Abstract
Post-translational modification of nucleocytoplasmic proteins by O-linked β-N-acetylglucosamine (O-GlcNAc) has for the last 25 years emerged as an essential glucose-sensing mechanism. The liver X receptors (LXRs) function as nutritional sensors for cholesterol-regulating lipid metabolism, glucose homeostasis, and inflammation. LXRs are shown to be post-translationally modified by phosphorylation, acetylation, and sumoylation, affecting their target gene specificity, stability, and transactivating and transrepressional activity, respectively. In the present study, we show for the first time that LXRα and LXRβ are targets for glucose-hexosamine-derived O-GlcNAc modification in human Huh7 cells. Furthermore, we observed increased hepatic LXRα O-GlcNAcylation in vivo in refed mice and in streptozotocin-induced refed diabetic mice. Importantly, induction of LXRα O-GlcNAcylation in both mouse models was concomitant with increased expression of the lipogenic gene SREBP-1c (sterol regulatory element-binding protein 1c). Furthermore, glucose increased LXR/retinoic acid receptor-dependent activation of luciferase reporter activity driven by the mouse SREBP-1c promoter via the hexosamine biosynthetic pathway in Huh7 cells. Altogether, our results suggest that O-GlcNAcylation of LXR is a novel mechanism by which LXR acts as a glucose sensor affecting LXR-dependent gene expression, substantiating the crucial role of LXR as a nutritional sensor in lipid and glucose metabolism.
Keywords: Diseases/Metabolic, Glycosylation, Metabolism/Glucose, Metabolism/Lipogenesis, Phosphorylation/Transcription factors, Protein/Post-translational Modification, Receptors/Nuclear, Signal Transduction
Introduction
Liver X receptor (LXR)4 α (NR1H3) and LXRβ (NR1H2) are ligand-activated transcription factors belonging to the nuclear receptor family and work as heterodimers with the retinoic X receptor (RXR). LXRα is expressed primarily in liver, macrophages, adipose tissue, and the interstitial epithelium, whereas LXRβ is ubiquitously expressed (1, 2). Both isoforms act as sterol sensors binding endogenous oxysterol ligands (3–5). LXRs are known to play a crucial role in lipid and glucose metabolism, partly through their regulation of sterol regulatory element-binding protein 1c (SREBP-1c), a key regulator of lipogenesis (6, 7). Furthermore, LXRs are reported to be involved in both insulin-mediated activation and lipid-mediated repression of SREBP-1c transcription (8–10), supporting a role of LXRs as nutritional sensors. In addition to the role of LXRs in fatty acid homeostasis, these nuclear receptors are also known to control gene expression linked to cholesterol homeostasis in response to oxidized cholesterol (7). Because LXRs play an important role in both cholesterol efflux from macrophages and act as a modulator of immune responses (11–13), activation of LXR signaling may have beneficial effects on atherosclerosis. However, LXR activation also has proatherogenic capacity by activating SREBP-1c and fatty acid synthase gene expression (6, 14). Whether a net outcome of LXR activation is pro- or antiatherogenic may depend on conditions regulating post-translational modifications on LXRs.
LXRs are shown to be post-translationally modified by phosphorylation (15–17), acetylation (18), and sumoylation (19), affecting their target gene specificity, stability, and transactivating and transrepressional activity, respectively. Recently, Mitro et al. (20) showed that physiological concentrations of glucose were able to activate LXR and induce expression of genes involved in both lipid and cholesterol homeostasis in liver. Moreover, they made the surprising discovery that glucose acts as an endogenous LXR ligand in human HepG2 cells. These findings are somewhat controversial because LXRs are known to be ligand-activated via a hydrophobic pocket in the C-terminal domain (21). This argues against a role for the highly hydrophilic glucose molecule acting as a ligand for LXRs and suggests that glucose exerts its effect on LXR via activation of downstream glucose signaling, potentially the hexosamine biosynthetic pathway involving O-linked glycosylation. Cytoplasmic and nuclear proteins can be dynamically modified by O-linked β-N-acetylglucosamine (O-GlcNAc) by the enzyme O-GlcNAc transferase using UDP-GlcNAc generated by the hexosamine biosynthetic pathway as substrate (22, 23). O-GlcNAc is attached to specific serine and threonine residues on nuclear and cytoplasmic proteins analogous to phosphorylation (22). 2–5% of glucose entering the cell is diverted into the hexosamine biosynthetic pathway. Because O-GlcNAc levels on proteins appear to be sensitive to flux through this pathway, O-GlcNAc transferase can be considered as a general sensor of glucose availability that modifies proteins according to changes in levels of UDP-GlcNAc. O-GlcNAcylation has been shown to be an important regulatory mechanism for a number of nutrient- and stress-responsive transcription factors, including p53, NF-κB, and FOXO1 (24–27).
Here, we report for the first time that LXRs are O-GlcNAcylated in human Huh7 liver cells and that in vivo O-GlcNAcylation of hepatic LXRα is induced in refed and streptozotocin (STZ)-induced diabetic mice concomitant with increased SREBP-1c expression. Moreover, we show that glucose increased LXR/RXR-dependent activation of the mouse SREBP-1c promoter in Huh7 cells via the hexosamine biosynthetic pathway. Our findings provide a novel mechanism for direct glucose response of LXRs, crucial for proper regulation of lipid and glucose metabolism.
EXPERIMENTAL PROCEDURES
Reagents
d-(+)-Glucose solution (G8769; Sigma), d-(+)-glucosamine hydrochloride (G4875; Sigma), d-(+)-galactose (G0750; Sigma), 6-diazo-5-oxo-l-norleucine (DON, D2141; Sigma), O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate (PUGNAc, A157250; Toronto Chemical), succinylated wheat germ agglutinin (sWGA)-agarose (AL-1023S; Vector Laboratories), N-acetyl-d-glucosamine (GlcNAc, 01140; Sigma-Aldrich), GlcNAc-thiazoline (provided by Dr. Spencer Knapp, Rutgers, NJ).
Plasmids
Construction of pFLAG-hLXRα (wild type (WT) (amino acids 1–447), amino acids 166–447, 259–447) and pFLAG-hLXRβ (WT (amino acids 1–461), amino acids 1–157, 157–461, and 273–461) was performed using PCR with human LXR cDNA as a template. The PCR fragments were inserted into pFLAG (modified from pcDNA3; Invitrogen) (28) using Xho1 (LXRα) and EcoRI (LXRβ) sites. Sequences were verified by direct DNA sequencing. Other plasmids were: pSG5 (Stratagene), pRL (Promega), and pSG5-hRXRα, pcDNA3-hLXRα, and pcDNA3-hLXRβ (provided by Krister Bamberg, AstraZeneca, Sweden). The following mouse SREBP-1c promoter constructs were provided by Dr. Nobuhiro Yamada, University of Tokyo, Japan: pGL2basic/−550mSREBP1cprom-Luc (pBP1c550-Luc), pGL2prom/mLXREab-Luc (pLXRE-Luc), pGL2prom/mLXREaM-Luc, pGL2prom/mLXREbM-Luc, and pGL2prom/mLXREabM-Luc (29). The LXR response elements LXREa and LXREb in the mouse SREBP-1c promoter are highly similar to LXRE1 and LXRE2 in the promoter of human SREBP-1c (30).
Cell Cultures and Transfections
The Huh7 liver hepatoma cell line was maintained in Dulbecco's modified Eagle's medium (DMEM) (D6546; Sigma) supplemented with 10% fetal bovine serum (F7524; Sigma), 4 mm l-glutamine (G7513; Sigma), and 1% penicillin-streptomycin (P4458; Sigma). The FLAG-hLXRα-expressing Flp-InTM293 cell line was generated using the Flp-In system (Invitrogen) in accordance with the manufacturer's instructions. The cells were maintained in DMEM containing 5 mm d-glucose (D6046; Sigma) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. All cells were kept at 37 °C in a humidified atmosphere containing 5% CO2. Huh7 cells were transfected using Lipofectamine 2000 (Invitrogen) as instructed by the manufacturer with the following details. Cells were seeded in DMEM containing 5 mm d-glucose 1 day before transfection. Transfection was performed in serum-free DMEM containing 1 mm d-glucose. 5 h after transfection, media were changed to serum-free DMEM with different glucose concentrations together with 0.2–5 mm glucosamine, 5 μm DON, 50 μm GlcNAc-thiazoline, or 50–100 μm PUGNAc for 24 h as specified in each experiment.
Preparation of Protein Extract
Cells were scraped in phosphate-buffered saline and centrifuged for 3500 rpm for 3 min, and pellets were resuspended in radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris-Cl, pH 8, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 2 mm EDTA, 2.5 mm sodium pyrophosphate, 1 mm NaF, 1 mm Na3VO4, 1 mm β-glycerophosphate, 1 μm O-GlcNAcase inhibitor GlcNAc-thiazoline, and CompleteTM protease inhibitors (Roche Applied Science). For preparation of total protein extract from mice liver, ∼25 mg of liver tissue was homogenized in 500 μl of lysis buffer (phosphate-buffered saline containing 0.1% Nonidet P-40, 0.1% SDS and CompleteTM protease inhibitor). Tissue and cold lysis buffer were combined in tubes containing ceramic beads (Recellys CK28; Bertin Technologies) and homogenized in Precellys®24 homogenizer (Bertin Technologies) 2 × 15 s at 5000 rpm.
Immunoprecipitation and sWGA Pulldown
Protein extracts (300–500 μg) were incubated with protein A/G-agarose beads (sc-2003; Santa Cruz Biotechnology) for 1 h at 4 °C to minimize unspecific binding. Extracts were transferred to clean tubes and incubated further with 1 μg of LXR antibody, 2 μg of rabbit FLAG antibody, or IgG control (mouse gamma globulin, 015-000-002; Jackson ImmunoResearch Laboratories) together with protein G-agarose beads (sc-2002; Santa Cruz Biotechnology) overnight at 4 °C. For anti-mouse FLAG immunoprecipitation, EZview Red anti-FLAG M2 affinity gel (F2426; Sigma) was used according to the manufacturer's manual. Succinylated WGA pulldown was performed as immunoprecipitation except 40 μl of sWGA-agarose was added after preincubation with protein A/G-agarose beads.
Western Blotting
O-GlcNAc levels were determined by SDS-PAGE and blotting with mouse monoclonal anti-O- GlcNAc antibodies RL2 (MA1-072, 1:1000; Affinity BioReagents), CTD110.6 (MMS-248R, 1:1000; Covance), or the GlcNAc-binding lectin sWGA conjugated to horseradish peroxidase (HRP), sWGA-HRP (H-2102-1, 1:10,000; EY Laboratories). Ectopically expressed FLAG-tagged LXRs were detected with mouse monoclonal anti-human LXR antibodies (PP-K8607-00 (LXRα) from R&D Systems or PP-K8917-00 (LXRβ), 1:500) or rabbit polyclonal anti-FLAG antibody (F7425, 1:5000; Sigma,), whereas a rabbit polyclonal anti-mouse LXR antibody (1:1000, provided by Knut Steffensen, Karolinska Institutet, Sweden) (31) was used for detecting LXRα in mice liver. Secondary light chain-specific antibodies from Jackson ImmunoResearch Laboratories were used at 1:10,000 dilutions (anti-mouse, 115-035-174; anti-rabbit, 211-032-171). Anti-mouse IgM (A8786; Sigma) was used at a 1:5000 dilution.
In Vitro Translation and GlcNAcylation of LXRα and LXRβ in Rabbit Reticulocyte Lysate
35S-Labeled LXRα and LXRβ proteins were in vitro translated from pcDNA3-hLXRα and pcDNA3-hLXRβ or different pFLAG-hLXRα/pFLAG-hLXRβ deletion constructs using the Promega TNT T7 Quick Coupled Transcription-Translation system (Promega, Madison, WI) and [35S]methionine (PerkinElmer Life Sciences), as instructed by the manufacturer. 40 μl of the reaction mixtures were diluted in 200 μl of 0.5 m HEPES, pH 7.5, containing 0.1 m NaCl and incubated with 50 μl of sWGA-agarose (equilibrated in HEPES-NaCl buffer) overnight at 4 °C. After extensive washing in HEPES-NaCl buffer containing 0.2% Nonidet P-40, GlcNAcylated LXRs were batch-eluted three times in 200 μl of 0.5 m GlcNAc following elution in 400 μl of 0.5 m galactose. GlcNAc-eluted proteins were subjected to SDS-PAGE together with an aliquot of the TNT reaction mixture. The gel was analyzed by fluorography using En3Hance reagent (PerkinElmer Life Sciences).
Promoter Activity Assays
Huh7 cells were seeded at a density of 8 × 104 cells/well in 24-well plates. The next day, cells were co-transfected with 400 ng of luciferase reporter construct, 20 ng of Renilla luciferase control plasmid, pRL, 100 ng of pSG5-hRXRα with 100 ng of pcDNA3-hLXRα, pcDNA3-hLXRβ, or empty vector pSG5 as control. After 24 h, the cells were harvested in 100 μl of reporter lysis buffer (Promega). Luciferase activities were measured using the Dual- Luciferase reporter assay system (Promega) in 96-well plates on a Synergy 2 Multi-Probe Microplate Reader (BioTek Instruments) according to the manufacturer's protocol.
Animals
All use of animals was approved and registered by the Norwegian Animal Research authority and the regional ethical committee for animal experiments in Sweden. The mice (mixed genetic background based on 129/Sv and C57BL/6J strains, backcrossed in C57BL/6J for at least six generations) were housed in a temperature-controlled (22 °C) facility with a strict 12-h light/dark cycle with free access to water during experiments. The mice were killed by cervical dislocation; tissues were excised, rapidly frozen in liquid nitrogen, and stored at −80 °C until further analysis. They were fed an R36 Lactamin diet containing 55.7% carbohydrates, 18.5% protein, and 4% fat (Lactamin AB; Stockholm, Sweden). Mice used in the STZ experiment were of the C57BL/6J strain and fed a diet containing 64% carbohydrates, 31.5% protein, and 4.5% fat (SDS RM no.1 maintenance, Special Diets Services, UK).
Fasting-Refeeding Experiments
Male mice were fasted and refed for 24 h and 12 h, respectively. All mice were killed at the end of the dark period.
STZ Treatment
Male mice were pretreated with two intraperitoneal injections of STZ (100 mg/kg) (S0130; Sigma) with 1 day between injections. STZ was freshly made in sodium citrate buffer (50 mmol/liter, pH 4.5) immediately before injections. Seven days after the first STZ injection, mice were included in fasting-refeeding experiment as described above.
Quantitative RT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen). cDNA was synthesized using a High Capacity cDNA Archive Kit (Applied Biosystems). Analysis of mRNA expression was done by quantitative reverse transcription-PCR on 7900HT (Applied Biosystems), and subsequent data analysis was done using SDS 2.3 software. Gene expression was normalized against the expression of the ribosomal protein 36B4. Primer sequences are available upon request.
Statistical Analysis
Data are expressed as mean ± S.D. Statistical significance between groups was assessed by a two-tailed t test, equal variances not assumed.
RESULTS
LXRα and LXRβ Are Modified by O-GlcNAc in Huh7 Cells
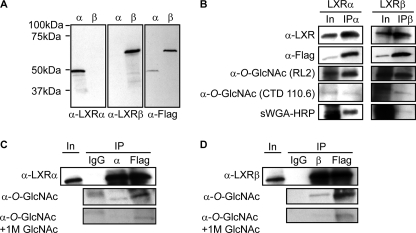
To confirm specificity of the LXR antibodies used in this study, FLAG-tagged human LXRα and LXRβ were transiently transfected into Huh7 liver cells followed by SDS-PAGE and immunoblotting with monoclonal anti-human LXRα, anti-human LXRβ, and anti-FLAG antibodies (Fig. 1A). Additionally, FLAG-tagged mouse LXRα was similarly tested using polyclonal anti-LXR antibody and anti-FLAG antibodies in immunoblotting experiments (data not shown). In agreement with a previous report (15), LXRα migrated as one major and one minor band around 50 kDa, whereas LXRβ migrated as a single band of ∼60 kDa. Because of recent reports debating how, and if, LXRs are glucose-responsive nuclear receptors in liver (20, 32–34) and the fact that O-GlcNAc functions as a nutrient sensor, we investigated whether LXRα and LXRβ are modified by O-GlcNAc in Huh7 cells cultured under high glucose conditions. Cells transfected with FLAG-tagged LXRα or LXRβ were immunoprecipitated with antibodies against LXRα (Fig. 1B, left panel) and LXRβ (Fig. 1B, right panel), and immunoblots were incubated with two different anti-O-GlcNAc antibodies (RL2 and CTD110.6) and the terminal GlcNAc-specific lectin, sWGA, which binds to N- and O-GlcNAcylated proteins. Enrichment of GlcNAc-containing glycoproteins using this lectin dramatically enrich for CTD110.6- and RL2-reactive proteins (35) (see Fig. 3A, lower panel). All three detection methods showed O-GlcNAcylation of both LXRα and LXRβ determined by the observation of RL2-, CTD110.6-, and sWGA-reactive bands of ∼50 kDa (LXRα) and 60 kDa (LXRβ). Specificity of LXR O-GlcNAcylation was confirmed by similar RL2 antibody reactivity in anti-FLAG immunoprecipitates, but not in anti-IgG immunoprecipitates and by reduced reactivity in the presence of free GlcNAc (Fig. 1, C and D).
FIGURE 1.
LXRα and LXRβ are modified by O-GlcNAc in Huh7 cells. A, Huh7 cells transfected with FLAG-hLXRα (α) or FLAG-hLXRβ (β) (25 mm glucose, 2.5% serum) were subjected to SDS-PAGE and blotted using anti-LXRα, anti-LXRβ, or anti-FLAG antibodies. B, FLAG-hLXRα and FLAG-hLXRβ were immunoprecipitated (IP) from Huh7 cells (25 mm glucose, 10% serum) using anti-LXRα or anti-LXRβ antibodies. Input (In, 8%) and immunoprecipitated proteins were subjected to SDS-PAGE and blotted with anti-O-GlcNAc antibodies CTD110.6 or RL2, or sWGA-HRP to verify O-GlcNAc modification. Input shows total amount of O-GlcNAc-modified proteins. Anti-LXRα (left panel), anti-LXRβ (right panel), and anti-FLAG antibodies were used to detect LXRs. C and D, FLAG-hLXRα (C) or FLAG-hLXRβ (D) was immunoprecipitated from Huh7 cells (25 mm glucose, 10% serum) using anti-LXRα or anti-LXRβ antibodies or anti-FLAG M2 affinity gel. Mouse IgG immunoprecipitation was performed as control. Input (8%) and immunoprecipitated proteins were subjected to SDS-PAGE and blotted with anti-LXRα, anti-LXRβ, or anti-O-GlcNAc (RL2) antibodies. Specificity was confirmed by GlcNAc competition (bottom panels).
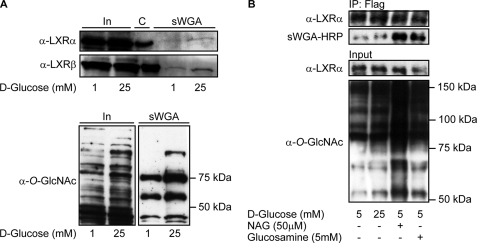
FIGURE 3.
LXR GlcNAcylation is elevated by glucose in Huh7 cells and in stably transfected FLAG-hLXRα FlpInTM293 cells. A, GlcNAc-modified proteins from FLAG-hLXRα or FLAG-hLXRβ-overexpressing Huh7 cells (1 mm and 25 mm glucose) were absorbed on sWGA-agarose beads (total amount of proteins loaded onto sWGA beads was the same in all lanes). Input (In, 10%) and sWGA-precipitated proteins (sWGA) were subjected to SDS-PAGE and blotted using anti-LXRα or anti-LXRβ antibodies. Immunoprecipitated LXRs are loaded as size control (C) (upper panel). O-GlcNAc-modified proteins from input (10%, left panel) and sWGA precipitation (right panel) were detected using anti-O-GlcNAc (CTD110.6) antibody (lower panel). B, FLAG-hLXRα was immunoprecipitated (IP) from FLAG-hLXRα stably transfected FlpInTM293 cells (5 mm glucose, 25 mm glucose, 5 mm glucose + GlcNAc (NAG), 5 mm glucose + glucosamine) using rabbit anti-FLAG antibody. Input (2%) and immunoprecipitated proteins were subjected to SDS-PAGE and blotted with anti-LXRα antibody and sWGA-HRP or anti-O-GlcNAc (CTD110.6) antibody to verify GlcNAc modification.
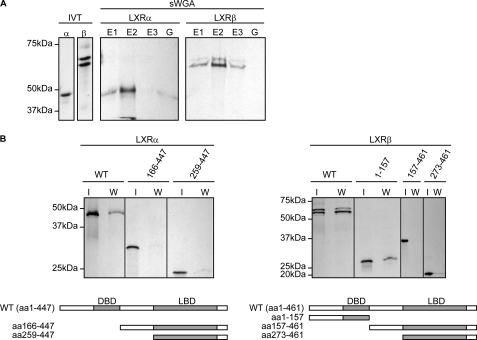
In Vitro GlcNAcylation of LXRα and LXRβ
In vitro translation was used to generate 35S-labeled LXRα and LXRβ in rabbit reticulocyte lysate to test for their ability to bind sWGA-agarose because proteins expressed in rabbit reticulocyte lysate are modified by O-GlcNAc (36). When the translation mixtures were added to sWGA-agarose beads, 35S-labeled LXRα and LXRβ were retained on the lectin beads and eluted with 0.5 m GlcNAc (Fig. 2A). We next performed in vitro translation on deletion constructs of LXRα and LXRβ to elucidate which domain(s) contain O- GlcNAc site(s). Fig. 2B shows that sWGA binding of 35S-labeled LXRα and LXRβ depends on the N-terminal region containing the activation function-1 and DNA-binding domains, suggesting that this region contains putative O-GlcNAc binding site(s).
FIGURE 2.
In vitro GlcNAcylation of LXRα and LXRβ. A, in vitro translated (IVT) 35S-labeled hLXRα or 35S-labeled hLXRβ proteins in rabbit reticulocyte lysate were absorbed on sWGA-agarose beads. Beads were eluted once with 0.5 m galactose (G) followed by three times elution with 0.5 m GlcNAc (E1, E2, E3). In vitro translated lysates (8%) and sWGA eluates (sWGA; E1, E2, E3, G) were subjected to SDS-PAGE and analyzed by fluorescence autoradiography. B, in vitro translated 35S-FLAG-hLXRα or 35S-FLAG-hLXRβ full-length and truncated proteins were absorbed on sWGA-agarose beads. In vitro translated lysates (I, 8%) and sWGA eluates (W) were subjected to SDS-PAGE and analyzed by fluorescence autoradiography. Schematic figures of the LXR proteins are shown. DBD, DNA-binding domain; LBD, ligand-binding domain.
LXR GlcNAcylation Is Elevated by Glucose
To determine the effect of glucose on O-GlcNAcylation of LXRα and LXRβ, LXR-transfected Huh7 cells were grown in serum-free medium containing 1 mm glucose or 25 mm glucose for 24 h. GlcNAcylated proteins were absorbed on sWGA-agarose beads and analyzed by immunoblotting using anti-LXR antibodies (Fig. 3A, upper panel). We observed increased association of LXRα and LXRβ with the beads in high glucose-treated cells and in cells treated with 5 mm glucose (data not shown). This was not seen in cells transfected with empty vector (data not shown). LXRα appear to have weaker affinity for sWGA than LXRβ judged by weaker LXRα immunoreactive bands observed in sWGA eluates and longer exposure time needed for sWGA-retained 35S-labeled LXRα in the in vitro GlcNAcylation assay (Fig. 2). This can be explained by fewer O- GlcNAcylation sites on LXRα and/or the position of the site(s) because WGA has high affinity for oligomeric and terminal GlcNAc (37). High glucose did not affect the expression level of transfected LXR proteins, suggestive of a glucose-dependent regulation of LXR O-GlcNAcylation in these cells. As expected, we observed increased recovery of total O-GlcNAcylated proteins on sWGA beads in cells treated with high glucose (Fig. 3A, lower panel). To verify that glucose via the hexosamine pathway directly modifies O-GlcNAc levels on LXR, we performed Western blotting using HRP-conjugated sWGA on immunoprecipitated FLAG-LXRα stably expressed in FlpInTM293 cells (Fig. 3B, upper panel). Because of single-copy integration via FlpIn recombination, these cells are a good model system for targeted integration of expression vectors ensuing high levels of expression. In these cells, we observed increased O-GlcNAc modification of LXRα in high glucose-, GlcNAc-thiazoline-, and glucosamine-treated cells concomitant with an overall increase in whole cell protein O-GlcNAcylation (Fig. 3B, lower panel). Similarly, we observed increased O-GlcNAc antibody reactivity in Western blots of endogenously expressed LXRβ immunoprecipitated from THP-1 macrophages treated with increasing glucose concentrations (data not shown).
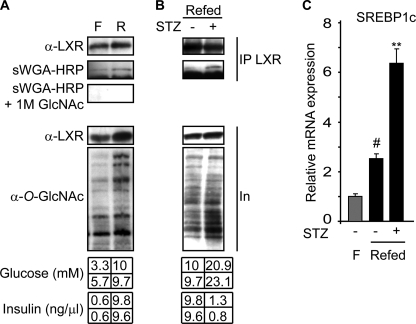
In Vivo Hepatic LXR O-GlcNAcylation Is Induced by High Glucose and STZ Treatment
We next analyzed in vivo GlcNAcylation of LXR in livers isolated from fasted and refed mice. The mean plasma glucose and insulin concentrations in fasted mice were 4.5 mm and 0.6 ng/μl, respectively, compared with 9.9 mm and 9.7 ng/μl after refeeding. Using HRP-conjugated sWGA in Western blotting experiments, we found that liver LXRα was more O-GlcNAcylated in refed compared with fasted liver. Specificity was confirmed by inhibited sWGA-HRP reactivity in the presence of free GlcNAc (Fig. 4A). Furthermore, the effect of hyperglycemia independent of insulin was studied in diabetic mice (refed + STZ) with mean blood glucose and insulin levels at 22 mm and 1.05 ng/μl, respectively. Interestingly, we observed higher relative levels of O-GlcNAcylated LXR in refed STZ-treated mice compared with refed control mice (Fig. 4B).
FIGURE 4.
In vivo hepatic LXR O-GlcNAcylation is induced by refeeding and STZ treatment. A, mice were fasted for 24 h (F) or fasted for 24 h then refed for 12 h (R) (n = 2/lane). B, mice were treated with STZ for 1 week before they were fasted for 24 h then refed for 12 h (STZ-Refed). Controls (Refed) were not treated with STZ (n = 2/lane). A and B, LXRα was immunoprecipitated (IP) using anti-LXR antibody. Input (In, 12%) and immunoprecipitated proteins were subjected to SDS-PAGE and blotted with anti-LXR or sWGA-HRP. Specificity was confirmed by GlcNAc competition (A). Plasma glucose and insulin levels for each animal are shown in the bottom panel. C, hepatic gene expression of SREBP-1c was analyzed by quantitative reverse transcription-PCR and normalized against expression of the ribosomal protein 36B4 (n = 4/group for fasted (gray bar), n = 4–6/group for refed (black bars)). Values are given as mean ± S.D. (error bars), and the expression in the fasted controls is set as 1. **, p < 0.01 STZ versus control; #, p < 0.01 fasted versus refed.
To elucidate the physiological role of increased O-GlcNAc modification on LXRα in the regulation of hepatic lipogenesis, we isolated RNA from all three mouse models discussed above and studied hepatic mRNA expression of LXR-responsive SREBP-1c. Fasting to refeeding resulted in a 2.5-fold increase in SREBP-1c mRNA expression, and a further 2.5-fold increase was seen in diabetic mice (Fig. 4C). Moreover, and in agreement with a previous report (38), we observed increased SREBP-1c mRNA expression in STZ-treated fasted mice (data not shown). Because the induction of SREBP-1c mRNA expression in all mouse models is blunted in LXR α/β double knock-out mice (32),5 our in vivo data argue for a role of O-GlcNAcylated LXR in regulating SREBP-1c mRNA expression.
O-GlcNAc Regulates LXR Transactivation of the SREBP-1c Promoter
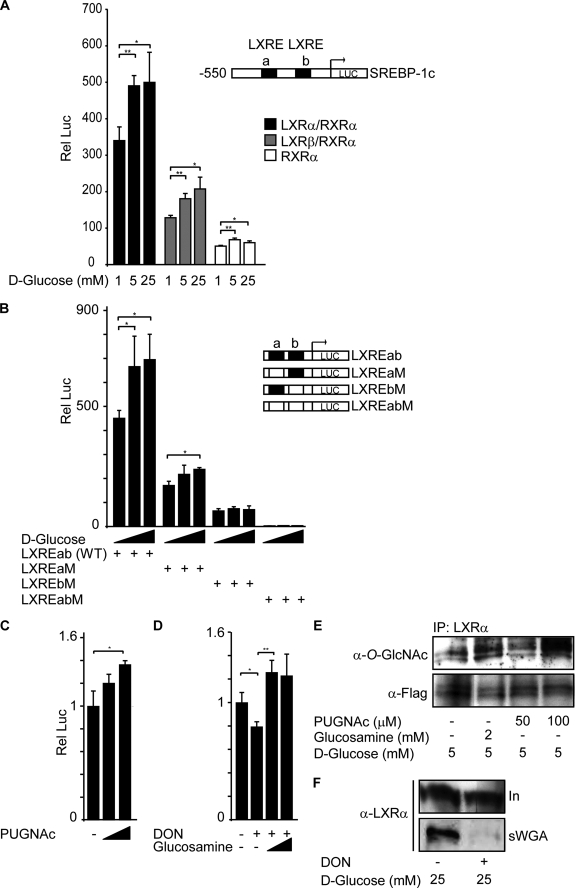
To examine a role of O-GlcNAc on LXR activity, we asked whether glucose could regulate the SREBP-1c promoter through LXRs. Huh7 cells were co-transfected with RXRα and LXRα or LXRβ expression vectors together with a luciferase reporter construct containing 550 bp of the proximal 5′-flanking region of the mouse SREBP-1c promoter (pBP1c550-Luc) (Fig. 5A). In control cells transfected with RXRα and the empty vector pSG5, there were only small effects on luciferase activity probably because of endogenous LXR in Huh7 cells (white bars). Co-transfection with LXRα/RXRα (black bars) resulted in ∼7-fold higher basal luciferase activities (1 mm glucose), and LXRβ/RXRα (gray bars) resulted in 2.5-fold higher luciferase activities compared with RXRα alone, suggesting that LXRα is the major transactivator of SREBP-1c transcription in Huh7 cells. However, glucose-induced luciferase activities increased in a similar manner in LXRα- and LXRβ-transfected cells (40–60%; similar between 5 and 25 mm glucose).
FIGURE 5.
O-GlcNAc regulates LXR transactivation of the SREBP-1c promoter. A, Huh7 cells were transfected with pBP1c550-Luc reporter, Renilla luciferase control plasmid (pRL), and RXRα expression vector together with LXRα (black bars), LXRβ (gray bars), or empty vector pSG5 (white bars). 5 h after transfection, cells were stimulated with different glucose concentrations (1, 5, or 25 mm) for 24 h. A schematic figure of the reporter construct including LXRE elements a and b is shown. B, Huh7 cells were transfected with different pBP1cLXRE-Luc reporter constructs (LXREab, LXREaM, LXREbM, or LXREabM), pRL LXRα, and RXRα expression vectors. 5 h after transfection, cells were stimulated with different glucose concentrations (1, 5, or 25 mm) for 24 h. A schematic figure of the LXRE reporter constructs is shown. C, Huh7 cells were transfected with pBP1cLXREab-Luc reporter, pRL LXRα, and RXRα expression vectors. 5 h after transfection, cells were stimulated with 5 mm glucose in the absence or presence of PUGNAc (50 μm and 100 μm) for 24 h. The luciferase value at 5 mm glucose without stimulation is set as 1. D, Huh7 cells were transfected with pBP1cLXREab-Luc reporter, pRL LXRα, and RXRα expression vectors. 5 h after transfection, cells were stimulated with 25 mm glucose in the absence or presence of DON (5 μm) and glucosamine (0.2 mm and 1 mm) for 24 h. The luciferase value at 25 mm glucose without stimulation is set as 1. All luciferase data are presented as one representative experiment of three or more independent experiments performed in triplicate ± S.D. (error bars). *, p < 0.05; **, p < 0.01. E, LXRα was immunoprecipitated (IP) from FLAG-hLXRα-overexpressing Huh7 cells treated with 5 mm glucose for 24 h in the absence or presence of 2 mm glucosamine or 100 μm PUGNAc. Immunoprecipitated proteins were subjected to SDS-PAGE and blotted with anti-O-GlcNAc (RL2) or anti-FLAG antibodies. F, O-GlcNAc-modified proteins from FLAG-LXRα-overexpressing Huh7 cells cultured for 24 h in 25 mm d-glucose in the absence or presence of DON (5 μm) were absorbed on sWGA-agarose beads. Input (In, 10%) and sWGA pulled-down proteins (sWGA) were subjected to SDS-PAGE and blotted with anti-LXRα antibody.
To show that LXR transactivation of the SREBP-1c promoter depends on the two LXRE elements of the promoter, we co-transfected LXRα and RXRα with the pBP1cLXRE-Luc promoter/enhancer construct (LXREab-Luc) or mutated versions of the LXRE elements (LXRaM-Luc, LXREbM-Luc, and LXREa+bM-Luc) (29) (Fig. 5B). The glucose responsiveness of the LXREab-Luc enhancer/promoter was similar as the endogenous −550 bp SREBP-1c promoter and relative luciferase activities dropped by ∼60% in cells transfected with the LXREaM-Luc construct in which the upstream LXREa site was mutated. However, glucose responsiveness remained the same, suggesting that the downstream LXREb site is capable of conferring responsiveness to glucose and O-GlcNAcylated LXRα, but that both sites are necessary for maximal responses. Mutation of the downstream LXREb site led to an 85% reduction in relative luciferase activities, and the glucose responsiveness appeared to be lost in cells co-transfected with this construct. Mutations of both LXREa+b sites resulted in only background luciferase activities.
To confirm that the hexosamine biosynthetic pathway is involved in glucose-mediated induction of the LXREab enhancer, we treated cells co-transfected with LXRα/RXRα with PUGNAc, an inhibitor of O-GlcNAcase (Fig. 5C). PUGNAc treatment (100 μm) resulted in 37% increase in relative luciferase activities driven by the LXREab enhancer. Furthermore, DON treatment reduced LXREab-luciferase activities by 20%, and luciferase activities increased by 26% above basal levels in cells treated with glucosamine and DON (Fig. 5D). In line with these observations, immunoblotting showed increased LXRα O-GlcNAcylation in glucosamine- or PUGNAc-treated cells (Fig. 5E), whereas a decrease in sWGA-precipitated LXRα was seen in cells treated with DON (Fig. 5F). Because PUGNAc treatment increases protein O-GlcNAcylation and glucosamine feeds into the hexosamine biosynthetic pathway downstream of the rate-limiting enzyme, glutamine:fructose-6-phosphate amidotransferase, these data suggest that the hexosamine pathway via LXR O-GlcNAcylation is involved in glucose-dependent LXRE/SREBP-1c promoter activation.
DISCUSSION
In the present study we demonstrate that LXR is a target for glucose-regulated O-linked GlcNAc modification and that O-GlcNAcylated LXR is involved in activation of SREBP-1c transcription both in vitro and in vivo. Modulation of post-translational modifications of LXR, such as Ser/Thr O-GlcNAcylation and O-phosphorylation may occur in both the absence and presence of natural ligand-tuning LXR activities in a cell- and gene-specific manner (39), depending on the nutritional stimuli.
LXR is a major regulator of SREBP-1c expression and an insulin-mediating factor in liver (8, 9). Moreover, glucose has been shown to activate the mouse SREBP-1c promoter independently of insulin (5, 7, 29, 40), possibly via a direct activation of LXR by glucose (20). However, LXR acting as a glucose sensor has lately been debated (32–34). With this in mind, it is important to distinguish between the effects seen by insulin and glucose when studying LXR activation of SREBP-1c. From our in vivo STZ studies, we found that hyperglycemia was able to induce the level of SREBP-1c mRNA expression more than 6.5-fold compared with the fasted situation. This induction could not be explained by insulin because the levels of insulin were approximately equal between STZ-treated and fasted control mice. Hasty et al. (40) have previously shown that SREBP-1c is regulated by glucose at the transcriptional level and that the hexosamine biosynthetic pathway may be involved in this process because treatment with azaserine, an inhibitor of glutamine:fructose-6-phosphate amidotransferase, inhibited the glucose-induced up-regulation of both precursor and mature forms of SREBP-1c. Furthermore, the regulation of SREBP-1c expression seen in our mouse models was not seen in LXR α/β double knock-out mice,5 indicating that the effect of glucose previously reported (5, 7, 29, 40) works through the hexosamine pathway and LXR and is an important contribution to the regulation of SREBP-1c.
Yamamoto et al. (16) recently reported that protein kinase A-induced phosphorylation of LXRα inhibited the expression of SREBP-1c in mice liver via reduced DNA binding and co-activator recruitment. Because glucagon/cAMP/protein kinase A signaling may, at least in part, explain down-regulation of SREBP-1 expression in response to fasting, it is likely that PKA-mediated phosphorylation of LXR contributes to the fasting signal on SREBP-1c. In the present study, we report increased hepatic levels of O-GlcNAcylated LXRα in both refed and STZ-induced diabetic mice concomitant with increased expression of SREBP-1c. Moreover, we show that glucose via the hexosamine biosynthetic pathway increased LXR/RXR-driven SREBP-1c promoter activity in Huh7 cells. We hypothesize that nutrient-activated hexosamine signaling promotes LXR O- GlcNAcylation, contributing to increased SREBP-1c expression in the liver.
Human LXRα has been shown to be phosphorylated on serine 198 (15, 17), affecting transcription in a gene-specific manner in macrophages (17). Because other serine or threonine residues may be phosphorylated upon protein kinase A signaling, it is currently not known whether Ser198 is the site of protein kinase A phosphorylation on human LXR in the liver. In our in vitro studies, we observed only modest LXR/RXR transactivation of the SREBP-1c promoter in high glucose-treated cells. This might be explained by constitutive phosphorylation competing for the same site(s) as GlcNAc on LXR and/or inhibitory phosphorylation occurring on adjacent GlcNAc sites. Recently, Housley et al. (26) reported elevated O-GlcNAc on FOXO1 by high glucose and a subsequent reduction by insulin. They further showed that O-GlcNAc modification increased substantially on the insulin-insensitive mutant FOXO1 lacking three AKT phosphorylation sites (T24A, S256A, S319A), resulting in increased FOXO1-dependent luciferase reporter activity. These observations implied overlapping and/or adjacent O-phosphorylation and O-GlcNAc sites on FOXO1, and the authors further identified several O-GlcNAc sites on FOXO1, one of which is adjacent to an AKT phosphorylation site (Thr317). We currently do not know whether mechanisms similar to those for FOXO1 apply to LXR. Because a switch between phosphorylation and glycosylation on LXR may be an important mechanism for nutritional regulation of SREBP-1c transcription, it will be interesting to study whether O-GlcNAc and O-phosphate compete for the same sites or are situated at different serines and/or threonines on LXR. Judging from our in vitro GlcNAcylation results (Fig. 2B), we believe that the major O-GlcNAc site(s) on LXRα and LXRβ resides in the N-terminal region containing the activation function-1 and DNA-binding domains. A more detailed mapping of the GlcNAc sites on LXR and site-directed mutagenesis are subjects for future investigations in our laboratory.
In the present study, our main objective was to examine whether LXR is a target for O-GlcNAcylation in response to glucose independently of ligand. However, we cannot exclude that LXR O-GlcNAcylation may be positively or negatively regulated by LXR and/or RXR ligand because a recent study by Torra et al. (17) reported that Ser198 phosphorylation of LXRα in RAW macrophages was induced by both synthetic and natural oxysterol LXR ligands and reduced by the RXR ligand 9-cis-retinoc acid. Additionally, in our study we see a robust induction of hepatic SREBP-1c mRNA expression by glucose in vivo not observed in LXR α/β double knock-out mice (Fig. 4C).5 This suggests that LXR is essential for SREBP-1c gene expression and that endogenous ligands may potentiate O-GlcNAcylation of LXR.
In conclusion, this present study reports O-GlcNAcylation of LXR in response to physiological glucose concentrations in liver, suggesting that this post-translational modification is instrumental in the glucose responses of LXR. Because protein O-GlcNAcylation of transcription factors has emerged as an essential glucose-sensing mechanism (41), it is important to study O-GlcNAc modification of LXR in more detail. Future investigations will reveal the functional consequences of this modification on LXR with respect to tissue and target gene specificity.
Acknowledgments
We thank Dr. Knut Steffensen (Department of Biosciences and Nutrition, Karolinska Institutet, Novum, Sweden) for providing the rabbit polyclonal LXR antibody and Dr. Hitoshi Shimano (Department of Internal Medicine, Endocrinology and Metabolism, University of Tsukuba, Japan) for providing the mouse SREBP-1c promoter constructs.
This work was supported by the Norwegian Research Council, the Novo Nordic Foundation, Anders Jahres Foundation, Joh H. Andresens Medical Foundation, Aktieselskabet Freia Chocolade Fabriks Medical Foundation, Johan Throne Holst Foundation, and the European Union-granted project 6th framework, CRESCENDO.
S. Holm, S. M. Ulven, K. Bamberg, V. H. Telle-Hansen, C. Bindesboll, K. R. Steffensen, Y. Qin, K. T. Dalen, J.-A. Gustaffson, and N. I. Nebb, unpublished data.
- LXR
- liver X receptor
- LXRE
- LXR response element
- RXR
- retinoic X receptor
- SREBP-1c
- sterol regulatory element-binding protein 1c
- O-GlcNAc
- O-linked β-N-acetylglucosamine
- STZ
- streptozotocin
- DON
- 6-diazo-5-oxo-l-norleucine
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate
- sWGA
- succinylated wheat germ agglutinin
- WT
- wild type
- DMEM
- Dulbecco's modified Eagle's medium
- HRP
- horseradish peroxidase.
REFERENCES
- 1.Apfel R., Benbrook D., Lernhardt E., Ortiz M. A., Salbert G., Pfahl M. (1994) Mol. Cell. Biol. 14, 7025–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teboul M., Enmark E., Li Q., Wikström A. C., Pelto-Huikko M., Gustafsson J. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2096–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peet D. J., Janowski B. A., Mangelsdorf D. J. (1998) Curr. Opin. Genet. Dev. 8, 571–575 [DOI] [PubMed] [Google Scholar]
- 4.Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. (2000) Genes Dev. 14, 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joseph S. B., Laffitte B. A., Patel P. H., Watson M. A., Matsukuma K. E., Walczak R., Collins J. L., Osborne T. F., Tontonoz P. (2002) J. Biol. Chem. 277, 11019–11025 [DOI] [PubMed] [Google Scholar]
- 7.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., Lustig K. D., Shan B. (2000) Genes Dev. 14, 2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobin K. A., Ulven S. M., Schuster G. U., Steineger H. H., Andresen S. M., Gustafsson J. A., Nebb H. I. (2002) J. Biol. Chem. 277, 10691–10697 [DOI] [PubMed] [Google Scholar]
- 9.Chen G., Liang G., Ou J., Goldstein J. L., Brown M. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11245–11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshikawa T., Shimano H., Yahagi N., Ide T., Amemiya-Kudo M., Matsuzaka T., Nakakuki M., Tomita S., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Takahashi A., Sone H., Osuga J., Gotoda T., Ishibashi S., Yamada N. (2002) J. Biol. Chem. 277, 1705–1711 [DOI] [PubMed] [Google Scholar]
- 11.Joseph S. B., Castrillo A., Laffitte B. A., Mangelsdorf D. J., Tontonoz P. (2003) Nat. Med. 9, 213–219 [DOI] [PubMed] [Google Scholar]
- 12.Walcher D., Kümmel A., Kehrle B., Bach H., Grüb M., Durst R., Hombach V., Marx N. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1022–1028 [DOI] [PubMed] [Google Scholar]
- 13.Myhre A. E., Agren J., Dahle M. K., Tamburstuen M. V., Lyngstadaas S. P., Collins A. J., Foster S. J., Thiemermann C., Aasen A. O., Wang J. E. (2008) Shock 29, 468–474 [DOI] [PubMed] [Google Scholar]
- 14.Liang G., Yang J., Horton J. D., Hammer R. E., Goldstein J. L., Brown M. S. (2002) J. Biol. Chem. 277, 9520–9528 [DOI] [PubMed] [Google Scholar]
- 15.Chen M., Bradley M. N., Beaven S. W., Tontonoz P. (2006) FEBS Lett. 580, 4835–4841 [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto T., Shimano H., Inoue N., Nakagawa Y., Matsuzaka T., Takahashi A., Yahagi N., Sone H., Suzuki H., Toyoshima H., Yamada N. (2007) J. Biol. Chem. 282, 11687–11695 [DOI] [PubMed] [Google Scholar]
- 17.Torra I. P., Ismaili N., Feig J. E., Xu C. F., Cavasotto C., Pancratov R., Rogatsky I., Neubert T. A., Fisher E. A., Garabedian M. J. (2008) Mol. Cell. Biol. 28, 2626–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007) Mol. Cell 28, 91–106 [DOI] [PubMed] [Google Scholar]
- 19.Ghisletti S., Huang W., Ogawa S., Pascual G., Lin M. E., Willson T. M., Rosenfeld M. G., Glass C. K. (2007) Mol. Cell 25, 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitro N., Mak P. A., Vargas L., Godio C., Hampton E., Molteni V., Kreusch A., Saez E. (2007) Nature 445, 219–223 [DOI] [PubMed] [Google Scholar]
- 21.Williams S., Bledsoe R. K., Collins J. L., Boggs S., Lambert M. H., Miller A. B., Moore J., McKee D. D., Moore L., Nichols J., Parks D., Watson M., Wisely B., Willson T. M. (2003) J. Biol. Chem. 278, 27138–27143 [DOI] [PubMed] [Google Scholar]
- 22.Hart G. W., Housley M. P., Slawson C. (2007) Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 23.Love D. C., Hanover J. A. (2005) Sci. STKE 312, re13. [DOI] [PubMed] [Google Scholar]
- 24.Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. (2006) Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 25.Yang W. H., Park S. Y., Nam H. W., Kim D. H., Kang J. G., Kang E. S., Kim Y. S., Lee H. C., Kim K. S., Cho J. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Housley M. P., Rodgers J. T., Udeshi N. D., Kelly T. J., Shabanowitz J., Hunt D. F., Puigserver P., Hart G. W. (2008) J. Biol. Chem. 283, 16283–16292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo M., Zilberfarb V., Gangneux N., Christeff N., Issad T. (2008) FEBS Lett. 582, 829–834 [DOI] [PubMed] [Google Scholar]
- 28.Båvner A., Matthews J., Sanyal S., Gustafsson J. A., Treuter E. (2005) Nucleic Acids Res. 33, 3561–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Osuga J., Harada K., Gotoda T., Kimura S., Ishibashi S., Yamada N. (2001) Mol. Cell. Biol. 21, 2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberkofler H., Schraml E., Krempler F., Patsch W. (2004) Biochem. J. 381, 357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakobsson T., Venteclef N., Toresson G., Damdimopoulos A. E., Ehrlund A., Lou X., Sanyal S., Steffensen K. R., Gustafsson J. A., Treuter E. (2009) Mol. Cell 34, 510–518 [DOI] [PubMed] [Google Scholar]
- 32.Lazar M. A., Willson T. M. (2007) Cell Metab. 5, 159–161 [DOI] [PubMed] [Google Scholar]
- 33.Denechaud P. D., Bossard P., Lobaccaro J. M., Millatt L., Staels B., Girard J., Postic C. (2008) J. Clin. Invest. 118, 956–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiota M., Magnuson M. A. (2008) J. Clin. Invest. 118, 841–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comer F. I., Vosseller K., Wells L., Accavitti M. A., Hart G. W. (2001) Anal. Biochem. 293, 169–177 [DOI] [PubMed] [Google Scholar]
- 36.Starr C. M., Hanover J. A. (1990) J. Biol. Chem. 265, 6868–6873 [PubMed] [Google Scholar]
- 37.Monsigny M., Roche A. C., Sene C., Maget-Dana R., Delmotte F. (1980) Eur. J. Biochem. 104, 147–153 [DOI] [PubMed] [Google Scholar]
- 38.Matsuzaka T., Shimano H., Yahagi N., Amemiya-Kudo M., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Tomita S., Sekiya M., Hasty A., Nakagawa Y., Sone H., Toyoshima H., Ishibashi S., Osuga J., Yamada N. (2004) Diabetes 53, 560–569 [DOI] [PubMed] [Google Scholar]
- 39.Rosenfeld M. G., Lunyak V. V., Glass C. K. (2006) Genes Dev. 20, 1405–1428 [DOI] [PubMed] [Google Scholar]
- 40.Hasty A. H., Shimano H., Yahagi N., Amemiya-Kudo M., Perrey S., Yoshikawa T., Osuga J., Okazaki H., Tamura Y., Iizuka Y., Shionoiri F., Ohashi K., Harada K., Gotoda T., Nagai R., Ishibashi S., Yamada N. (2000) J. Biol. Chem. 275, 31069–31077 [DOI] [PubMed] [Google Scholar]
- 41.Issad T., Kuo M. (2008) Trends Endocrinol. Metab. 19, 380–389 [DOI] [PubMed] [Google Scholar]