Abstract
The epidermal growth factor repeats of the Notch receptor are extensively glycosylated with three different O-glycans. O-Fucosylation and elongation by the glycosyltransferase Fringe have been well studied and shown to be essential for proper Notch signaling. In contrast, biosynthesis of O-glucose and O-N-acetylglucosamine is less well understood. Recently, the isolation of the Drosophila mutant rumi has shown that absence of O-glucose impairs Notch function. O-Glucose is further extended by two contiguous α1,3-linked xylose residues. We have identified two enzymes of the human glycosyltransferase 8 family, now named GXYLT1 and GXYLT2 (glucoside xylosyltransferase), as UDP-d-xylose:β-d-glucoside α1,3-d-xylosyltransferases adding the first xylose. The enzymes are specific for β-glucose-terminating acceptors and UDP-xylose as donor substrate. Generation of the α1,3-linkage was confirmed by nuclear magnetic resonance. Activity on a natural acceptor could be shown by in vitro xylosylation of a Notch fragment expressed in a UDP-xylose-deficient cell line and in vivo by co-expression of the enzymes and the Notch fragment in insect cells followed by mass spectrometric analysis of peptide fragments.
Keywords: Cell/Cell-Cell Interaction, Glycoproteins/Biosynthesis, Glycosylation, Receptors, Signal Transduction, Subcellular Organelles/Golgi, Signal Transduction/Notch
Introduction
More than half of the 29–36 epidermal growth factor (EGF)2 repeats of the mammalian Notch receptors are modified with O-fucose (Fuc), O-glucose (Glc), or O-N-acetylglucosamine (GlcNAc) glycans. These O-glycans are linked to specific serine or threonine residues in distinct consensus sequences (1, 2), which are also found on EGF repeats of various other proteins that include blood coagulation factor VII and Notch ligands Delta and Serrate/Jagged (3).
Although further modifications of O-GlcNAc have not been identified yet, O-Fuc added by the protein-O-fucosyltransferase 1 (Pofut1) can be elongated to the tetrasaccharide Siaα2,3/6Galβ1,4GlcNAcβ1,3Fucα1-O-Ser/Thr. O-Fucosylation is clearly essential for Notch function, and Pofut1 null mice exhibit an embryonic lethal phenotype (4–7). The enzyme has a chaperone function that, at least in Drosophila, seems more critical than its fucosyltransferase activity to functionalize Notch. Elongation of O-Fuc by the GlcNAc-transferase Fringe in Drosophila (8, 9) or its three mammalian homologs (10) modulates relative signaling intensities by different Notch ligands. In Drosophila, Fringe enhances signaling of Delta but reduces that of Serrate (11). Although more complicated in mammals, Fringe modification of Notch alters interactions with its ligands as well (3, 8, 12).
The O-Glc glycan typically exists as the trisaccharide Xylα1,3Xylα1,3Glcβ1-O-Ser in mammals (1, 13). On factor VII, this glycan is bound to serine 52, and mutation of this amino acid reduces the coagulant activity in a clotting assay (14). The gene encoding the protein O-glucosyltransferase (Poglut) has recently been identified via the Drosophila mutant rumi that showed a temperature-dependent Notch phenotype (15). Based on the role of O-Fuc modifications, it is anticipated that xylosylation of O-Glc is playing a similar role in Notch signaling as glycosylation by Fringe. Although enzymatic activities of the two α1,3-xylosyltransferases have been detected in mammalian cells (16–18), the genes encoding the xylosyltransferases have not yet been identified, hampering further functional studies of O-Glc modifications. Here we describe the identification of two genes, now named GXYLT1 and GXYLT2 (glucoside xylosyltransferase), encoding enzymes able to transfer xylose to the O-Glc residue bound to Notch EGF repeats both in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Enzyme Expression
Protein A fusions of human GLT8D1, GLT8D2, GXYLT1, and GXYLT2 (Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC)), for which constructs are described in the supplemental Experimental Procedures, were expressed in Sf9 insect cells grown in Insect-Xpress medium (Lonza) by baculovirus infection (Bac-to-Bac®; Invitrogen). After 72 h of incubation, secreted proteins were purified using 100 μl of IgG-Sepharose-6 Fast Flow (GE Healthcare) per 100 ml of culture supernatant for 12 h at 4 °C, washed according to the manufacturer's guidelines, and stored at −20 °C in 50% glycerol, 2 mm MnCl2, 10 mm MOPS, pH 7.
Xylosyltransferase Activity Assays
Assays were performed in 50 μl of reaction buffer (100 mm MOPS, pH 7.5, 10 mm MnCl2, 10 mm ATP). Acceptors were applied to a final concentration of 100 μm, and UDP-[6-3H]Gal, UDP-[1-3H]Glc (GE Healthcare), or UDP-[U-14C]Xyl (PerkinElmer Life Sciences) was diluted with cold UDP sugars to obtain 5 μm final concentration with a specific activity of 4 kBq/nmol for 3H sugars and 0.75 kBq/nmol for [14C]Xyl. Reactions were started by the addition of 10 μl of bead-coupled enzyme, incubated at 37 °C for 1 h, and terminated with 1 ml of ice-cold H2O. Radioactivity associated to the acceptors was determined after purification on C18 columns (Sep-PakR Vac 3cc; Waters Corp.) as described (19) and counted by liquid scintillation (LS 6500; Beckman Coulter). Enzymatic activity was expressed per μg of protein as calculated from Coomassie Blue staining using protein A standard as reference (see Fig. 2E). Additionally, factor VII EGF1 (20) and Glc-R (in which R is a hydrophobic aglycon) as reference were tested at a final concentration of 2 μm using 5 μm UDP-[14C]Xyl (7.5 kBq/nmol). Incubation time was increased to 2 h, and washing was performed with 0.5 m acidic acid, pH 3.4, to disassociate bead-coupled enzyme-product complexes.
FIGURE 2.
In vitro enzymatic activity. A, activity of GLT8D1, GLT8D2, GXYLT1, and GXYLT2 with acceptors mimicking the two naturally occurring xylosyltransferase substrates on EGF domains using UDP-Xyl as donor substrate. no Acc., no acceptor. B, donor substrate specificity using Glc-R as substrate. C, acceptor substrate specificity of GXYLT1 and GXYLT2 using para-nitrophenol (pNP)-linked monosaccharides as acceptor substrates. D, glucosylated EGF1 of factor VII when compared with unglucosylated EGF1 and Glc-R as acceptor, now measured at 2 μm acceptor concentration (when compared with 100 μm in A–C) to adapt to the low availability of factor VII EGF1. E, quantification of enzyme bound to 0.5 μl of IgG beads using a protein A standard and Coomassie Blue staining, of which 10 μl was used per assay in A–D.
Expression and Purification of Notch EGF1–5 and in Vitro Xylosylation
His/Myc-tagged mouse Notch EGF1–5 was expressed in two 175-cm2 flasks of pgsI-208 and control Chinese hamster ovary (CHO) cells as described previously (21). Media (80 ml) containing secreted proteins were loaded on nickel affinity columns (HisTrap HP, 1 ml; GE Healthcare), washed with 10 volumes of binding buffer (20 mm Tris-HCl, pH 8; 150 mm NaCl), and eluted with 20 mm Tris-HCl, pH 8; 150 mm NaCl; 500 mm imidazole using a 10-ml linear gradient. Protein fractions were pooled, desalted (HiPrep 26/10; GE Healthcare) with 10 mm MOPS, pH 7, and finally concentrated to 100 μl using Amicon Ultra-4 centrifugal devices (Millipore). Purification was confirmed by SDS-PAGE followed by Coomassie Blue staining and immunoblotting with monoclonal antibody 9E10 (anti-Myc). Ten μl of purified Notch EGF1–5 was incubated under standard assay conditions in the presence of 100 μm cold UDP-Xyl as donor and enzyme for 4 h, separated by SDS-PAGE, and analyzed by mass spectrometry.
In Vivo Xylosyltransferase Activity
Sf9 cells were co-infected with baculovirus encoding Notch EGF1–5 and GXYLT1, GXYLT2, or GLT8D1 for 72 h. Purification and further processing of secreted EGF1–5 was performed as described for EGF1–5 from CHO cells.
Liquid Chromatography-MS/MS
Protein bands were excised from gels and trypsinized, and peptides were recovered as described (22). Reverse-phase chromatography using acetonitrile as an eluent was performed on a Waters nanoACQUITY UPLC device equipped with an analytical column (Waters, BEH130 C18, 100 μm × 100 mm, 1.7-μm particle size) coupled online to an electrospray mass ionization Q-TOF Ultima (Waters). Spectra were recorded in positive ion mode, and peptides were automatically subjected to fragmentation (MS/MS). Alternatively, MALDI-TOF MS was carried out on a VoyagerDE Pro (Applied Biosystems). Peptide solution was mixed with matrix (α-cyano-4-hydroxy-cinnamic acid (Bruker Daltonics), 5 mg/ml in 50% acetonitrile with 0.1% trifluoroacetic acid) and then spotted on the target plate. Spectra were acquired in positive ion mode, averaging about 1000 laser shots.
RESULTS
Selection of Potential Xylosyltransferases
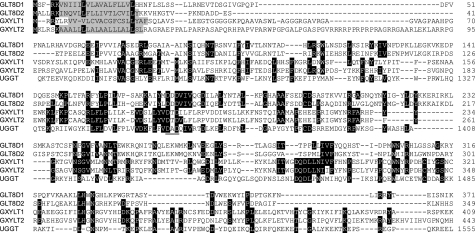
UDP-glucose:glycoprotein glucosyltransferase (UGGT) catalyzes the transfer of glucose in α1,3-linkage to N-glycans of unfolded protein. Because the generated linkage is often conserved within glycosyltransferase gene families and considering that the only difference between xylose and glucose is the absence of the C6 primary alcohol group in xylose, we hypothesized that the xylosyltransferases involved in the generation of the Xylα1,3Xylα1,3Glcβ1 trisaccharide occurring on Notch might be homologous to UGGT. The latter enzyme is much larger than typical glycosyltransferases, having a domain potentially involved in the recognition of unfolded proteins followed by a C-terminal catalytic domain. Using this catalytic domain in a position-specific iterated (PSI)-BLAST, a group of four human genes with unknown function was identified. The genes, which were at that time named GLT8D1 to GLT8D4 (glycosyltransferase 8 domain-containing), belong to glycosyltransferase gene family 8 of the Carbohydrate-Active EnZymes (CAZy) data base (23) and possess the glycosyltransferase DXD motif involved in metal binding. GLT8D1 and GLT8D2 as well as GLT8D3 and GLT8D4, which are now renamed GXYLT1 and GXYLT2 (HGNC), showed ∼50% identity with each other at the amino acid level, but between the GLT8D1–2 and GXYLT1–2 pairs and with UGGT, identity drops to about 20%. All four subfamily members show the typical type-II architecture of Golgi glycosyltransferases with the prediction of a small N-terminal cytoplasmic domain, a transmembrane domain followed by a stem region, and a C-terminal catalytic domain that aligns with UGGT (Fig. 1).
FIGURE 1.
Sequence alignment. The complete amino acid sequence of four human GLT8 family members is aligned with the N-terminal catalytic domain of UGGT starting at amino acid 1243. Sequence conservation is shown when three out of five amino acids are identical. Putative transmembrane domains are shown shaded, and the conserved DXD motif is underlined.
GXYLT1 and GXYLT2 Are α1,3-Xylosyltransferases
To assess the enzymatic activity of the four putative glycosyltransferases, constructs for expression of a secreted soluble form of the enzymes in Sf9 insect cells were generated. All enzymes were expressed with an N-terminal protein A tag and lacked the predicted cytoplasmic and transmembrane domains. The enzymes secreted in the medium at similar levels (as seen by Western blotting) were captured on IgG beads by the protein A tag and used in bound form for in vitro glycosyltransferase assays. Initial assays were carried out using the synthetic acceptors Glc-R and Xyl-Glc-R (24). These compounds mimic the natural acceptors of the two different xylosyltransferases involved in the synthesis of the Xylα1,3Xylα1,3Glcβ1 trisaccharide on EGF domains and allow separation of reaction products from unincorporated donor by reverse-phase chromatography. Assays carried out with UDP-[14C]Xyl as donor substrate revealed the xylosyltransferase activity of GXYLT1 and GXYLT2 on Glc-R but not on Xyl-Glc-R. In contrast, GLT8D1 and GLT8D2 were inactive with both acceptors (Fig. 2A). Specificity for both the donor substrate UDP-Xyl and the β-linked glucose acceptor was demonstrated by a series of assays using different donor substrates and a panel of acceptor sugars linked to para-nitrophenol (Fig. 2, B and C). In addition, the linkage formed by GXYLT1 was investigated. The product obtained from a reaction with 1 mg of Glc-R was purified on a C18 cartridge and analyzed by high performance liquid chromatography and 1H-NMR. This showed that the product was identical to the synthetic Xyl-Glc-R (24), which confirmed the generation of an α1,3-linkage (supplemental Fig. S1).
The α1,3-Xylosyltransferases Act on EGF Repeats
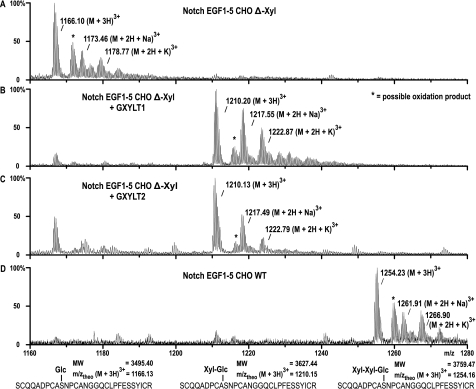
The activity of GXYLT1 and GXYLT2 on two different EGF repeat-containing substrates was subsequently tested. Using a radioactive assay similar to the one described above, we first demonstrated that recombinant factor VII EGF repeat 1, produced in Escherichia coli and in vitro glucosylated using Poglut (15, 20), was functioning as an acceptor, but not the unglucosylated EGF repeat (Fig. 2D). Similarly, activity of both enzymes on a protein containing the first five EGF repeats (EGF1–5) of mouse Notch1 (25) was established. For this purpose, the Notch acceptor structure was expressed in CHO pgsI-208 cells, which are deficient in UDP-xylose synthase. Although in wild type CHO cells the dominant glycan structure found on EGF1–5 is Xyl-Xyl-Glc-O, expression of EGF1–5 in pgsI-208 resulted in the exclusive substitution by Glc-O (21). After purification, the glucosylated EGF1–5 was incubated with the IgG bead-linked enzymes in the presence of UDP-Xyl, and the reaction products were analyzed by mass spectrometry after in-gel trypsinization (Fig. 3). EGF1–5 contains two O-Glc consensus sequences on EGF repeats 2 and 4. Fig. 3 shows the analysis of the glucosylated peptide derived from repeat 4, but repeat 2 is identically modified by the xylosyltransferases. The dominant peak at m/z 1210 demonstrated the addition of a pentose residue to the O-glucosylated peptide by GXYLT1 and GXYLT2 (Fig. 3, B and C). In contrast, only the initial acceptor visible at m/z 1166 is found in the negative control (Fig. 3A) and is not changed by incubation with GLT8D1 or GLT8D2 (data not shown).
FIGURE 3.
Xylosyltransferase activity using Notch EGF1–5 as acceptor. EGF1–5 produced in UDP-Xyl synthase-negative cells (A) was incubated with GXYLT1 or GXYLT2 (B and C), and tryptic peptides were analyzed by liquid chromatography-electrospray ionization-TOF mass spectrometry. Shown is the O-glucosylated peptide of EGF4. GXYLT1 and GXYLT2 were able to extend the O-glucosylated peptide by one xylose but not to produce the full trisaccharide that is produced in wild type CHO cells (D). To confirm that the m/z values shown represent the differentially glycosylated peptides of EGF4, sequencing was done by tandem mass spectrometry and shown in supplemental Fig. S2. MW, molecular weight.
In vivo activity was shown using a similar approach in insect cells. Sf9 cells express Poglut (20) and, based on the observed structures, the first xylosyltransferase. Upon transfection with an EGF1–5 construct, a mixture of peptides modified with Glc-O and Xyl-Glc-O in an approximate 2:1 ratio is produced (not shown but identical to Fig. 4A, where GLT8D1 is co-expressed). Co-expression of EGF1–5 and GXYLT1 or GXYLT2 resulted, however, in a considerable increase of the xylosylated peptide, now analyzed by more quantitative MALDI mass spectrometry (Fig. 4).
FIGURE 4.
In vivo xylosyltransferase activity. Shown are the MALDI-mass spectra of insect cell-produced EGF1–5 that was co-infected with GLT8D1 as a negative control (A), GXYLT1 (B), or GXYLT2 (C). The glucosylated peptide with the calculated nominal mass of 3498 Da is most dominant in A but is reduced in favor of the Xyl-Glc-peptide (3630 Da) after incubation with GXYLT1 or GXYLT2. As for Fig. 3, peptide sequencing was performed and is presented in supplemental Fig. S2.
DISCUSSION
Considering that similar glycosyltransferase reactions are often catalyzed by homologous glycosyltransferases, we hypothesized that the xylosyltransferases acting on EGF domains and UGGT might show a significant degree of homology. UGGT uses UDP-glucose as donor substrate, a nucleotide sugar very similar to UDP-xylose, and transfers the substrate in an α1,3-linkage. Moreover, it possesses a protein binding domain as expected for the EGF-specific xylosyltransferases. In the CAZy data base, the glycosyltransferase family 24 (GLT24) contains exclusively UGGTs from different species (23). In contrast, the candidate xylosyltransferases selected by their homology to UGGT belong to the huge GLT8 family. Their putative activity was difficult to predict from this affiliation because the GLT8 family contains enzymes of various origin transferring a broad variety of sugars. Other mammalian members of the GLT8 family are LARGE, a putative glycosyltransferase acting on α-dystroglycan (26), and glycogenin, a protein catalyzing its own oligo-glucosylation in α1,4-linkage to prime glycogen biosynthesis (27). Despite their limited homology to UGGT, we could, however, demonstrate that GXYLT1 and GXYLT2 are xylosyltransferases catalyzing a similar reaction as UGGT.
The activity of GXYLT1 and GXYLT2 as α1,3-xylosyltransferases was unambiguously demonstrated using both synthetic and natural acceptor substrates. Assays with synthetic acceptors first provided evidence for the xylose donor specificity and β-glucose acceptor specificity. Subsequently, activity on EGF repeats was explicitly established by taking advantage of the availability of the UDP-xylose synthase-deficient CHO cell line pgsI-208 (21). This cell line is devoid of UDP-Xyl and allowed the production of EGF repeats exclusively modified with unsubstituted O-Glc and thus representing the natural substrate for the identified xylosyltransferases. The elongation of this substrate by GXYLT1 and GXYLT2 was then displayed by mass spectrometry. Finally, the enzymes were shown to act in vivo in Sf9 insect cells.
In evolution, GXYLT1 and GXYLT2 seem to have split only in the vertebrates. Orthologs of both xylosyltransferases are found in zebrafish, but the cephalochordate Branchiostoma floridae and other sequenced eumetazoans until the sea anemone Nematostella vectensis (28) only present a single gene. Caenorhabditis elegans seems to have lost the gene, and no ortholog is found in the unicellular choanoflagellate Monosiga brevicollis, which is the closest living relative of metazoans. Emerging of the xylosyltransferase therefore appears to match that of Notch signaling (29).
Amino acid sequence differences between GXYLT1 and GXYLT2 are most apparent in the putative stem regions where GXYLT2 exhibits an arginine-rich and a proline-rich insertion. Although GXYLT1 appears more active than GXYLT2 in the assays performed in this study, no differences in specificity could be observed. As only three different EGF domains have been utilized, it cannot be excluded that there is preference for specific EGF domains. However, such differences have not been revealed for the three different mammalian Fringe GlcNAc transferases acting on O-fucose (10). The enzymatic activity of the two xylosyltransferases could thus as well be redundant and the enzymes only differentially expressed.
Extension of O-Fuc glycans by the enzyme Fringe has been shown to play a key role in modulation of Notch signaling, but the role of xylosylation of O-Glc could until now only be speculated about. Identification of two human genes encoding xylosyltransferases of the EGF O-glucosylation pathway will now enable functional studies that resolve the role of O-Glc xylosylation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM061126 (to R. S. H.). This work was also supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) (to R. G.-S.) for the Junior Research Group “Stem Cell Glycans” established under the roof of REBIRTH, a DFG cluster of Excellence at Hannover Medical School.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Table S1, and Figs. S1 and S2.
- EGF
- epidermal growth factor
- GLT8
- glycosyltransferase 8 family
- GXYLT
- glucosyl xylosyltransferase
- Poglut
- protein O-glucosyltransferase
- UGGT
- UDP-glucose:glycoprotein glucosyltransferase
- CHO
- Chinese hamster ovary
- MS
- mass spectrometry
- MS/MS
- tandem MS
- MALDI
- matrix-assisted laser desorption/ionization
- TOF
- time of flight.
REFERENCES
- 1.Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., Haltiwanger R. S. (2000) J. Biol. Chem. 275, 9604–9611 [DOI] [PubMed] [Google Scholar]
- 2.Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., Okajima T. (2008) J. Biol. Chem. 283, 35486–35495 [DOI] [PubMed] [Google Scholar]
- 3.Luther K. B., Haltiwanger R. S. (2009) Int. J. Biochem. Cell Biol. 41, 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamura Y., Saga Y. (2008) Mech. Dev. 125, 663–673 [DOI] [PubMed] [Google Scholar]
- 5.Shi S., Stanley P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stahl M., Uemura K., Ge C., Shi S., Tashima Y., Stanley P. (2008) J. Biol. Chem. 283, 13638–13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okajima T., Xu A., Lei L., Irvine K. D. (2005) Science 307, 1599–1603 [DOI] [PubMed] [Google Scholar]
- 8.Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., Vogt T. F. (2000) Nature 406, 369–375 [DOI] [PubMed] [Google Scholar]
- 9.Brückner K., Perez L., Clausen H., Cohen S. (2000) Nature 406, 411–415 [DOI] [PubMed] [Google Scholar]
- 10.Rampal R., Li A. S., Moloney D. J., Georgiou S. A., Luther K. B., Nita-Lazar A., Haltiwanger R. S. (2005) J. Biol. Chem. 280, 42454–42463 [DOI] [PubMed] [Google Scholar]
- 11.Xu A., Haines N., Dlugosz M., Rana N. A., Takeuchi H., Haltiwanger R. S., Irvine K. D. (2007) J. Biol. Chem. 282, 35153–35162 [DOI] [PubMed] [Google Scholar]
- 12.Hicks C., Johnston S. H., diSibio G., Collazo A., Vogt T. F., Weinmaster G. (2000) Nat. Cell Biol. 2, 515–520 [DOI] [PubMed] [Google Scholar]
- 13.Hase S., Nishimura H., Kawabata S., Iwanaga S., Ikenaka T. (1990) J. Biol. Chem. 265, 1858–1861 [PubMed] [Google Scholar]
- 14.Bjoern S., Foster D. C., Thim L., Wiberg F. C., Christensen M., Komiyama Y., Pedersen A. H., Kisiel W. (1991) J. Biol. Chem. 266, 11051–11057 [PubMed] [Google Scholar]
- 15.Acar M., Jafar-Nejad H., Takeuchi H., Rajan A., Ibrani D., Rana N. A., Pan H., Haltiwanger R. S., Bellen H. J. (2008) Cell 132, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishimizu T., Sano K., Uchida T., Teshima H., Omichi K., Hojo H., Nakahara Y., Hase S. (2007) J. Biochem. 141, 593–600 [DOI] [PubMed] [Google Scholar]
- 17.Omichi K., Aoki K., Minamida S., Hase S. (1997) Eur. J. Biochem. 245, 143–146 [DOI] [PubMed] [Google Scholar]
- 18.Minamida S., Aoki K., Natsuka S., Omichi K., Fukase K., Kusumoto S., Hase S. (1996) J. Biochem. 120, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 19.Palcic M. M., Heerze L. D., Pierce M., Hindsgaul O. (1988) Glycoconj. J. 5, 49–63 [Google Scholar]
- 20.Shao L., Luo Y., Moloney D. J., Haltiwanger R. (2002) Glycobiology 12, 763–770 [DOI] [PubMed] [Google Scholar]
- 21.Bakker H., Oka T., Ashikov A., Yadav A., Berger M., Rana N. A., Bai X., Jigami Y., Haltiwanger R. S., Esko J. D., Gerardy-Schahn R. (2009) J. Biol. Chem. 284, 2576–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 23.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krylov V., Ustyuzhanina N., Grachev A., Bakker H., Nifantiev N. (2007) Synthesis 2007, 3147–3154 [Google Scholar]
- 25.Shao L., Moloney D. J., Haltiwanger R. (2003) J. Biol. Chem. 278, 7775–7782 [DOI] [PubMed] [Google Scholar]
- 26.Kanagawa M., Saito F., Kunz S., Yoshida-Moriguchi T., Barresi R., Kobayashi Y. M., Muschler J., Dumanski J. P., Michele D. E., Oldstone M. B., Campbell K. P. (2004) Cell 117, 953–964 [DOI] [PubMed] [Google Scholar]
- 27.Pitcher J., Smythe C., Cohen P. (1988) Eur. J. Biochem. 176, 391–395 [DOI] [PubMed] [Google Scholar]
- 28.Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., Terry A., Shapiro H., Lindquist E., Kapitonov V. V., Jurka J., Genikhovich G., Grigoriev I. V., Lucas S. M., Steele R. E., Finnerty J. R., Technau U., Martindale M. Q., Rokhsar D. S. (2007) Science 317, 86–94 [DOI] [PubMed] [Google Scholar]
- 29.King N., Westbrook M. J., Young S. L., Kuo A., Abedin M., Chapman J., Fairclough S., Hellsten U., Isogai Y., Letunic I., Marr M., Pincus D., Putnam N., Rokas A., Wright K. J., Zuzow R., Dirks W., Good M., Goodstein D., Lemons D., Li W., Lyons J. B., Morris A., Nichols S., Richter D. J., Salamov A., Sequencing J. G., Bork P., Lim W. A., Manning G., Miller W. T., McGinnis W., Shapiro H., Tjian R., Grigoriev I. V., Rokhsar D. (2008) Nature 451, 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.