout mice, normalized to those of Histone H3 and reported in a histogram as delta of FMR1 knockout vs wildtype value. Error bars represent standard error. (c) The level of PSD–95 mRNA in hippocampal or cortical neuronal cell culture was estimated by quantitative RT–PCR, normalizing the values to Histone mRNA. The histogram represents the delta of FMR1 knockout vs wildtype value and the bars represent the standard errors of three independent measurements. (d) Protein extracts from cortex, hippocampus or cerebellum of four wildtype and four FMR1 knockout mice were analyzed for PSD–95 and eIF4E proteins and reported in a histogram with standard error (right panel). Western blot from one of the four independent mice analyzed for PSD–95 and eIF4E is shown. *, p<0.05 and **, p<0.01 for knockout versus wildtype by Student’s t test in all panels.
These data suggest that either transcription or stability of the PSD–95 mRNA is regulated by FMRP in the hippocampus. Critically, the hippocampus is important for learning processes altered in FXS patients40 and loss of PSD–95 results in hippocampal–dependent learning defects20.
Activity–dependent FMRP control of PSD–95 mRNA stability
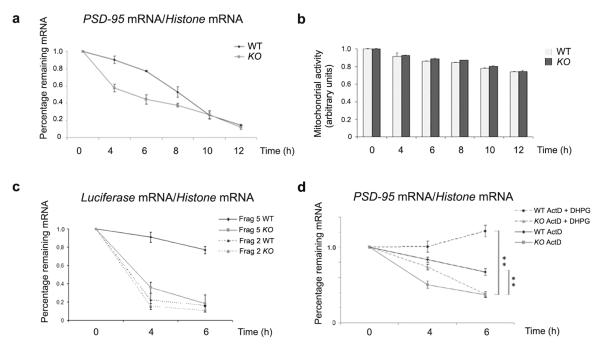
To directly assess whether this change in mRNA level was due to altered transcription or mRNA stability, we examined the half–life of the PSD–95 message in cortical and hippocampal primary cultured neurons. Interestingly, after transcriptional blockade with Actinomycin D, PSD–95 mRNA abundance was significantly and selectively reduced in hippocampal cultures in the absence of FMRP (Fig. 8a and Supplemental Fig. 5). Stability of PSD–95 mRNA was unaffected in FMR1 knockout cortical cultures (Supplementary Fig. 6) in agreement with prior results (Fig. 6a, 7b and 7c). These results were not due to non–specific cell death effects as the morphology of hippocampal cells from both wildtype and FMR1 knockout mice were the same (Supplementary Fig. 7) and cell survival was the same in both genotypes (Fig. 8b) although we did note that after 12 hours both the wildtype and FMR1 knockout neurons showed some increase in cell death (≈25%). Combined, these results suggest that FMRP functions to stabilize the PSD–95 mRNA specifically in the hippocampus. Furthermore, the stability of a reporter (Renilla luciferase) RNA carrying the FMRP–interacting portion of the PSD–95 3′UTR (fragment 5), was also more stable when transfected into wildtype versus FMR1 knockout hippocampal neurons (Fig. 8c), while a reporter RNA containing another PSD–95 3′UTR that does not bind FMRP (fragment 2) was equally unstable in both cultures (Fig. 8c). These data strongly suggest that a direct interaction between FMRP and the PSD–95 3′UTR is necessary to confer mRNA stabilization.
Figure 8. FMRP regulates the stability of PSD–95 mRNA in hippocampal cells through an activity-dependent mechanism.
(a) RNA was isolated at the indicated times after Actinomycin D application to hippocampal neurons from wildtype or FMR1 knockout mice and the stability of PSD–95 mRNA was normalized to the values of Histone H3 mRNA. (b) MTT assay performed on wildtype and FMR1 knockout hippocampal cells during the Actinomycin D treatments. Standard error of three measurements for each time point is reported. (c) Stability of a chimeric mRNA containing the Renilla luciferase reporter and either fragment 2 or 5 of PSD–95 3′UTR transfected into wildtype or FMR1 knockout hippocampal neurons. mRNA levels were measured at the indicated times after Actinomycin D application by quantitative RT–PCR, normalizing the values to Histone H3 mRNA. (d) mRNA was isolated at the indicated times after Actinomycin D or Actinomycin D + DHPG application to hippocampal neurons from wildtype or FMR1 knockout mice. The stability of PSD–95 mRNA in wildtype or FMR1 knockout hippocampal cells was measured by quantitative RT–PCR. **, p<0.01 for knockout versus wildtype by Student’s t test.
As FMRP has not been previously shown to regulate mRNA stability, we also assessed the stability of eleven other FMRP targets and two synaptic scaffolding proteins whose mRNAs are localized in dendrites (Homer 1a and Shank 1). Of these mRNAs (Supplementarly Table 1) only myelin basic protein mRNA (MBP) changed its stability. MBP mRNA is a target of FMRP regulation41, present only in glia cells, which also express FMRP8,41. We detected the MBP mRNA because our primary neurons were cocultured with glial cells. Importantly, while this list is clearly not exhaustive, our analyses suggest that FMRP–mediated mRNA stabilization is a highly selective mechanism with respect to both cell type and target mRNA and that it works in both neurons and glia.
As FMRP is regulated by mGluR activation (e.g.29,42,43) we also investigated whether mGluR stimulation would alter FMRP-dependent PSD–95 mRNA stabilization. Using two independent protocols (see Methods for details), we found that the presence of DHPG further stabilized PSD–95 mRNA in wildtype cells at both time points measured (Fig. 8d). In FMR1 knockout cells, the addition of DHPG provided only transient stabilization that did not persist at the later time point, suggesting that DHPG might also have a transient, FMRP–independent effect on mRNA half–life. Quantification of three independent experiments indicated that there was a significant DHPG–dependent stabilization effect only in the wildtype neurons and this effect is mostly lost in FMR1 knockout hippocampal cells (Fig. 8d). Combined, the data suggest that there is a long–lasting FMRP–dependent stabilization effect via mGluR–specific neuronal activity.
DISCUSSION
In this paper we have shown that FMRP interacts directly with the 3′UTR of PSD–95 mRNA. However, we find that PSD–95 mRNA polysomal association remains the same in wildtype and FMR1 knockout mice and that the PSD–95 mRNA is still localized in FMR1 knockout neurons. While translation of the PSD–95 mRNA may decrease due to post–initiation mechanisms (i.e. as in the case of some miRNAs37), that we cannot detect with the current assay, this translation mechanism would be different from that previously documented for other FMRP targets (e.g. Arc).
Interestingly, we find that the FMRP:PSD–95 mRNA interaction resulted in a stabilization of the PSD–95 message that can be further increased via mGluR stimulation. In FMR1 knockout mice the PSD–95 message is less stable, resulting in a reduction in the levels of this critical synaptic protein. These observations are consistent with previous circumstantial evidence suggesting that FMRP could potentially control mRNA levels. A microarray study identified 113 FMRP–associated mRNAs whose level decreased in Fragile X cell lines, yet whose polysome profile remained unchanged16. Another study found decreases in the levels of some FMRP–target mRNAs in the absence of FMRP17. While neither group examined these mRNAs further, it is possible that reduced levels of these mRNAs actually reflect a loss of mRNA stability in the absence of FMRP. Our finding that at least one other mRNA (MBP) is destabilized in the absence of FMRP lends support to this idea.
Surprisingly, we find that stabilization of the PSD–95 message is dependent on the area of the brain examined. The effect is most prominent in the hippocampus, present to a minor extent in cerebellum, and not detected in the cortex. This lack of a cortical effect is consistent with previous findings that PSD–95 protein levels are the same in wildtype and FMR1 knockout cortical cells29. Those authors also observed an FMRP–dependent increase in PSD–95 protein, in cortical cells, shortly after DHPG treatment but found that protein levels fall back to baseline by 4 hours29, suggesting a transient surge in PSD–95 expression. We observe, in hippocampal neurons, that the relative level of the PSD–95 mRNA rises slightly after 4 to 6 hours of DHPG exposure, suggesting an additional activity–dependent increase in RNA stability. Combined, these data suggest that FMRP can regulate, according to the physiological state (DHPG–treated or not) and cell type (cortical or hippocampal), both a rapid rise in PSD–95 translation (cortex) and a more prolonged rise in PSD–95 mRNA levels due to an increase in stability (hippocampus), and suggests that FMRP could have multiple independent roles.
We have mapped the binding site of FMRP to a G–rich element that is flanked by two AU–rich elements (AREs), well known cis–acting mRNA elements that regulate mRNA half–life. Several trans–acting factors that aid in both stabilization and destabilization of target mRNAs are known to bind to AREs44. Interestingly, regulation of HuD, a member of the Hu class of ARE–binding proteins44, during neuronal development results in temporal regulation of GAP-4345. Similarly, regulation of mRNA stability is often the result of competition between stabilizing and destabilizing factors44. It is therefore plausible that the region specific regulation of the PSD–95 message is a result of interference between the stabilizing role of FMRP and stabilizing and destabilizing functions of other binding factors. In support of this notion, we find that the hippocampus and cortex display different forms of Hu–family proteins (Supplementary Fig. 8). Combinatorial models are an emerging theme explaining RNA:protein binding specificity (reviewed in46,47), and in our case may explain why FMRP does not stabilize all of its known binding targets (Supplementary Table 1).
We also found that the PSD–95 mRNA is localized in dendrites in vivo but that its localization is not dependent upon FMRP, further highlighting the complexity surrounding FMRPs many roles in the cytoplasm. Several factors are known to bind to FMRP and are presumed to aid FMRP in these cytoplasmic regulatory functions. However, to date only one of these interactions has also been shown to aid FMRP function. Cooperative binding between FMRP and the BC1 RNA leads to translational repression of a subset of mRNAs and BC1 functions as a repressor of translational initiation in rabbit reticulocite assays3and references therein. We expect further binding partners to collaborate with FMRP to aid translational repression, mRNP localization, and this newly identified role in mRNA stabilization.
While a large number of putative FMRP target mRNAs have been isolated in the past five years, relatively few are known to be involved in regulating synapse structure and function. Our results strengthen the idea that FMRP function is extremely important for the correct formation of the postsynaptic compartment. The results also support the notion that the underlying cause of FXS, and potentially other forms of mental retardation, may be through direct interference with synaptic signaling leading to spine dysmorphogenesis and ultimately to memory defects1. Interestingly, the mRNA encoding a PSD–95 associated protein, SAPAP4, has also been shown to be in a complex with FMRP16. In addition, PSD-95, SAPAP4, Arc and are all components of the large scale NMDA receptor signaling complex that also links NMDA receptors to the mGluR signaling pathway23, and disruption of PSD–9520, Arc48 and α–CaMKII22 all result in impairments in learning. This is of interest in light of the evidence suggesting that alterations in glutamate receptor signaling via mGluRs might underlie a number of the cognitive deficits associated with FXS23. Furthermore, various other cases of neurological deficits also result in a decrease in PSD–95 expression (e.g.49,50) suggesting that strict regulation of PSD–95 expression is required for proper brain function. PSD–95 is important in both behavioral memory and dendritic spine morphology27 both features of FXS. Combined these results suggest that FMRP may regulate NMDA and mGluR receptor signaling through several proteins, including PSD–95, and that the cognitive and anatomical defects in FXS may arise by disruption of this complex.
METHODS
Animals Treatment
Animal care conformed to institutional guidelines in compliance with national and international laws and policies. All animals were 3 week old males (C57/BL6 wildtype and two FMR1 knockout strains C57/BL6 and FVB background).
Western Blots
We used standard methodologies with an FMRP monoclonal antibody (MAB2160) from Chemicon and a polyclonal (rAM2) produced in our laboratory8. The PSD–95 antibody was from Upstate (1:1000) and the eIF4E antibody from Cell Signalling (1:10000). All secondary antibodies were from Promega. The proteins were revealed using ECL plus (Amersham) and a phosphoimager (Amersham).
cDNA constructs
We obtained a mouse PSD–95 cDNA construct with the 3′UTR from the IMAGE consortium (ID 10318) and also isolated PSD–95 coding and 3′UTR fragments via RT–PCR from mouse brain extract and T/A cloning (Promega Easy T/A cloning kit; pT/A–Fragment 1–5). Details of constructs and mutagenesis are reported in Supplementary Information. FMRP protein domains were previously reported34.
EMSA
We performed binding reactions using full length human FMRP protein in binding buffer (300 mM KCl, 5 mM MgCl2, 2 mM DTT, 0.5% glycerol, 20 mM HEPES pH 7.5 and 300 ng/□l tRNA), incubating at 25 °C or 4 °C for 30 minutes. We added heparin (0.3 □g) for 5 minutes before separation on a 6% native polyacrylamide gel. We performed binding reactions with FMRP domains in the same buffer plus 100 or 300 mM KCl and 50–100 ng recombinant protein.
In vitro transcription
We performed these reactions using standard protocols (Ambion SP6/T7 Mega-Script) with [α32P]UTP, [α35S]UTP and UTP-Cy5 for EMSA, Northern blot, in situ hybridization respectively.
Primary cultures
We prepared primary cortical and hippocampal neuronal cultures from embryonic mice (E15 – cortical, E19 – hippocampal) using standard protocols.
Neuronal transfection
We transfected hippocampal neurons at 14 DIV using a standard Ca2+ phosphate precipitation protocol. We washed the precipitate using Hanks’ balanced saline (HBSS) and performing Actinomycin D experiments 48 hours later.
FISH, Immunofluorescence and Immunohistochemistry
We fixed primary hippocampal and cortical neurons at room temperature for 15 minutes (4% paraformaldehyde, 2 mM MgCl2, 5 mM EGTA in PBS 1X) then UV irradiated and permeabilized the cells (1X PBS containing 0.1% Triton X–100). We prehybridized neurons (50% formamide, 2X SSC, 10 mM NaH2PO3) then hybridized at 42 °C (30% formamide, 10 mM NaH2PO3, 10% dextran sulfate, 2X SSC, 0.2% BSA, yeast tRNA and salmon sperm DNA 500 □g/ml, and in vitro synthesized Cy5–labeled riboprobe). We performed immunofluorescence and immunohistochemistry preincubation in 2% donkey serum, 0.2% Triton X–100, then incubation in 1% BSA with anti–FMRP antibodies8 then Cy3–labeled secondary anti–rabbit IgG (Jackson ImmunoResearch). We analyzed neurons by confocal scanning microscopy (Zeiss LSM 510).
Radioactive in situ hybridization
We cryostat sectioned, fixed (4% paraformaldehyde), permeabilized (Proteinase ) and acetylated (0.25% of acetic anhydride in 0.1 M triethanolamine pH 8.0) brains prior to prehybridization and hybridization using standard protocols (55 °C in 50% formamide, 1X Denhardt’s solution, 10% dextran sulphate, 0.3 M NaCl, 5 mM EDTA, 0.5 mg/ml yeast tRNA, 20 mM Tris HCl pH 8.0, 50 mM DTT and 105 cpm/μl of [α35S]UTP-riboprobe). Slides were emulsified (Kodak autoradiography emulsion NTB2) and developed (ILFORD PQ developer) after 7-15 days of exposition. We analyzed sections by ZEISS axioskop microscopy (1.25X or 5X objectives), acquired images with a Canon S50 digital camera and quantified the signal using two ImageQuant and ImageJ.
Immunoprecipitation and RT–PCR
Whole brain was homogenized in 10 mM HEPES pH 7.4; 200 mM NaCl, 0.5% Triton X–100, 30 mM EDTA, protease inhibitors (Sigma–Aldrich) and 30 U/ml Rnasin. We preblocked 20 μl protein A sepharose (0.1 □g/ml each BSA, yeast tRNA, glycogen) for 1 hr then immunoprecipitated with specific FMRP antibodies8. DNase I (RNase–free, Amersham Pharmacia Biotech) was added during washes. We treated the immunoprecipitate with 50 μg proteinase K (Sigma–Aldrich) prior to RNA extraction and precipitation. First–strand synthesis was performed using p(dN)6 and 100 U of M–MLV RTase (Invitrogen). RT–PCR was performed as described in8. Radioactive semiquantitative RT–PCR reactions were performed in nonsaturating conditions in the presence of [α32P]dCTP, 1 mM dCTP and 10 mM dATP, dGTP and dTTP and analyzed on a 5% polyacrylamide gel.
Reversible crosslinking
We performed experiments as previously described32. Briefly, we washed hippocampal neurons at 10 DIV with Neurobasal containing 2% B27 and crosslinked in 0.5% formaldehyde (J.T. Baker) for 30 min at room temperature and quenched with 0.25 M glycine (Biorad). We harvested cells by centrifugation, PBS wash and resuspension in RIPA buffer. We immunoprecipitated crosslinked complexes with an FMRP antibody8. Prior to RT–PCR, we reversed crosslinking by treatment at 70 °C.
Polysomal analysis and RT–PCR
We analyzed cytoplasmic brain extract (total brain and hippocampi) as previously described8. See Supplementary Information for details.
mRNA stability assay
We treated primary cortical or hippocampal cultures (10 DIV) from time 0 with Actinomycin D (10 μg/ml) for the indicated times. We washed cultures in PBS, extracted RNA with Trizol and analyzed RNA by quantitative RT–PCR. We used a NIKON C1 with plan–neofluar 20X to analyze both wildtype and FMR1 knockout cultures for morphology. We assessed mRNA stability after DHPG treatment in two different ways. First, we added DHPG (100 μM) to cultures pre-exposed to Actinomycin D for 3,30 hours or 5,30 hours. After 30 minutes of DHPG treatment, we collected mRNA for quantitative RT–PCR analysis. Second, we added DHPG (50 μM) and Actinomycin D jointly to cells at time 0 and collected RNA 4 or 6 hours later for quantitative RT–PCR analysis.
Quantitative RT–PCR
We performed reactions with MoMLV–reverse transcriptase (Invitrogen) and the TaqMan Universal PCR Master Mix (ABI 4304437) using dual–labeled TaqMan probes (Applied Biosystems). We detected mouse PSD–95 mRNA using the Pre–Developed TaqMan probe Mm00492193_m1 and compared with the endogenous control mRNA (mouse H3f3b mRNA Pre–Developed TaqMan probe Mm00787223_s1). Cycle parameters were as suggested by the manufacturer. Relative PSD–95 mRNA levels, normalized to H3f3b, were calculated as follows: 2−[□eltaCt(treated) − □eltaCt(untreated)] = 2−□eltaCt, where DeltaCt equals Ct(PSD–95) – Ct(H3f3b). β–Actin mRNA was detected with Pre–Developed TaqMan probe Mm00607939_s1, and Renilla luciferase mRNA was detected using primers specifically designed by the Applied Biosystems (See Supplementary information).
Primers used
We provide a table containing the primers used in this study in Supplementary Information.
Neuronal cell survival (Mitochondrial Activity)
We measured the mitochondrial activity using the colorimetric MTT assay by incubating hppocampal cultures for 30 minutes at 37 °C with 1 ml of Locke’s buffer (154 mM NaCl, 5.6 mM KCl, 3.6 mM NaHCO3, 2.3 mM CaCl2, 1.2 mM MgCl2, 5.6 mM glucose, 5 mM Hepes pH 7.4) containing ([3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium]) (Sigma). We then dissolved cultures with 700 μl of DMSO and tested viable neurons by production of the purple MTT cleavage product, formazan. We took three independent measurements of sample optical density using a VICTOR 3V 1420 Multilabel Counter at 490 nm and reported the mean with standard deviation. The value of each culture is divided by the reference value (control culture at time 0).
Northern blot analysis
2 μg of poly(A)+ RNA from the entire brain or 20 μg of total RNA were probed using a mouse [α32P]UTP PSD–95 mRNA antisense probe to fragment 5 (nucleotide 2820–3061 of BC014807), the entire 3′UTR (nucleotide 2227–3061 from BC014807) or the coding region (nucleotide 61–2226 of BC014807). We probed the same membrane with a cDNA antisense fragment (nucleotide 258–837 of X03672) and quantified radioactive signals with a phosphoimager (Amersham).
ACKNOWLEDGEMENTS
We thank B.A. Oostra for the FMR1 knockout mice, N.K. Gray and T. Achsel for their critical evaluation of the manuscript and O. Steward for precious suggestions and reagents. We thank M.A. Kiebler for advice on the neuronal transfection protocol.
This research was funded by an European Molecular Biology Organization short term fellowship, a Royal Society of Edinburgh SEELLD fellowship and a Biotechnology and Biological Sciences Research Council grant (C19143) to KSD, by Telethon (GGP05269), Ministero della Salute, Ministero della Università (FIRB) to CB and by Wellcome Trust grant number 056523 and the Wellcome Trust Genes to Cognition Programe to SGNG. FZ was supported by the Associazione Italiana Sindrome dell’X Fragile.
References
- 1.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat. Rev. Neurosci. 2005;6:376–87. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005;4:341–9. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 3.Zalfa F, Achsel T, Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr Opin Neurobiol. 2006;16:265–9. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–99. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 5.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. Embo J. 2001;20:4803–13. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. Rna. 2003;9:1198–207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Yun SW, Seto J, Liu W, Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–17. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- 8.Zalfa F, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–27. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 9.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–37. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zalfa F, et al. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J. Biol. Chem. 2005;280:33403–10. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 11.Johnson EM, et al. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res. 2006;83:929–43. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- 12.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–53. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 13.Darnell JC, et al. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005 doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Zhong N, Ju W, Nelson D, Dobkin C, Brown WT. Reduced mRNA for G3BP in fragile X cells: evidence of FMR1 gene regulation. Am J Med Genet. 1999;84:268–71. doi: 10.1002/(sici)1096-8628(19990528)84:3<268::aid-ajmg20>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 17.Miyashiro KY, et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–31. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 18.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–50. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonegawa S, et al. Hippocampal CA1-region-restricted knockout of NMDAR1 gene disrupts synaptic plasticity, place fields, and spatial learning. Cold Spring Harb Symp Quant Biol. 1996;61:225–38. [PubMed] [Google Scholar]
- 20.Migaud M, et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–9. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 21.Fagiolini M, et al. Separable features of visual cortical plasticity revealed by N-methyl-D-aspartate receptor 2A signaling. Proc Natl Acad Sci U S A. 2003;100:2854–9. doi: 10.1073/pnas.0536089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–11. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 23.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–9. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 24.Sheng M, Kim MJ. Postsynaptic signaling and plasticity mechanisms. Science. 2002;298:776–80. doi: 10.1126/science.1075333. [DOI] [PubMed] [Google Scholar]
- 25.Cuthbert PC, Stanford LE, Coba MP, Ainger JA, Fink AE, Opazo P, Delgado JY, Komiyama NH, O’Dell TJ, Grant SGN. SAP102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J. Neurosci. 2007;27:2673–2682. doi: 10.1523/JNEUROSCI.4457-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarpey P, et al. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:318–24. doi: 10.1086/422703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vickers CA, et al. Neurone specific regulation of dendritic spines in vivo by post synaptic density 95 protein (PSD-95) Brain Res. 2006 doi: 10.1016/j.brainres.2006.03.075. [DOI] [PubMed] [Google Scholar]
- 28.Reiss AL, Lee J, Freund L. Neuroanatomy of fragile X syndrome: the temporal lobe. Neurology. 1994;44:1317–24. doi: 10.1212/wnl.44.7.1317. [DOI] [PubMed] [Google Scholar]
- 29.Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A. 2003;100:14374–8. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YQ, et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 31.Lu R, et al. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl. Acad. Sci. USA. 2004;101:15201–6. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–90. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 33.Bence M, Arbuckle MI, Dickson KS, Grant SG. Analyses of murine postsynaptic density-95 identify novel isoforms and potential translational control elements. Brain Res Mol Brain Res. 2005;133:143–52. doi: 10.1016/j.molbrainres.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 34.Adinolfi S, et al. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. Rna. 1999;5:1248–58. doi: 10.1017/s1355838299990647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–80. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 36.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–42. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 37.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2006 doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–59. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- 39.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 40.Loesch DZ, Huggins RM, Hagerman RJ. Phenotypic variation and FMRP levels in fragile X. Ment Retard Dev Disabil Res Rev. 2004;10:31–41. doi: 10.1002/mrdd.20006. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, et al. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Hum Mol Genet. 2004;13:79–89. doi: 10.1093/hmg/ddh009. [DOI] [PubMed] [Google Scholar]
- 42.Weiler IJ, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. USA. 1997;94:5395–400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–55. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith CL, et al. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J Neurobiol. 2004;61:222–35. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- 46.Singh R, Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol. 2005;12:645–53. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 47.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–10. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Plath N, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–44. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 49.Gylys KH, et al. Synaptic changes in Alzheimer’s disease: increased amyloid-beta and gliosis in surviving terminals is accompanied by decreased PSD-95 fluorescence. Am J Pathol. 2004;165:1809–17. doi: 10.1016/s0002-9440(10)63436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toro C, Deakin JF. NMDA receptor subunit NRI and postsynaptic protein PSD-95 in hippocampus and orbitofrontal cortex in schizophrenia and mood disorder. Schizophr Res. 2005;80:323–30. doi: 10.1016/j.schres.2005.07.003. [DOI] [PubMed] [Google Scholar]