Abstract
The early growth response gene product Egr-1 has been shown to have great impact on growth, proliferation, and differentiation in a wide variety of cells, including T cells. In this study, we show that Egr-1 is rapidly induced upon T cell stimulation and is expressed predominantly in T helper type 2 (Th2) compared with type 1 (Th1) cells. We further investigate the role of Egr-1 in regulation of the Th2 cytokine interleukin-4 (IL-4) expression. IL-4 is a key Th2 cytokine that regulates humoral immunity and also causes allergic inflammation. Regulation of IL-4 gene transcription in Th2 cells has been shown to be controlled by multiple T cell receptor (TCR)-induced transcription factors. However, only a few transcription factors were shown to be selectively induced in differentiated Th2 cells in response to TCR stimulation. Chromatin immunoprecipitation analysis demonstrates that Egr-1 binds to the IL-4 promoter in vivo upon T cell stimulation. Ectopic expression of Egr-1 enhances endogenous IL-4 mRNA expression and elevates IL-4 promoter activity. We also show that Egr-1, nuclear factor of activated T cell, and NF-κB cooperatively bind to an NFAT/NF-κB-overlapping IL-4 enhancer element and activate the IL-4 promoter synergistically. Furthermore, we show that antisense oligonucleotides that knock down Egr-1 expression attenuate IL-4 transcription. Our study provides the first evidence that Egr-1 protein is differentially expressed in Th1 and Th2 cells and is involved in the acute phase of the IL-4 transcription in response to TCR stimulation.
Keywords: Cell/Blood, Cytokines, Cytokines/Interleukins, Gene/Regulation, Immunology, RNA, Transcription, Transcription/General Factors
Introduction
Interleukin (IL)2-4 plays a pivotal role in the differentiation of T helper type 2 (Th2) cells that secrete IL-4, IL-5, and IL-13 and in the development of humoral immunity (1–3). IL-4 also plays a central role in the pathogenesis of allergic inflammatory diseases (4, 5). Expression of the IL-4 gene by T cells has been documented to occur at two distinct steps: an initial step of differentiation of naïve CD4 T cells into effector Th2 cells and the acute induction of the IL-4 gene expression in differentiated Th2 cells (6–9).
To date, seven transcription factors, STAT6, GATA-3, RBPJκ, c-Maf, NFAT, IRF4, and the AP-1 family protein JunB, have been implicated in Th2-specific regulation of IL-4 transcription (6, 8, 10–14). Among them, only a few transcription factors, such as JunB (but not the other Jun family members), were shown to be selectively activated in Th2 cells during differentiation by T cell receptor (TCR) engagement (11). The NFAT families of transcription factors, which encompass five evolutionary related proteins, play an important role in expression of many cytokine genes (15). Mature T cells express predominantly NFATp and NFATc, and both have been shown to activate the IL-4 gene in response to TCR stimulation (16, 17). Although NFATp and NFATc are expressed in both Th1 and Th2 cells, NFATp was shown to bind to the IL-4 enhancer and the IL-4 promoter only in stimulated Th2 cells, whereas the same transcription factor binds to the interferon (IFN)-γ promoter only in stimulated Th1 cells (12). The molecular mechanisms for the cell type-restricted binding of NFATp are still obscure.
Previously, a comparison study of expression profiles of Th1 and Th2 mRNA libraries reviewed that the early growth response protein (Egr)-1 mRNA was overexpressed in Th2 cells (18). Egr-1 is a zinc finger transcription factor discovered independently by several laboratories searching for genes essential for growth, proliferation, or differentiation (19–23). To date, four closely related Egr proteins, Egr-1, Egr-2, Egr-3, and Egr-4, have been identified (24). All four Egr proteins recognize the consensus sequence GCG(G/C/T)GGGCG but bind to distinct target sequences with different binding affinities (25, 26). Many environmental signals, including growth factors, mitogens, hormones, and neurotransmitters, induce Egr-1 expression (27). In T cells, expression of Egr-1, Egr-2, and Egr-3 can be induced through TCR stimulation (28). In contrast to Egr-1, expressions of Egr-2 and Egr-3 are dependent on NFAT activation, and therefore, their expression is considered to be a secondary response to T cell activation (28–30). The importance of Egr-1 in T cell biology has been documented by its role during T cell development in the thymus (30–32). Egr-1-deficient mice show defects in positive selection resulting in a reduced percentage of CD4+ and CD8+ single-positive mature T cells in the thymus (33). In contrast, Egr-1 overexpression in the thymus allowed positive selection of thymocytes (31). Egr-1 has also been shown to control survival of mature thymocytes and newly emigrated thymocytes (34). The survival role of Egr-1 in thymocyte development can be explained by its function during activation of the T cell survival cytokine IL-2 and its receptor (35–37). In contrast, Egr-2 and Egr-3 were shown to contribute to a negative regulation of T cell activation and to be involved in T cell anergy (28).
The observation that Egr-1 mRNA is expressed preferentially in Th2 cells prompted us to investigate further whether the Egr-1 protein is expressed preferentially in Th2 cells and, if so, what role Egr-1 plays in the regulation of Th2 cytokine gene expression. In this study, we show that the Egr-1 protein is induced rapidly upon TCR stimulation and is expressed predominantly in Th2 cells during differentiation. We further demonstrate that Egr-1 activates IL-4 promoter activity in vitro and binds to the human IL-4 promoter in vivo. Finally, we identify an Egr-1-binding site within the human IL-4 promoter and show that Egr-1 cooperates with NFAT and NF-κB to activate human IL-4 transcription.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
The cell lines used in this study were the human T cell leukemia cell line Jurkat, the mouse Th1 clones Cl29 and B10BI, and the mouse Th2 clones D10G4.1 and L1/1. Jurkat T cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum, 50 μg/ml gentamicin (Invitrogen), or 100 units/ml penicillin and 100 units/ml streptomycin, 6 mm HEPES (1 m solution; Invitrogen), and 2 mm l-glutamine (200 mm solution; Invitrogen) at 37 °C in a humidified atmosphere containing 5% CO2. Th1 and Th2 clones were cultured as above supplemented with 2 units/ml recombinant human IL-1, 25 units/ml recombinant human IL-2.
Purification of Primary Human and Mouse T Lymphocytes
Human primary T cells were prepared from peripheral blood of healthy donors. The mononuclear cells were isolated over Ficoll-Hypaque (Biochrom, Berlin, Germany), washed three times with phosphate-buffered saline, and incubated in Dulbecco's modified Eagle's medium for 1 h in plastic tissue culture dishes at 37 °C in a 5% CO2 incubator. Nonadherent lymphocytes cells were collected from the plastic dishes. T cells were purified from the mixed lymphocyte population up to 90% purity by rosetting with 2-amino-ethylisothyo-uronium-bromide-treated sheep red blood cells. Naïve CD4+CD62L+ T cells were isolated from mouse spleen and purified by CD4CD62L MicroBeads (MACS Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions.
In Vitro Th1/Th2 Differentiation
In vitro differentiation of mouse Th1/Th2 cells was carried out by the established method (38). Briefly, purified CD4+CD62L+ naïve cells (1 × 106/ml) were cultured on plates precoated with α-CD3 (1 μg/ml) and in the presence of soluble α-CD28 (5 μg/ml). Th1 conditions were established by using IL-12 (3.4 ng/ml), IL-2 (20 units/ml), and α-IL-4 antibody (2 μg/ml) (BD Transduction Laboratories). Th2 conditions were established by using IL-4 (3000 units/ml), IL-2 (20 units/ml), and α-IFN-γ antibody (2 μg/ml) (BD Transduction Laboratories). 48 h after starting the culture, cells were replated to fresh medium containing the above polarizing cytokines and anti-cytokine antibodies plus IL-2 (5 units/ml). Cells were cultured further for another 2 days and then washed and restimulated (1 × 106/ml) with plate-bound α-CD3 for the indicated times.
Western Blot Analysis
1 × 107 cells were sedimented and lysed for 20 min in ice-cold lysis buffer (50 mm Tris-HCl, pH 8.0, 120 mm NaCl, 1% Nonidet P-40, 0.5% sodium desoxycholate, 0.1% SDS, 2 mm EDTA, 25 mm NaF, 0.2 mm NaVO4, 1 mm dithiothreitol, and the complete protease inhibitor mixture from Roche Applied Science). After removing the cell debris by centrifugation at 13,000 rpm for 30 min, equal amounts of proteins were separated by SDS-PAGE, blotted onto a nitrocellulose membrane (Amersham Biosciences), and blocked with 5% bovine serum albumin in 0.05% Tween 20/phosphate-buffered saline. The following antibodies were used: α-Egr-1, α-Egr-3, α-YY-1, α-p65 (rabbit, sc-109), α-T-bet mAb 4B10 (Santa Cruz Biotechnology), α-Egr-2 (Lifespan), α-NFATc (7A6) (Alexis Biochemicals), α-NFATp (Dianova, Hamburg), and α-tubulin (Sigma). For stripping, blots were incubated for 30 min in a buffer containing 62.5 mm Tris-HCl, pH 6.8, 2% SDS, and 100 mm β-mercaptoethanol at 56 °C. The blots were washed six times for 10 min in phosphate-buffered saline/Tween and blocked again in 5% bovine serum albumin.
Reverse Transcription-PCR and Quantitative Real Time PCR
RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer's instructions. 1 μg of total RNA was reverse-transcribed using the PerkinElmer GeneAmp RNA PCR kit. Aliquots of cDNA were PCR-amplified in a 50-μl reaction buffer with 2.5 units of Taq polymerase containing 10 mm human IL-4 primer pair (456-bp PCR product; Stratagene) and 2.5 μm human β-actin primer pair (661-bp PCR product; Stratagene). 35 cycles were carried out, each consisting of the following conditions: 94 °C 1 min, 56 °C 1 min, and 72 °C 1 min for human IL-4 and 20 cycles for β-actin. Amplification products were separated by electrophoresis on 1.2% agarose gels. The sequences of primers and fluorescent-labeled probes for TaqMan quantitative real time PCR are as follows. Human IL-4 primers were: forward, 5′-ACA GCC TCA CAG AGC AGA AGA CT-3′; reverse, 5′-TGT GTT CTT GGA GGC AGC AA-3′; and probe, 5′-TGT GCA CCG AGT TGA CCG TAA CAG ACA T-3′. Human β-actin primers were: forward, 5′-ACC CAC ACT GTG CCC ATC TAC GA-3′; reverse, 5′-CAG CGG AAC CGC TCA TTG CCA ATG G-3′; and probe, 5′-ATG CCC TCC CCC ATG CCA TCC TGC GT-3′. PCR was performed in a 12.5-μl reaction mixture (PCR kit from Eurogentech, Belgium) that contained 0.08 μg of reverse-transcribed cDNA, 7.5 pm forward primers, 22.5 pm reverse primers, and 5 pm probe. For each sample, three PCRs were performed. The resulting relative increase in reporter fluorescent dye emission was monitored by the TaqMan system (GeneAmp 5700 sequence detection system and software; PerkinElmer). The level of IL-4 mRNA, relative to β-actin, was calculated using the formula: Relative mRNA expression = 2−(Ct of IL-4 − Ct of β-actin), where Ct is the respective threshold cycle value.
Chromatin Immunoprecipitation (ChIP) Assays
PMA- (5 ng/ml) and ionomycin- (0.5 μm) stimulated Jurkat T cells (2 × 107) or α-CD3- (coated with 30 μg/ml) and α-CD28- (5 μg/ml)-stimulated human peripheral blood T cells (1 × 107) were mixed with formaldehyde (final concentration, 1%). The cells were lysed and sonificated. Protein concentrations of the final products were measured by the Bio-Rad assay. Equal amounts of proteins (100 μg) were taken for ChIPs performed with 2 μg of α-Egr-1, α-p65, α-NFATc, or with α-rabbit IgG or α-mouse IgG as controls for 2 h at 4 °C. Following deproteination and reversal of cross-links as described previously (39), the presence of selected DNA sequences was assessed by PCR. PCR primers specific for the human IL-4 promoter −336 to −40 were (−336)-5′-CAACAAATTCGGACACCTG-3′ and (−40)-5′-GCTGAAACCGAGGGAAAAT-3′ (yielding a 296-bp product); the −112 primer was (−112)-5′-TCAGCACCTCTCTTCCAGGAG-3′; the primers for −644 to −434 were (′644)-5′-CCTACCTTGCCAAGGGCTTC-3′ and (−434)-5′-GAAACTGTCCTGTCATGGAAAAGAT-3′.
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear extracts were prepared from nonstimulated and stimulated cells as described previously (40). The human IL-4 enhancer element probe P1 and the EMSA protocol were described previously (41). Briefly, the DNA-binding reactions between protein and 32P-labeled probe were performed on ice for 30 min. DNA·protein complexes were separated by electrophoresis on native 5% polyacrylamide gels in 0.5 ×TBS buffer (1 × TBS buffer contains 89 mm Tris, pH 8.2, 89 mm boric acid, and 2 mm EDTA). For supershifting or inhibition assays, antibodies were preincubated with nuclear extracts on ice for 20 min prior to the addition of the labeled probe.
Plasmid Constructs
The luciferase reporter plasmid pLuc-IL4 containing bp −269/+11 of the human IL-4 promoter, and the luciferase constructs containing four copies of either the DNA sequence of human IL-4 P1 or the NFAT-binding sequence in the human IL-2 promoter were constructed previously (39). The Egr-1 expression vector (pCB6-Egr-1) containing the rat Egr-1 was kindly supplied by J. Milbrandt (Washington University, St. Louis) (42). The p65 (NF-κB) expression plasmid (pCMV-p65) was kindly supplied by L. Schmitz (University of Giessen, Germany) (43). The human NFATc expression vector pSH107c was kindly supplied by G. R. Crabtree (Stanford University, Stanford, CA) (44). The NFATp expression vector was kindly supplied by A. Rao (Harvard Medical School, Boston) (45).
DNA Transfection
4 × 106/ml Jurkat T cells in RPMI 1640 medium, 8 μg of the reporter plasmid, and 2–6 μg of each indicated expression plasmid were electroporated using a Bio-Rad Gene Pulser set at 960 microfarads, 240 V. The transfected cells were allowed to recover overnight and were stimulated with PMA (5 ng/ml) and ionomycin (0.5 μm) for 8 h. Equal transfection efficiency was controlled by co-transfection with 0.5 μg of pCMVEGFPspectrin and assessed by fluorescence-activated cell sorter. Luciferase activity was determined in 10 μl of cell extract using the luciferase assay substrate (Promega) with a Duolumat LB9507 luminometer (Berthold, Germany).
Egr-1 Antisense Oligonucleotide Knockdown Experiments
To inhibit the expression of endogenous Egr-1, we used an antisense approach. The sequences of the antisense were described previously (46), and the control scrambled oligonucleotides were ggaatctcattcgatgcatac. The antisense (10 μg) and the control scrambled oligonuleotides (10 μg) were transfected into Jurkat T cells using the Amaxa T cell NucleofectorTM kit V (Amaxa) according to the manufacturer's instructions. The transfection efficiency was more than 80% as controlled by transfection with an expression vector containing the green fluorescent protein (pCMVEGFPspectrin) and assessed by fluorescence-activated cell sorter. After transfection (overnight), cells were treated with PMA (5 ng/ml) and ionomycin (0.5 μm) for the indicated times. Cells were then harvested for further analysis.
RESULTS
Egr-1 Is Preferentially Expressed in Activated Th2 Cells
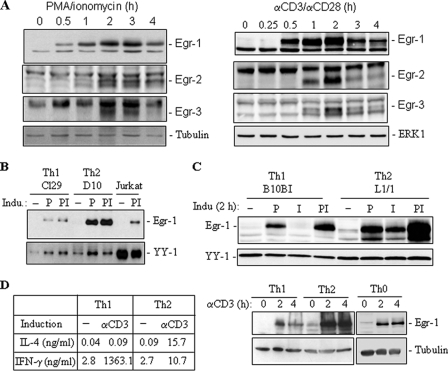
To gain an overall view of the Egr-1 expression pattern in T cells, we carried out a kinetic analysis of Jurkat T cells stimulated through their TCR by α-CD3 (in the presence of α-CD28) or by PMA/ionomycin to mimic TCR stimulation. Total cell lysates were prepared and subjected to Western blot analysis. Consistent with an earlier observation (28), Egr-1 protein was rapidly induced upon T cell stimulation with a peak at 2 h (Fig. 1A). In contrast, Egr-2 and Egr-3 whose expressions depend on NFAT activation (28–30), were induced much slower and expressed during a shorter time period than Egr-1 (Fig. 1A). Because Egr-1 mRNA was observed to be expressed preferentially in differentiated Th2 cells (18), we wondered whether this published Egr-1 mRNA expression pattern correlates with Egr-1 protein expression levels. Thus, we first compared two pairs of murine Th1/Th2 (Cl29/D10G4.1 and B10BI/AuA4) cell lines for their Egr-1 protein expression profiles. The Th1 and Th2 cell lines Cl29 and D10G4.1 were stimulated with PMA alone or with PMA plus ionomycin for 2 h, and the lysates were analyzed by Western blotting. Egr-1 was not detectable in the nonstimulated cells and was expressed upon T cell stimulation (Fig. 1B). Consistent with the mRNA expression data (18), Egr-1 protein was shown to be expressed predominantly in the Th2 cell line D10G4.1 compared with the Th1 cell line Cl29. To investigate signals required for the preferential expression of Egr-1 in Th2 cells further, the Th1 and Th2 cell lines B10BI and L1/1 were stimulated with PMA and ionomycin alone or in a combination of PMA and ionomycin for 2 h. Similar to Cl29 and D10G4.1, Egr-1was not detectable in the nonstimulated cells and was expressed upon T cell stimulation by PMA (Fig. 1C). Interestingly, ionomycin alone could not induce Egr-1 expression in the Th1 B10BI cells, but could in the Th2 L1/1 cells. Significant increase in Egr-1 expression was observed in the Th2 L1/1 cells when stimulated with PMA and ionomycin together (Fig. 1C). A slightly enhanced Egr-1 expression was also observed in Th1 B10BI cells but at a much smaller level compared with Th2 L1/1 cells.
FIGURE 1.
Egr-1 is expressed rapidly upon T cell activation and is expressed preferentially in Th2 cells. A, Egr-1 is induced rapidly upon T cell stimulation. Jurkat T cells were stimulated with either α-CD3/α-CD28 or PMA/ionomycin for the indicated times. Cell lysates were subjected to Western blot analysis with antibodies against Egr-1, Egr-2, and Egr-3. B–D, Egr-1 is expressed preferentially in Th2 cells. B, murine Th1 clone Cl29 and Th2 clone D10G4.1 cells were stimulated with PMA alone (P) or PMA plus ionomycin (PI) for 2 h. Nuclear extracts were analyzed by Western blotting for Egr-1 expression. Jurkat T cells were used as a positive control. The constitutive transcription factor YY-1 was used for control of equal loading. C, murine Th1 clone B10BI and L1/1 cells were stimulated with PMA or ionomycin alone or in combination for 2 h. Cell lysates were subjected to Western blot analysis. D, naïve CD4+ T cells (Th0) were differentiated under either Th1 or Th2 culture conditions. The differentiated cells were restimulated with α-CD3 for the indicated times. The cytokine expression profiles of differentiated cells were analyzed in supernatants by enzyme-linked immunosorbent assay. Total cell lysates were analyzed for Egr-1 expression by Western blotting. Tubulin was used to control equal loading. All results are representative of at least two independent experiments.
To confirm the above observations further, freshly isolated naïve murine CD4+CD62L+ T cells were subjected to differentiation in vitro under either Th1 or Th2 culture conditions. The differentiation status was controlled in supernatants of restimulated cells by enzyme-linked immunosorbent assay for the expression profiles of the Th1 cytokine IFN-γ and the Th2 cytokine IL-4 (Fig. 1D, left panel). Examination of the freshly differentiated Th1 and Th2 cells confirmed that Egr-1 protein was preferentially expressed in Th2 cells (Fig. 1D, right, lower panels).
Egr-1 Binds to the IL-4 Promoter in Vivo and Activates IL-4 Transcription
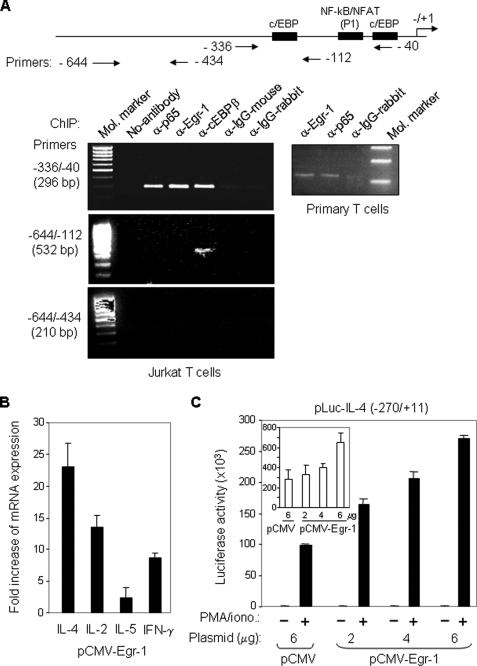
Because Egr-1 is expressed predominantly in Th2 cells, we asked whether Egr-1 promotes expression of the Th2 cytokine IL-4. We first examined whether Egr-1 interacts with the IL-4 promoter in vivo. Jurkat and freshly isolated human peripheral blood T cells were stimulated with PMA/ionomycin or α-CD3/α-CD28 for 3 h and 4 h, respectively. The activated cells were subjected to a ChIP analysis. PCR primers specific for the human IL-4 promoter (−336 to −40) were designed. The experiments showed that the 296-bp IL-4 promoter PCR product was produced in DNA precipitated by antibodies against Egr-1 in activated Jurkat and peripheral blood T cells (Fig. 2A). In contrast, use of the IgG control antibodies did not result in the generation of the 296-bp IL-4 promoter product. Because NF-κB and C/EBP are known IL-4 promoter-binding factors, we also included antibodies against C/EBP and the NF-κB subunit p65 in the ChIP analysis in parallel. The binding intensity of Egr-1 to the IL-4 promoter was comparable with that of NF-κB (p65) and C/EBP (Fig. 2A). The specific binding of Egr-1 to the P1 region was confirmed further by the analysis using primers from −644 to −112 and −644 to −434. Only a single 532-bp PCR product was detected in the α-C/EBP immunoprecipitate by the primers −644 to −112, and no PCR products were detected by the primers −644 to −434 in any of the immunoprecipitates. This experiment demonstrates that Egr-1 interacts with the human IL-4 promoter in vivo.
FIGURE 2.
Egr-1 binds to the IL-4 promoter in vivo and activates IL-4 transcription. A, Egr-1 interacts with the human IL-4 promoter in vivo. Jurkat (left) and freshly isolated human peripheral blood T cells (right) were stimulated with PMA (10 ng/ml)/ionomycin (1 μm) or α-CD3 (30 μg/ml)/α-CD28 (5 μg/ml), respectively. After 3 h (Jurkat) and 4 h (primary T cells) of stimulation, cells were subjected to ChIP analysis with the indicated antibodies. A map of the IL-4 promoter showing the location of the primers used for ChIP is also presented. B, ectopic expression of Egr-1 enhances endogenous cytokine expression. Jurkat T cells were transfected with an Egr-1 expression plasmid (pCMV-Egr-1) or with the empty plasmid (pCMV). After overnight recovery of the cells, RNA was prepared, and the expression levels of IL-4, IL-2, IL-5, and IFN-γ were determined by quantitative real time PCR. The results are presented as fold increase in mRNA levels of Egr-1-transfected cells versus empty vector-transfected cells. Results are representative of two independent experiments assayed in triplicate (error bars denote the S.D.). C, ectopic expression of Egr-1 enhances IL-4 promoter activity. The human IL-4 promoter-luciferase reporter construct was co-transfected with either the Egr-1-containing (2–6 μg) or the empty (6 μg) expression vector into Jurkat T cells. After overnight recovery, the transfected cells were split and stimulated with PMA/ionomycin (PMA/iono.) or left unstimulated for 8 h. The promoter activities are given as luciferase activity. Inset, promoter activities in unstimulated conditions are shown magnified. One representative of two independent experiments is shown.
We next investigated whether Egr-1 activates IL-4 transcription. Jurkat T cells were transiently transfected with an Egr-1 expression plasmid or the empty control plasmid. Jurkat T cells are known to produce both Th1 and Th2 cytokines, including IL-2, IFN-γ, IL-4, and IL-5 (35, 41, 47–49). The effects of Egr-1 on mRNA expression of these cytokines were analyzed by real time PCR. The experiment showed that ectopic expression of Egr-1 in Jurkat T cells led to an ∼23-fold increase in endogenous IL-4 mRNA expression (Fig. 2B). Egr-1 was previously shown to activate the IL-2 promoter (35). In support of this report, ectopic expression of Egr-1 also resulted in an increase in IL-2 mRNA expression by ∼12-fold. Egr-1 showed less of an effect on IL-5 expression, where only a 2.5-fold increase in the IL-5 mRNA expression was seen. The mRNA expression level of IFN-γ was also increased upon overexpression of Egr-1 but at a substantially smaller level than the IL-4 mRNA (Fig. 2B). These data indicate that Egr-1 may promote IL-4 transcription. To investigate further the role of Egr-1 in regulation of IL-4 transcription, a luciferase reporter plasmid containing the human IL-4 promoter was co-transfected with different amounts of the Egr-1 expression plasmid into Jurkat T cells. Overexpression of Egr-1 was shown to enhance the transcriptional activity of the IL-4 promoter in a dose-dependent manner (Fig. 2C). These experiments demonstrate that Egr-1 can promote IL-4 expression at the transcriptional level.
Egr-1, NFAT, and NF-κB Co-bind to the IL-4 Promoter Enhancer Element P1
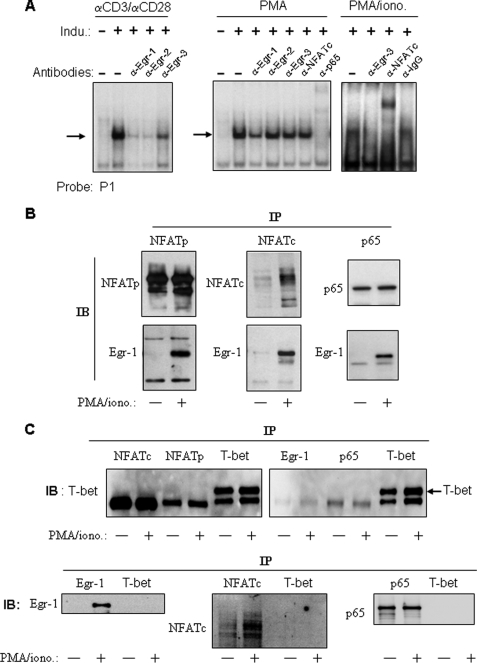
The human IL-4 promoter contains five TCR-inducible regulatory elements that coordinately regulate the acute IL-4 transcription in response to T cell stimulation (9). Three of these, P0, P1 and P4, have been implicated in Th2 specific protein/DNA interactions (40, 50–52). Therefore, we carried out EMSAs to examine whether any of these elements are binding sites for Egr-1. In stimulated cells, we detected an interaction of the P1 element with Egr proteins (Fig. 3A). In the unstimulated (resting) stage, no protein bindings could be detected by the P1 probe (Fig. 3A). However, inducible binding appeared after stimulation with α-CD3·α-CD28, PMA, or PMA plus ionomycin. Formation of the α-CD3·αCD28 inducible complex was inhibited by antibodies against all three Egr (Egr-1, Egr-2, and Egr-3) proteins, demonstrating that Egr proteins were involved in the formation of the P1·protein complex (Fig. 3A, left). Egr-1 expression can be induced by PMA alone (Fig. 1B), whereas expression of Egr-2 and Egr-3 depends on NFAT activation, which requires calcium signals (28–30). In accordance, the complex induced by PMA alone was only diminished by the antibody against Egr-1 but not by antibodies against Egr-2 and Egr-3 (Fig. 3A, center). Binding of Egr-3 to the P1 probe occurred after stimulation of the cells with PMA plus ionomycin (which provides a calcium signal) as shown by inhibition with the antibody against Egr-3 (Fig. 3A, right). The complex formed by the P1 probe was also supershifted by antibodies against NFAT and p65 (Fig. 3A, center and right), indicating that the complex also contained NFAT and NF-κB. Again, supershifting of NFAT was only seen in PMA/ionomycin-stimulated cells but not in cells stimulated with PMA alone.
FIGURE 3.
Egr-1 binds to the IL-4 P1 enhancer element and interacts with NFAT and NF-κB. A, Egr-1 binds to the human IL-4 P1 enhancer element upon stimulation with PMA or α-CD3/α-CD28. The 32P-labeled IL-4 P1 probe was incubated with nuclear extracts (10 μg) prepared from Jurkat T cells nonstimulated (−) or stimulated (+) for 2 h with α-CD3 (30 μg/ml)/α-CD28 (5 μg/ml), PMA (10 ng/ml), or PMA/ionomycin (1 μm) (PMA/iono.) in the absence or presence of the indicated antibodies. After 30 min of incubation, the samples were subjected to EMSA. The inducible complexes are indicated by arrows. Results are representative of two independent experiments. B, Egr-1 co-immunoprecipitates with NFAT and NF-κB. Total lysates from Jurkat cells either nonstimulated (−) or stimulated (+) with PMA/ionomycin for 2 h were immunoprecipitated (IP) with antibodies against NFATp, NFATc, or NF-κB subunit p65 and then immunoblotted (IB) with α-Egr-1 antibodies or the indicated control antibodies. Results are representative of three independent experiments. C, unrelated antibody, α-T-bet, was used as control and showed that NFAT, p65, and Egr-1 did not co-precipitate with T-bet.
Because three inducible transcription factors (Egr-1, NFAT, and NF-κB) were found in the P1 complex, we assumed that these proteins might physically interact with each other. Previous studies using a glutathione S-transferase “pulldown” technique have shown that Egr-1 forms heterodimers with NFAT and NF-κB (53, 54). To confirm the physical interactions among Egr-1, NFAT, and NF-κB p65, we carried out an immunoprecipitation experiment followed by Western blot analysis. The experiment confirmed that Egr-1 co-precipitates with NFATp, NFATc, and p65. Co-precipitation occurred only in stimulated but not in unstimulated cells (Fig. 3B). As a control, T-bet does not co-precipitate with Egr-1, NFAT, and p65; and Egr-1, NFAT, and p65 do not co-precipitate with T-bet (Fig. 3C). Altogether, these experiments demonstrate that Egr-1 can interact with NFAT and NF-κB to co-bind to the P1 enhancer element. This result is confirmed by the ChIP analysis presented in Fig. 2A which showed binding of Egr-1 only to an area of the IL-4 promoter containing the P1 element.
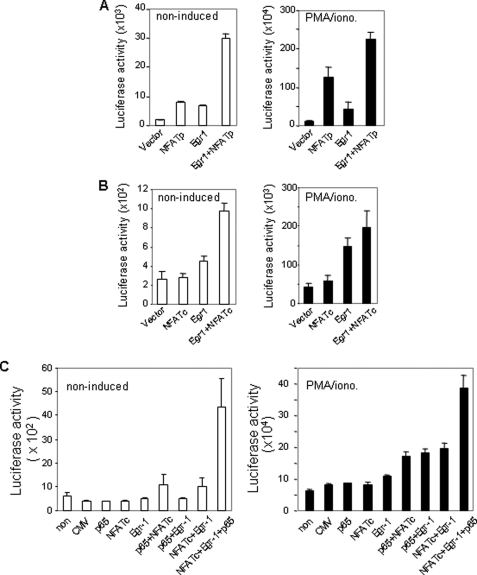
Egr-1, NFAT, and NF-κB Synergistically Activate the IL-4 P1 Element
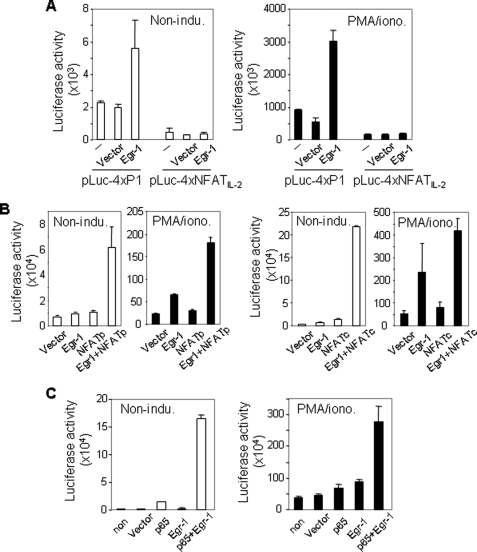
To confirm that the P1 element is a relevant target of Egr-1, a luciferase reporter plasmid containing four copies of the IL-4 P1 element was co-transfected with the Egr-1 expression plasmid into Jurkat T cells. As a negative control, a luciferase reporter plasmid containing four copies of the NFAT-binding element of the human IL-2 promoter was co-transfected in parallel. Overexpression of Egr-1 was shown to enhance transcriptional activity of the P1 element in both noninduced and PMA/ionomycin-induced Jurkat T cells significantly (Fig. 4A). In contrast, no elevation of the transcriptional activity was seen in cells co-tranfected with the IL-2 NFAT reporter plasmid (Fig. 4A). Because Egr-1, NFAT, and NF-κB co-bound to the P1 element, we asked whether these transcription factors could cooperate with each other to activate the P1 element. To answer this question, the P1-luciferase reporter plasmid was co-tranfected with the NFATp, NFATc, and Egr-1 expression plasmid either alone or in combination into Jurkat T cells. To optimize the condition, the expression plasmids were titrated to a level that alone had only little effect on P1 activity. These experiments showed that Egr-1 synergized with NFAT to activate P1-mediated transcription (Fig. 4B). The synergistic effect was also observed by a combination of Egr-1 with NF-κB (Fig. 4C). A less pronounced synergy was seen in cells stimulated with PMA/ionomycin, which may be because PMA/ionomycin stimulation increased the endogenous Egr-1, NFAT, and NF-κB levels, which may alter the optimal combination.
FIGURE 4.
Egr-1 enhances the transcriptional activity mediated by the P1 enhancer element. A, Egr-1 enhances the transcriptional activity of the IL-4 P1 but not the IL-2 NFAT-binding element. Luciferase (Luc) reporter constructs containing four copies of either the human IL-4 P1 or the human IL-2 NFAT-binding element were co-transfected with the Egr-1 (4 μg) or the empty (4 μg) expression vector into Jurkat T cells. After overnight recovery, cells were split and either left unstimulated or stimulated with PMA (10 ng/ml)/ionomycin (1 μm) (PMA/iono.) for 8 h, followed by determining luciferase activity. B, Egr-1 cooperates with NFAT to enhance the transcriptional activity of the P1 element synergistically. The P1 luciferase reporter construct was co-transfected with Egr-1 (1 μg), NFATp (1 μg), NFATc (1 μg), or empty expression vector (1 μg) alone or in combinations into Jurkat T cells. After overnight recovery, cells were split and stimulated as in A. C, Egr-1 cooperates with NF-κB to enhance the transcriptional activity of the P1 element synergistically. The P1 luciferase reporter construct was co-transfected with Egr-1 (1 μg), p65 (1 μg), or empty expression vector alone or in combination into Jurkat T cells. After overnight recovery, cells were split and stimulated as in B. Results are representative of two (A) or three (B and C) independent experiments, each with triplicate transfections (error bars indicate S.D.).
Egr-1, NF-AT, and NF-κB Cooperatively Activate the IL-4 Promoter
To investigate further the effect of Egr-1 on IL-4 transcription, the IL-4 promoter-luciferase reporter plasmid described in Fig. 2C (which contains the P1 element) was co-transfected with the Egr-1, NFAT, and NF-κB expression plasmids alone or in combinations. Co-transfection of the IL-4 promoter construct at a suboptimal dose of the NFAT or Egr-1 expression plasmid alone showed only a slightly increased IL-4 promoter activity. However, combinations of Egr-1 with NFATp (Fig. 5A) or NFATc (Fig. 5B) significantly enhanced expression of the reporter gene. Further enhanced IL-4 promoter activity was seen after combined ectopic expression of Egr-1, NFAT, and NF-κB (p65) together in Jurkat T cells (Fig. 5C). Again, high synergy was noted especially in the absence of stimulation. Less synergy in stimulated cells is most likely due to induction of the endogenous respective transcription factors leading to less optimal conditions for the demonstration of synergy. These experiments demonstrate that Egr-1 cooperates with NFAT and NF-κB to activate IL-4 transcription.
FIGURE 5.
Egr-1 cooperates with NFAT and NF-κB to activate the transcriptional activity of the IL-4 promoter. A and B, Egr-1 cooperates with NFAT to enhance the transcriptional activity of the IL-4 promoter. The human IL-4 luciferase reporter construct was co-transfected with either Egr-1 (2 μg), NFATp (2 μg), NFATc (2 μg), or the empty expression vector (2 μg) alone or in combination into Jurkat T cells. After overnight recovery, the cells were treated as described for Fig. 4. C, Egr-1 cooperates with NFAT and NF-κB to enhance the transcriptional activity of the IL-4 promoter. The human IL-4 luciferase reporter construct was co-transfected with 1 μg of each expression plasmid alone or in combination as indicated. After overnight recovery, cells were split and then stimulated as in A and B. All results are representative of three independent experiments, each with triplicate transfections (error bars indicate S.D.).
Egr-1 Antisense Oligonucleotides Down-regulate IL-4 mRNA Expression
Activation of IL-4 transcription is regulated by multiple inducible transcription factors (9). To determine the role of Egr-1 in activation of the IL-4 transcription, we carried out an antisense approach to knock down endogenous Egr-1. Antisense Egr-1 oligodeoxynucleotides were shown to inhibit inducible expression of Egr-1 protein completely (Fig. 6A). Knockdown of Egr-1 resulted in down-regulation of the IL-4 mRNA expression by ∼78% assessed by quantitative real time PCR (Fig. 6B). The experiment demonstrates that Egr-1 is involved in optimal activation of IL-4 transcription.
FIGURE 6.
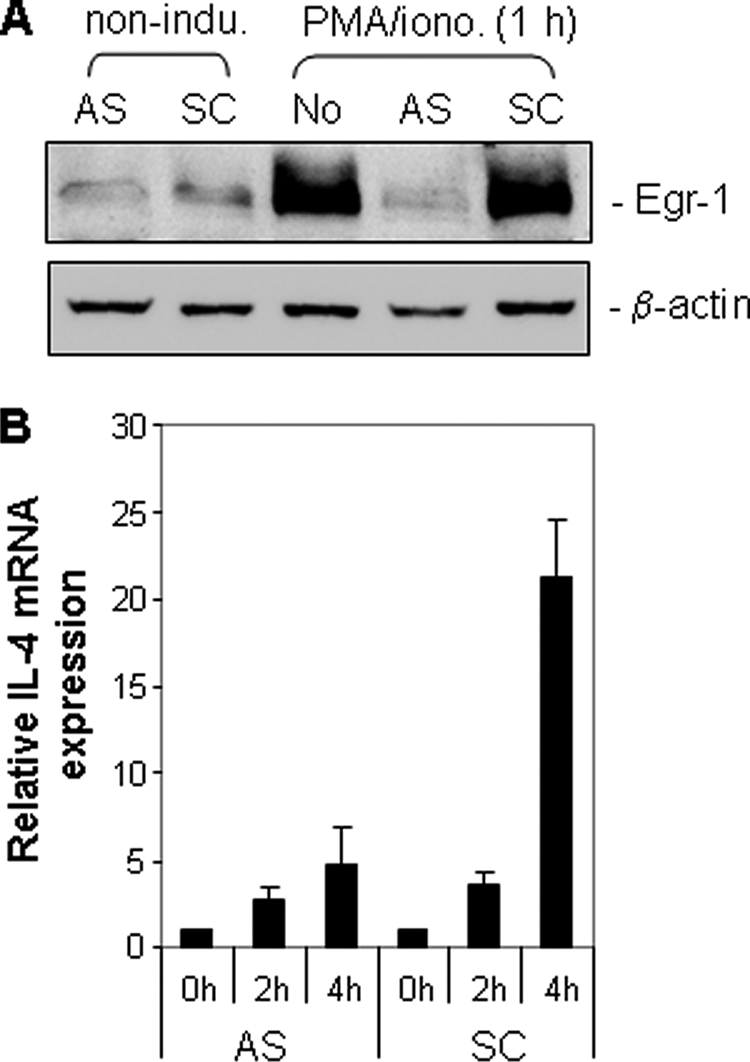
Egr-1 antisense oligonucleotides down-regulate endogenous IL-4 mRNA expression. A, Egr-1 antisense oligonucleotides down-regulate Egr-1 expression. Jurkat T cells were transfected with either Egr-1 antisense (AS) or control scrambled (CS) oligonucleotides as described under “Experimental Procedures.” After overnight recovery, cells were split and either untreated or treated with PMA (5 ng/ml) and isonomycin (0.5 μm) (PMA/iono.) for 1 h. The expression levels of Egr-1 were checked by Western blotting. B, Egr-1 antisense oligonucleotides down-regulate endogenous IL-4 mRNA expression. The endogenous IL-4 mRNA expression level of the antisense or the control scrambled oligonucleotide transfected cells was assessed after 2- and 4-h stimulation with PMA and ionomycin by quantitative real time PCR. Results are representative of two independent experiments with triplicate assays (error bars indicate S.D.).
DISCUSSION
Egr-1, known as an early growth response gene, has been shown to have great impact on growth, proliferation, and differentiation in a wide variety of cells (19–23). In T cells, Egr-1 can be induced by TCR engagement (30–37). Consistent with the previous studies, we show that Egr-1 is induced rapidly upon T cell stimulation. However, Egr-1 is expressed at a significantly higher level in differentiated Th2 compared with Th1 cells (Fig. 1). Interestingly, we found that in Th2 cells, Egr-1 expression can be induced by a calcium signal and that the calcium signal synergizes with the PMA-induced signal to enhance Egr-1 expression (Fig. 1C). In contrast, Egr-1 expression could not be induced by a calcium signal alone in Th1 cells. Thus, the preferential expression of Egr-1 in Th2 cells may involve a calcium-mediated mechanism.
Th1 and Th2 cells are characterized by expression of distinct sets of cytokines, typically IFN-γ for Th1 and IL-4 for Th2 cells. Predominant expression of Egr-1 in Th2 cells indicates that Egr-1 may be a driving force for early expression of some Th2 genes. Indeed, we found that Egr-1 interacts with the IL-4 promoter in vivo (Fig. 2A) and activates IL-4 gene expression at the transcriptional level. This is demonstrated by the facts that (i) ectopic expression of Egr-1 enhances endogenous expression of IL-4 mRNA and activates IL-4 promoter activity in vitro (Fig. 2B and 2C) and (ii) antisense RNA against Egr-1 down-regulates IL-4 transcription (Fig. 6).
In this study, we further characterized the potential binding site for Egr-1 in the human IL-4 promoter. The human IL-4 promoter contains five inducible regulatory elements (9) of which P1 is one of the most important elements and binds to NFAT and NF-κB upon TCR stimulation (41, 51, 55). Herein, we show that the P1 element is also a target site of Egr-1 (Fig. 3A). The P1 element was shown previously to bind nuclear proteins preferentially in activated Th2 but not in activated Th1 cells (51). Therefore, this site was proposed to be a Th2-specific protein·DNA-interacting site (51). However, NFAT and NF-κB (p50/p65) are not Th2-specific. Nevertheless, NFATp, which accounts for 80–90% of the total level of NFAT proteins, was shown to bind to the IL-4 promoter only in stimulated Th2 cells, whereas the same transcription factor binds to the IFN-γ promoter only in stimulated Th1 cells (12, 51). Because NFATp is expressed in both Th1 and Th2 cells, it is difficult to interpret the Th2-specific bindings of transcription factors on the P1 element. One explanation would be that Egr-1 forms heterodimers with NFAT and NF-κB to bind to the P1 site. This possibility is supported by our co-immunoprecipitation experiment (Fig. 3B) and also by the results of other investigators which used a glutathione S-transferase pulldown technique to demonstrate interactions of Egr-1, NF-κB, and NFAT with each other to form heterodimers (53, 54).
Besides the P1 element, the P4 and P0 element were also shown to interact differentially with nuclear proteins in Th1 and Th2 cells (40, 52). However, we did not detect bindings of the P4 and P0 element to the Egr-1 protein, although both elements are binding sites for NF-AT. Apparently, formation of the Egr-1/NFAT heterodimer alone is not sufficient to bring Egr-1 to the NFAT-binding site as also demonstrated in our experiment showing that the IL-2 NFAT element did not respond to the Egr-1-mediated transcriptional activation (Fig. 4A). The binding site for NFAT usually contains an AP1-binding sequence. Because our experiments showed that the IL-2 NFAT binding element, which contains an AP-1 binding sequence, did not respond to Egr-1, AP1 is unlikely to play a role in supporting binding of Egr-1 to the NFAT site. The IL-4 P1 NFAT-binding motif differs from the IL-2 NFAT-binding motif in that the IL-4 P1 NFAT-binding sequence also interacts with NF-κB (41). Thus, NFAT and NF-κB together may support binding of Egr-1 to the P1 region of the IL-4 promoter. Because JunB, which has been shown to be selectively induced in Th2 cell in response to TCR stimulation (11), was detected in the inducible P0 DNA/protein complex (40), JunB may account for the observed Th2-specific interaction with the P0 element.
The IL-4 gene, like many inducible genes, is under the control of a large array of cis-acting and enhancer elements, which act together to achieve a fine degree of control over the transcriptional activity of the gene (9). To date, seven transcription factors, STAT6, GATA-3, RBPJk, c-Maf, NFAT, IRF4, and JunB, have been shown to participate in Th2-specific regulation of IL-4 transcription (6, 8, 10–14). Among them, only JunB was shown to be selectively activated in Th2 cells after TCR stimulation (11). In this study, we show that Egr-1 is also preferentially expressed in Th2 cells in response to T cell stimulation. We show that Egr-1 forms a DNA-binding complex with NFAT and NF-κB on the IL-4 P1 site and cooperates with NFAT and NF-κB to enhance transcription of the IL-4 gene. Silencing Egr-1 expression resulted in 78% reduction of IL-4 transcription, indicating that Egr-1 may play a significant role in transcriptional activation of the IL-4 gene in Th2 polarized cells.
Taken together, we have shown that the early growth response gene product Egr-1 is expressed preferentially in Th2 cells and is involved in transcriptional activation of the key Th2 cytokine gene IL-4. Because Egr-1 is expressed predominantly in Th2 cells, we assume that Egr-1 may play an important role in the effector function of Th2 cells.
This work was supported by the Spitzencluster Initiative with Apogenix BioRN-INA-10 (to M. L.-W.) and by Deutsche Forschungsgemeinschaft Grant SFB TA22 (to M. L.).
- IL
- interleukin
- Th1 and Th2
- T helper types 1 and 2, respectively
- TCR
- T cell receptor
- IFN
- interferon
- Egr
- early growth response
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- PMA
- phorbol 12-myristate 13-acetate
- NFAT
- nuclear factor of activated T cell
- T-bet
- T box transcription factor
- C/EBP
- CCAAT/enhancer-binding proteins.
REFERENCES
- 1.Seder R. A., Paul W. E. (1994) Annu. Rev. Immunol. 12, 635–673 [DOI] [PubMed] [Google Scholar]
- 2.Brown M. A., Hural J. (1997) Crit. Rev. Immunol. 17, 1–32 [DOI] [PubMed] [Google Scholar]
- 3.Jankovic D., Sher A., Yap G. (2001) Curr. Opin. Immunol. 13, 403–409 [DOI] [PubMed] [Google Scholar]
- 4.Tepper R. I., Levinson D. A., Stanger B. Z., Campos-Torres J., Abbas A. K., Leder P. (1990) Cell 62, 457–467 [DOI] [PubMed] [Google Scholar]
- 5.Ricci M., Matucci A., Rossi O. (1997) J. Investig. Allergol. Clin. Immunol. 7, 144–150 [PubMed] [Google Scholar]
- 6.Agarwal S., Rao A. (1998) Immunity 9, 765–775 [DOI] [PubMed] [Google Scholar]
- 7.Murphy K. M., Reiner S. L. (2002) Nat. Rev. Immunol. 2, 933–944 [DOI] [PubMed] [Google Scholar]
- 8.Amsen D., Blander J. M., Lee G. R., Tanigaki K., Honjo T., Flavell R. A. (2004) Cell 117, 515–526 [DOI] [PubMed] [Google Scholar]
- 9.Li-Weber M., Krammer P. H. (2003) Nat. Rev. Immunol. 3, 534–543 [DOI] [PubMed] [Google Scholar]
- 10.Zheng W., Flavell R. A. (1997) Cell 89, 587–596 [DOI] [PubMed] [Google Scholar]
- 11.Li B., Tournier C., Davis R. J., Flavell R. A. (1999) EMBO J. 18, 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwal S., Avni O., Rao A. (2000) Immunity 12, 643–652 [DOI] [PubMed] [Google Scholar]
- 13.Lohoff M., Mittrücker H. W., Prechtl S., Bischof S., Sommer F., Kock S., Ferrick D. A., Duncan G. S., Gessner A., Mak T. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11808–11812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rengarajan J., Mowen K. A., McBride K. D., Smith E. D., Singh H., Glimcher L. H. (2002) J. Exp. Med. 195, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabtree G. R., Olson E. N. (2002) Cell 109, S67–S79 [DOI] [PubMed] [Google Scholar]
- 16.Monticelli S., Rao A. (2002) Eur. J. Immunol. 32, 2971–2978 [DOI] [PubMed] [Google Scholar]
- 17.Peng S. L., Gerth A. J., Ranger A. M., Glimcher L. H. (2001) Immunity 14, 13–20 [DOI] [PubMed] [Google Scholar]
- 18.Zelenika D., Adams E., Humm S., Graca L., Thompson S., Cobbold S. P., Waldmann H. (2002) J. Immunol. 168, 1069–1079 [DOI] [PubMed] [Google Scholar]
- 19.Milbrandt J. (1987) Science 238, 797–799 [DOI] [PubMed] [Google Scholar]
- 20.Lim R. W., Varnum B. C., Herschman H. R. (1987) Oncogene 1, 263–270 [PubMed] [Google Scholar]
- 21.Sukhatme V. P., Cao X. M., Chang L. C., Tsai-Morris C. H., Stamenkovich D., Ferreira P. C., Cohen D. R., Edwards S. A., Shows T. B., Curran T., LeBeau M. M., Adamson E. D. (1988) Cell 53, 37–43 [DOI] [PubMed] [Google Scholar]
- 22.Christy B. A., Lau L. F., Nathans D. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 7857–7861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemaire P., Revelant O., Bravo R., Charnay P. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4691–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckmann A. M., Wilce P. A. (1997) Neurochem. Int. 31, 477–510; discussion 517–526 [DOI] [PubMed] [Google Scholar]
- 25.Swirnoff A. H., Milbrandt J. (1995) Mol. Cell. Biol. 15, 2275–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zipfel P. F., Decker E. L., Holst C., Skerka C. (1997) Biochim. Biophys. Acta 1354, 134–144 [DOI] [PubMed] [Google Scholar]
- 27.Gashler A., Sukhatme V. P. (1995) Prog. Nucleic Acids Res. Mol. Biol. 50, 191–224 [DOI] [PubMed] [Google Scholar]
- 28.Safford M., Collins S., Lutz M. A., Allen A., Huang C. T., Kowalski J., Blackford A., Horton M. R., Drake C., Schwartz R. H., Powell J. D. (2005) Nat. Immunol. 6, 472–480 [DOI] [PubMed] [Google Scholar]
- 29.Mages H. W., Stamminger T., Rilke O., Bravo R., Kroczek R. A. (1993) Int. Immunol. 5, 63–70 [DOI] [PubMed] [Google Scholar]
- 30.Shao H., Kono D. H., Chen L. Y., Rubin E. M., Kaye J. (1997) J. Exp. Med. 185, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki T., Lemonnier F. A. (1998) J. Exp. Med. 188, 715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basson M. A., Wilson T. J., Legname G. A., Sarner N., Tomlinson P. D., Tybulewicz V. L., Zamoyska R. (2000) J. Immunol. 165, 2444–2450 [DOI] [PubMed] [Google Scholar]
- 33.Bettini M., Xi H., Milbrandt J., Kersh G. J. (2002) J. Immunol. 169, 1713–1720 [DOI] [PubMed] [Google Scholar]
- 34.Schnell F. J., Kersh G. J. (2005) J. Immunol. 175, 2270–2277 [DOI] [PubMed] [Google Scholar]
- 35.Skerka C., Decker E. L., Zipfel P. F. (1995) J. Biol. Chem. 270, 22500–22506 [DOI] [PubMed] [Google Scholar]
- 36.Lin J. X., Leonard W. J. (1997) Mol. Cell. Biol. 17, 3714–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decker E. L., Skerka C., Zipfel P. F. (1998) J. Biol. Chem. 273, 26923–26930 [DOI] [PubMed] [Google Scholar]
- 38.Lohoff M., Duncan G. S., Ferrick D., Mittrücker H. W., Bischof S., Prechtl S., Röllinghoff M., Schmitt E., Pahl A., Mak T. W. (2000) J. Exp. Med. 192, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li-Weber M., Giaisi M., Baumann S., Pálfi K., Krammer P. H. (2004) Eur. J. Immunol. 34, 1111–1118 [DOI] [PubMed] [Google Scholar]
- 40.Li-Weber M., Salgame P., Hu C., Davydov I. V., Laur O., Klevenz S., Krammer P. H. (1998) J. Immunol. 161, 1380–1389 [PubMed] [Google Scholar]
- 41.Li-Weber M., Giasi M., Krammer P. H. (1998) J. Biol. Chem. 273, 32460–32466 [DOI] [PubMed] [Google Scholar]
- 42.Russo M. W., Matheny C., Milbrandt J. (1993) Mol. Cell. Biol. 13, 6858–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz M. L., Baeuerle P. A. (1991) EMBO J. 10, 3805–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Northrop J. P., Ho S. N., Chen L., Thomas D. J., Timmerman L. A., Nolan G. P., Admon A., Crabtree G. R. (1994) Nature 369, 497–502 [DOI] [PubMed] [Google Scholar]
- 45.Luo C., Burgeon E., Carew J. A., McCaffrey P. G., Badalian T. M., Lane W. S., Hogan P. G., Rao A. (1996) Mol. Cell. Biol. 16, 3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worden B., Yang X. P., Lee T. L., Bagain L., Yeh N. T., Cohen J. G., Van Waes C., Chen Z. (2005) Cancer Res. 65, 7071–7080 [DOI] [PubMed] [Google Scholar]
- 47.Neumann M., Grieshammer T., Chuvpilo S., Kneitz B., Lohoff M., Schimpl A., Franza B. R., Jr., Serfling E. (1995) EMBO J. 14, 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proksch P., Giaisi M., Treiber M. K., Pálfi K., Merling A., Spring H., Krammer P. H., Li-Weber M. (2005) J. Immunol. 174, 7075–7084 [DOI] [PubMed] [Google Scholar]
- 49.Li-Weber M., Giaisi M., Krammer P. H. (2001) Eur. J. Immunol. 31, 3694–3703 [DOI] [PubMed] [Google Scholar]
- 50.Bruhn K. W., Nelms K., Boulay J. L., Paul W. E., Lenardo M. J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9707–9711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenner C. A., Szabo S. J., Murphy K. M. (1997) J. Immunol. 158, 765–773 [PubMed] [Google Scholar]
- 52.Li-Weber M., Salgame P., Hu C., Davydov I. V., Krammer P. H. (1997) J. Immunol. 158, 1194–1200 [PubMed] [Google Scholar]
- 53.Chapman N. R., Perkins N. D. (2000) J. Biol. Chem. 275, 4719–4725 [DOI] [PubMed] [Google Scholar]
- 54.Decker E. L., Nehmann N., Kampen E., Eibel H., Zipfel P. F., Skerka C. (2003) Nucleic Acids Res. 31, 911–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abe E., De Waal Malefyt R., Matsuda I., Arai K., Arai N. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2864–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]