Abstract
Using sensitive sequence profile analysis, we identify a hitherto uncharacterized cysteine-rich, transmembrane (TM) module, CYSTM, found in a wide range of tail-anchored membrane proteins across eukaryotes. This superfamily includes Schizosaccharomyces Uvi15, Arabidopsis PCC1, Digtaria CDT1 and Saccharomyces proteins YDL012C and YDR210W, which have all been implicated in resistance/response to stress or pathogens. Based on the pattern of conserved cysteines and data from different chemical genetics studies, we suggest that CYSTM proteins might have critical role in responding to deleterious compounds at the plasma membrane via chelation or redox-based mechanisms. Thus, CYSTM proteins are likely to be part of a novel cellular protective mechanism that is widely active in eukaryotes, including humans.
Contact: aravind@ncbi.nih.gov
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
High-throughput genetic screens in model systems such as Saccharomyces cerevisiae have yielded a wealth of data on the cellular apparatus and biochemical processes involved in tolerance to environmental stresses (Wuster and Madan Babu, 2008). The basic assumption in these studies is that if a gene is important for normal growth in presence of a given stress its partial or complete loss would uncover a stress–gene interaction, which can be measured as compromised fitness. In particular, natural resistance to chemical stresses has been intensely studied to obtain a handle on the pharmacology, mode of action and off-target effects of various medically and commercially relevant substances (Ericson et al., 2008; Giaever et al., 2002; Hillenmeyer et al., 2008; Parsons et al., 2004). These studies have shown that cellular defenses against deleterious substances work at a variety of levels ranging from direct interaction and expulsion to more subtle effects involving direct and indirect backups that act as intrinsic buffers in the system (Venancio et al., 2009). Nevertheless, given the magnitude of the information that has accumulated over the past few years, there are still several concealed aspects of chemical resistance that remain to be discovered in this data. We were especially interested in uncovering previously unknown resistance mechanisms that might be widely conserved across eukaryotes. We recently compiled a comprehensive collection of chemical genetics datasets from 34 different studies, including homozygous and heterozygous mutants and covering 425 compounds [(Venancio et al., 2009) and T.M. Venancio et al., manuscript in preparation]. This data can be represented as a bimodal network, i.e. the chemical-phenotype (CP) network, which links genes to the respective compounds against which they provide natural resistance. Genes in this network can be further connected to each other, based on statistically significant overlaps in their interactions with chemicals, to generate an undirected network (the shared chemical phenotype or SCP network; T.M. Venancio et al., manuscript in preparation). Highly connected genes or hubs in the SCP network functionally cooperate with a wide range of genes in providing chemical tolerance and could hence be critical nodal points in the phenomenon of natural resistance. Among these hubs, we recovered two paralogous proteins, YDL012C and YDR210W, which were believed to be yeast-specific proteins with no previously characterized domains (Smith et al., 1999). A third paralog YBR016W was also recovered in the SCP network albeit only with a moderate number of connections. Using sensitive sequence profile analysis, we present evidence that these proteins define a novel superfamily of transmembrane (TM) domains that are widely distributed across eukaryotes and have a potential conserved role in resistance to various chemical/environmental stresses.
2 METHODS
Using data from 34 independent studies (Supplementary Material), we extracted every reported case in which a homozygous or heterozygous gene deletion resulted in a growth defect (decreased fitness) in the presence of a chemical. Every such chemical–gene interaction contributed a single edge in the CP network. By simulations using degree-preserving random networks, we created the SCP network by retaining only the significant interactions (P ≤ 0.001; T.M. Venancio et al., manuscript in preparation). Profile searches against the NR database were performed using the PSI-BLAST (Altschul et al., 1997) and the HMMSEARCH and JACKHMMER programs of the recently developed HMMER3 package (Eddy, 2008). For PSI-BLAST, parameters were adjusted for short sequences in the initial run (inclusion threshold = 0.01). Sequences detected in PSI-BLAST searches were used to create an initial profile for HMMSEARCH and new sequences were added to it as they were detected. The JACKHMMER inclusion threshold was set as 0.001. Multiple alignments were constructed using the KALIGN program (Lassmann and Sonnhammer, 2005) followed by manual editing using the HMMSEARCH HSPs. Transmembrane helices and membrane topology were predicted using the TMHMM program (Krogh et al., 2001). The multiple alignment was used to predict protein secondary structure using the JPRED program (Cuff and Barton, 2000).
3 RESULTS AND DISCUSSION
3.1 Identification and characterization of the CYSTM module
Examination of the sequences of YDL012C, YDR210W and YBR016W revealed that they are all small proteins (<150 aa) sharing a peculiar structure: they possess a variable N-terminal segment, which is non-globular and enriched in proline and glutamine, followed by a conserved C-terminal region (35–40 aa) comprising a distinct module, which includes a single TM helix (Fig. 1). This suggested that they are tail-anchored cell membrane proteins and this has been experimentally confirmed in the case of the three above proteins (Beilharz et al., 2003; Huh et al., 2003) (Fig. 2). The C-terminal TM helix in these proteins differed from all other previously characterized TM helices in having a unique pattern of conserved residues suggesting that it might have functional significance for these proteins (Fig. 1). To better understand the evolutionary affinities and functions of this conserved module in the yeast proteins, we initiated transitive sequence profile searches using the PSI-BLAST program against the NR database. These searches recovered homologous proteins from several other fungi and also a fourth paralog from yeast YDR034W-B, which has also been shown to be a membrane-anchored protein (Huh et al., 2003). Given that short sequences with distinctive compositions fare poorly in PSI-BLAST searches, we also initiated HMM searches with HMMSEARCH and JACKHMMER (Eddy, 2008). These searches recovered several other proteins with significant expect values from various animals, choanoflagellates, fungi, plants, chlorophytes and the alveolate Perkinsus. These included previously studied proteins such as the Schizosaccharomyces pombe stress and chemical response protein Uvi15 (gi: 19112015; e = 10−5, iteration 3 in a JACKHMMER search seeded with YDL012C C-terminal region), the plant pathogen resistance protein PCC1 (gi: 18403306; e = 10−6, iteration 4) and the plant heavy metal resistance proteins such as CDT1 (gi: 197927011; e = 10−4, iteration 5). Further, reciprocal JACKHMMER searches with selected examples from the newly detected sequences also recovered the original yeast proteins with significant e-values. These observations indicated that rather being yeast specific, this conserved region defines a novel superfamily of modules that is widely distributed across eukaryotes.
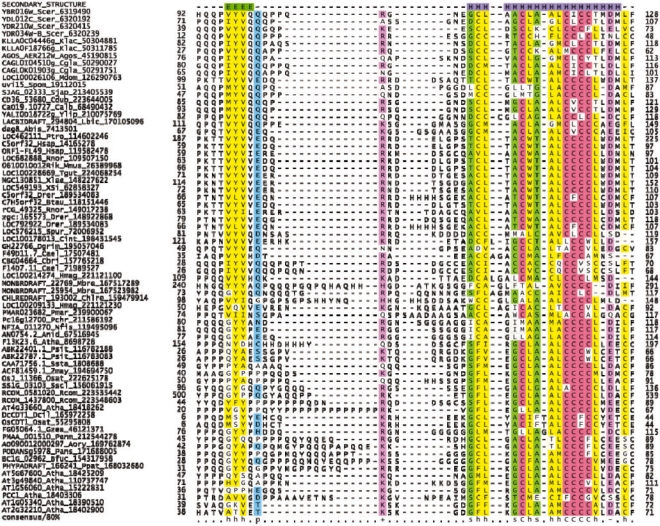
Fig. 1.
Multiple sequence alignment of the CYSTM domain superfamily. The columns were colored according to the consensus shown below the alignment, and the predicted secondary structure is shown on top. The sequences are labeled using the gene name, species abbreviation and GenBank gi number and sequence identifiers. The species abbreviations are—Abis: Agaricus bisporus; Atha: Arabidopsis thaliana; Agos: Ashbya gossypii; Anid: Aspergillus nidulans; Aory: Aspergillus oryzae; Btau: Bos taurus; Bfuc: Botryotinia fuckeliana; Cbri: Caenorhabditis briggsae; Cele: Caenorhabditis elegans; Calb: Candida albicans; Cdub: Candida dubliniensis; Cgla: Candida glabrata; Crei: Chlamydomonas reinhardtii; Cint: Ciona intestinalis; Drer: Danio rerio; Dcil: D.ciliaris; Dgri: Drosophila grimshawi; Gzea: Gibberella zeae; Hsap: Homo sapiens; Hmag: Hydra magnipapillata; Klac: Kluyveromyces lactis; Lbic: Laccaria bicolor; Mdom: Monodelphis domestica; Mbre: Monosiga brevicollis MX1; Mmus: Mus musculus; Nfis: Neosartorya fischeri; Osat: O.sativa; Ptro: Pan troglodytes; Pchr: Penicillium chrysogenum; Pmar: Penicillium marneffei; Pmar: Perkinsus marinus; Ppat: Physcomitrella patens; Psit: Picea sitchensis; Pans: Podospora anserina; Rnor: Rattus norvegicus; Rcom: Ricinus communis; Scer: S.cerevisiae; Sjap: Schizosaccharomyces japonicus; Spom: S.pombe; Sscl: Sclerotinia sclerotiorum; Ssta: Sporobolus stapfianus; Spur: Strongylocentrotus purpuratus; Tgut: Taeniopygia guttata; Xtro: Xenopus tropicalis; Xlae: Xenopus laevis; Ylip: Yarrowia lipolytica; Zmay: Zea mays.
Fig. 2.

A speculative model of the CYSTM module-anchored plasma in the membrane. Lipids are represented with black tail and yellow head groups. The protein transmembrane regions colored in red, with cysteines are represented as brown dashes. The intracellular unstructured regions are shown as random coil. The conserved acidic position (usually Asp) is shown binding the extracellular lipid head.
A multiple alignment of this domain (Fig. 1) showed that the features found in the above yeast proteins are widely conserved throughout the family and found in no other membrane protein family. These include: (i) An N-terminal cytoplasmic element that is predicted to adopt an extended conformation (β-strand) connected by a highly variable linker to (ii) a C-terminal TM helix with 5–6 cysteines followed by an acidic residue. Of these, 3–4 cysteines occur consecutively to constitute a conserved cysteine patch that is a hallmark of this superfamily (Fig. 1). Hence, we named this module CYSTM after this CYS-rich TM element. The CYSTM module is always present at the extreme C-terminus of the protein in which it is present. Furthermore, like the yeast prototypes, majority of these proteins also possess a proline/glutamine-rich segment upstream of the CYSTM module that is likely to form a polar, disordered head in the cytoplasm (Fig. 1). Consistent with this, in addition to the four yeast paralogs, two representatives from plants namely PCC1 and CDT1 have also been experimentally shown to be membrane proteins (Kuramata et al., 2009; Sauerbrunn and Schlaich, 2004). Based on these features, a number of predictions can be made regarding the structure and interactions of the CYSTM module. The presence of a solitary β-strand with conserved hydrophobic residues in the N-terminal cytoplasmic part indicates that it is likely to homo- or hetero-dimerize via this element (e.g. as in the case of the p53 tetramerization domain or the MetJ/Arc-like transcription factors; Figs 1 and 2). Further, presence of an atypical well-conserved acidic residue at the C-terminal end of the TM helix suggests that it might interact with a positively charged moiety in the lipid head group (Fig. 1) (von Heijne, 2007). This raises the possibility of potential specific association with zwitterionic lipids with available positive charges, such as phosphatidylethanolamine or phosphatidylcholine, which are abundant in membranes of all organisms with CYSTM proteins (Vance and Vance, 2008). Modeling the TM segment as an α-helix shows that up to 3–4 of the cysteines characteristic of this domain could approximately localize to the same face of the helix with the other cysteines probably oriented away from them (Fig. 2). This could potentially result in a prominent ridge of sulfhydryl groups on the face bearing the majority of the cysteines.
3.2 Possible functions of the CYSTM module
The CP network suggests that the yeast proteins YDL012C, YDR210W and YBR016W are together involved in resistance against a diverse set of substances that include DNA-damaging agents such as mitomycin C, the replication inhibitor methotrexate, the oxidizing agent hydrogen peroxide and the potential membrane destabilizing agent 1,8-nonadiene (Supplementary Material). In particular, YDL012C and YDR210W significantly overlap in the chemicals against which they provide resistance, suggesting that they might function together as a complex. Representatives of this superfamily from other organisms have also been independently implicated in stress responses. These include Uvi15 from the distantly related yeast S.pombe, which has been shown to be induced by heat shock and chemical stresses such as DNA-damaging agents and the amino acid analog canavanine. Further, deletion of Uvi15 resulted in loss of viability under stress conditions and stationary phase (Lee et al., 1995). The Arabidopsis representative of the superfamily, PCC1, was induced via the salicylic acid-dependent pathway upon pathogen exposure and its overexpression conferred resistance to oomycetes (Sauerbrunn and Schlaich, 2004). A group of plant CYSTM proteins typified by CDT1 was shown in Digitaria ciliaris and Oryza sativa to confer tolerance to heavy metals such as cadmium and copper (Kuramata et al., 2009). Heterologous expression of CDT1 in yeast showed that it conferred metal resistance by preventing uptake of the metal into the cell. Thus, consistently across eukaryotes different versions of the CYSTM module appears to have a role in stress response or tolerance, and more specifically in resistance to deleterious substances, implying that this might be a general function of the superfamily.
Typically, single TM protein modules are poorly conserved across diverse organisms unless they have a specialized function [e.g. the KASH module in nuclear membrane proteins (Fischer et al., 2004)]. Moreover, conservation of specific residues inside a TM, rather than an overall conservation of hydrophobic character, is atypical of membrane proteins unless they serve a ligand interaction or enzymatic role (von Heijne, 2007). These observations, taken together with the widespread evidence for a role in stress/chemical resistance response, suggest that the peculiar pattern of conserved cysteines and the acidic residue in the CYSTM domain are directly responsible for this function. Further, as suggested above the conserved acidic residue could allow tight association with certain types of lipids and alter the permeability of the plasma membrane to deleterious substances. Such membrane alterations could also explain the role of CYSTM proteins such as PCC1 in pathogen resistance, namely in blocking invasion of intracellular pathogens that form an interface with the host membrane. The peculiar arrangement of sulfhydryl groups within the membrane could also alter the redox potential of the membrane or potentially directly chelate metal ions. This proposal is consistent with the observation that the plant CDT1 excludes heavy metal ions (Kuramata et al., 2009).
The alteration of redox potential of the membrane by CYSTM proteins might also affect the uptake of certain compounds and also allow quenching of potentially damaging radicals. If this were the case, the CYSTM family could be seen as a membrane-associated analog of the metallothionein-like system against redox stresses (Deneke, 2000). Additionally, the cytoplasmic polar disordered head seen in majority of members of this superfamily is comparable in residue composition and organization to the ‘prion-like’ proteins, which assume multiple alternative conformational states (Perrett and Jones, 2008). Hence, it is conceivable that this cytoplasmic head could under certain circumstances assume some degree of structure. It particular, in some members of the superfamily the short repeats between the TM helix and the first well-predicted strand could potentially form additional small strands. It remains to be seen if such conformational changes might have any role in the stress-response functions of this superfamily.
4 CONCLUSIONS
By combining information from chemical genetics studies and sequence profile analysis, we uncover a previously unrecognized conserved module that could mediate tolerance and response to a wide range of stresses at the level of the plasma membrane. The unique pattern of conserved cysteines in the CYSTM module could be central to its protective function. Its conservation across eukaryotes, including humans, indicates that further studies on this module might be useful in uncovering hitherto unrecognized defensive strategies against environmental insults.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lakshminarayan Iyer for assistance early in the project.
Funding: Intramural Research Program of the National Institutes of Health, USA.
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz T, et al. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Deneke SM. Thiol-based antioxidants. Curr. Top. Cell. Regul. 2000;36:151–180. doi: 10.1016/s0070-2137(01)80007-8. [DOI] [PubMed] [Google Scholar]
- Eddy SR. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 2008;4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson E, et al. Off-target effects of psychoactive drugs revealed by genome-wide assays in yeast. PLoS Genet. 2008;4:e1000151. doi: 10.1371/journal.pgen.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, et al. Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye. Genetics. 2004;168:1385–1393. doi: 10.1534/genetics.104.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Krogh A, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuramata M, et al. Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant Cell Physiol. 2009;50:106–117. doi: 10.1093/pcp/pcn175. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, et al. Characterization of uvi15+, a stress-inducible gene from Schizosaccharomyces pombe. Mol. Gen. Genet. 1995;246:663–670. doi: 10.1007/BF00290711. [DOI] [PubMed] [Google Scholar]
- Parsons AB, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- Perrett S, Jones GW. Insights into the mechanism of prion propagation. Curr. Opin. Struct. Biol. 2008;18:52–59. doi: 10.1016/j.sbi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Sauerbrunn N, Schlaich NL. PCC1: a merging point for pathogen defence and circadian signalling in Arabidopsis. Planta. 2004;218:552–561. doi: 10.1007/s00425-003-1143-z. [DOI] [PubMed] [Google Scholar]
- Smith KN, et al. Disruption and functional analysis of seven ORFs on chromosome IV: YDL057w, YDL012c, YDL010w, YDL009c, YDL008w (APC11), YDL005c (MED2) and YDL003w (MCD1) Yeast. 1999;15:1255–1267. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1255::AID-YEA451>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins, and Membranes. Amsterdam, Boston: Elsevier; 2008. [Google Scholar]
- Venancio TM, et al. High-confidence mapping of chemical compounds and protein complexes reveals novel aspects of chemical stress response in yeast. Mol. Biosyst. 2009 doi: 10.1039/b911821g. [Epub ahead of print, doi:10.1039/b911821g, 27 August, 2009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. The membrane protein universe: what's out there and why bother? J. Intern. Med. 2007;261:543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- Wuster A, Madan Babu M. Chemogenomics and biotechnology. Trends Biotechnol. 2008;26:252–258. doi: 10.1016/j.tibtech.2008.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.