Abstract
The amyloid-β (Aβ) peptide, widely known as the causative molecule of Alzheimer disease (AD), is generated by the sequential cleavage of amyloid precursor protein (APP) by the aspartyl proteases BACE1/β-secretase and presenilin/γ-secretase. Inhibition of BACE1, therefore, is a promising strategy for preventing the progression of AD. However, β-secretase inhibitors (BSIs) exhibit unexpectedly low potency in cells expressing “Swedish mutant” APP (APPswe) and in the transgenic mouse Tg2576, an AD model overexpressing APPswe. The Swedish mutation dramatically accelerates β-cleavage of APP and hence the generation of Aβ; this acceleration has been assumed to underlie the poor inhibitory activity of BSI against APPswe processing. Here, we studied the mechanism by which the Swedish mutation causes this BSI potency decrease. Surprisingly, decreased BSI potency was not observed in an in vitro assay using purified BACE1 and substrates, indicating that the accelerated β-cleavage resulting from the Swedish mutation is not its underlying cause. By focusing on differences between the cell-based and in vitro assays, we have demonstrated here that the potency decrease is caused by the aberrant subcellular localization of APPswe processing and not by accelerated β-cleavage or the accumulation of the C-terminal fragment of β-cleaved APP. Because most patients with sporadic AD express wild type APP, our findings suggest that the wild type mouse is superior to the Tg2576 mouse as a model for determining the effective dose of BSI for AD patients. This work provides novel insights into the potency decrease of BSI and valuable suggestions for its development as a disease-modifying agent.
Keywords: Cell, Cell/Neuron, Cell/Secretion, Diseases/Alzheimer Disease, Diseases/Amyloid, Enzymes/Inhibitors, Neurochemistry, Proteases/Aspartyl Protease
Introduction
Alzheimer disease (AD)2 is the most common type of dementia associated with neurodegeneration. Amyloid β (Aβ) peptides accumulate in the brains of AD patients and are deposited as insoluble plaques, the hallmarks of AD pathophysiology (1). Aβ is produced by sequential cleavage of amyloid precursor protein (APP) by the aspartyl proteases BACE1/β-secretase and presenilin/γ-secretase. Growing evidence indicates that the acceleration of Aβ generation can trigger the cognitive dysfunction characteristic of AD (2–4). In fact, many risk factors for AD, including higher levels of β-secretase and γ-secretase expression, oxidative stress, and insulin dysfunction, promote the generation of Aβ (5–11). Therefore, the inhibition of Aβ production is one of the most promising therapeutic approaches for preventing the progression of AD (12–16).
BACE1/β-secretase inhibitors (BSIs) have been investigated as AD-modifying agents since the gene encoding the BACE1 enzyme was cloned in 1999 (17, 18). BACE1-deficient mice are viable, and the dramatic decrease in Aβ levels caused by the genetic deletion of BACE1 in AD model mice can ameliorate AD phenotypes such as memory impairment (19–21). However, BSI is less able to reduce Aβ in an AD model mouse (Tg2576) than in wild type mice (22–24), raising doubts about the potential clinically effective dose of BSI and thus raising concerns about the safety of the treatment and its cost in clinical trials.
Tg2576 is a transgenic mouse expressing “Swedish mutant” APP (APPswe) (25). This two-amino acid mutation, which was discovered in Swedish familial AD patients (26), dramatically accelerates β-site processing of APP. Therefore, the weakening of BSI potency in the Tg2576 mouse appears to be attributable to the Swedish mutation. Several reports have shown that BSIs are less potent against Aβ generation in cells stably transfected with the APPswe variant than in those transfected with wild type APP (APPwt) (22–24, 27), but the mechanism underlying this reduction in BSI inhibitory activity has not yet been elucidated. If the poor potency of BSIs in Tg2576 mice arises from differences between AD and non-AD that are unrelated to the Swedish mutation, then a high dose of BSI would be required to effectively prevent AD progression in both sporadic and Swedish type AD. Therefore, to accurately predict the clinically effective dose of BSI, we must elucidate the mechanism by which the Swedish mutation affects BSI potency.
In this study, in vitro BSI assays using purified BACE1 and substrate peptides showed that, in contrast to previous results from cell-based assays, BSI is as potent a cleavage inhibitor for APPswe as it is for APPwt. This finding suggests that differences between the cell-based and in vitro enzymatic assays might underlie the apparent effect of the Swedish mutation on BSI potency. Our analysis of these differences demonstrates that the potency decrease is caused by the aberrant subcellular localization of APPswe processing and not by accelerated β-cleavage or by the accumulation of the C-terminal fragment of β-cleaved APP (βCTF). Our findings suggest that the abnormal subcellular site of APPswe processing is responsible for the weakened inhibitory activity of BSIs against Aβ production in APPswe-expressing cells.
EXPERIMENTAL PROCEDURES
In Vitro BACE1 Activity Assay
In vitro BACE1 activity assays were performed using substrate peptides from the American Peptide Company, Inc. (Sunnyvale, CA), recombinant human BACE1 from R & D Systems (Minneapolis, MN), and BSI OM99–2 (28) or β-secretase Inhibitor IV from Calbiochem (29). The substrate peptide sequences were SEVKMDAEFRHDSGYEK-biotin (wild type; wt) and SEVNLDAEFRHDSGYEK-biotin (Swedish; swe). Peptides and inhibitors were dissolved in dimethyl sulfoxide (DMSO), and dissolved peptides were stored at −20 °C.
The standard reaction buffer was 50 mm sodium acetate, pH 4.5, containing 0.25 mg/ml bovine serum albumin (BSA). In experiments to check the pH dependence of IC50 values, citrate-phosphate buffer was used because of its broad buffering range. The reactions were carried by mixing 89 μl of substrate solution, 1 μl of inhibitor solution or DMSO, and 10 μl of BACE1 in each well of a 96-well plate and incubating the plate under the conditions described in Fig. 1 and Table 1. The reactions were terminated by the addition of 30 μl of 1 m Tris-HCl, pH 7.6.
FIGURE 1.
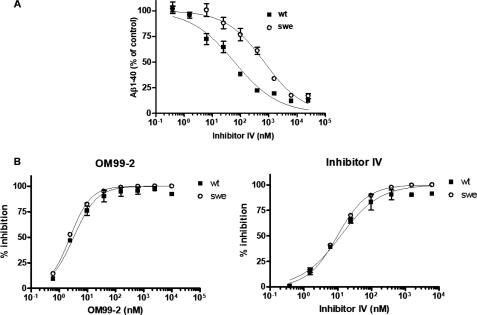
The β-cleavage of wild type and “Swedish mutant” APP substrates is inhibited with equal efficiency by BSIs in an in vitro BACE1 activity assay. A, SH-SY5Y cells stably transfected with wild type APP (APPwt; ■) or “Swedish mutant” APP (APPswe; ○) were treated with Inhibitor IV at the indicated concentrations for 24 h, and the conditioned medium was analyzed for the amount of Aβ1–40 as described under “Experimental Procedures.” Aβ1–40 quantification data are expressed relative to those for DMSO-treated control cultures (defined as 100%). The IC50 values for APPwt and APPswe are 61 and 694 nm, respectively. B, an in vitro BACE1 assay was performed using 0.45 nm purified human BACE1, 4 μm substrate peptide (a greater than 100-fold excess of substrate), and the indicated concentrations of BSI OM99–2 or Inhibitor IV. The inhibition data are expressed relative to control reaction mixtures lacking BACE1 enzyme (defined as 100%). However, 0% inhibition is defined as that obtained for a control solution treated with DMSO (no inhibitor).
TABLE 1.
Summary of IC50 values for BSIs in an in vitro BACE1 assay under various conditions
| Parameter | IC50 ± S.D. |
IC50 ratio (swe:wt) | |
|---|---|---|---|
| Wt | Swe | ||
| nm | |||
| β-Secretase inhibitor compound | |||
| OM99–2 | 3.3 ± 0.3 | 2.4 ± 0.1 | 0.7 |
| Inhibitor IV | 12.4 ± 2.3 | 9.8 ± 0.9 | 0.8 |
| Reaction timea | |||
| 3 h | 3.5 ± 0.5 | 4.1 ± 1.0 | 1.2 |
| 6 h | 5.4 ± 1.9 | 8.2 ± 1.4 | 1.5 |
| 24 h | 6.0 ± 1.1 | 9.4 ± 0.5 | 1.5 |
| Substrate/enzyme ratioa | |||
| 200:1 | 19 ± 2 | 23 ± 6 | 1.2 |
| 50:4 | 21 ± 3 | 28 ± 6 | 1.3 |
| 25:8 | 24 ± 5 | 32 ± 6 | 1.4 |
| 16:16 | 26 ± 6 | 46 ± 10 | 1.7 |
| pHa | |||
| 4.6 | 5.6 ± 1.0 | 5.0 ± 1.1 | 0.9 |
| 6.2 | 5.6 ± 0.4 | 4.6 ± 1.7 | 0.8 |
a These experiments were performed using Inhibitor IV.
Enzyme-linked immunosorbent assays (ELISAs) were used to measure the products of BACE1 enzymatic cleavage. The reaction mixtures were appropriately diluted in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST) and 1% BSA and transferred to a detection plate coated with a monoclonal antibody specific for the N-terminal end generated by BACE1 cleavage (82E1; IBL Co., Ltd., Gunma, Japan). The plate was incubated overnight at 4 °C and then washed five times with TBST. Neutravidin-horseradish peroxidase (Thermo Scientific, Inc., Rockford, IL) was diluted 1:10,000 in sample dilution buffer, and 100 μl of this diluted solution was added to each well. The plate was incubated for 1 h at room temperature, washed five times with TBST, and developed using SuperSignal ELISA Pico chemiluminescent substrate (Thermo Scientific, Inc.). Luminescence counts were measured using an ARVO MX plate reader (PerkinElmer Life Sciences).
Cell-based Aβ Production Activity Assay
SH-SY5Y human neuroblastoma cells stably transfected with APP isoform 695 (APP695) were maintained in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum (4.5 g/liter) and 100 μg/ml hygromycin B. Cells transfected with wild type or Swedish mutant APP695 (APPwt or APPswe, respectively) were designated SHwt cells and SHswe cells, respectively. For inhibitor treatments, the cells were seeded in 96-well plates at a density of 8 × 105 cells/ml (150 μl of growth medium/well), incubated for 2 h, and then treated with 2 μl of inhibitor diluted in DMSO. The final DMSO concentration was 1%. The cells were then incubated for 24 h at 37 °C in a humidified 5% CO2 atmosphere. To measure secreted Aβ, the conditioned medium was transferred to a 96-well plate, which was stored at 4 °C until use.
For quantification of βCTF, treated cells were lysed in TBS containing Complete protease inhibitor mixture (Roche Applied Science) and 1% Triton X-100 for 2 h at 4 °C. The amount of βCTF in the lysate was determined using a βCTF ELISA.
Cell Surface Biotinylation
Five milliliters of SHwt or SHswe cells were seeded into 6-cm dishes (2 × 106 cells/dish). After 24 h, the cells were washed three times with Hanks' balanced salt solution (HBSS) and then biotinylated with Sulfo-NHS-LC-Biotin (0.5 mg/ml; Thermo Scientific, Inc.) in cold HBSS for 1 h at 4 °C. The cultures were washed with 100 mm glycine in cold HBSS and rinsed twice with cold HBSS. The cells were harvested in cold phosphate-buffered saline (PBS) and collected by centrifugation. Cell pellets were suspended in PBS containing 1% Nonidet P-40 and Complete protease inhibitor mixture (Roche Applied Science), sonicated, and centrifuged at 16,000 × g for 10 min. The protein concentrations of the supernatant fractions were determined using a BCA assay kit (Thermo Scientific, Inc.) and normalized to the controls.
The normalized SHwt and SHswe cell lysates were incubated with streptavidin beads (Thermo Scientific, Inc.) overnight. The biotinylated molecules were eluted by heating at 95 °C for 10 min in LDS sample buffer (Invitrogen). The eluates were analyzed by Western blotting using anti-APP antibody 6E10 (Covance, Princeton, NJ).
Immunofluorescence and Image Acquisition
SHwt or SHswe cells cultured on plastic discs in 24-well plates were rinsed with HBSS, incubated with antibody 6E10 (1:200) to label APP on the cell surface, and washed three times with cold HBSS. Either immediately thereafter or after a 15-min incubation at 37 °C to allow endocytosis to occur, the cells were fixed for 15 min at room temperature in 4% paraformaldehyde. The fixed cells were permeabilized, blocked with PBS containing 0.1% Triton X-100 and 3% normal goat serum for 15 min, and incubated with a rabbit polyclonal antibody specific for the C-terminal region of APP (AB5352; Millipore, Billerica, MA) overnight at 4 °C. The primary antibodies were labeled with secondary antibodies conjugated to Alexa Fluor 488 (for AB5352) or 594 (for 6E10) (Invitrogen). The images were acquired using an Eclipse FN1 microscope (Nikon, Tokyo, Japan) equipped with a 40× objective. Exposure time and gain remained constant for all of the images.
Cell-free β-Secretase Assay
SHwt and SHswe cell lysates were assayed for β-secretase in a cell-free system. First, SHwt and SHswe cells were cultured in 150-mm dishes, washed with PBS, suspended with trypsin-EDTA, diluted with growth medium, and centrifuged. The resulting cell pellets were washed twice with PBS and quickly frozen in liquid N2. Frozen cells were homogenized in a buffer containing 50 mm MES, pH 5.5, Complete protease inhibitor mixture (Roche Applied Science), the aspartic protease inhibitor pepstatin A (10 μm; Roche Applied Science), and the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester (10 μm; Calbiochem) using 30 strokes of a tight fitting Dounce homogenizer. The lysis buffer was detergent-free to avoid disruption of the conformation of the APP substrate and BACE1 enzyme.
The cell lysates were mixed in 96-well plates with reaction buffer containing various concentrations of Inhibitor IV and incubated for 1 h at 25 °C with shaking. The reactions were terminated by the addition of 1 m Tris-HCl, pH 7.6, containing 3% Triton X-100 and 50 μm Inhibitor IV. Solubilized βCTF was quantified using an ELISA.
Quantification of Aβ1–40, Aβ1-x, and βCTF
An Aβ1–40 homogenous time resolved fluoresence kit was purchased from Nihon Schering (Osaka, Japan). Briefly, antibody-EuK (55 ng/ml), antibody-XL665 (400 ng/ml), and phosphate buffer (50 mm; pH 7.4) containing 0.2% BSA and 0.5 m KF were added into each well of a 384-well plate. Samples of conditioned cell culture medium or synthetic peptide standards were added to yield a total assay volume of 20 μl/well. After mixing, the reaction mixture was incubated at 4 °C to reach equilibrium binding and then read on an ARVO multilabel counter (PerkinElmer Life Sciences).
An Aβ1-x ELISA was established using the commercially available antibodies 82E1 (IBL Co., Ltd.) and 4G8 (Covance). First, a Maxisorp plate (Nunc, Rochester, NY) was coated with 82E1 (0.5 μg/ml) in 50 mm Tris-HCl, pH 8, overnight at 4 °C and then blocked with TBST containing 0.5% BSA. Next, conditioned medium was appropriately diluted with sample dilution buffer (TBST containing 1% BSA), added to the 82E1-coated wells, and incubated overnight at 4 °C. After four washes, horseradish peroxidase-conjugated 4G8 (0.05 μg/ml in sample dilution buffer) was added, and the mixture was incubated for 1 h at room temperature. The peptides sandwiched with both 82E1 and 4G8 were quantified as luminescence counts (see “In Vitro BACE1 Activity Assay”). βCTF was quantified using a βCTF ELISA kit (IBL Co., Ltd.) according to the manufacturer's protocol.
Data Analysis
GraphPad Prism (GraphPad Software, Inc., San Diego, CA) was used to graph and analyze data. All of the titration curves were fitted to a sigmoidal dose-response equation to determine the IC50 values of the tested compounds.
RESULTS
BSIs Are Equally Potent Inhibitors of BACE1 Cleavage of APPwt and APPswe in Vitro
It has been widely observed that, in cells, BSIs are much less effective inhibitors of β-cleavage of APPswe than of β-cleavage of APPwt (22–24, 27). In the present study, we assessed the relative effectiveness of Inhibitor IV at reducing the production of Aβ in SH-SY5Y cells stably expressing APPwt versus APPswe. As shown in Fig. 1A, Inhibitor IV was about 10-fold less potent against β-cleavage of APPswe in SH-SY5Y cells than against the β-cleavage of APPwt reported previously.
The Swedish mutation dramatically accelerates β-cleavage of APP in both in vitro enzymatic assays and in cell-based assays. Therefore, we investigated whether it would cause a similar decrease in BSI potency in vitro using purified BACE1 and substrate peptides. The IC50 values of inhibitors of BACE1 activity are summarized in Table 1. The representative BACE1 inhibitors OM99–2 and Inhibitor IV were equally potent inhibitors of cleavage of the wild type and Swedish type substrates.
The APPswe-induced Decrease in BSI Potency Observed in Cells Does Not Occur in Vitro under Various Cell-like Conditions
The discrepancy between in vitro and cell-based assay data suggests that the decreased potency of BSI in cells expressing APPswe might result from differences between the two assays. Not only do the assays differ in reaction conditions, such as the enzyme: substrate ratio, reaction time, and pH, but they also differ in that βCTF accumulates in cells expressing APPswe and in the subcellular compartmentalization of the β-cleavage reaction.
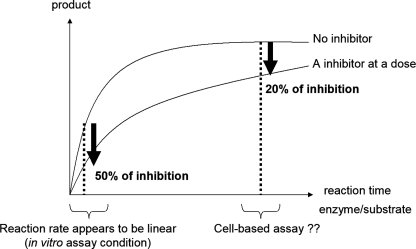
We first examined the influence of reaction conditions on the IC50 value of BSI. Generally, an enzymatic reaction proceeds linearly during the early phase and then reaches a plateau; the percentage of inhibition generally appears to be lower at the plateau than in the early phase (Fig. 2). Because cell-based assays usually use longer reaction times than those in vitro, the BACE1 cleavage reaction might be saturated under cell-based assay conditions. In addition, because the kcat/Km of APPswe is much higher than that of APPwt, the β-cleavage reaction may reach a plateau much earlier for APPswe than for APPwt. We speculated that in cells, the saturation of BACE1 cleavage of APPswe might decrease the potency of BSI. Therefore, we performed in vitro BACE1 activity assays for various lengths of time and compared the resulting IC50 values for the processing of the Swedish and wild type peptides. Unexpectedly, the ratio of IC50 values for APPswe and APPwt (IC50 swe:wt) did not increase with longer reaction times (Table 1). Even after 24 h, the IC50 value of swe:wt was much less than 10, and it was the same as that in the cell-based assay.
FIGURE 2.
Schematic model of the “saturation hypothesis.” In this hypothetical scheme, inhibitors are less powerful under product-saturated conditions arising from lengthy reaction times or an excessively high enzyme: substrate ratio.
Next, to examine the saturation hypothesis (see the schematic in Fig. 2), we incubated BACE1:substrate mixtures of various ratios for 24 h to bring the reactions closer to plateau. Even when BACE1 and its substrate were present in equal amounts, IC50 swe:wt was still much less than 10 (Table 1), which is inconsistent with the saturation hypothesis.
We next examined whether pH conditions might affect BSI potency. Although APP was previously thought to be cleaved by BACE1 in acidic cell compartments of pH 4.5, the optimal pH for BACE1, β-cleavage of APP occurs in early endosomes with a pH of ∼6 (30). We therefore examined whether the presence of a higher pH in the cellular compartments in which β-cleavage occurs might explain the decreased potency. We measured BACE1 activities at pH 4.5 and 6.2 and plotted the percentage of inhibition at each dose of Inhibitor IV (Table 1). Both the IC50 value and IC50 swe:wt remained unaffected by pH, indicating that the Swedish mutation-linked BSI potency decrease is not caused by a higher pH in the cellular compartment in which β-cleavage of APP occurs.
βCTF Accumulation Is Not Involved in the BSI Potency Decrease
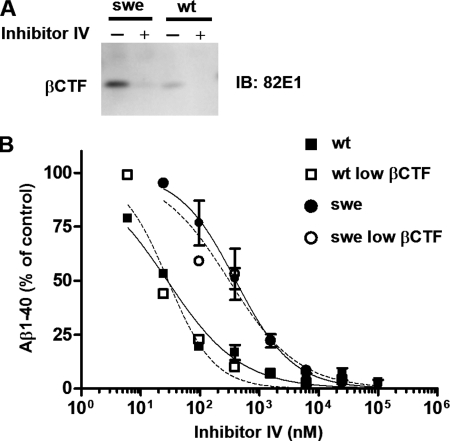
βCTF has been reported to accumulate robustly in cells stably expressing APPswe (22, 31), suggesting that the inhibition of βCTF production by BSI may not lower the amount of Aβ secreted from SHswe cells. Specifically, the βCTF that accumulates in SHswe cells can be cleaved by γ-secretase to release Aβ into the medium, which might then lead to the BSI potency decrease. In this case, inhibiting the accumulation of βCTF before assaying the BSI assay would abolish the apparent potency decrease. To examine this possibility, we performed cell-based assays with Inhibitor IV pretreatment to prevent the accumulation of βCTF. After 24 h of pretreatment (Fig. 3A), we exposed SHwt and SHswe cells to a range of Inhibitor IV concentrations and quantified the Aβ1–40 secreted into the media. As shown in Fig. 3, the IC50 values for SHswe cells were about 10 times higher than those for SHwt cells, regardless of pretreatment conditions, indicating that βCTF accumulation in SHswe cells is not the cause of the reduced potency of BSI.
FIGURE 3.
βCTF accumulation is not the cause of the Swedish mutation-linked reduction in BSI Aβ-reducing potency in APPswe-expressing SH-SY5Y cells. A, Western blot demonstrating robust accumulation of C-terminal fragments of β-cleaved APP (βCTF) in SH-SY5Y cells stably transfected with APPswe (SHswe cells) but not in SH-SY5Y cells stably expressing APPwt (SHwt cells). Pretreatment with Inhibitor IV (25 μm) for 24 h reduced the level of βCTF in pretreated SHswe cells to a level below that in SHwt cells pretreated with DMSO. B, SHwt and SHswe cells pretreated with Inhibitor IV or DMSO were treated again with Inhibitor IV at the indicated concentrations for 24 h, and the amount of Aβ1–40 in the conditioned medium was measured as described under “Experimental Procedures.” The data for Aβ1–40 levels in the culture medium are expressed relative to the data for medium from DMSO-treated control cultures (defined as 100%). The IC50 values with and without pretreatment were as follows: SHwt, 28 nm; pretreated SHwt (low βCTF), 29 nm; SHswe, 400 nm; pretreated SHswe (low βCTF), 311 nm. IB, immunoblot.
The Swedish Mutation Decreases BSI Potency against βCTF Production in a Cell-based Assay
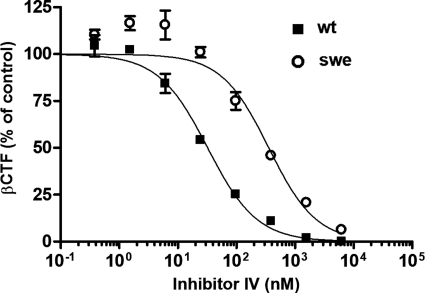
To confirm that the reduced effectiveness of BSIs against APPswe processing is independent of βCTF accumulation, we investigated whether Inhibitor IV equally prevents βCTF generation in SHwt and SHswe cells. βCTF was quantified using a βCTF ELISA kit after the cells were exposed to various Inhibitor IV concentrations for 24 h. As shown in Fig. 4, Inhibitor IV exhibited the usual decrease in inhibitory activity in these experiments. In addition, the IC50 values for βCTF production were comparable with those for Aβ production. These data suggest that no association exists between the accumulation of βCTF and the decreased effectiveness of BSI.
FIGURE 4.
BSI inhibits the generation of βCTF in SH-SY5Y cells stably expressing Swedish type APP (SHswe) less potently than in SH-SY5Y cells stably expressing wild type APP (SHwt). SHwt and SHswe cells were treated with Inhibitor IV at the indicated concentrations for 24 h and then lysed in Tris-buffered saline containing 1% Triton X-100. The amount of βCTF in the cell lysate was measured using a βCTF enzyme-linked immunosorbent assay kit (ELISA). The βCTF quantification data are expressed relative to those for the DMSO-treated control cultures (defined as 100%). The IC50 values in SHwt and SHswe cells are 32 and 356 nm, respectively.
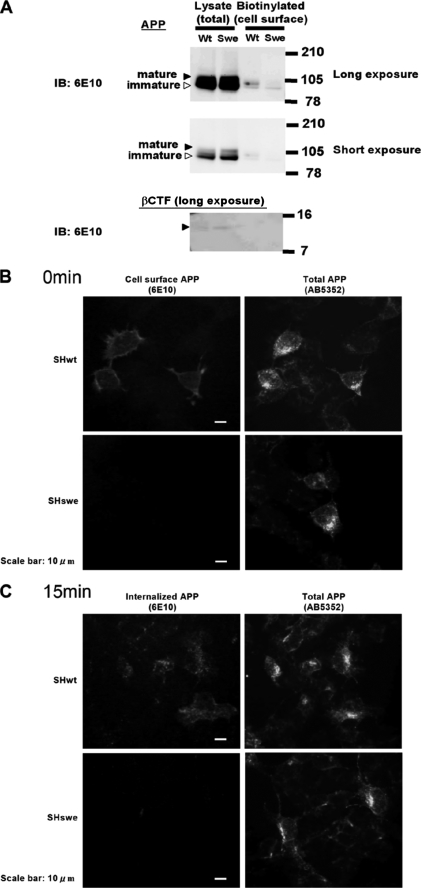
The Swedish Type APP Was Not Exposed to the Plasma Membrane, whereas Wild Type Was
The aberrant subcellular localization of APPswe processing by BACE1 has been reported by several groups (31, 34, 35, 40) with the specific consensus that although APPwt is cleaved by BACE1 in the early endosomes, the β-cleavage of APPswe occurs primarily within the secretory pathway. However, few studies have directly compared the localization of BACE1 processing for APPwt versus APPswe, particularly in human neuronal cells. Therefore, to confirm that the subcellular localization of APP processing by BACE1 is altered by the Swedish mutation, we compared the amounts of APPwt and APPswe reaching the plasma membrane without first undergoing β-cleavage in the secretory vesicles.
First, we labeled cell surface APP with biotin, precipitated the biotin-labeled APP using streptavidin beads, and quantified the biotin-labeled and total APP by Western blotting. As shown in Fig. 5A, the level of total APP in SHswe cells was similar to or higher than that in SHwt cells. In contrast, the level of biotin-labeled APP in SHswe cells was much lower than that in SHwt cells. In particular, mature, post-translationally modified APP was only minimally biotinylated in SHswe cells, suggesting that mature APPswe ready for processing by proteases is almost entirely cleaved by BACE1 before it appears on the cell surface.
FIGURE 5.
The Swedish type APP is not exposed to the plasma membrane, whereas wild type is. A, SHwt and SHswe cell surfaces were biotinylated for 1 h at 4 °C, conditions under which no endocytosis occurs. The biotinylated cell surface APP was precipitated with streptavidin-coated beads, and the precipitated and total APP were subjected to Western blot analysis with anti-APP antibody 6E10. Black arrowheads indicate mature APP; white arrowheads indicate immature APP. βCTF was barely detectable in the SHswe lysate lane. B, APP on the surface of SHwt and SHswe cells was labeled with antibody 6E10 for 45 min at 4 °C. After washing, the cells were immediately fixed with 4% paraformaldehyde. Scale bar, 10 μm. C, same as B, except that the washed cells were incubated for 15 min at 37 °C before fixing with 4% paraformaldehyde. Scale bar, 10 μm. IB, immunoblot.
Second, to demonstrate that most APPswe is neither exposed to the cell surface nor internalized by endocytosis, we conducted an immuno-uptake assay. APP on the SHwt and SHswe cell surfaces was labeled with the anti-APP antibody 6E10 and traced during its uptake by endocytosis. Although antibody 6E10 recognizes βCTF in addition to APP, the amount of βCTF was so much lower than that of APP (Fig. 5A) that we concluded that the observed 6E10 immunoreactivity was attributable to APP. As shown in Fig. 5B, the surfaces of SHwt cells, but not of SHswe cells, were substantially stained with 6E10. After a 15-min incubation at 37 °C to promote endocytosis, APP-bound 6E10 produced a fine granular staining pattern in SHwt cells but not in SHswe cells (Fig. 5C). In contrast, the anti-APP antibody AB5352 yielded comparable staining of total APP in SHwt and SHswe cells (Fig. 5, B and C, right panels). Taken together, these results indicate that APPswe does not reach the plasma membrane and is not endocytosed, unlike APPwt, suggesting that APPswe is mostly β-cleaved before it reaches the plasma membrane, whereas APPwt is β-cleaved after it reaches the plasma membrane.
BSI Equally Inhibits the Processing of APPwt and APPswe in a Cell-free Assay
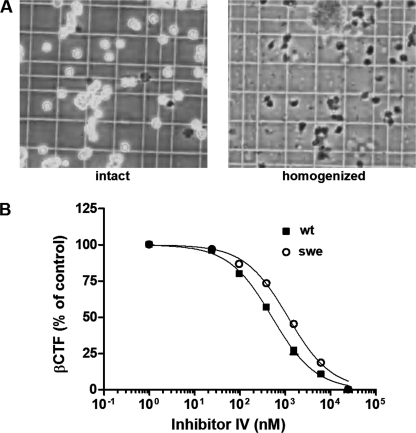
Finally, we evaluated whether the subcellular compartment in which APP processing occurs influences the potency of BSI. Although APPwt is predominantly cleaved in early endosomes by BACE1 (32), the processing of APPswe occurs within the secretory pathway (33). Therefore, we investigated whether the distinct subcellular localization of β-cleavage leads to differences in the APPwt and APPswe processing inhibition by BSI. In our cell-free assay, cellular compartments were thoroughly disrupted by homogenization, osmotic shock, and a freeze-thaw process (Fig. 6A). The resulting lysates were prepared in detergent-free lysis buffer to maintain protein conformations and protein-protein interactions. The SHwt and SHswe cell lysates were incubated in reaction buffer containing various concentrations of Inhibitor IV, and the percentage of inhibition by BSI relative to the DMSO control was plotted (Fig. 6B). Under these conditions, the IC50 values for APPswe and APPwt processing were comparable, suggesting that cellular partitioning is involved in the BSI potency decrease caused by the Swedish mutation.
FIGURE 6.
In a cell-free assay with disrupted subcellular compartmentalization, BSI inhibits the processing of APPswe as effectively as APPwt. A, intact or homogenized cells were suspended in phosphate-buffered saline containing 10% trypan blue and visualized under a microscope at 40× magnification. B, cells homogenized as in A were treated with Inhibitor IV at the indicated concentrations for 1 h at 25 °C, and the membranes were disrupted with 1% Triton X-100 to solubilize membrane-anchored βCTF. βCTF levels were determined using an ELISA; the βCTF quantification data are expressed relative to those for DMSO-treated control cell lysates (defined as 100%). The IC50 values for APPwt and APPswe are 514 and 1158 nm, respectively.
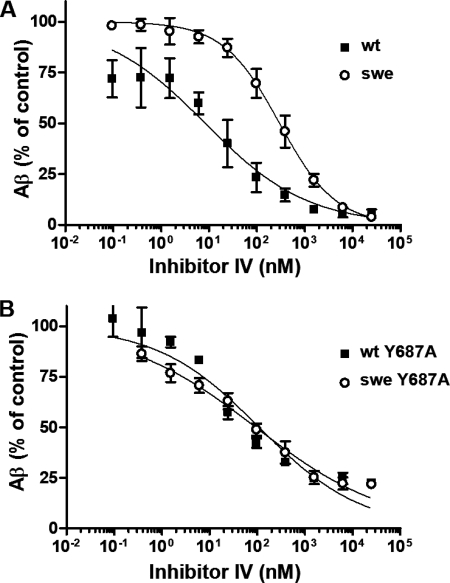
In a Cell-based Assay, the Y687A Mutation Suppresses the Negative Effect of the Swedish Mutation on the Potency of BSI
The results of our cell-free assay suggested that the inhibitory potency of BSI depends on the subcellular location of APP β-cleavage. We therefore expected that restricting the subcellular site of β-cleavage would abolish the influence of the Swedish mutation on BSI potency. The trafficking and metabolism of APP is known to be regulated by its C-terminal region, which has the amino acid sequence YENPTY (34–38). Phosphorylation of the tyrosine residue at position 687 in the C-terminal domain is essential for the localization of APP on the plasma membrane. In fact, the Y687A mutation of APP695 dramatically reduces the amount of APP on the cell surface (38). Furthermore, although the Y687A and wild type peptides are cleaved equally well by the purified α-secretase TACE in a cell-free assay, Y687A is poorly processed by cell surface α-secretase in a cell-based assay (38). Thus, full-length Y687A APP does not reach the plasma membrane and is strictly confined to the secretory vesicles even after its maturation in the Golgi apparatus. On the other hand, another study reported that the amount of secreted Aβ was unaffected by the Y687A mutation (36), indicating that, unlike the nonamyloidogenic α-secretase-dependent cleavage of APP, the amyloidogenic cleavage of APP by β- and γ-secretases could occur without reaching the cell surface.
Hence, to clarify the mechanism by which the site of β-cleavage influences BSI potency, we inserted the Y687A mutation into APPwt and APPswe and analyzed the effect of Inhibitor IV on Aβ production in SH-SY5Y cells stably expressing these mutated proteins (APPY687A and APPsweY687A, respectively). Aβ production curves were produced by plotting the relative amount of Aβ1-x at each dose of Inhibitor IV. As shown in Fig. 7B, the curves for APPsweY687A and APPY687A were nearly identical, whereas the fitted curve for APPswe was shifted to the right relative to that for APPwt (Fig. 7A). Thus, the ability of Inhibitor IV to prevent β-cleavage of Y687A-containing APP was independent of the β-cleavage site sequence, wild or Swedish type, supporting the hypothesis that the decreased effectiveness of BSI against APPswe cleavage is a result of differences in the subcellular sites of β-cleavage in APPwt- and APPswe-expressing cells.
FIGURE 7.
BSI inhibits Aβ production in SH-SY5Y cells expressing the Y687A mutants of APPswe (APPsweY687A) and APPwt (APPY687A) with equal potency. A, SHwt (■) and SHswe cells (○) were treated with Inhibitor IV at the indicated concentrations for 24 h, and the amount of Aβ in the conditioned medium was measured using an Aβ1-x ELISA. Aβ1-x quantification data are expressed relative to those for DMSO-treated control cultures (defined as 100%). The IC50 values for SHwt and SHswe are 19 and 298 nm, respectively. B, same as A, except that experiment was performed using SH-SY5Y cells stably expressing APPY687A (■) or APPsweY687A mutants (○). The IC50 values for APPY687A and APPsweY687A are 103 and 93 nm, respectively.
DISCUSSION
Inhibition of β-secretase activity is the most promising strategy for modifying the course of AD, and many companies have long been attempting to develop BSIs for this purpose. To develop medicines with sufficient clinical efficacy, the preclinical data must allow accurate prediction of the clinically effective dose. Therefore, it is important to determine which of the model systems (wild type or Tg2576 mice) more accurately reflects AD patients in terms of the Aβ-lowering effectiveness of BSI and to elucidate the mechanism by which the Swedish mutation weakens the inhibitory potency of BSI.
We first attempted to recreate the BSI potency-decreasing effect of the Swedish mutant in vitro using purified BACE1 and substrates but were unable to adequately mimic cellular conditions. We then redirected our focus to a search for cellular conditions that would abolish the potency- decreasing effect of the Swedish mutant, i.e. we created a cell-based assay that was closer to in vitro conditions. By examining the ability of BSIs to reduce Aβ secretion from cells with no βCTF accumulation and their ability to inhibit the generation of βCTF, we unambiguously determined that βCTF accumulation does not underlie the potency-decreasing effect of the Swedish mutant, despite previous assertions to the contrary (22). Data from perturbation and alteration of the subcellular APP processing site suggest that the BSI potency decrease is, instead, a result of the anomalous subcellular localization of APPswe β-cleavage.
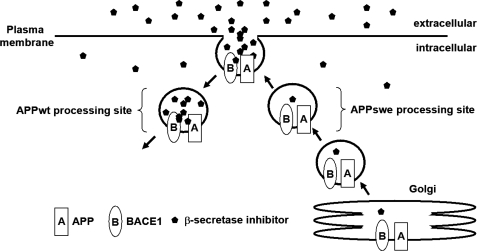
Why is the efficacy of BSI reduced by this change in the subcellular site of β-cleavage? Our findings give rise to two different speculations. First, the location of β-cleavage of APPswe might be generally difficult for chemicals to reach, probably because the place must be usually protected from exogenous enemies for cell survival. In fact, certain anti-cancer agents accumulate in certain organelles, thereby reducing their potency (39, 40), and Rajendran et al. (41) recently demonstrated that efficient targeting of BSI to the β-cleavage site can dramatically improve its inhibitory power against cellular Aβ production. These findings clearly indicate that drugs do not always spread into a cell uniformly and that their distribution patterns greatly influence their efficacies. Therefore, the notion that the BSI potency decrease is caused by a greatly decreased concentration of inhibitors at the subcellular site of APPswe β-cleavage is very reasonable. On the other hand, if this idea is indeed correct, then the potency decrease linked to the Swedish mutation would appear to be compound-specific rather than an example of a general phenomenon, considering that the distribution patterns of exogenous compounds are determined by particular physical characteristics. However, other groups have observed Swedish mutation-linked decreases in the potencies of several BSI series in cell-based assays (22, 24, 27). Moreover, we have confirmed that, in addition to Inhibitor IV, some compounds described in patents also exhibit the BSI potency decrease and that this decrease is abolished by the Y687A mutation (data not shown). These data suggest that most BSIs are distributed similarly in cells, probably because compounds with high affinities for the BACE1 active site share some physical properties.
A second, simpler explanation is that β-cleavage of APPswe occurs before it reaches the plasma membrane (31, 33), whereas APPwt is processed in an early endosome originating at the cell surface (32, 36). Both BACE1 and APP are transported from the Golgi apparatus to the plasma membrane and then to endosomes. The active site of BACE1 on the plasma membrane is exposed and more easily accessible to inhibitors than that of intracellular BACE1. BACE1 that cleaves APPwt is sometimes bound to BSI on the cell surface prior to APP processing, but the enzyme that processes APPswe is not. For either (or both) of the reasons described above (Fig. 8), the aberrant localization of APPswe processing may lower the potency of BSIs.
FIGURE 8.
Schematic diagram of the proposed model for increased BSI concentrations in early endosomes. BSIs added to the medium are taken up by endocytosis and diffuse into compartments at various rates depending on their physical properties (such as cell permeability). Compounds bound to or adjacent to BACE1 on the cell surface are efficiently taken up into early endosomes, so that BSIs may be at higher concentrations in early endosomes than in secretory vesicles.
This work is important for the accurate estimation of clinically effective doses of BSIs. According to our results, the Aβ-lowering potency of BSI in sporadic AD patients may be better modeled by the wild type mouse than by the Tg2576 mouse. Almost all AD patients express wild type APP, suggesting that β-cleavage takes place during endocytosis, as in wild type mice. However, we cannot rule out the possibility that the location of APP processing is aberrant in sporadic AD patients, although no direct evidence supports this hypothesis. It has been reported that the trafficking and metabolism of APP are affected by the phosphorylation of its C terminus and by its interaction with X11, Fe65, LRP1, and others (10, 34, 42–47). Moreover, phosphorylation of APP has been observed in postmortem human brains (48). However, the pathophysiological role of APP phosphorylation remains controversial (49). In the near future, clinical data for BSI efficacy in AD patients, in combination with the results of this study, will enable us to infer the precise subcellular site of APP processing.
Acknowledgments
We thank all of the members of the Pain & Neurology Laboratory, particularly Dr. A. Kato, Dr. G. Sakaguchi, Dr. H. Ito, K. Nishitomi, A. Mikami, I. Nanchi, and M. Hosono (Shionogi & Co., Ltd.) for helpful discussions and technical suggestions.
Footnotes
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- BSI
- β-secretase inhibitor
- βCTF
- C-terminal fragment of β-cleaved APP
- TBS
- Tris-buffered saline
- wt
- wild type
- Swe
- Swedish type
- SHwt
- SH-SY5Y stably expressing APPwt
- SHswe
- SH-SY5Y stably expressing APPswe
- Aβ
- amyloid β
- DMSO
- dimethyl sulfoxide
- BSA
- bovine serum albumin
- ELISA
- enzyme-linked immunosorbent assay
- HBSS
- Hanks' balanced salt solution
- PBS
- phosphate-buffered saline
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Selkoe D. J., Bell D. S., Podlisny M. B., Price D. L., Cork L. C. (1987) Science 235, 873–877 [DOI] [PubMed] [Google Scholar]
- 2.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 3.Klyubin I., Walsh D. M., Lemere C. A., Cullen W. K., Shankar G. M., Betts V., Spooner E. T., Jiang L., Anwyl R., Selkoe D. J., Rowan M. J. (2005) Nat. Med. 11, 556–561 [DOI] [PubMed] [Google Scholar]
- 4.Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L. B., Lindholm K., Yan R., Citron M., Xia W., Yang X. L., Beach T., Sue L., Wong P., Price D., Li R., Shen Y. (2003) Nat. Med. 9, 3–4 [DOI] [PubMed] [Google Scholar]
- 6.He W., Lu Y., Qahwash I., Hu X. Y., Chang A., Yan R. (2004) Nat. Med. 10, 959–965 [DOI] [PubMed] [Google Scholar]
- 7.Dodson S. E., Gearing M., Lippa C. F., Montine T. J., Levey A. I., Lah J. J. (2006) J. Neuropathol. Exp. Neurol. 65, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada H., Tamaoka A., Ishii K., Shoji S., Kametaka S., Kametani F., Saito Y., Murayama S. (2006) Neurosci. Res. 54, 24–29 [DOI] [PubMed] [Google Scholar]
- 9.Majercak J., Ray W. J., Espeseth A., Simon A., Shi X. P., Wolffe C., Getty K., Marine S., Stec E., Ferrer M., Strulovici B., Bartz S., Gates A., Xu M., Huang Q., Ma L., Shughrue P., Burchard J., Colussi D., Pietrak B., Kahana J., Beher D., Rosahl T., Shearman M., Hazuda D., Sachs A. B., Koblan K. S., Seabrook G. R., Stone D. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17967–17972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Q. L., Galasko D. R., Ringman J. M., Vinters H. V., Edland S. D., Pomakian J., Ubeda O. J., Rosario E. R., Teter B., Frautschy S. A., Cole G. M. (2009) Arch Neurol. 66, 448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhiyou C., Yong Y., Shanquan S., Jun Z., Liangguo H., Ling Y., Jieying L. (2009) Neurochem. Res. 34, 1226–1235 [DOI] [PubMed] [Google Scholar]
- 12.Nishitomi K., Sakaguchi G., Horikoshi Y., Gray A. J., Maeda M., Hirata-Fukae C., Becker A. G., Hosono M., Sakaguchi I., Minami S. S., Nakajima Y., Li H. F., Takeyama C., Kihara T., Ota A., Wong P. C., Aisen P. S., Kato A., Kinoshita N., Matsuoka Y. (2006) J. Neurochem. 99, 1555–1563 [DOI] [PubMed] [Google Scholar]
- 13.Meredith J. E., Jr., Thompson L. A., Toyn J. H., Marcin L., Barten D. M., Marcinkeviciene J., Kopcho L., Kim Y., Lin A., Guss V., Burton C., Iben L., Polson C., Cantone J., Ford M., Drexler D., Fiedler T., Lentz K. A., Grace J. E., Jr., Kolb J., Corsa J., Pierdomenico M., Jones K., Olson R. E., Macor J. E., Albright C. F. (2008) J. Pharmacol. Exp. Ther. 326, 502–513 [DOI] [PubMed] [Google Scholar]
- 14.Sankaranarayanan S., Price E. A., Wu G., Crouthamel M. C., Shi X. P., Tugusheva K., Tyler K. X., Kahana J., Ellis J., Jin L., Steele T., Stachel S., Coburn C., Simon A. J. (2008) J. Pharmacol. Exp. Ther. 324, 957–969 [DOI] [PubMed] [Google Scholar]
- 15.Sankaranarayanan S., Holahan M. A., Colussi D., Crouthamel M. C., Devanarayan V., Ellis J., Espeseth A., Gates A. T., Graham S. L., Gregro A. R., Hazuda D., Hochman J. H., Holloway K., Jin L., Kahana J., Lai M. T., Lineberger J., McGaughey G., Moore K. P., Nantermet P., Pietrak B., Price E. A., Rajapakse H., Stauffer S., Steinbeiser M. A., Seabrook G., Selnick H. G., Shi X. P., Stanton M. G., Swestock J., Tugusheva K., Tyler K. X., Vacca J. P., Wong J., Wu G., Xu M., Cook J. J., Simon A. J. (2009) J. Pharmacol. Exp. Ther. 328, 131–140 [DOI] [PubMed] [Google Scholar]
- 16.Asai M., Hattori C., Iwata N., Saido T. C., Sasagawa N., Szabó B., Hashimoto Y., Maruyama K., Tanuma S., Kiso Y., Ishiura S. (2006) J. Neurochem. 96, 533–540 [DOI] [PubMed] [Google Scholar]
- 17.Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlogue L., John V. (1999) Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 18.Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 19.Ohno M., Sametsky E. A., Younkin L. H., Oakley H., Younkin S. G., Citron M., Vassar R., Disterhoft J. F. (2004) Neuron 41, 27–33 [DOI] [PubMed] [Google Scholar]
- 20.McConlogue L., Buttini M., Anderson J. P., Brigham E. F., Chen K. S., Freedman S. B., Games D., Johnson-Wood K., Lee M., Zeller M., Liu W., Motter R., Sinha S. (2007) J. Biol. Chem. 282, 26326–26334 [DOI] [PubMed] [Google Scholar]
- 21.Ohno M., Cole S. L., Yasvoina M., Zhao J., Citron M., Berry R., Disterhoft J. F., Vassar R. (2007) Neurobiol. Dis. 26, 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riddell D., Atchison K., Yu D., Turner J., Wang E., Gonzales C., Warwick H., Kim J., Malamas M., Wagner E., Aschmies S., Albinson K., Erdel J., Gunawan I., Revilla-Sanchez R., Takano K., Warren N., Fang K., Bard J., Hirst W., Pangalos M., Moss S., Haydon P., Robichaud A., Reinhart P. (2009) in International Conference on Alzheimer's Disease, Vienna, Austria: on July 15, 2009 [Google Scholar]
- 23.Shitaka Y., Yarimizu J., Nagakura A., Mitani Y., TAkami S., Miyake A., Hamakawa N., Hey J., Koelsch G., Bilcer G., Matsuoka N. (2008) in International Conference on Alzheimer's Disease, Chicago, IL: on July 28, 2008 [Google Scholar]
- 24.Malamas M., Erdei J., Gunawan I., Robichaud A., Turner J., Hu Y., Wagner E., Aschmies S., Comery T., Fan K. (2008) in International Conference on Alzheimer's Disease, Chicago, IL: on July 28, 2008, 4, 515–515 [Google Scholar]
- 25.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Science 274, 99–102 [DOI] [PubMed] [Google Scholar]
- 26.Citron M., Oltersdorf T., Haass C., McConlogue L., Hung A. Y., Seubert P., Vigo-Pelfrey C., Lieberburg I., Selkoe D. J. (1992) Nature 360, 672–674 [DOI] [PubMed] [Google Scholar]
- 27.Hussain I., Hawkins J., Harrison D., Hille C., Wayne G., Cutler L., Buck T., Walter D., Demont E., Howes C., Naylor A., Jeffrey P., Gonzalez M. I., Dingwall C., Michel A., Redshaw S., Davis J. B. (2007) J. Neurochem. 100, 802–809 [DOI] [PubMed] [Google Scholar]
- 28.Hong L., Koelsch G., Lin X., Wu S., Terzyan S., Ghosh A. K., Zhang X. C., Tang J. (2000) Science 290, 150–153 [DOI] [PubMed] [Google Scholar]
- 29.Pietrak B. L., Crouthamel M. C., Tugusheva K., Lineberger J. E., Xu M., DiMuzio J. M., Steele T., Espeseth A. S., Stachel S. J., Coburn C. A., Graham S. L., Vacca J. P., Shi X. P., Simon A. J., Hazuda D. J., Lai M. T. (2005) Anal. Biochem. 342, 144–151 [DOI] [PubMed] [Google Scholar]
- 30.Stachel S. J., Coburn C. A., Rush D., Jones K. L., Zhu H., Rajapakse H., Graham S. L., Simon A., Katharine Holloway M., Allison T. J., Munshi S. K., Espeseth A. S., Zuck P., Colussi D., Wolfe A., Pietrak B. L., Lai M. T., Vacca J. P. (2009) Bioorg Med. Chem. Lett. 19, 2977–2980 [DOI] [PubMed] [Google Scholar]
- 31.Thinakaran G., Teplow D. B., Siman R., Greenberg B., Sisodia S. S. (1996) J. Biol. Chem. 271, 9390–9397 [DOI] [PubMed] [Google Scholar]
- 32.Koo E. H., Squazzo S. L. (1994) J. Biol. Chem. 269, 17386–17389 [PubMed] [Google Scholar]
- 33.Haass C., Lemere C. A., Capell A., Citron M., Seubert P., Schenk D., Lannfelt L., Selkoe D. J. (1995) Nat. Med. 1, 1291–1296 [DOI] [PubMed] [Google Scholar]
- 34.King G. D., Perez R. G., Steinhilb M. L., Gaut J. R., Turner R. S. (2003) Neuroscience 120, 143–154 [DOI] [PubMed] [Google Scholar]
- 35.Ono Y., Kinouchi T., Sorimachi H., Ishiura S., Suzuki K. (1997) J. Biochem. 121, 585–590 [DOI] [PubMed] [Google Scholar]
- 36.Perez R. G., Soriano S., Hayes J. D., Ostaszewski B., Xia W., Selkoe D. J., Chen X., Stokin G. B., Koo E. H. (1999) J. Biol. Chem. 274, 18851–18856 [DOI] [PubMed] [Google Scholar]
- 37.Steinhilb M. L., Turner R. S., Gaut J. R. (2002) J. Neurochem. 80, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K., Niidome T., Akaike A., Kihara T., Sugimoto H. (2008) Biochem. Biophys. Res. Commun. 377, 544–549 [DOI] [PubMed] [Google Scholar]
- 39.Lee C. M., Tannock I. F. (2006) Br J. Cancer 94, 863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon S. M., Schindler M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3497–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajendran L., Schneider A., Schlechtingen G., Weidlich S., Ries J., Braxmeier T., Schwille P., Schulz J. B., Schroeder C., Simons M., Jennings G., Knölker H. J., Simons K. (2008) Science 320, 520–523 [DOI] [PubMed] [Google Scholar]
- 42.Yoon I. S., Pietrzik C. U., Kang D. E., Koo E. H. (2005) J. Biol. Chem. 280, 20140–20147 [DOI] [PubMed] [Google Scholar]
- 43.Cam J. A., Bu G. (2006) Mol. Neurodegener. 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spoelgen R., von Arnim C. A., Thomas A. V., Peltan I. D., Koker M., Deng A., Irizarry M. C., Andersen O. M., Willnow T. E., Hyman B. T. (2006) J. Neurosci. 26, 418–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt V., Sporbert A., Rohe M., Reimer T., Rehm A., Andersen O. M., Willnow T. E. (2007) J. Biol. Chem. 282, 32956–32964 [DOI] [PubMed] [Google Scholar]
- 46.Marzolo M. P., Bu G. (2009) Semin Cell Dev. Biol. 20, 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borg J. P., Ooi J., Levy E., Margolis B. (1996) Mol. Cell. Biol. 16, 6229–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee M. S., Kao S. C., Lemere C. A., Xia W., Tseng H. C., Zhou Y., Neve R., Ahlijanian M. K., Tsai L. H. (2003) J. Cell Biol. 163, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sano Y., Nakaya T., Pedrini S., Takeda S., Iijima-Ando K., Iijima K., Mathews P. M., Itohara S., Gandy S., Suzuki T. (2006) PLoS One 1, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]