Abstract
Motivation: The Nearest Alignment Space Termination (NAST) tool is commonly used in sequence-based microbial ecology community analysis, but due to the limited portability of the original implementation, it has not been as widely adopted as possible. Python Nearest Alignment Space Termination (PyNAST) is a complete reimplementation of NAST, which includes three convenient interfaces: a Mac OS X GUI, a command-line interface and a simple application programming interface (API).
Results: The availability of PyNAST will make the popular NAST algorithm more portable and thereby applicable to datasets orders of magnitude larger by allowing users to install PyNAST on their own hardware. Additionally because users can align to arbitrary template alignments, a feature not available via the original NAST web interface, the NAST algorithm will be readily applicable to novel tasks outside of microbial community analysis.
Availability: PyNAST is available at http://pynast.sourceforge.net.
Contact: rob.knight@colorado.edu
1 INTRODUCTION
The Nearest Alignment Space Termination (NAST) tool (DeSantis et al., 2006b) was developed to efficiently align thousands of 16S rRNA genes using an alignment compression algorithm to create multiple sequence alignments (MSAs) with a set number of columns. The Greengenes ribosomal database (DeSantis et al., 2006a) has oriented the 400 000 known near full length 16S rRNA genes with NAST, and this aligner has become an integral tool in microbial community analysis. Additionally, users have created project-specific MSAs of this scale via the web interface at http://greengenes.lbl.gov/NAST. While NAST has been available to a wide audience as a web application, the difficulty of installing it locally has limited the applicability of NAST for many users.
Here, we present Python Nearest Alignment Space Termination (PyNAST), a complete reimplementation of the NAST algorithm using the PyCogent toolkit (Knight et al., 2007). Several key features have been added in PyNAST representing significant enhancements. New features include:
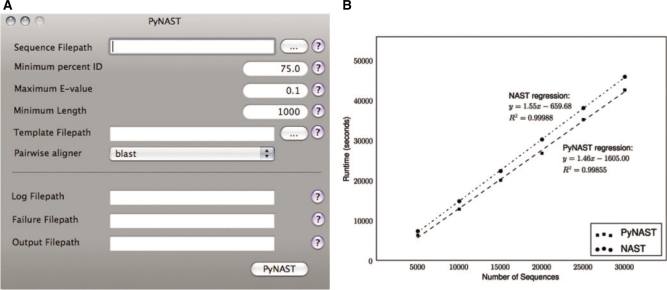
three convenient interfaces: a Mac OS X GUI (Fig. 1a), a command-line interface and a simple application programming interface (API);
parameterized algorithms at key steps of the analysis [e.g. pairwise alignment can be performed with BLAST, MUSCLE (Edgar, 2004), MAFFT (Katoh et al., 2005), ClustalW (Thompson et al., 1994), or the PyCogent pairwise hidden Markov model (HMM) aligner, and is extensible to incorporate new pairwise aligners];
an open source software package with minimal dependencies (Python, NumPy and BLAST), designed for easy installation on single machines or in a cluster environment;
ability to align an arbitrary sequence against an arbitrary template alignment, rather than only 16S sequences.
Fig. 1.
(A) Screenshot of the PyNAST graphical user interface for Mac OS X. (B) Runtime of PyNAST is compared with that of NAST, each running on a single processor. PyNAST has a slightly shorter per sequence runtime (slope). The candidate sequences used in this evaluation ranged from 917 to 1343 bases, with a median length of 1294. The template alignment was a Greengenes core set (dated November 8, 2007) with 7682 positions and 4938 sequences.
2 ALGORITHM
The NAST algorithm aligns a candidate sequence to a template alignment. The output, the aligned candidate sequence, is guaranteed to be the same length as the input template alignment. In the original NAST implementation, each user-submitted candidate sequence is aligned to the Greengenes ‘Core Set’ (a template alignment), comprising approximately 5000 aligned non-chimeric sequences representative of the currently recognized diversity among bacteria and archaea. In PyNAST, the user can specify an arbitrary template alignment in a standard fasta alignment file to which candidate sequences should be aligned.
The NAST algorithm is applied to a candidate sequence and template alignment as follows. First, the sequence most similar to the candidate sequence in the template alignment (the template sequence) is identified using BLAST (Altschul et al., 1990). Gaps are removed from the template sequence, and it is then pairwise aligned with the candidate sequence. Next, the gap spacing from the template alignment is reintroduced into the pairwise alignment yielding an alignment that may be longer than the template alignment. To reduce the length of the pairwise alignment to that of the template alignment, gaps must be removed from the pairwise alignment. New gaps which were introduced in the aligned template sequence during pairwise alignment are removed, and to maintain the alignment, the nearest gap character in the aligned candidate sequence is also removed. Thus by introducing local misalignments, the candidate sequence is globally aligned to the template alignment without disrupting the length of the template alignment.
3 SPEED BENCHMARK
The runtime of PyNAST was compared against the runtime of NAST on a collection of 30 000 16S rRNA sequences, and subsets of this collection containing 5000, 10 000, 15 000, 20 000 and 25 000 sequences each. The command-line implementation of NAST was used in this study, and compared directly with command-line PyNAST. BLAST 2.2.16 was used for the database search and pairwise alignment steps in both applications. PyNAST runs faster than the original NAST implementation, requiring 1.46 s/sequence versus 1.55 s/sequence (Fig. 1b).
The rate-limiting step in PyNAST is the pairwise alignment, and the algorithm is therefore of complexity corresponding to the pairwise alignment algorithm. When Blast is used for the pairwise alignment, the complexity is O(nm) where n and m refer to the lengths of the candidate and template sequences.
4 CONCLUSIONS
The key advantage of the NAST algorithm is that it aligns relatively conserved regions well, and avoids expanding the alignment with new gaps in non-conserved regions. Because new sequences are aligned to the same length as the template alignment, newly acquired sequences can be analyzed in the context of existing alignments which may have been developed through costly processes such as manual alignment. Users are thus, for example, able to calculate distance matrices for diversity estimates and taxonomically classify organisms in microbiome samples based on existing high-quality alignments.
PyNAST is a reimplementation of NAST, introducing new features that increase its portability and flexibility. Its availability as an open source application with three convenient interfaces will allow the application of the NAST algorithm on a wider basis, to larger datasets, and in novel domains.
Funding: This work was funded in part by grants T15LM009451 to JGC; a Bill and Melinda Gates Foundation Mal-ED Network Discovery Project to William Petri; 1U01HG004866-01 to Owen White; Human Microbiome Demonstration project grant UH2DK083981 to FDB, James Lewis, and Gary Wu. This work was also partially supported by grant UH2CA140233 from Human Microbiome Project of the NIH Roadmap Initiative and National Cancer Institute to Zhiheng Pei.
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, et al. Greengenes, a chimera-checked 16s rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006a;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, et al. NAST: a multiple sequence alignment server for comparative analysis of 16s rRNA genes. Nucleic Acids Res. 2006b;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, et al. Mafft version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, et al. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]