Abstract
Cannabinoids have been shown to exert anti-inflammatory activities in various in vivo and in vitro experimental models as well as ameliorate various inflammatory degenerative diseases. However, the mechanisms of these effects are not completely understood. Using the BV-2 mouse microglial cell line and lipopolysaccharide (LPS) to induce an inflammatory response, we studied the signaling pathways engaged in the anti-inflammatory effects of cannabinoids as well as their influence on the expression of several genes known to be involved in inflammation. We found that the two major cannabinoids present in marijuana, Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD), decrease the production and release of proinflammatory cytokines, including interleukin-1β, interleukin-6, and interferon (IFN)β, from LPS-activated microglial cells. The cannabinoid anti-inflammatory action does not seem to involve the CB1 and CB2 cannabinoid receptors or the abn-CBD-sensitive receptors. In addition, we found that THC and CBD act through different, although partially overlapping, mechanisms. CBD, but not THC, reduces the activity of the NF-κB pathway, a primary pathway regulating the expression of proinflammatory genes. Moreover, CBD, but not THC, up-regulates the activation of the STAT3 transcription factor, an element of homeostatic mechanism(s) inducing anti-inflammatory events. Following CBD treatment, but less so with THC, we observed a decreased level of mRNA for the Socs3 gene, a main negative regulator of STATs and particularly of STAT3. However, both CBD and THC decreased the activation of the LPS-induced STAT1 transcription factor, a key player in IFNβ-dependent proinflammatory processes. In summary, our observations show that CBD and THC vary in their effects on the anti-inflammatory pathways, including the NF-κB and IFNβ-dependent pathways.
Keywords: Cytokines/Chemokines, Transcription/NF-κB, NF-κB, STAT, Cannabidiol, Cannabinoids, Microglia, Neuroinflammation
Introduction
Δ9-Tetrahydrocannabinol (THC)3 is a major constituent of Cannabis and serves as an agonist of the cannabinoid receptors CB1 (located mainly in neural cells) and CB2 (located mainly on immune cells). The second major constituent of Cannabis extract is cannabidiol (CBD), which is virtually inactive at the CB1 and CB2 receptors (1). Thus, because of its negligible activity at the CB1 receptor, CBD lacks the psychoactive effects that accompany the use of THC. Moreover, CBD was demonstrated to antagonize some undesirable effects of THC, including intoxication, sedation, and tachycardia, while sharing neuroprotective, anti-oxidative, anti-emetic, and anti-carcinogenic properties (2–4). Both THC and CBD have been shown to exert anti-inflammatory properties and to modulate the function of immune cells, including suppression of humoral response, immune cell proliferation, maturation, and migration, and antigen presentation (5–9). Despite increasing amounts of such observations, the molecular mechanisms involved in these cannabinoid-mediated effects are not yet fully understood.
Microglial cells are resident macrophages of the central nervous system and serve as early host defense against pathogens. Activation of microglial cells leads to the release of proinflammatory and neurotoxic factors and serves as part of the neuroinflammatory process (10). The BV-2 murine microglial cell line is known to retain morphological, phenotypic, and functional properties associated with freshly isolated microglia such as expression of nonspecific esterase activity, phagocytic ability, and the absence of peroxidase activity (11, 12). Furthermore, these cells release lysozyme and, when stimulated, interleukin (IL)-1 and tumor necrosis factor α (11, 12). Close similarities between BV-2 and primary microglia in mechanisms mediating microglial stimulations, e.g. by lipopolysaccharide (LPS), S100B, or β-amyloid, were reported (13). These properties make BV-2 cells an appropriate model for studying the activation of microglia in vitro. It has recently been shown that BV-2 cells express elements of the cannabinoid signaling systems, including the presence of endocannabinoids, i.e. anandamide and 2-arachidonoylglycerol, and cannabinoid or cannabinoid-like receptors such as CB2, GPR55, and abnormal cannabidiol (abn-CBD)-sensitive receptors but very little CB1 cannabinoid receptor (14–16).
In this study, we used the BV-2 microglial cell line and assessed the effects of THC and CBD on the LPS-activated microglial secretion of proinflammatory cytokines such as interleukin IL-1β, IL-6, and of interferon β (IFNβ). LPS signaling through TLR4 (toll-like receptor 4) is known to activate several intracellular pathways and to induce broad changes in gene expression, eventually inducing the release of various proinflammatory cytokines and neurotoxic factors (17). LPS activates two basic intracellular pathways via specific adaptor proteins. The first is the myeloid differentiation factor 88 (MyD88)-adaptor protein-dependent pathway that leads to activation of NF-κB-dependent transcription. The second pathway (the MyD88-independent pathway) is dependent on the toll-interleukin-1 receptor (TIR) domain-containing adaptor-inducing interferon-β (TRIF) protein. Its activation turns on the interferon-regulated factor 3 (IRF3)-dependent pathway that enhances the production of IFNβ (18). IFNβ, in an autocrine way, acts via the type I interferon receptor and via signal transducers and activators of transcription (STAT)-dependent pathways and activates a second wave of gene expression including chemokines such as chemokine 2 (CCL2 (C-C motif ligand 2)). We studied the effects of THC and CBD on these two pathways. In addition, we studied the effect of these materials on the expression of several genes, belonging to suppressors of cytokine signaling (SOCS) family, that are involved in the negative regulation of proinflammatory events.
We found that although both THC and CBD exert inhibitory effects on the production of inflammatory cytokines in activated microglial cells in culture, their activities seem to involve both different and overlapping intracellular pathways. These effects are not mediated via CB1, CB2, nor abn-CBD-sensitive receptors.
EXPERIMENTAL PROCEDURES
Reagents
LPS (Escherichia coli serotype 055:B5) and propidium iodide (PI) were purchased from Sigma. THC and CBD were obtained from the National Institute on Drug Abuse (Baltimore, MD). SR141716, SR144528, and abn-CBD were obtained from Tocris (Ellisville, MO). Stocks of these materials in ethanol or DMSO were kept at −80 °C and diluted into medium just prior to experiments. Final concentration of ethanol or DMSO in culture medium was 0.1%. At this concentration, ethanol or DMSO did not show any significant effect on the investigated parameters.
Microglial Cell Culture
The BV-2 murine microglial cell line, originally generated by E. Blasi (University of Perugia, Perugia, Italy (see Ref. 11)), was kindly provided by Prof. E. J. Choi from the Korea University (Seoul, Korea). The BV-2 cells were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2 in high glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% heat-inactivated fetal bovine serum, streptomycin (100 μg/ml), and penicillin (100 units/ml) (Biological Industries Ltd., Kibbutz Beit Haemek, Israel).
Cell Viability Test
BV-2 cell viability was measured by fluorescence-activated cell sorting analysis using PI staining. Microglial cells (1 × 106 cells in 100-mm plates) were pretreated with THC or CBD (both at 10 μm in growth medium) and 2 h later stimulated with 100 ng/ml LPS. The cells were collected 4 h after LPS stimulation and spun down for 5 min at 2000 rpm; the cell pellets were washed twice with Dulbecco's PBS without Ca2+/Mg2+, pH 7.4, fixed in 70% ethanol at −20 °C overnight, followed by incubation with RNase (0.2 mg/ml) at 37 °C, PBS rinsing, and staining with PI (50 μg/ml) for 15 min on ice. The single cell fluorescence of 20,000 cells (for each sample) was measured using a flow cytometer (FACSCalibur, BD Biosciences). The PI emission was detected in the FL2 channel using an emission filter of 585 nm. The data were analyzed using the CellQuest software. The apoptotic cells were defined as cells in sub-G0/G1 phase with hypodiploid DNA content (19).
Enzyme-linked Immunosorbent Assay (ELISA)
Microglial cells were pretreated with cannabinoids for 2 h and stimulated with LPS (100 ng/ml in growth medium) for 4 h. The cultured media were then collected and spun down for 5 min at 2000 rpm, and the concentrations of IL-1β, IL-6, and IFNβ in the medium were determined by ELISA using specific monoclonal antibodies and the procedures recommended by the suppliers (R&D Systems, Minneapolis, MN and PBL Interferon Source, Piscataway, NJ). The serum in the culture media did not interfere with the assays. Whenever CB1 or CB2 receptor antagonists (SR141716 and SR144528, respectively) or abn-CBD were applied, they were added 30 min before the beginning of the THC or CBD treatment.
Western Blot Analysis
To examine the levels of IL-1 receptor-associated kinase 1 (IRAK-1) and of IκB proteins and of the phosphorylated form of the p65 NF-κB subunit, BV-2 cells were incubated with THC or CBD at 1, 5, or 10 μm. Two h later the cells were stimulated for 15 min with 100 ng/ml LPS. The cells were then rinsed twice with ice-cold PBS and lysed with RIPA buffer (140 mm NaCl, 20 mm Tris, pH 7.4, 10% glycerol, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and leupeptin at 20 μg/ml). Lysates were centrifuged at 4 °C (10 min, 14,000 rpm) and pellets discarded, and the supernatants were aliquoted and stored at −20 °C for further analysis.
Aliquots of 25 μg of proteins (as measured with the Bradford protein assay) from each sample were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk in 10 mm Tris-HCl, pH 7.6, containing 150 mm NaCl and 0.5% Tween 20 (TBST). The blots were incubated overnight at 4 °C with primary antibodies, including rabbit anti-IRAK-1, rabbit anti-IκBα (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-phospho-p65 (Ser-536) (Cell Signaling, Danvers, MA), or general rabbit anti-p65 subunit of NF-κB (Santa Cruz Biotechnology). After extensive wash with TBST, horseradish peroxidase-conjugated secondary goat anti-rabbit antibody (Jackson ImmunoResearch, West Grove, PA) was applied for 1 h at room temperature, and the blots were extensively washed and visualized using an enhanced chemiluminescence detection kit (EZ-ECL Biological Industries). The blots were scanned and quantified with NIH Image 1.63. The intensity of the staining of β-actin (using anti-β-actin monoclonal antibody, Santa Cruz Biotechnology) and the total amount of each of the studied proteins were used as loading controls for data normalization.
The immunoblotting assay served also to detect the activation level of STAT1 and of STAT3 after LPS stimulation in the presence or absence of cannabinoids. BV-2 cells were pretreated with THC or CBD for 2 h, and 100 ng/ml LPS was then added for 2 h (for STAT1) or for 2 or 4 h (for STAT3). Cells were then rinsed twice with ice-cold PBS, lysed with RIPA buffer, and centrifuged, and aliquots of the supernatants were applied to immunoblotting analysis as described above. Membranes were probed with rabbit polyclonal antibodies against phospho-STAT1 (Tyr-701) or phospho-STAT3 (Tyr-705) or against the general forms of STAT1 or STAT3 (Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch).
RNA Extraction
BV-2 cells were pretreated with cannabinoids for 2 h and then stimulated with LPS (100 ng/ml) in the presence of cannabinoids for an additional 4 h (same conditions as those used for the ELISA). Total RNA was extracted using the VersageneTM RNA purification kit and the manufacturer's instructions (Gentra Systems, Minneapolis, MN). Purity of extracted RNA was determined by absorbance ratio at 260/280 nm using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA integrity was assessed by electrophoresis on ethidium bromide-stained 1.2% agarose gels.
Quantitative Real Time PCR (qPCR)
cDNA was synthesized using the QuantiTect reverse transcription kit according to the manufacturer's instructions (Qiagen AG, Basel, Switzerland). qPCR was carried out as reported earlier (20). The cDNA of each specific gene was amplified with a pair of specific primers. The primers were designed using the PrimerQuest online tool and synthesized by Metabion International (Planegg-Martinsried, Germany). GenBankTM accession numbers for the different genes and the primer sequences used for amplification are given in Table 1. Each qPCR mixture (in 20 μl) contained cDNA samples (in 3 μl), 125 nm of each forward and reverse primers, and 10 μl of AbsoluteTM Blue QPCR SYBR Green ROX Mix (Thermo Fisher Scientific, Epsom, Surrey, UK). For each of the analyzed gene products, we ran normal and mock reverse-transcribed samples (in the absence of reverse transcriptase) as well as no template control (total mix without cDNA). qPCR was carried out using the Rotor-Gene 3000 qPCR instrument (Corbett Research, Sydney, Australia). RNA expression level was expressed as fold change using the calculation method described by Pfaffl (21). Quantification was performed by “the comparative cycle of threshold method,” with β2-microglobulin (B2m) gene product for normalization, as B2M mRNA expression was found not to be affected by the various treatments. The qPCR runnings were repeated 3 times using mRNA preparations from independent experiments.
TABLE 1.
Sequences of primers used for qPCR amplification of selected gene products
| Gene | Accession no. | Forward | Reverse |
|---|---|---|---|
| B2m | NM_009735 | AGTTCCACCCGCCTCACATTGAAA | TCGGCCATACTGGCATGCTTAACT |
| Il1b | NM_008361 | GCAACTGTTCCTGAACTC | CTCGGAGCCTGTAGTGCA |
| Ifnb1 | NM_010510 | TGCCATCCAAGAGATGCTCCAGAA | AGAAACACTGTCTGCTGGTGGAGT |
| Socs3 | NM_007707 | AGCAGATGGAGGGTTCTGCTTTGT | ATTGGCTGTGTTTGGCTCCTTGTG |
| Cish | NM_009895 | TGGGCCCAAAGTAGTCCTGAATGT | AGAAGAGTGGGAGCCCTTGTGTTT |
| Ccl2 | NM_011333 | CATGCTTCTGGGCCTGCTGTTC | CATGCTTCTGGGCCTGCTGTTC |
Statistical Analysis
The Graph Pad Prism program was used for statistical analysis. The data are expressed as the mean ± S.E. and analyzed for statistical significance using one-way analysis of variance (ANOVA), followed by Bonferroni or Dunnett's post-hoc tests. p < 0.05 was considered significant.
RESULTS
Cell Viability
The PI incorporation assay followed by fluorescence-activated cell sorting analysis was used to test BV-2 cell viability. Under control conditions there were 2.6 ± 0.9% of dead cells. Neither 4 h of stimulation with LPS alone (3.1 ± 0.5% of dead cells) nor the addition of LPS after 2 h of preincubation with 10 μm THC (2.4 ± 1.1%) or 10 μm CBD (5.0 ± 1.8%) significantly affected the viability of the BV-2 microglial cells. A 6-h THC treatment without LPS resulted in 1.6 ± 0.4% of dead cells, although incubation with CBD alone resulted in 7.3 ± 1.5% of dead cells. One-way ANOVA F(5,20) = 3.75, p < 0.05, Bonferroni post hoc test did not reveal a significant effect of these treatments versus control, n = 3–5.
THC and CBD Decrease the Release of Cytokines from LPS-stimulated BV-2 Microglial Cells
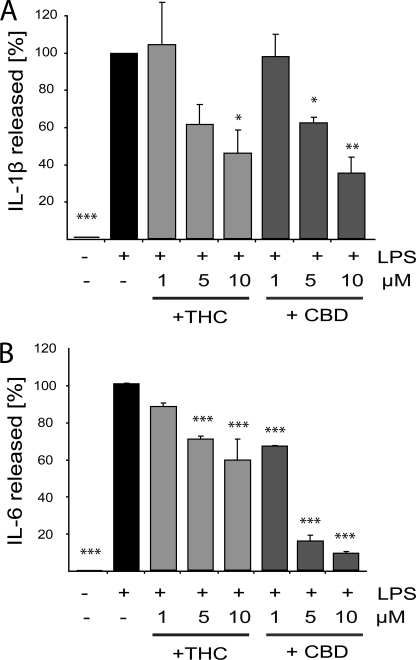
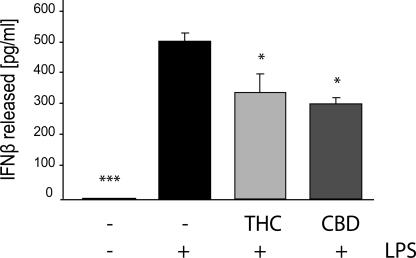
LPS stimulation induces the activation of several intracellular pathways involved in innate immune response. Indeed, as revealed by ELISA, a 4-h LPS stimulation of BV-2 microglial cells led to release of IL-1β, IL-6, and IFNβ proinflammatory cytokines (Figs. 1 and 2). Pretreatment with THC or CBD (at 1, 5, or 10 μm) significantly and dose-dependently decreased the amount of released IL-1β and of released IL-6 (Fig. 1, A and B, respectively). At a 10 μm dose, THC and CBD inhibited the LPS-induced IL-1β release by 54 ± 13 and 64 ± 9%, respectively (Fig. 1A). Regarding IL-6, THC at 5 μm decreased its release by 30 ± 2% and at 10 μm by 41 ± 11%, in comparison with LPS alone. The release of IL-6 was more strongly inhibited by CBD than by THC. The lowest dose of CBD used (1 μm) reduced the release of IL-6 from LPS-activated microglia by ∼25% (an effect comparable with that achieved with 5 μm THC), whereas 5 and 10 μm reduced the release of IL-6 by 85 ± 2 and 91 ± 1%, respectively (Fig. 1B). Both cannabinoids decreased the level of LPS-induced release of IFNβ. At 10 μm, THC and CBD reduced the LPS-induced release of IFNβ by 34 ± 12 and 37 ± 7%, respectively (Fig. 2).
FIGURE 1.
THC and CBD inhibit the LPS-induced release of IL-1β and IL-6 from BV-2 cells. Cells were preincubated for 2 h with the indicated concentrations of THC or CBD and then activated for 4 h with 100 ng/ml LPS. Cell-free media were then collected, and the release of IL-1β (A) and IL-6 (B) was measured using ELISA. The percentage compared with LPS applied alone (marked as 100%) is expressed as the mean ± S.E. of three independent experiments. One-way ANOVA was used as follows: IL-1β, F(7,16) = 10.21, p < 0.001 and IL-6 F(7,16) = 81.8, p < 0.0001. Bonferroni post hoc test: *, p < 0.05; **, p < 0.01; ***, p < 0.001 show significant differences from LPS-treated cells.
FIGURE 2.
THC and CBD decrease the LPS-induced release of IFNβ from BV-2 cells. Cells were pretreated for 2 h with THC or CBD (both at 10 μm) and then activated for 4 h with 100 ng/ml LPS. Cell-free media were then collected and subjected to ELISA for IFNβ. Each bar represents the mean (in pg/ml) ± S.E. from three independent experiments. One-way ANOVA was used as follows: F(3,8) = 35.4, p < 0.001; Bonferroni post hoc test: *, p < 0.05; ***, p < 0.001 versus LPS-treated BV-2 cells.
Unstimulated BV-2 microglial cells did not release detectable amounts of either IL-1β, IL-6, or IFNβ. Moreover, application of cannabinoids for 6 h at the highest concentration tested (10 μm) did not have any effect on cytokine release from unstimulated BV-2 cells (data not shown). Thus, the cannabinoid-induced inhibition of the release of IL-1β, IL-6, and IFNβ could be observed only when the microglial cells were activated.
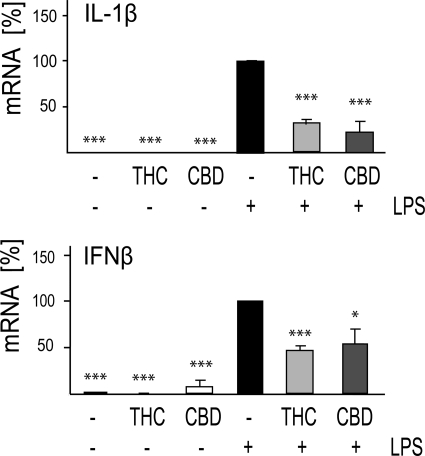
qPCR analysis revealed up-regulated levels of IL-1β and IFNβ mRNA following LPS stimulation of BV-2 microglial cells. THC and CBD at 10 μm decreased the expression of Il1b transcripts by 69 and 78%, respectively. Similarly, IFNβ mRNA level was decreased by 54% by 10 μm THC and by 46% by 10 μm CBD (Fig. 3). Thus, the decrease in release seems to be due to the cannabinoid effect on the mRNA expression of these molecules. However, we cannot rule out additional effects on the release per se.
FIGURE 3.
CBD and THC decrease the mRNA levels of LPS-up-regulated IL-1β and IFNβ. Cells were treated for 2 h with 10 μm THC or CBD. LPS (100 ng/ml) was then added, and 4 h later the cells were harvested, and RNA was extracted for qPCR analysis. The bar graphs present the percent of mRNA expression (average ± S.E. from three independent experiments) versus LPS-only treated samples (taken as 100%). One-way ANOVA was used as follows: IL-1β F(5,12) = 57.2, p < 0.001; IFNβ F(5,10) = 25.16, p < 0.001; Dunnett's post hoc tests: *, p < 0.05, ***, p < 0.001 versus LPS.
CB1 and CB2 Receptors and abn-CBD-sensitive Receptors Do Not Mediate the THC and CBD Inhibitory Effects on Activated BV-2 Cells
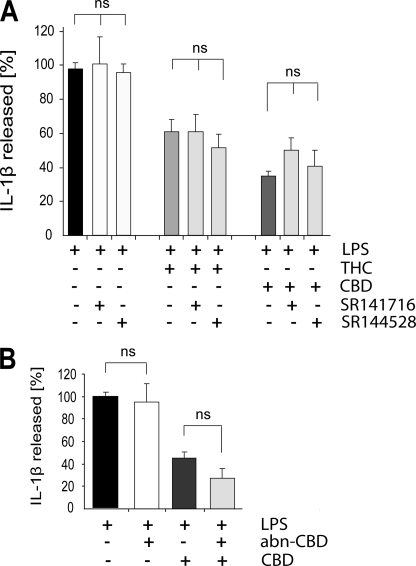
In search for possible receptor targets for THC and CBD that could mediate these immunomodulating effects, we applied selective antagonists of CB1 (SR141716) and CB2 (SR144528) receptors. Because previous observations indicated that CBD is able to antagonize the abn-CBD-induced migration of BV-2 microglial cells (14, 22), we tested the effect of abn-CBD by itself as well as its effect in the presence of CBD.
As shown in Fig. 4A, neither 0.5 μm SR141716 nor 0.5 μm SR144528 given 30 min before CBD or THC affected the inhibitory effect of either THC or CBD (both given at 10 μm) on IL-1β release. Fig. 4B shows that 1 μm abn-CBD (a concentration that induces migration of BV-2 cells (14)) did not affect the CBD inhibition of LPS-induced IL-1β release. Neither 0.5 μm SR141716, SR144528, nor 1 μm abn-CBD affected the LPS effect by themselves. In addition, SR141716, SR144528, and abn-CBD when given alone (without LPS) did not affect the basal level of IL-1β release (data not shown). These results suggest that CB1 and CB2 receptors as well as abn-CBD-sensitive receptors are not involved in the anti-inflammatory effects of THC and CBD in this model of microglial activation.
FIGURE 4.
CB1 and CB2 receptor antagonists as well as abn-CBD do not affect the THC- and CBD-induced inhibition of IL-1β release from LPS-stimulated BV-2 cells. Cells were pretreated for 30 min with SR141716 or SR144528 (both at 0.5 μm) (A) or abn-CBD (1 μm) (B), followed by the addition of 10 μm THC or CBD and 2 h later of LPS (100 ng/ml). Cell-free media were collected 4 h later and assayed for released IL-1β by ELISA. The data are expressed as percentage of released IL-1β ±S.E. from three to four independent experiments. The amount released with LPS alone is represented as 100%. A, one-way ANOVA was used as follows: F(6,14) = 6.58, p < 0.01. Bonferroni post hoc analysis showed that neither SR141716 nor SR144528 affected THC or CBD inhibition of IL-1β release. B, one-way ANOVA was used as follows: F(3,8) = 14.34, p < 0.01. Bonferroni post hoc analysis showed that pretreatment with abn-CBD did not change the effect of CBD on LPS stimulated IL-1β release. ns, not significant.
CBD but Not THC Inhibits the NF-κB-dependent Pathway
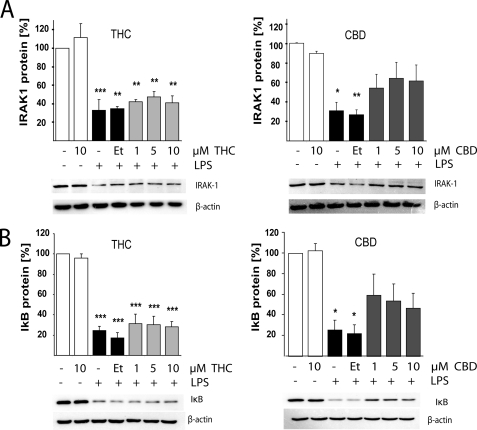
The NF-κB p65-p50 protein complex is present in an inactive form in the cytoplasm while bound to its inhibitory protein IκB. It has been shown that LPS activation of TLR4 leads to IκB inactivation via IRAK-1 kinase-dependent phosphorylation of IκB, which is followed by ubiquitin-dependent degradation of both IRAK-1 and IκB. This action allows the NF-κB p65 subunit to become phosphorylated and to be translocated to the nucleus (23).
As shown in Fig. 5, 15 min of LPS (100 ng/ml) stimulation leads to degradation of IRAK-1 (Fig. 5A) and of IκB proteins (Fig. 5B) in BV-2 microglial cells. LPS-activated cells contain ∼30% of IRAK-1 protein levels as compared with the control level (non activated samples). Pretreatment with CBD partially prevented the LPS-induced reduction in IRAK-1 protein level achieving ∼60% of control levels for both 5 and 10 μm CBD, demonstrating that CBD decreases IRAK-1 degradation. Interestingly, pretreatment with THC did not have any significant effect on the degradation of IRAK-1. None of the THC pretreated samples differed statistically from LPS-only treated BV-2 cells.
FIGURE 5.
CBD, but less so THC, partially reverses the LPS-induced degradation of IRAK-1 and of IκB in LPS-stimulated BV-2 cells. Cells were pretreated for 2 h with THC or CBD at the indicated concentrations followed by 15 min of incubation with LPS (100 ng/ml) and lysed in RIPA buffer, and 20 μg of protein aliquots were subjected to Western blot analysis for IRAK-1 (A) and IκB (B). β-Actin served as a loading control. Bars show the average results of three repetitions with 100% representing the amounts of proteins in control cells. One-way ANOVA was used as follows for IRAK-1 expression: THC-treated cells F(6,14) = 16.79, p < 0.0001, and for CBD-treated cells F(6,14) = 6.00, p < 0.01. One-way ANOVA was used as follows for IκB protein expression: THC-treated samples F(6,14) = 36.59, p < 0.0001, and for CBD-treated samples F(6,14) = 6.39, p = 0.01, followed by Bonferroni post hoc test. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control. Cannabinoid vehicle (0.1% ethanol; Et) did not affect the LPS-induced IRAK-1 or IκB degradation.
In parallel to the effect on IRAK-1, LPS treatment reduced the amount of IκB by 80% in comparison to nonstimulated cells. CBD partially reversed the LPS effect and decreased IκB degradation. Thus, in cells preincubated for 2 h with CBD prior to the LPS application, IκB was present at a much higher level reaching 50–55% of the control (non-LPS) level. On the other hand, THC had no effect on IκB level at all concentrations tested. As an important control, we show that neither THC nor CBD at 10 μm affects the level of IRAK-1 and of IκB proteins when added to the cells in the absence of LPS.
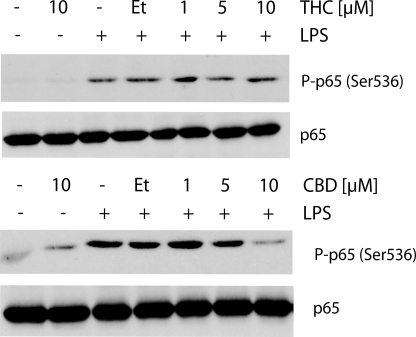
In agreement with these results, LPS activation for 15 min resulted in profound phosphorylation of the p65 NF-κB subunit, and this activation was decreased following pretreatment with 10 μm CBD (and to a lesser extent by 5 μm CBD) but not following THC treatment at any of the concentrations applied (Fig. 6). The 0.1% ethanol used as cannabinoid vehicle did not affect the level of phosphorylated p65. CBD or THC applied without LPS had no effect. Altogether, these observations suggest that CBD, but not THC, inhibits the LPS activation of the pathway leading to NF-κB phosphorylation.
FIGURE 6.
CBD, but not THC, reduces LPS-induced NF-κB p65 subunit phosphorylation. Cells were pretreated for 2 h with CBD or THC at the indicated concentrations followed by LPS (100 ng/ml) for 15 min. Cell homogenates (20 μg of protein aliquots) were subjected to Western blot analysis using antibodies against the phospho-form of NF-κB p65 (Ser-536) subunit. The amount of total p65 served as a loading control. Et, ethanol.
Both CBD and THC Regulate the Activity of the IFNβ Pathway
As described above, the level of released IFNβ protein was significantly lowered when BV-2 cells were pretreated for 2 h with CBD or THC prior to LPS stimulation. It was therefore of interest to study the effect of LPS on IFNβ signaling (activated via the MyD88-independent pathway) and to determine the effects of THC and CBD on this cascade. At the first step, we studied the effects of the cannabinoids on the LPS/IFNβ-induced activation of the transcription factors STAT1 and STAT3, the major mediators of IFNβ signaling (24, 25).
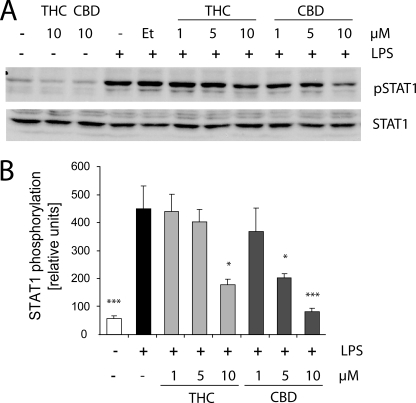
Evaluation of the phosphorylation kinetics of STAT1α (at Tyr-701) revealed maximal STAT1α activation following 2 h with LPS (100 ng/ml) (data not shown). A 2-h pretreatment with 10 μm THC (but less so with 1 or 5 μm) significantly decreased the phosphorylation of STAT1α (Fig. 7). CBD applied 2 h before LPS decreased STAT1α activation already at 5 μm and showed a very strong inhibition at 10 μm. Neither THC nor CBD given alone at 10 μm affected the basal STAT1α phosphorylation level (Fig. 7A). Similarly, 0.1% ethanol (the vehicle for the applied cannabinoids) did not affect the level of LPS-induced STAT1α phosphorylation. The total level of STAT1α remained unaffected by any of the cannabinoid treatments (Fig. 7A).
FIGURE 7.
THC and CBD decrease LPS-induced STAT1 phosphorylation. Cells were pretreated for 2 h with various concentrations of either THC or CBD prior to the addition of LPS for an additional 2 h. A shows representative Western blots for pSTAT1 (Tyr-701) and the general form of STAT1; B, bar graph shows the level of phosphorylation of STAT1 from three independent experiments. One-way ANOVA was used s follows: F(8,18) = 8.1, p < 0.001, followed by Bonferroni post hoc test. *, p < 0.05, ***, p < 0.001 versus ethanol (Et) + LPS.
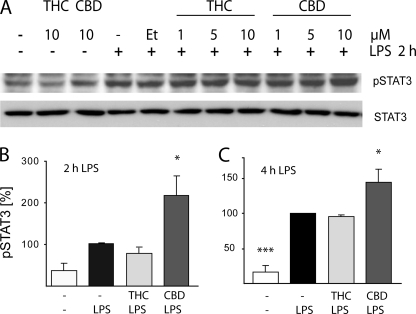
At the next step, we determined the level of activation of STAT3 following LPS stimulation. As shown at Fig. 8A, 2 h with LPS stimulates STAT3 phosphorylation, and this phosphorylation was potentiated when LPS-stimulated cells were preincubated with CBD at 10 μm (but less so with 1 or 5 μm). THC did not affect the level of STAT3 activation at any of the concentrations applied (1, 5, or 10 μm). Neither THC nor CBD given alone (10 μm) affected the basal level of STAT3 phosphorylation. In addition, neither of the treatments had any significant effect on the total amount of STAT3 (Fig. 8A). A time-response experiment (2 and 4 h with LPS) showed increased STAT3 phosphorylation at 2 h and stronger phosphorylation at 4 h (Fig. 8, B and C). The potentiation by 10 μm CBD was observed for both 2 and 4 h of LPS stimulation, whereas THC had no effect at both time points tested (Fig. 8, B and C).
FIGURE 8.
CBD increases LPS-induced STAT3 phosphorylation. A, cells were pretreated for 2 h with various concentrations of either THC or CBD prior to the addition of LPS for additional 2 h. Representative Western blots for pSTAT3 (Tyr-705) and for the general form of STAT3 are shown. B and C show the phosphorylation of STAT3 in bar graphs presenting the results from three independent experiments on cells stimulated with LPS for 2 h (B) or 4 h (C). B, one-way ANOVA was used as follows: F(3,11) = 8.32, p < 0.01, followed by Dunnett's post hoc test. *, p < 0.05 versus LPS. C, ANOVA was used as follows: F(3,8) = 27.09, p < 0.001; *, p < 0.05; ***, p < 0.001 versus LPS alone. Et, ethanol.
These results indicate that the inhibition exerted by THC and CBD on the release of LPS-induced IFNβ from BV-2 cells is in correlation with the lowered level of STAT1 phosphorylation. On the other hand CBD (but not THC) increases the level of STAT3 phosphorylation in LPS stimulated BV-2 microglial cells.
CBD and THC Affect the Level of Negative Regulators of IFNβ/STAT Pathway
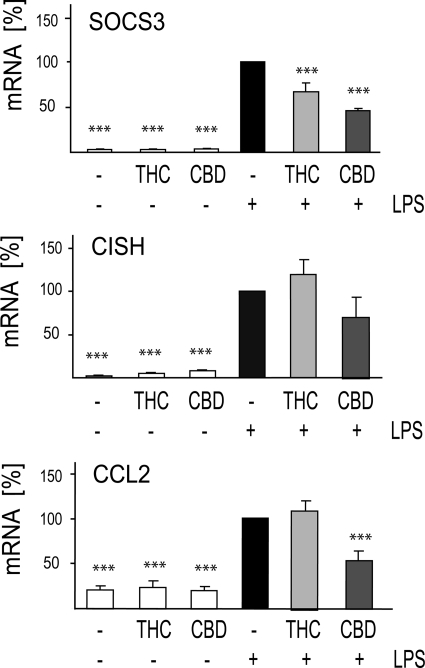
Transcripts for negative regulators of the IFNβ/STAT pathway, including Socs3 and Cish as well as of the Ccl2 chemokine whose transcription is dependent on IFNβ release and is induced via STAT1 pathway, were selected for evaluation by means of qPCR, and the results are displayed in Fig. 9.
FIGURE 9.
LPS up-regulated SOCS3, CISH, and CCL2 mRNAs are differently modulated by CBD and THC. Cells were treated for 2 h with 10 μm THC or CBD. LPS (100 ng/ml) was then added, and 4 h later the cells were harvested, and RNA was extracted for qPCR analysis. The bar graphs present the percent of mRNA expression (average ± S.E. from three to four independent experiments) versus LPS-only treated samples (taken as 100%). One-way ANOVA was used as follows: for SOCS3 F(5,12) = 100.5, p < 0.001; for CISH F(5,12) = 20.7, p < 0.001; for CCL2 F(5,12) = 32.81, p < 0.0001. Dunnett's post hoc test: ***, p < 0.001 versus LPS.
We found that LPS-up-regulated the transcription of both Socs3 and Cish genes. The amount of LPS induced SOCS3 mRNAs were partially reduced by CBD. The amount of LPS-induced CISH mRNA was not significantly affected by either THC or CBD.
LPS stimulation resulted in increased mRNA for CCL2, an IFNβ dependent chemokine. This LPS-induced stimulation was decreased by CBD by 58% but completely unaffected by THC.
DISCUSSION
In this study, we activated BV-2 microglial cells with LPS and observed vast release of IL-1β, IL-6, and IFNβ cytokines, all well recognized as key mediators of inflammatory responses (26). Cannabinoid treatment by either THC or CBD strongly reduced the LPS-induced release of IL-1β, IL-6, and IFNβ. The inhibitory effects of these two cannabinoids on the release of IL-1β and IFNβ were similar. On the other hand, the release of IL-6 was inhibited to a much stronger extent by CBD than by THC.
It is well known that THC and CBD differ in their pharmacology toward the currently known cannabinoid receptors. THC is a CB1 and CB2 receptor partial agonist, whereas CBD exhibits a very low affinity toward both receptors (27, 28). Because CB2 receptors are expressed on various immune cells (29), including primary microglia and the BV-2 microglial cell line (14, 16), they seem to be primary candidates to mediate cannabinoid immunomodulation. Immunosuppressive effects of THC were shown to be predominantly CB2 mediated as revealed by using knock-out systems (30). Accordingly, CB2 agonists were shown to be immunosuppressive in rat primary microglial cultures (31). Rat primary microglial cells were also shown to express CB1 receptors whose activation leads to induction of NO production (32). To determine whether CB1 or CB2 receptors mediate the THC immunosuppression in our system, we applied the respective antagonists before THC treatment. We found that neither CB1 nor CB2 receptor antagonists interfered with the THC inhibitory effect on LPS-induced IL-1β release, suggesting that other receptor(s) or nonreceptor targets seem to be involved. The exact reasons for these different results are not clear. One possibility is that THC is a very weak agonist of the CB2 (33) and that the CB1 is almost absent in BV-2 cells (16). However, we cannot rule out that different inflammatory models may involve somewhat different mechanisms.
As for CBD, whereas binding assays show negligible affinity of CBD toward CB1 and CB2 receptors (27), several functional studies showed that some of the CBD effects may involve cannabinoid receptors. Thus, the CB1 antagonist AM251 reversed the slowing of gastrointestinal motility by CBD in LPS-treated septic mice (34). Moreover, Sacerdote et al. (35) showed that CB1 and CB2 receptor antagonists reversed the CBD modulation of IL-12 and IL-10 release in mouse peritoneal macrophages in vitro and showed that CBD decreased the formyl-methionyl-leucyl-phenylalanine-induced chemotaxis of macrophages in a CB2-dependent manner. However, using our model of microglial activation and applying CB1 and CB2 antagonists prior to CBD treatment, we found that neither of these antagonists affected CBD diminution of IL-1β release suggesting that this CBD activity is not CB1/CB2-mediated.
CBD may act as a partial agonist toward abn-CBD-sensitive receptors. These are putative new members of the cannabinoid receptor family, not yet cloned, but pinpointed pharmacologically in mice lacking CB1 and CB2 receptors (36). abn-CBD-sensitive receptors were shown to be present in microglial cells and regulate their migration (14, 22). However, in our hands abn-CBD pretreatment did not have any effect by itself and did not interfere with the CBD suppression of LPS-induced IL-1β release, thus excluding the involvement of abn-CBD-sensitive receptors in these CBD mediated effects.
Several studies pointed out that cannabinoids could have CB1/CB2 receptor-independent mechanisms of action (37–41). This observation is in agreement with our current findings and with the results of Kaplan et al. (42) showing that CBD and WIN 55212-3 exhibit immunosuppressive effects via non-CB1 and non-CB2 mechanisms. Moreover, Kaplan et al. (42) showed that both cannabinol (a weak CB2 agonist) and CBD inhibit IL-2 release from T lymphocytes, although for cannabinol the effect is CB2-dependent but not for CBD. Altogether, these results indicate that the immunoregulating effects of cannabinoids can be both CB2 receptor- and nonreceptor-mediated even in the same system.
Despite the increasing data on the immune regulatory effects of cannabinoids in vitro (39, 43, 44) and in vivo (45–47), the signaling pathways responsible for these effects are for the most part unknown. Moreover, the modulation of IFNβ-related processes by cannabinoids has been only poorly addressed.
The expression of IL-1β, IL-6, and IFNβ gene products are tightly regulated, and their promoter regions possess binding sites for specific inducible transcription factors. NF-κB is a primary (although not the only one) regulator of IL-1β and IL-6 cytokines, whereas IFNβ is expressed in response to IRF-3 factor activation (48).
The NF-κB pathway is a primary intracellular pathway controlling the transcription of many inflammatory genes (23). The NF-κB p65-p50 protein complex is present in the cytoplasm through association with the inhibitor protein, IκB, which masks the nuclear localization signal present within the NF-κB p65 subunit. The activation of NF-κB by extracellular inducers (including LPS) depends on the rapid phosphorylation of IκB by upstream IRAK-1 followed by ubiquitination and targeting for proteosome degradation of both proteins, IRAK-1 and IκB. The NF-κB p65 subunit then undergoes phosphorylation, followed by translocation to the nucleus where it regulates the expression of various inflammatory genes, including IL-1β and IL-6 (48).
Our data provide several indications for the effects of CBD in decreasing the activity of the NF-κB signaling pathway. The first includes the partial reversal of the LPS-induced degradation of IRAK-1 intermediate kinase; the second is the reversal of IκB degradation, and the third is the lowering of the NF-κB p65 subunit phosphorylation. Interestingly, treatment with THC did not significantly affect this cascade at any of these steps, thus questioning the involvement of the NF-κB pathway in the THC-mediated effects in BV-2 microglia. Several previous studies suggested the involvement of the NF-κB pathway in cannabinoid-induced immunosuppression in macrophages (49), thymocytes (50), monocytes (51), and granulomatous tissue (52). In all of these cases, the effects were shown to be CB2 receptor-mediated. It is interesting to note that 2-arachidonoylglycerol endocannabinoid was shown to decrease NF-κB activation in injured murine brain via CB1 receptors (53). However, in our microglial system neither the CB1 (which is present in a low concentration if at all (16)) nor the CB2 cannabinoid receptors seem to be involved. Interestingly, the non-CB1/CB2-mediated anti-inflammatory effects of cannabinoids mediated via NF-κB and other pathways were also observed in several nonimmune cells, including astrocytes and neuronal PC12 cells (54, 55).
Other pathways could be affected by cannabinoids promoting anti-inflammatory activities in microglial cells. For example, the released IL-1β could promote activation of the NF-κB pathway, whereas IFNβ promotes the interferon-stimulated response element (ISRE) pathway, and IL-6 induces the NF-κB as well as several other pathways (e.g. STAT- and ISRE-dependent) (17). Both THC and CBD decrease LPS-induced IFNβ production and release. These cannabinoids exert their inhibitory activity upstream of IFNβ synthesis, e.g. at the level of the MyD88-independent pathway that is leading to the activation of IRF-3. The IRF-3 pathway is activated following its phosphorylation by TBK1 (TANK-binding kinase 1) associated with the TRIF adaptor protein of TLR4 receptors. The activated IRF-3 binds the ISRE DNA sequence inducing the production of the IFNβ cytokine (17). IFNβ expression activates a second wave of gene expression (including chemokines such as CXCL10, CCL5, and CCL2) via the IFN receptor and the Janus tyrosine kinase/STAT pathways. Briefly, the released IFNβ binds to IFN receptor and induces phosphorylation of the Janus tyrosine kinase family members leading to the activation of STAT multifamily proteins. Upon activation, the members of the STAT family induce the expression of pro- as well as of anti-inflammatory genes through binding to the various ISRE as well as to some IFN-γ-activated sequence promoter sites to induce expression of interferon-stimulated genes (24). The major mediators of IFNβ signaling are STAT1 and STAT3 (24, 25). Indeed, we observed profound activation of STAT1 and STAT3 following LPS stimulation.
STAT1 and STAT3 have similar structures; both are phosphorylated on tyrosine residues upon cytokine stimulation, and both form homo- or heterodimers through the reciprocal Src homology 2 domain/phosphotyrosine interactions, move to the nucleus, bind to respective sequences on promoter sites, and activate transcription of a large number of genes. Various STAT1 and STAT3 dimers bind selectively to very similar but not identical elements and thus activate different but to some extent overlapping genes. This is likely to account for their different biological effects. For example, STAT1 homodimers exert proinflammatory effects via binding to ISRE and IFN-γ-activated sequence elements, inducing the expression of many chemokines (e.g. CCL2, ICAM1, and CXCL10), which regulate the migration or adhesion of immune cells (24). In contrast, STAT3 exerts anti-inflammatory effects via the increased synthesis of IL-10 (an anti-inflammatory interleukin) or via direct binding to consensus elements of various IL-10-inducible genes (56). Several reports have revealed mechanisms responsible for STAT3-mediated attenuation of immune responses. For example, activated STAT3 was shown to suppress LPS-induced IL-6, tumor necrosis factor α, and IL-12 gene expression in macrophages and in dendritic cells (57, 58). STAT3 deficiency (or inactivation) makes the mutant mice highly susceptible to LPS shock and results in increased production of inflammatory cytokines such as tumor necrosis factor α, IL-1, and IFNγ from macrophages or neutrophils (59, 60). In addition, studies on STAT3-deficient cells revealed the existence of reciprocal STAT1/STAT3 regulatory mechanisms and explained the increase in proinflammatory STAT1 activity in the absence/inactivation of STAT3 (61–63). Indeed, the balance between the proinflammatory STAT1 and the anti-inflammatory STAT3 seems to determine the final outcome of cell activation, i.e. immune tolerance versus chronic inflammatory state (24, 25). Thus, STAT3 forms a feedback loop that is switched on by LPS and serves as a counterbalance mechanism to reduce the risk of chronic inflammation.
In our experiments, we observed that although both cannabinoids reduce the activation of the proinflammatory STAT1, CBD (but not THC) strengthens the activation of STAT3. Thus, CBD seems to decrease the ongoing pro-inflammatory processes as well as intensify events counteracting inflammation. Moreover, we observed that LPS-induced STAT1-dependent expression of CCL2 mRNA was down-regulated following CBD (but not THC) pretreatment. CCL2 was shown to be up-regulated in STAT3 knock-out macrophages (64) pointing to a tight regulation of CCL2 levels by STAT3. Indeed, we found that the effect of CBD on the down-regulation of CCL2 mRNA parallels the up-regulation of STAT3 activation and STAT1 down-regulation and further indicates that these STAT molecules have a role in CBD-induced anti-inflammatory effects.
The IFNβ-inducible Janus tyrosine kinase/STAT pathway is under the control of feedback inhibitors belonging to the SOCS family that include SOCS1–SOCS7 and CISH (65, 66). The transcription of several of these inhibitors is up-regulated as a feedback response to immune activation by a variety of immune cytokines and by LPS (67). Indeed, our qPCR measurement of mRNAs for SOCS3 and CISH shows that LPS profoundly up-regulates the expression of SOCS3 and of CISH in BV-2 microglial cells. CBD, and to a lesser extent THC, significantly suppressed the expression of SOCS3, but not of CISH, suggesting the involvement of particular negative regulators in the anti-inflammatory activity of the cannabinoids. Because SOCS3 has been shown to decrease STAT3 activity (68, 69) and the absence of SOCS3 (e.g. in SOCS3−/− macrophages) led to increased STAT3-mediated anti-inflammatory effects (69), the inducible effect of CBD on the activation of STAT3 could be mediated via its effect on SOCS3 expression. This mode of regulation is in line with the CBD anti-inflammatory activity in LPS-activated microglial cells.
The NF-κB pathway can also be regulated by STAT-dependent molecules. Nishinakamura et al. (70) showed that activated STAT3 (STAT3C, a modified form of STAT3) reduced LPS-induced NF-κB transcription through αCP-1 (an RNA-binding protein that contains a K-homology domain with specificity for C-rich pyrimidine tracts) without affecting the TLR4 signal transduction, meaning without affecting phosphorylation of IκB and without affecting the DNA binding activity of NF-κB. We hypothesize that this regulation may be responsible at least in part for the diminution of IL-6 release by CBD.
As for THC, it did not affect STAT3 phosphorylation and had a reduced effect on NF-κB. This could explain its reduced effect on the LPS-induced release of IL-6, in comparison with the effects of CBD. As for its effects on IL-1β, this might be due to the effect of THC on the release of IFNβ and the concomitant reduction in STAT1 phosphorylation. Although we did not observe a direct effect of THC on the NF-κB pathway, an increasing number of genome-wide analyses indicate that modulation of IFNβ pathway activity results in diminished transcription of NF-κB-dependent genes (71, 72). This reciprocal regulation may be involved in THC-exerted anti-inflammatory effects.
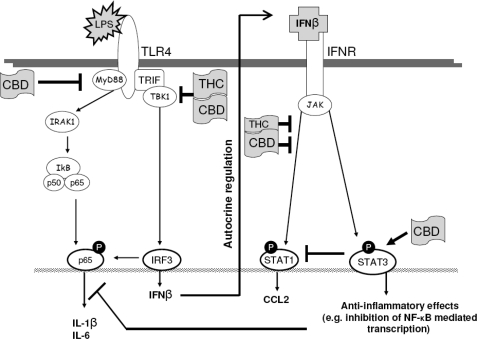
In summary, our results show that although both THC and CBD exert anti-inflammatory effects, the two compounds engage different, although to some extent overlapping, intracellular pathways. Both THC and CBD decrease the activation of proinflammatory signaling by interfering with the TRIF/IFNβ/STAT pathway (see Scheme 1). CBD additionally suppresses the activity of the NF-κB pathway and potentiates an anti-inflammatory negative feedback process via STAT3. It is well known that NF-κB, IRF-3, and the STAT factors are induced by a broad spectrum of endogenous signals whose level is increased in response to cytotoxic changes. These include mitogens, cytokines, and neurotoxic factors (73, 74). The cannabinoids by moderating or disrupting these signaling networks may show promise as anti-inflammatory agents.
SCHEME 1.
Schematic figure showing possible sites for the anti-inflammatory activities of THC and CBD in LPS-activated BV-2 microglia. Pointed arrows indicate activation, and blunt arrows indicate inhibition.
This work was supported in part by the Dr. Miriam and Sheldon G. Adelson Center for the Biology of Addictive Diseases, by the Adelsons' Program for Nerve Regeneration and Repair, by the Nella and Leon Benoziyo Center for Neurosciences, and by the Israeli Ministry of Health (to Z. V.).
This work is dedicated to the memory of Maciej Pietr.
- THC
- Δ9-tetrahydrocannabinol
- CBD
- cannabidiol
- abn-CBD
- abnormal cannabidiol
- STAT
- signal transducers and activators of transcription
- IL
- interleukin
- IFN
- interferon
- LPS
- lipopolysaccharide
- PI
- propidium iodide
- PBS
- phosphate-buffered saline
- ELISA
- enzyme-linked immunosorbent assay
- qPCR
- quantitative real time PCR
- ANOVA
- analysis of variance
- IRF3
- interferon-regulated factor 3
- ISRE
- interferon-stimulated response element.
REFERENCES
- 1.Thomas B. F., Gilliam A. F., Burch D. F., Roche M. J., Seltzman H. H. (1998) J. Pharmacol. Exp. Ther. 285, 285–292 [PubMed] [Google Scholar]
- 2.Mechoulam R., Peters M., Murillo-Rodriguez E., Hanus L. O. (2007) Chem. Biodivers 4, 1678–1692 [DOI] [PubMed] [Google Scholar]
- 3.Izzo A. A., Borrelli F., Capasso R., Di Marzo V., Mechoulam R. (2009) Trends Pharmacol. Sci. 30, 515–527 [DOI] [PubMed] [Google Scholar]
- 4.Zuardi A. W. (2008) Rev. Bras. Psiquiatr. 30, 271–280 [DOI] [PubMed] [Google Scholar]
- 5.Malfait A. M., Gallily R., Sumariwalla P. F., Malik A. S., Andreakos E., Mechoulam R., Feldmann M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9561–9566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Shabat S., Hanus L. O., Katzavian G., Gallily R. (2006) J. Med. Chem. 49, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 7.Klein T. W., Cabral G. A. (2006) J. Neuroimmune Pharmacol. 1, 50–64 [DOI] [PubMed] [Google Scholar]
- 8.Baker D., Jackson S. J., Pryce G. (2007) Br. J. Pharmacol. 152, 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borrelli F., Aviello G., Romano B., Orlando P., Capasso R., Maiello F., Guadagno F., Petrosino S., Capasso F., Di Marzo V., Izzo A. A. (2009) J. Mol. Med. 87, 1111–1121 [DOI] [PubMed] [Google Scholar]
- 10.Dheen S. T., Kaur C., Ling E. A. (2007) Curr. Med. Chem. 14, 1189–1197 [DOI] [PubMed] [Google Scholar]
- 11.Blasi E., Barluzzi R., Bocchini V., Mazzolla R., Bistoni F. (1990) J. Neuroimmunol. 27, 229–237 [DOI] [PubMed] [Google Scholar]
- 12.Bocchini V., Mazzolla R., Barluzzi R., Blasi E., Sick P., Kettenmann H. (1992) J. Neurosci. Res. 31, 616–621 [DOI] [PubMed] [Google Scholar]
- 13.Kim S. H., Smith C. J., Van Eldik L. J. (2004) Neurobiol. Aging 25, 431–439 [DOI] [PubMed] [Google Scholar]
- 14.Walter L., Franklin A., Witting A., Wade C., Xie Y., Kunos G., Mackie K., Stella N. (2003) J. Neurosci. 23, 1398–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrier E. J., Kearn C. S., Barkmeier A. J., Breese N. M., Yang W., Nithipatikom K., Pfister S. L., Campbell W. B., Hillard C. J. (2004) Mol. Pharmacol. 65, 999–1007 [DOI] [PubMed] [Google Scholar]
- 16.Pietr M., Kozela E., Levy R., Rimmerman N., Lin Y. H., Stella N., Vogel Z., Juknat A. (2009) FEBS Lett. 583, 2071–2076 [DOI] [PubMed] [Google Scholar]
- 17.Gay N. J., Gangloff M. (2007) Annu. Rev. Biochem. 76, 141–165 [DOI] [PubMed] [Google Scholar]
- 18.Kawai T., Takeuchi O., Fujita T., Inoue J., Mühlradt P. F., Sato S., Hoshino K., Akira S. (2001) J. Immunol. 167, 5887–5894 [DOI] [PubMed] [Google Scholar]
- 19.Nicoletti I., Migliorati G., Pagliacci M. C., Grignani F., Riccardi C. (1991) J. Immunol. Methods 139, 271–279 [DOI] [PubMed] [Google Scholar]
- 20.Butovsky E., Juknat A., Elbaz J., Shabat-Simon M., Eilam R., Zangen A., Altstein M., Vogel Z. (2006) Mol. Cell. Neurosci. 31, 795–804 [DOI] [PubMed] [Google Scholar]
- 21.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franklin A., Stella N. (2003) Eur. J. Pharmacol. 474, 195–198 [DOI] [PubMed] [Google Scholar]
- 23.Rothwarf D. M., Karin M. (1999) Sci. STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 24.Wesoly J., Szweykowska-Kulinska Z., Bluyssen H. A. (2007) Acta Biochim. Pol. 54, 27–38 [PubMed] [Google Scholar]
- 25.Regis G., Pensa S., Boselli D., Novelli F., Poli V. (2008) Semin. Cell Dev. Biol. 19, 351–359 [DOI] [PubMed] [Google Scholar]
- 26.Suzumura A., Takeuchi H., Zhang G., Kuno R., Mizuno T. (2006) Ann. N.Y. Acad. Sci. 1088, 219–229 [DOI] [PubMed] [Google Scholar]
- 27.Showalter V. M., Compton D. R., Martin B. R., Abood M. E. (1996) J. Pharmacol. Exp. Ther. 278, 989–999 [PubMed] [Google Scholar]
- 28.Pertwee R. G. (2008) Br. J. Pharmacol. 153, 199–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackie K. (2008) J. Neuroendocrinol. 20, Suppl. 1, 10–14 [DOI] [PubMed] [Google Scholar]
- 30.Buckley N. E., McCoy K. L., Mezey E., Bonner T., Zimmer A., Felder C. C., Glass M., Zimmer A. (2000) Eur. J. Pharmacol. 396, 141–149 [DOI] [PubMed] [Google Scholar]
- 31.Romero-Sandoval E. A., Horvath R., Landry R. P., DeLeo J. A. (2009) Mol. Pain 5, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabral G. A., Harmon K. N., Carlisle S. J. (2001) Adv. Exp. Med. Biol. 493, 207–214 [DOI] [PubMed] [Google Scholar]
- 33.Bayewitch M., Rhee M. H., Avidor-Reiss T., Breuer A., Mechoulam R., Vogel Z. (1996) J. Biol. Chem. 271, 9902–9905 [DOI] [PubMed] [Google Scholar]
- 34.de Filippis D., Iuvone T., d'amico A., Esposito G., Steardo L., Herman A. G., Pelckmans P. A., de Winter B. Y., de Man J. G. (2008) Neurogastroenterol. Motil. 20, 919–927 [DOI] [PubMed] [Google Scholar]
- 35.Sacerdote P., Martucci C., Vaccani A., Bariselli F., Panerai A. E., Colombo A., Parolaro D., Massi P. (2005) J. Neuroimmunol. 159, 97–105 [DOI] [PubMed] [Google Scholar]
- 36.Járai Z., Wagner J. A., Varga K., Lake K. D., Compton D. R., Martin B. R., Zimmer A. M., Bonner T. I., Buckley N. E., Mezey E., Razdan R. K., Zimmer A., Kunos G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14136–14141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makriyannis A., Yang D. P., Griffin R. G., Das Gupta S. K. (1990) Biochim. Biophys. Acta 1028, 31–42 [DOI] [PubMed] [Google Scholar]
- 38.Felder C. C., Veluz J. S., Williams H. L., Briley E. M., Matsuda L. A. (1992) Mol. Pharmacol. 42, 838–845 [PubMed] [Google Scholar]
- 39.Puffenbarger R. A., Boothe A. C., Cabral G. A. (2000) Glia 29, 58–69 [PubMed] [Google Scholar]
- 40.Berdyshev E. V., Schmid P. C., Krebsbach R. J., Hillard C. J., Huang C., Chen N., Dong Z., Schmid H. H. (2001) Biochem. J. 360, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price T. J., Patwardhan A., Akopian A. N., Hargreaves K. M., Flores C. M. (2004) Br. J. Pharmacol. 142, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan B. L., Rockwell C. E., Kaminski N. E. (2003) J. Pharmacol. Exp. Ther. 306, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 43.Watzl B., Scuderi P., Watson R. R. (1991) Int. J. Immunopharmacol. 13, 1091–1097 [DOI] [PubMed] [Google Scholar]
- 44.Srivastava M. D., Srivastava B. I., Brouhard B. (1998) Immunopharmacology 40, 179–185 [DOI] [PubMed] [Google Scholar]
- 45.Gallily R., Yamin A., Waksmann Y., Ovadia H., Weidenfeld J., Bar-Joseph A., Biegon A., Mechoulam R., Shohami E. (1997) J. Pharmacol. Exp. Ther. 283, 918–924 [PubMed] [Google Scholar]
- 46.Smith S. R., Terminelli C., Denhardt G. (2000) J. Pharmacol. Exp. Ther. 293, 136–150 [PubMed] [Google Scholar]
- 47.Roche M., Diamond M., Kelly J. P., Finn D. P. (2006) J. Neuroimmunol. 181, 57–67 [DOI] [PubMed] [Google Scholar]
- 48.Covert M. W., Leung T. H., Gaston J. E., Baltimore D. (2005) Science 309, 1854–1857 [DOI] [PubMed] [Google Scholar]
- 49.Jeon Y. J., Yang K. H., Pulaski J. T., Kaminski N. E. (1996) Mol. Pharmacol. 50, 334–341 [PubMed] [Google Scholar]
- 50.Herring A. C., Kaminski N. E. (1999) J. Pharmacol. Exp. Ther. 291, 1156–1163 [PubMed] [Google Scholar]
- 51.Rajesh M., Mukhopadhyay P., Haskó G., Huffman J. W., Mackie K., Pacher P. (2008) Br. J. Pharmacol. 153, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Filippis D., Russo A., De Stefano D., Maiuri M. C., Esposito G., Cinelli M. P., Pietropaolo C., Carnuccio R., Russo G., Iuvone T. (2007) J. Mol. Med. 85, 635–645 [DOI] [PubMed] [Google Scholar]
- 53.Panikashvili D., Mechoulam R., Beni S. M., Alexandrovich A., Shohami E. (2005) J. Cereb. Blood Flow Metab. 25, 477–484 [DOI] [PubMed] [Google Scholar]
- 54.Esposito G., De Filippis D., Maiuri M. C., De Stefano D., Carnuccio R., Iuvone T. (2006) Neurosci. Lett. 399, 91–95 [DOI] [PubMed] [Google Scholar]
- 55.Curran N. M., Griffin B. D., O'Toole D., Brady K. J., Fitzgerald S. N., Moynagh P. N. (2005) J. Biol. Chem. 280, 35797–35806 [DOI] [PubMed] [Google Scholar]
- 56.Murray P. J. (2006) Biochem. Soc. Trans. 34, 1028–1031 [DOI] [PubMed] [Google Scholar]
- 57.Butcher B. A., Kim L., Panopoulos A. D., Watowich S. S., Murray P. J., Denkers E. Y. (2005) J. Immunol. 174, 3148–3152 [DOI] [PubMed] [Google Scholar]
- 58.Barton B. E. (2006) Expert Opin. Ther. Targets 10, 459–470 [DOI] [PubMed] [Google Scholar]
- 59.Takeda K., Clausen B. E., Kaisho T., Tsujimura T., Terada N., Förster I., Akira S. (1999) Immunity 10, 39–49 [DOI] [PubMed] [Google Scholar]
- 60.Akira S. (2000) Oncogene 19, 2607–2611 [DOI] [PubMed] [Google Scholar]
- 61.Costa-Pereira A. P., Tininini S., Strobl B., Alonzi T., Schlaak J. F., Is'harc H., Gesualdo I., Newman S. J., Kerr I. M., Poli V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8043–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qing Y., Stark G. R. (2004) J. Biol. Chem. 279, 41679–41685 [DOI] [PubMed] [Google Scholar]
- 63.Tanabe Y., Nishibori T., Su L., Arduini R. M., Baker D. P., David M. (2005) J. Immunol. 174, 609–613 [DOI] [PubMed] [Google Scholar]
- 64.Matsukawa A., Kudo S., Maeda T., Numata K., Watanabe H., Takeda K., Akira S., Ito T. (2005) J. Immunol. 175, 3354–3359 [DOI] [PubMed] [Google Scholar]
- 65.Jin H. J., Shao J. Z., Xiang L. X., Wang H., Sun L. L. (2008) Mol. Immunol. 45, 1258–1268 [DOI] [PubMed] [Google Scholar]
- 66.Croker B. A., Kiu H., Nicholson S. E. (2008) Semin. Cell Dev. Biol. 19, 414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wormald S., Hilton D. J. (2007) Curr. Opin. Hematol. 14, 9–15 [DOI] [PubMed] [Google Scholar]
- 68.Hong F., Jaruga B., Kim W. H., Radaeva S., El-Assal O. N., Tian Z., Nguyen V. A., Gao B. (2002) J. Clin. Invest. 110, 1503–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yasukawa H., Ohishi M., Mori H., Murakami M., Chinen T., Aki D., Hanada T., Takeda K., Akira S., Hoshijima M., Hirano T., Chien K. R., Yoshimura A. (2003) Nat. Immunol. 4, 551–556 [DOI] [PubMed] [Google Scholar]
- 70.Nishinakamura H., Minoda Y., Saeki K., Koga K., Takaesu G., Onodera M., Yoshimura A., Kobayashi T. (2007) Int. Immunol. 19, 609–619 [DOI] [PubMed] [Google Scholar]
- 71.Ogawa S., Lozach J., Benner C., Pascual G., Tangirala R. K., Westin S., Hoffmann A., Subramaniam S., David M., Rosenfeld M. G., Glass C. K. (2005) Cell 122, 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lacaze P., Raza S., Sing G., Page D., Forster T., Storm P., Craigon M., Awad T., Ghazal P., Freeman T. C. (2009) BMC Genomics 10, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ock J., Jeong J., Choi W. S., Lee W. H., Kim S. H., Kim I. K., Suk K. (2007) J. Neurosci. Res. 85, 1989–1995 [DOI] [PubMed] [Google Scholar]
- 74.Pais T. F., Figueiredo C., Peixoto R., Braz M. H., Chatterjee S. (2008) J. Neuroinflammation 5, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]