Abstract
CXCR4-using human immunodeficiency virus, type 1 (HIV-1) variants emerge late in the course of infection in >40% of individuals infected with clade B HIV-1 but are described less commonly with clade C isolates. Tat is secreted by HIV-1-infected cells where it acts on both uninfected bystander cells and infected cells. In this study, we show that clade B Tat, but not clade C Tat, increases CXCR4 surface expression on resting CD4+ T cells through a CCR2b-dependent mechanism that does not involve de novo protein synthesis. The expression of plectin, a cytolinker protein that plays an important role as a scaffolding platform for proteins involved in cellular signaling including CXCR4 signaling and trafficking, was found to be significantly increased following B Tat but not C Tat treatment. Knockdown of plectin using RNA interference showed that plectin is essential for the B Tat-induced translocation of CXCR4 to the surface of resting CD4+ T cells. The increased surface CXCR4 expression following B Tat treatment led to increased function of CXCR4 including increased chemoattraction toward CXCR4-using-gp120. Moreover, increased CXCR4 surface expression rendered resting CD4+ T cells more permissive to X4 but not R5 HIV-1 infection. However, neither B Tat nor C Tat was able to up-regulate surface expression of CXCR4 on activated CD4+ T cells, and both proteins inhibited the infection of activated CD4+ T cells with X4 but not R5 HIV-1. Thus, B Tat, but not C Tat, has the capacity to render resting, but not activated, CD4+ T cells more susceptible to X4 HIV-1 infection.
Keywords: Cell/Chemotaxis, Receptors/Chemokine, Signal Transduction/G Proteins, Viruses/HIV, RNA interference, CCR2, CXCR4, Plectin, Tat
Introduction
Human immunodeficiency virus, type 1 (HIV-1)2 exhibits high genetic variability, with strains divided into three main groups: major (M), which are the cause of most HIV-1 infections worldwide, outlier (O), and new (N; non-M and non-O; 1). Within group M, nine clades are recognized, designated by the letters A–D, F–H, J, and K. In addition, circulating recombinant forms (CRF) have also been identified (1). Globally, over 50% of all infections are caused by clade C, whereas clade B, the most studied clade, represents just 10% of all infections but is dominant in both Europe and North America. Recent research has shown that the different clades and CRF of HIV-1 have biological differences with respect to transmission (2), replication (3), and disease progression (4, 5). Moreover, the HIV-1 proteins gp120 (6), Nef (7, 8), Vif, Vpr, Vpu (9, 10), and Tat (11–19) show clade and isotype-specific properties at both the molecular and biological levels. Furthermore, the HIV-1 long terminal repeat (LTR) also displays clade-specific properties (20). Therefore, generalizations of HIV-1 transmission, pathogenesis, and tissue involvement based on clade B studies may be misleading.
HIV-1 isolates are distinguishable by the chemokine receptor they use for cell entry (21). Chemokine (CXC motif) receptor 4 (CXCR4)-using isolates are termed X4, chemokine (C-C motif) receptor 5 (CCR5)-using isolates are termed R5, and isolates that use both CXCR4 and CCR5 are termed R5X4. R5 viruses predominate early in HIV-1 infection for all clades, whereas X4 or R5X4 viruses emerge in about 40% of late clade B HIV-1 infection (22). The precise mechanism by which this emergence occurs is poorly understood, but it coincides with both significantly lower levels of CD4+ T cells and a lower thymic production of new T cells than in individuals with R5 variants irrespective of viral load (23–25). It is possible that the R5 to X4 shift is also a cause of the CD4+ T cell decline, because X4 variants target the naïve cell pool (26). In clade C HIV-1 infection, R5 variants dominate all stages of disease, including late stage AIDS, with only limited X4 or R5X4 variants described or characterized (27–29), suggesting that factors limiting the development of X4 clade C viruses exist, although it is unclear whether they are host or virological constraints.
CXCR4 is a seven-transmembrane receptor coupled to a pertussis toxin (PTX)-sensitive heterotrimeric Gi protein, which modulates the levels of intracellular cAMP by inhibiting the activity of adenylate cyclase. It also links to phosphoinositide 3-kinase, which is intrinsically linked to cell motility by promoting reorganization of the actin cytoskeleton (30). CXCR4 also activates Src kinase, which in conjunction with other protein tyrosine kinases of the Syk, Tec, and focal adhesion kinase families promotes the activation of mitogen-activated protein kinases (MAPK) and Ras-related GTPases, which change the transcriptional profile of the cell and promote actin remodeling (30). The natural ligand for CXCR4 is CXCL12, which is a chemoattractant for CD4+ T cells.
HIV-1 clade B Tat (B Tat) has been shown to up-regulate CXCR4 on resting CD4+ T cells (31, 32). Tat is an 86–101-residue regulatory protein (9–11 kDa) that is essential for productive and processive transcription from the HIV-1 LTR (33) and is secreted from unruptured, HIV-1-infected cells where it has a variety of effects on both uninfected and infected cells (reviewed in Refs. 34 and 35). The cysteine-rich and core domains of Tat are responsible for its trans-activational properties (33). Aligning this region of Tat with several β-chemokines indicates positioning and similarity of key residues critical for β-chemokine receptor binding and signal transduction (36), including a CC(F/Y) motif at positions 30–32. However, clade-specific variations occur, with more than 90% of HIV-1 clade C Tat proteins (C Tat) possessing a C31S mutation (12), and HIV-1 CRF_AE Tat proteins have an (F/Y)32W mutation (19). We have previously shown that the C31S variation found in C Tat results in its reduced ability to bind CCR2b and stimulate TNF and CCL2 production from monocytes (16). In this study, we show that B Tat but not C Tat up-regulates the surface expression of CXCR4 on resting CD4+ T cells, rendering a larger population of resting CD4+ T cells more susceptible to X4 HIV-1 infection.
EXPERIMENTAL PROCEDURES
CD4+ T Cells
Human peripheral blood mononuclear cells were isolated from the heparinized blood of healthy HIV-1 seronegative donors by density centrifugation over Ficoll-PaqueTM Premium (GE Healthcare). CD4+ T cells were isolated from peripheral blood mononuclear cells by negative selection using the CD4+ T cell isolation kit from Miltenyi Biotec (Auburn, CA) in accordance with the manufacturer's instructions. The final cultures of CD4+ T cells were always >95% pure, as determined by two-color flow cytometry analysis using monoclonal fluorescently tagged antibodies specific for CD4 and CD3 (BD Pharmingen). After purification, the cells were resuspended in AIM-V medium with human serum albumin (AIM-V; Invitrogen) at 1 × 106 CD4+ T cells/ml for 24 h to allow for maximal unstimulated surface expression of CXCR4 before use.
Synthesis of Tat Proteins
Full-length synthetic Tat proteins were synthesized in the solid phase using the method of Barany and Merrifield (37) with fast Fmoc (9-fluoenylmethoxy carbonyl) chemistry and 4-hydroxymethyl-phenoxymethyl-copolystyrene, 1% divinylbenzene preloaded resin (0.5 mmol; Applied Biosystems) on an automated ABI 433A synthesizer (Applied Biosystems) as previously described (11). Purification and analysis using high performance liquid chromatography was performed as previously described (11, 14). Amino acid analysis was performed on a model 6300 Beckman analyzer, and mass spectrometry was carried out using an Ettan matrix-assisted laser desorption ionization time-of-flight apparatus (Amersham Biosciences). The trans-activation activities of the synthetic Tat proteins were analyzed by monitoring the production of β-galactosidase after activation of lacZ expression in HeLa-CD4-LTR-β-galactosidase cells as previously described (16).
Viruses
The following viruses were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: HIV-1IIIB from Dr. Jean-Marie Bechet and Dr. Luc Montagnier (38, 39) and HIV-193In905 from Dr. Robert Bollinger and the UNAIDS Network for HIV Isolation and Characterization, and the Division of AIDS, NIAID, National Institutes of Health (40). The viruses were propagated on phytohemagglutinin (PHA) from Phaseolus vulgaris (Sigma) and interleukin (IL)-2 (Roche Applied Science)-activated peripheral blood mononuclear cells, treated with RNase-free DNase I (Invitrogen), and purified using Vivaspin 20 columns with a 300,000 molecular weight cut-off (Sartorius Stedim Biotech, Edgewood, NY). The 50% tissue culture infective dose (TCID50) was determined by serial dilution of the virus and calculated using the Spearman-Kärber method as described elsewhere (41). The HIV-1 p24 antigen concentration in culture supernatants was determined using the Alliance HIV-1 p24 antigen enzyme-linked immunosorbent assay kit (PerkinElmer Life Sciences).
Antagonists and Inhibitors
The CXCR4 antagonist AMD3100, the chemokine (C-C motif) receptor 2b (CCR2b) antagonist RS102895, the extracellular signal-regulated kinase (Erk) 1/2 inhibitor U0126, and pertussis toxin (PTX) from Bordetella pertussis were all purchased from Sigma. The cytotoxic effect of the different inhibitors and antagonists was tested by the trypan blue dye exclusion assay, and none was found to be cytotoxic (viability was >99%). Anti-CD3 antibody (clone HIT3a) was purchased from (eBioscience, San Diego, CA).
Flow Cytometry
All of the flow cytometry was performed on a FACSCalibur flow cytometer (BD Biosciences). All of the cytogram analysis was performed using CellQuest Pro® software (BD Biosciences).
Surface protein expression of CXCR4, CCR5, CD25, CD69, human leukocyte antigen-DR, and CD45RO on CD4+ T cells was investigated by flow cytometry using fluorescently labeled monoclonal antibodies CD4PerCp, CD3FITC, CXCR4PE, CD45ROAPC, CCR5APC, CD14APC, CD14FITC, CD25APC, CD69APC, and human leukocyte antigen-DRPE (where FITC is fluorescein isothiocyanate, PE is phycoerythrin, APC is allophycocyanin, and PerCp is peridin-chlorophyll protein) obtained from BD Pharmingen as described previously (42). The results are expressed as [mean fluorescence intensity in treated cells/mean fluorescence intensity in untreated cells] × 100. Total CXCR4 content was evaluated by surface staining for CXCR4, then permeabilizing with Cytofix/Cytoperm (BD Biosciences) and restaining for CXCR4. Nonspecific fluorescence was assessed using isotype- and fluorophore-matched controls. Receptor internalization was measured by the retention of anti-CXCR4 antibody binding following elimination of cell surface bound antibody via acid wash as described previously (42). Receptor internalization was quantified as the percentage of total anti-CXCR4 fluorescence intensity (pH 7 wash) retained following acid wash (pH 4).
For the quantification of Erk1/2 phosphorylation, Tat-conditioned cells or unconditioned controls were harvested and stimulated with gp120 for 2 min. The reaction was stopped by adding an equal volume of glacial Dulbecco's phosphate-buffered saline (DPBS) supplemented with 2% (w/v) paraformaldehyde. The cells were permeabilized in 90% glacial methanol overnight, washed twice in DPBS supplemented with 1% (w/v) bovine serum albumin and 0.1% (w/v) NaN3, and stained with the 20A Alexa Fluor 647-conjugated monoclonal anti-phospho-Erk1/2 (Thr202/Tyr204) obtained from BD Pharmingen. The specificity for phosphorylated Erk1/2 was assessed using 40 nm 12-O-tetradecanoylphorbol-13-acetate, which induced a significant increase in mean fluorescence over unstimulated control cells.
Filamentous actin (F-actin) polymerization following the addition of gp120 was assessed mainly as previously described (42, 43). Briefly, CD4+ T cells (107/ml) were incubated in RPMI 1640 medium containing 20 mm HEPES (both Invitrogen) and gp120MN (Immunodiagnostics, Woburn, MA) or gp120SF162 (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health). Every 15 s, 100 μl of cell suspension was added to 400 μl of DPBS supplemented with 4.5% (w/v) paraformaldehyde, 100 nm FITC-labeled phalloidin, and 125 μg/ml l-α-lysophosphatidylcholine (both Sigma). The results are expressed as [median fluorescence intensity after the addition of ligand/median fluorescence intensity before the addition of ligand] × 100.
Real Time PCR
CXCR4 mRNA and HIV-1 LTR DNA quantification were measured by real time PCR using a using the LightCycler System and the FastStart RNA Master SYBR Green I kit and the FastStart DNA Master SYBR Green I (all from Roche Applied Science), respectively, as previously described (42). All of the results are expressed as the ratio between the copy number of the target gene and the copy number of RNA polymerase II and normalized so that CXCR4 mRNA expression and HIV-1 LTR in unconditioned cells equals 1.00.
siRNA Transfection
A control duplex siRNA consisting of a scramble sequence that does not lead to the degradation of any cellular message and three plectin duplex siRNAs were purchased from Santa Cruz Biotechnology. RNA duplexes were transfected into primary resting CD4+ T cells using Lipofectamine RNAiMAX in Opti-MEM I (both Invitrogen) with a final siRNA concentration of 40 nm. The cells were then used after 48 h, and the extent of knockdown was quantified by immunoblotting as described below.
Immunoblotting
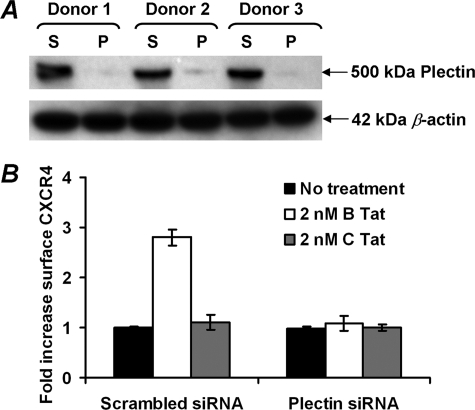
Plectin was quantified by electrophoresis and subsequent immunoblotting using anti-plectin C-20 (Santa Cruz Biotechnology) or E398P (Abcam) as described previously (42). The membranes were then stripped and reprobed for β-actin using a monoclonal antibody (clone AC-74; Sigma) The relative density of the plectin (target) band was compared with the β-actin (reference) band and analyzed using the freely available ImageJ (National Institutes of Health).
Total CXCR4 content and the phosphorylation of Erk1/2, p38, and stress-activated protein kinase/Jun amino-terminal kinase (SAPK/JNK) were determined by electrophoresis and subsequent immunoblotting. Briefly, the cells were washed and lysed in CelLytic M in the presence of phosphatase and protease inhibitors (all from Sigma) and subjected to centrifugation (15,000 × g, 15 min) to remove debris. 40 μg/sample was then heated to 100 °C for 10 min in SDS sample buffer containing reducing agent and separated by 4–12% bis-Tris-polyacrylamide gel (Invitrogen) and transferred to nitrocellulose membranes (Fisher). The membranes were blocked overnight with DPBS supplemented with 0.1% (v/v) polysorbate 20 (Sigma) and 5% (w/v) dried nonfat milk (Genesee Scientific, San Diego, CA) and probed with an antibody against either CXCR4 (Abcam), phosphorylated p38 (Thr180/Tyr182), phosphorylated SAPK/JNK (Thr183/Tyr185), or phosphorylated Erk1/2 (Thr202/Tyr204) (from Cell Signaling Technology) followed by detection using the WesternBreeze chemiluminescence kit (Invitrogen). The membranes were subsequently stripped and reprobed with anti-β-actin (Sigma) in the case of CXCR4 or with the corresponding antibody against total p38, Erk1/2, or SAPK/JNK (all from Cell Signaling Technology) to confirm equal loading.
Chemotaxis Assays
The migration of CD4+ T cells in response to gp120 was evaluated using 24-well Transwell migration chambers with 5-μm-pore size polyvinylpyrrolidone-free polycarbonate filters (Corning) as previously described (44).
HIV-1 Infection Assay
After 4 h of conditioning with Tat, CD4+ T cells were infected with X4 HIV-1IIIB or the R5 HIV-193In905 at a multiplicity of infection of 0.01 for 3 h and then washed three times with DPBS. The cells were then cultured in RPMI 1640 supplemented with 15% (v/v) fetal bovine serum, 2 mm glutamine, and antibiotics (all from Invitrogen). After 14 h, either the cells were harvested and total DNA was prepared with the QIAamp DNA mini kit in accordance with the manufacturer's directions (Qiagen), or the medium was supplemented with 20 units/ml IL-2 and cells were cultured for a further 96 h. The supernatants were then collected and assayed using the Alliance HIV-1 p24 antigen enzyme-linked immunosorbent assay kit (PerkinElmer Life Sciences).
Statistics
The results are expressed as the means ± S.E. of three or more experiments performed in triplicate. All of the p values correspond to two-tailed t tests.
RESULTS
CXCR4 Antibody Binding Sites Are Increased on B Tat-treated, but Not on C Tat-treated, Resting CD4+ T Cells
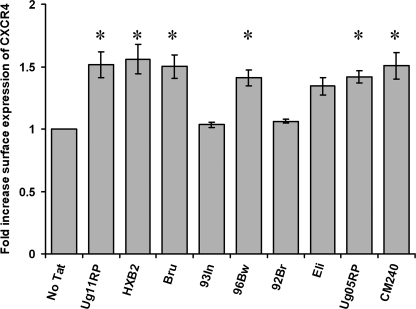
In this study, we initially used nine different Tat proteins from four different HIV-1 clades and one CRF: Clade A, Ug11RP (17); clade B, HXB2 (B Tat)(16) and Bru (11); clade C, 93In (C Tat)(16), 96Bw (46), and 92Br (45); clade D, Ug05RP (14), and Eli (45); and CRF_AE, CM240 (46). All of the Tat variants have similar trans-activational activity as described previously (16, 46).
We first examined whether the different variants of Tat have an effect on HIV-1 surface coreceptor expression on resting CD4+ T cells. CD4+ T cells were freshly isolated from healthy donors, cultured for 24 h as described under “Experimental Procedures,” and subsequently exposed to 2 nm Tat proteins for 4 h. The cells were then harvested, stained, and analyzed by flow cytometry. All of the Tat proteins except the clade C proteins 93In and 92Br induced a significant increase in the surface expression of CXCR4 (Fig. 1). We then investigated the dose-dependent response to B Tat and C Tat at 1, 10, and 50 nm. At all of the concentrations tested, B Tat significantly up-regulated CXCR4 surface expression, whereas C Tat did not (p < 0.0001; Fig. 2A). Flow cytometry demonstrated that neither B Tat nor C Tat altered the expression of the activation markers (human leukocyte antigen-DR, CD25, or CD69) and had no effect on the surface expression of CD4, CD3, CCR5, or CD45RO (data not shown).
FIGURE 1.
Tat proteins from different clades up-regulate CXCR4 expression of resting CD4+ T cells. Purified resting CD4+ T cells from HIV-negative subjects were incubated with 2 nm Tat from different clades. After 4 h, the cells were harvested and stained for surface CXCR4. All of the Tat proteins significantly up-regulated CXCR4 surface expression except the clade C Tat93In and Tat92Br. *, p < 0.001.
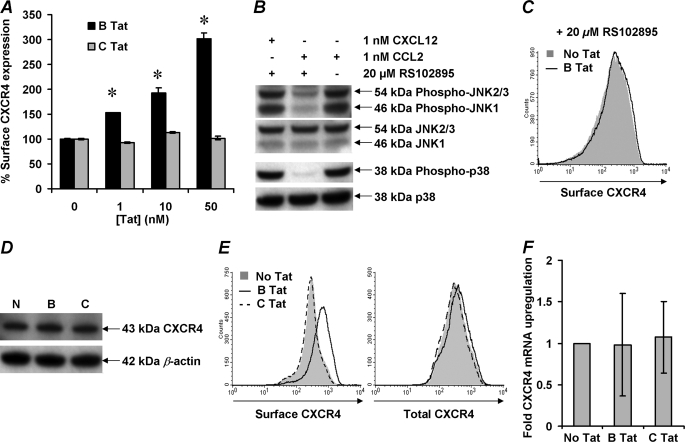
FIGURE 2.
B Tat induces up-regulation of surface CXCR4 expression on resting CD4+ T cells through a CCR2-dependent mechanism that does not require de novo protein synthesis. A, purified resting CD4+ T cells from HIV-negative subjects were incubated with increasing concentrations of Tat. After 4 h, the cells were harvested and stained for surface CXCR4. B Tat, but not C Tat, induced a dose-dependent increase in surface expression of CXCR4. The data are expressed as the means ± S.E. of the mean fluorescence intensity calculated from three independent experiments. B, purified resting CD4+ T cells were cultured in the presence or absence of 20 μm RS102895 and 1 nm CCL2 or CXCL12. After 4 h, the cells were harvested and analyzed for the phosphorylation of SAPK/JNK and p38 by immunoblotting. The blots were then stripped and reprobed for the expression of total SAPK/JNK or p38. RS102895 successfully inhibited the CCL2/CCR2-mediated phosphorylation events but not those through CXCL12/CXCR4, indicating CCR2 antagonism specificity. C, purified resting CD4+ T cells were pretreated with 20 μm RS102895 and then cultured for 4 h in the presence of 2 nm B Tat (black line histogram) or with vehicle control (solid gray histogram). The cells were then harvested and stained for surface CXCR4. RS102895 inhibited the up-regulation of surface CXCR4 expression. Histograms are shown from a representative donor. D–F, purified resting CD4+ T cells were incubated with 2 nm Tat for 4 h, lysed, subjected to electrophoresis, and immunoblotting (D). Alternatively, the cells were stained for surface CXCR4 or permeabilized and stained for total CXCR4 (E) or evaluated for CXCR4 mRNA content using real time PCR and summarized in F. The histograms and blots for a representative donor are shown. Neither Tat protein induced de novo protein synthesis. *, p < 0.001.
We have previously shown that both B Tat and C Tat bind CXCR4 with the same Kd but differ in their affinity for CCR2b (16). Therefore, we tested the hypothesis that B Tat triggers the up-regulation of surface CXCR4 expression through a CCR2b-dependent mechanism by incubating the cells with RS102895, a potent and specific spiropiperidine class inhibitor for CCR2b that has no significant inhibitory activity on other chemokine receptors (47). Indeed, RS102895 inhibits the downstream effects of the CCR2 ligand, CCL2, namely the phosphorylation of p38 and SAPK/JNK but not those of CXCL12 (Fig. 2B). Incubation of resting CD4+ T cells in the presence of RS102895 also resulted in the complete inhibition of surface CXCR4 up-regulation by B Tat (Fig. 2C). Importantly, the concentration of RS102895 used had no effect on the surface expression of CXCR4 (Fig. 2C).
A previous study using 200 μg/ml cycloheximide showed that the up-regulation of CXCR4 on resting CD4+ T cells by B Tat was due to de novo protein synthesis (32). However, we noticed that in their data and in our own experiments (not shown), treatment of resting CD4+ T cells with 200 μg/ml cycloheximide actually down-regulates CXCR4 expression. Therefore, to determine whether B Tat-induced up-regulation of surface CXCR4 on resting CD4+ T cells was due to de novo protein synthesis, CD4+ T cells were incubated with B Tat, and the expression of both surface and total CXCR4 was quantified by immunoblotting (Fig. 2D) and flow cytometry (Fig. 2E), and CXCR4 mRNA was quantified by real time PCR (Fig. 2F). Although cell surface expression of CXCR4 increases significantly (p < 0.0001), there was no increase in either total CXCR4 (p = 0.17; Fig. 2, D and E) or CXCR4 mRNA (p = 0. 93; Fig. 2F).
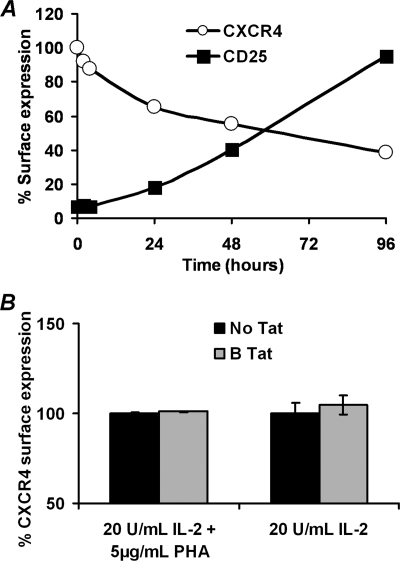
We then sought to examine the effect of Tat on the surface expression of CXCR4 on activated CD4+ T cells using CD25 as a marker of activation. We found that PHA-IL-2-stimulated CD4+ T cells have significantly lower levels of surface CXCR4 than resting CD4+ T cells after just 24 h of stimulation (Fig. 3A; p = 0.014). Moreover, B Tat had no effect on the surface expression of CXCR4 on IL-2 or IL-2/PHA-stimulated CD4+ T cells (p = 0.87 and 0.38 respectively; Fig. 3B).
FIGURE 3.
Expression of CXCR4 on IL-2 stimulated CD4+ T cells. A, surface expression of CXCR4 and CD25 (as an indicator of activation) was assessed over 96 h after stimulation with 20 units/ml IL-2 and 5 μg/ml PHA. As the cells became activated, CXCR4 expression was down-regulated. B, at 24 h post-IL-2 or IL2 plus PHA treatment, the ability of B Tat to up-regulate CXCR4 was assessed by flow cytometry. The data are expressed as the means ± S.E. of the mean fluorescence intensity calculated from three or more independent experiments performed in triplicate. B Tat had no effect on the CXCR4 surface expression of activated CD4+ T cells.
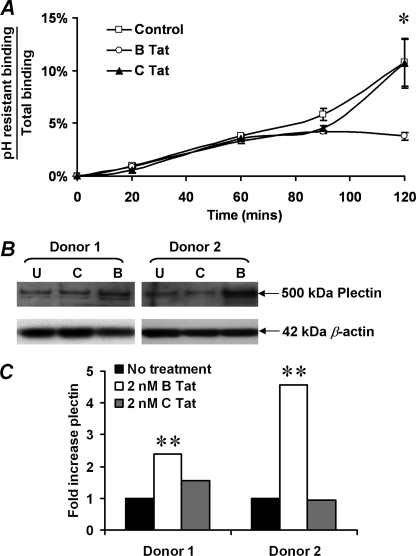
B Tat but Not C Tat Reduces CXCR4 Internalization
To determine whether altered receptor trafficking might play a role in B Tat up-regulation of CXCR4 cell surface expression, the receptor internalization rates were quantified by flow cytometric analysis of internalized anti-CXCR4 antibody following removal of surface-bound antibody by acid washing. Low pH washing efficiently removed cell surface-bound antibody as demonstrated by the abrogation of the fluorescent signal of both anti-CD4PerCp and anti-CXCR4PE (data not shown). Therefore, residual antibody binding reflects internalized receptors not exposed to extracellular low pH by acid washing at the time points shown (48). After 2 h of incubation at 37 °C in medium without Tat, the acid-resistant anti-CXCR4 antibody binding post-washing averaged 11% of total CXCR4 fluorescence intensity (Fig. 4A). Parallel incubation in the presence of 2 nm B Tat resulted in significantly decreased acid-resistant anti-CXCR4 antibody binding, reaching a 65% reduction at 120 min with an average of 3.6% total fluorescence intensity (p = 0.008). C Tat had no significant effect at all time points (p > 0.2). Thus, B Tat appears to up-regulate cell surface expression of CXCR4 in part by reducing receptor internalization rates rather than by increasing the size of the total CXCR4 receptor pool.
FIGURE 4.
Effect of B Tat on CXCR4 internalization and on plectin content. A, CXCR4 internalization was quantified by flow cytometric assessment of anti-CXCR4 antibody binding following acid stripping of cell surface-bound antibody. Acid-resistant binding (internalized CXCR4) is expressed as a fraction of total antibody binding over 2 h of incubation at 37 °C. B Tat reduced the CXCR4 internalization by 65% across four independent experiments (p = 0.008). B, effect of B Tat on plectin expression. Immunoblotting was used to quantify plectin levels in resting CD4+ T cells cultured for 4 h in the presence of 0 (U), 2 nm C Tat (C), or 2 nm B Tat (B). Two donors are shown. Parallel determination of β-actin verified equivalent protein loading. C, densitometric analysis of the immunoblots shown in B revealed that B Tat, but not C Tat, conditioning of resting CD4+ T cells significantly increased plectin content (p = 0.001). *, p = 0.008; **, p = 0.001.
In the absence of ligand binding, the precise mechanisms that regulate CXCR4 signaling and intracellular trafficking are not well understood. However, recent studies have suggested a role for plectin, a cytolinker protein that plays an important role as a scaffolding platform for proteins involved in cellular signaling and in CXCR4 signaling and trafficking (42, 49). Therefore, to determine whether B Tat has an effect on plectin expression, the plectin levels were quantified by immunoblotting (Fig. 4B). We found that B Tat significantly increased the expression of plectin in CD4+ T cells (mean fold increase 1 versus 3.5, p = 0.001; Fig. 4C). These data suggest that plectin may play a role in B Tat-dependent up-regulation of surface CXCR4 expression.
To understand the importance of the interaction between plectin, CXCR4, and CCR2b, we examined the effect of plectin RNA interference on B Tat-mediated CXCR4 up-regulation. We transfected primary CD4+ T cells with a scramble siRNA or with plectin-specific siRNA, treated these cells with 2 nm B Tat for 4 h, and determined surface CXCR4 expression by flow cytometry. Cells transfected with the scramble siRNA exhibited an increase in CXCR4 surface expression following B Tat treatment (Fig. 5). In contrast, cells transfected with plectin siRNA exhibited no increase in CXCR4 surface expression following B Tat treatment. These data indicate that plectin is essential for the increase in surface expression of CXCR4 following B Tat treatment.
FIGURE 5.
Effect of plectin RNA interference on B Tat-mediated CXCR4 surface up-regulation. Purified CD4+ T cells were transiently transfected with a scramble siRNA as a control (S) or plectin-specific siRNA (P) and subjected to immunoblot analysis as described above (A) or treated with B Tat, C Tat, or vehicle control (B). After 4 h, the cells were harvested and stained for surface CXCR4. Plectin siRNA almost completely abrogated the B Tat-mediated up-regulation of CXCR4 surface expression (p < 0.0001).
B Tat Increases CXCR4 Signaling, Leading to an Increase in CXCR4-tropic gp120-induced Actin Polymerization and Chemotaxis
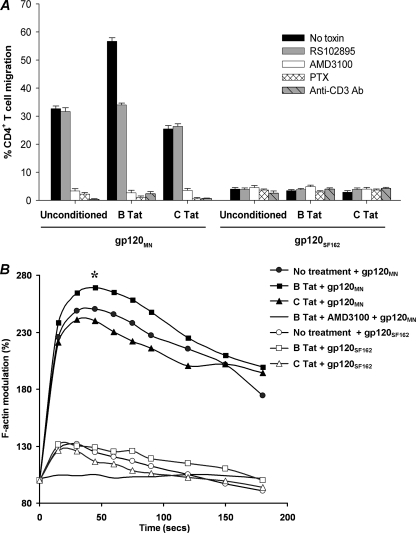
Using a Transwell assay, we tested the hypothesis that B Tat-induced CXCR4 surface up-regulation would enhance CD4+ T cell chemotaxis toward CXCR4-tropic gp120MN. We compared the percentage of unconditioned resting CD4+ T cells that migrated in response to gp120 with or without prior conditioning with Tat for 4 h. CD4+ T cell migration toward 200 nm gp120MN, the reported Kd of CXCR4-tropic gp120 for CXCR4 (50), was enhanced 73% by prior exposure to B Tat (p < 0.0001; Fig. 6A), whereas migration toward the CCR5-tropic gp120SF162 remained unaffected (Fig. 6A). Interestingly, prior incubation with C Tat inhibited the chemotactic effect of gp120MN by 22% (p < 0.0001; Fig. 6A). Preincubating CD4+ T cells with 20 μm RS102895 before B Tat exposure reduced migration toward gp120MN to 34%, the same as for non-B Tat-treated gp120MN-specific migration (Fig. 6A). All of the chemotactic activity in response to gp120MN was abrogated by pretreating the cells with 1 μm AMD3100, a specific CXCR4 antagonist, or 1 μg/ml PTX, which irreversibly inhibits the Gi subunits of heterotrimeric G proteins by ADP ribosylation (Fig. 6A), indicating a requirement for CXCR4 and Gαi proteins in CXCR4-tropic gp120MN-induced migration.
FIGURE 6.
B Tat, but not C Tat, augments CXCR4-tropic gp120MN chemotactic and F-actin response. A, CD4+ T cell migration induced by 200 nm gp120MN or gp120SF162 alone or with prior conditioning with 2 nm Tat and/or 20 μm RS102895, 1 μm AMD3100, 1 μg/ml PTX, or 10 μg/ml anti-CD3 antibody. The bars represent the means ± S.E. of the percentage of migrated cells from three independent experiments performed in triplicate. Chemokinesis was minimal when equal concentrations of gp120 was added to both the apical and basal chambers and has been subtracted from these histograms. CXCR4-tropic gp120MN induced chemotaxis of CD4+ T cells and was augmented by preconditioning with 2 nm B Tat but not C Tat. The augmentation was abrogated in the presence of RS102895 (p < 0.0001). Treatment with AMD3100, PTX, or anti-CD3 antibody inhibited gp120MN-induced chemotaxis. R5 gp120SF162 induced minimal chemotaxis and was unaffected by all inhibitor and stimulus treatments. B, effect of B Tat on CXCR4-tropic gp120MN-induced F-actin polymerization in resting CD4+ T cells. Using FITC-phalloidin as a probe for intracellular F-actin, the effects of Tat preconditioning on F-actin polymerization in resting CD4+ T cells was assessed by flow cytometry. The results show the kinetics of F-actin polymerization following stimulation of unconditioned cells with R5 gp120SF162 (open symbols) or X4 gp120MN (closed symbols) at time 0 with or without inhibitors. All of the data are representative of three independent experiments. *, p < 0.0001.
Previous studies have shown that CXCR4 physically interacts with the T cell receptor (TCR) in response to signaling via CXCL12 and uses TCR signaling machinery from signal initiation to signal termination. Therefore, to establish whether anti-CD3 stimulation would inhibit the migration of CD4+ T cells toward gp120MN, the response of CD4+ T cells pretreated with 10 μg/ml anti-CD3 antibody was evaluated. The response was reduced to 2% of cell migration in response to gp120MN without prior anti-CD3 treatment (p < 0.0001; Fig. 6A).
Cell polarization and motility are governed by actin cytoskeleton rearrangements. Actin turnover is also involved in HIV-1 entry, which requires disintegration and reconstitution of the membrane structure. Thus, by inducing F-actin polymerization, CXCR4 can regulate both X4 HIV-1 entry and chemotaxis. CD4+ T cells conditioned with B Tat for 4 h were treated with 200 nm gp120 and probed with FITC-phalloidin for F-actin polymerization at various time points. F-actin polymerization was rapidly induced in resting, untreated CD4+ T cells following the addition of 200 nm CXCR4-tropic gp120MN. Kinetics in all instances were rapid with the peak response achieved within 30 s post-stimulation and was completely abrogated in the presence of AMD3100, indicating a dependence on signaling through CXCR4 (Fig. 6B). Treating the cells with B Tat for 4 h prior to stimulation with gp120MN increased the F-actin polymerization response (p = 0.034; Fig. 6B). Conversely, conditioning the cells with C Tat slightly inhibited the polymerization of F-actin, although this was not significant (p = 0.44; Fig. 6B). The CCR5-tropic gp120SF162 also induced rapid polymerization of F-actin, although we observed no difference between the Tat-conditioned and nonconditioned cells (132% increase versus 130% increase, p = 0.87). The peak response achieved using gp120SF162 in nonconditioned cells was significantly less compared with gp120MN alone (2.5-fold increase versus 1.3-fold increase, p = 0.033; Fig. 6B).
B Tat Enhances CXCR4-tropic gp120-induced Erk1/2 Phosphorylation in Primary CD4+ T Cells
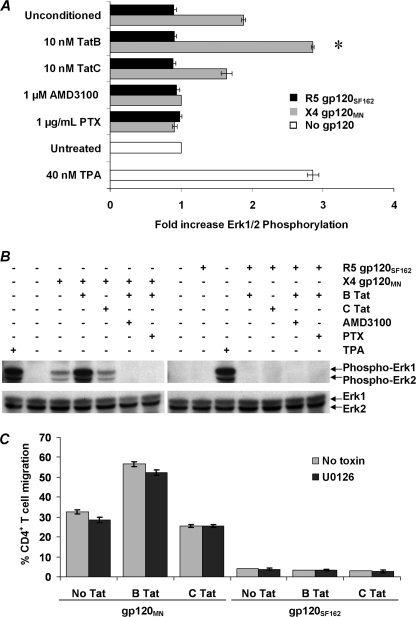
Erk1/2 has been implicated in regulating chemotaxis in some systems (51, 52). Therefore, we analyzed whether gp120 could induce the phosphorylation of Erk1/2 in resting unstimulated CD4+ T cells. At both 40 and 80 nm, we observed no effect of either the CXCR4-tropic gp120MN or the CCR5-tropic gp120SF162 (data not shown). However, at 200 nm, gp120MN induced significant phosphorylation of Erk1/2 (1.88-fold increase, p < 0.0001; Fig. 7, A and B). Furthermore, Erk1/2 phosphorylation was greater in cells conditioned with B Tat than in unconditioned cells (1.88 versus 2.85-fold increase, p < 0.0001; Fig. 7, A and B). Conversely, prior conditioning with C Tat had no effect on the extent of gp120MN-induced Erk1/2 phosphorylation. Pretreating the cells with AMD3100 or PTX abrogated gp120MN-induced phosphorylation of Erk1/2, indicating the inhibition of the Ras/MAPK pathway and a requirement for CXCR4 in gp120MN-induced Erk1/2 phosphorylation; conditioning with either Tat had no observable effect on this inhibition (Fig. 7, A and B). Conversely, the gp120SF162 had no effect on Erk1/2 phosphorylation at either the Kd for CCR5, 6 nm (53; data not shown), or at 200 nm (Fig. 7, A and B).
FIGURE 7.
Effect of B Tat on gp120 induced Erk1/2 phosphorylation in primary resting CD4+ T cells. A, primary resting CD4+ T cells pretreated with B Tat or C Tat for 4 h and then stimulated with gp120 for 2 min in the presence of 1 μm AMD3100, 1 μg/ml PTX, or vehicle control, fixed, permeabilized, and stained with a fluorescently tagged antibody specific for phosphorylated Erk1/2 and then analyzed by flow cytometry. The bars represent the means and S.E. of the relative change in fluorescence intensity indicative of Erk1/2 phosphorylation after stimulation with CXCR4-tropic gp120MN (black bars) or CCR5-tropic gp120SF162 (gray bars). White bars represent resting cells treated with 40 nm 12-O-tetradecanoylphorbol-13-acetate (TPA) or with vehicle control in the absence of gp120 stimulation. The change in Erk1/2 phosphorylation is expressed as the relative fold change in the mean fluorescence intensity, with the base-line fluorescence intensity before the addition of gp120 expressed as 1.00. Alternatively, cells were lysed, and Erk1/2 phosphorylation was determined through immunoblotting. B, B Tat, but not C Tat, preconditioning of resting CD4+ T cells significantly increased gp120MN-induced phosphorylation of Erk1/2 (p < 0.0001) but had no effect on gp120SF162-induced events. Both AMD3100 and PTX inhibited Erk1/2 phosphorylation by gp120MN. C, CD4+ T cell migration induced by 200 nm gp120MN or gp120SF162 after preconditioning with Tat and/or 10 μm U0126. The bars represent the means ± S.E. of the number of migrated cells from three independent experiments performed in triplicate. Chemokinesis was minimal when equal concentrations of gp120 were added to both the apical and basal chambers and has been subtracted from these histograms. *, p < 0.0001.
Although several studies have demonstrated activation of Erk1/2 in response to a CXCL12 or gp120 interaction with CXCR4, its role in chemokine-mediated chemotaxis is contentious (43, 54). Thus, although Erk1/2 is activated in response to gp120, the role it plays in gp120-mediated chemotaxis of resting CD4+ T cells is unknown. We therefore measured CD4+ T cell migration after preincubation with 10 μm U0126, an Erk1/2 inhibitor. U0126 had little effect on CD4+ T cell migration in response to 200 nm gp120MN (Fig. 7C). However, although U0126 had no effect on CD4+ T cell migration in response to gp120, it inhibits the productive infection of primary CD4+ T cells at a point before the reverse transcription step (42).
B Tat but Not C Tat Increases Entry of X4 HIV-1IIIB but Not R5 HIV-193In905 into Resting CD4+ T Cells
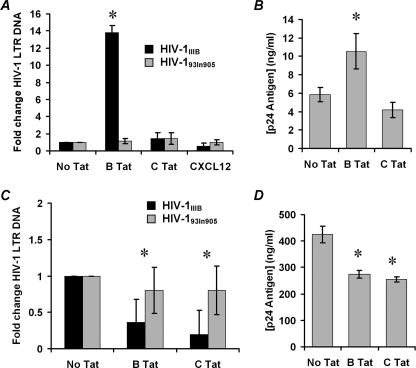
The previous results suggested that B Tat conditioning of resting CD4+ T cells selectively affects X4 HIV-1 gp120-CXCR4 interactions. Therefore, we tested the hypothesis that increased surface expression of CXCR4 will enhance X4 HIV-1 entry into resting CD4+ T cells while having no effect on R5 HIV-1 entry. To test this hypothesis directly, we measured viral entry by real time PCR. Highly purified resting CD4+ T cells were conditioned with B Tat, C Tat, or vehicle control for 4 h before infection with HIV-1 (multiplicity of infection of 0.01) for 3 h. After an additional 14 h culture in fresh medium, the samples were analyzed by real time PCR for the presence and quantity of strong stop HIV-1 DNA (with LTR R/U5 primers), an early product of reverse transcription. B Tat conditioning significantly enhanced X4 HIV-1IIIB infection 14-fold (p < 0.0001; Fig. 8A). Conversely, C Tat had no effect on HIV-1IIIB infection (p = 0.25). Neither B Tat nor C Tat had any effect on the R5 HIV-193In905 infection (p > 0.15).
FIGURE 8.
X4 HIV-1 infection of primary resting CD4+ T cells. A, purified resting CD4+ T cells were conditioned with 2 nm Tat, 1 nm CXCL12, or left unconditioned for 4 h before infection with X4 HIV-1IIIB (black bars) or the R5 HIV-193In905 (gray bars) at a multiplicity of infection of 0.01 for 3 h. The cells were then cultured for 14 h without further stimulation. Entry of HIV-1 was evaluated by real time PCR using primers specific for the LTR R/U5 region as described under “Experimental Procedures.” B, cells infected with HIV-1IIIB as described above were further cultured in the presence of 20 units/ml IL-2. After 96 h, the supernatants were collected and p24 quantified. C, purified CD4+ T cells were stimulated for 72 h in the presence of 20 units/ml IL-2, conditioned with 2 nm Tat for 4 h and then subjected to infection with HIV-1IIIB (black bars) or the HIV-193In905 (gray bars) at a multiplicity of infection of 0.01 for 3 h. The cells were then cultured for a further 14 h before the HIV-1 LTR DNA quantity was evaluated. D, cells infected with HIV-1IIIB from C were further stimulated with 20 units/ml IL-2 for 96 h before supernatants were collected, and p24 quantified. B Tat, but not C Tat, conditioning significantly augmented X4 HIV-1IIIB but not R5 HIV-193In905 infection of resting CD4+ T cells. Conversely, both B Tat and C Tat significantly inhibited X4 HIV-1IIIB but not R5 HIV-193In905 infection of activated CD4+ T cells. *, p < 0.05.
Virus replication was then induced using 20 units/ml IL-2 for a further 96 h, and p24 antigen was quantified at the end of this period. The increase in viral DNA found in B Tat-conditioned cells with the HIV-1IIIB correlated with a significant increase in p24 antigen concentration at 4 days post-infection (p = 0.034; Fig. 8B). C Tat conditioning resulted in no significant change in p24 at 4 days post-infection (p = 0.14).
Both B Tat and C Tat Inhibit the Entry of X4 HIV-1IIIB into PHA-activated CD4+ T Cells
In contrast to resting CD4+ T cells, PHA/IL-2-treated CD4+ T cells subjected to X4 HIV-1 infection resulted in significantly lower levels of viral LTR DNA copies in both clade B and clade C Tat-treated cells than in untreated cells (p = 0.003 and < 0.0001, respectively). Also, as in resting CD4+ T cells, there was no significant change in R5 HIV-1 viral LTR DNA copies found between either Tat treatment or the vehicle control (p = 0.63 and 0.58, respectively; Fig. 8C). This pattern extended to the supernatant p24 antigen levels at 96 h post-infection. Compared with unconditioned cells, there were significantly lower levels of p24 at 96 h in both B Tat- (274 ng/ml p24 versus 425 ng/ml p24, p = 0.005) and C Tat-treated cells (254 ng/ml p24 versus 425 ng/ml p24, p = 0.002; Fig. 8D).
DISCUSSION
Many different roles have been attributed to clade B Tat, including the induction of CD4+ T cell apoptosis (14–15, 35, 55–58), stimulation or inhibition of cytokine production, and as a monocyte chemoattractant (16, 36). The last role is thought to be a direct result of the ability of clade B Tat to bind CCR2b, inducing a transient calcium flux that activates a signaling module involving Gi and a hierarchy of MAPK family members that regulate chemotaxis. In a previous study, we showed that the clade C Tat used here was unable to bind CCR2b and failed to induce a transient calcium flux and activate signaling modules that regulate monocyte chemotaxis (16). Furthermore, although both Tat proteins bind CXCR4 with the same Kd, neither is able to induce a transient calcium flux in primary CD4+ T cells (16) or induce CD4+ T cell chemotaxis.3
In this study, we show that B Tat up-regulates the expression of CXCR4 on the surface of resting CD4+ T cells within 4 h through a dose- and CCR2b-dependent mechanism in a manner similar to CCL2 (42) that does not require de novo protein synthesis but involves the relocation of preexisting CXCR4 to the cell surface combined with a decrease in CXCR4 internalization. However, the mutation found in C Tat renders it unable to bind CCR2b (16) and subsequently unable to up-regulate CXCR4 expression on CD4+ T cells. Furthermore, because B Tat had no effect on CXCR4 expression in activated CD4+ T cells, the activation state of the cells appears to be important and may account for the discrepancies observed in the initial reports (31, 32, 59, 60).
The CXCR4-tropic gp120MN induced a CXCR4-specific signaling response in resting CD4+ T cells that led to F-actin polymerization, Erk1/2 phosphorylation, and chemotaxis. Treating CD4+ T cells with anti-CD3 antibody induced a marginal down-regulation of CXCR4 surface expression (data not shown), probably mediated via protein kinase C, which phosphorylates CXCR4 followed by receptor internalization (61). However, it is unlikely that this down-regulation accounted for the almost complete inhibition of gp120MN-induced chemotaxis we observed. Previous studies have shown that CXCR4 physically interacts with the TCR in response to signaling via CXCL12 and uses TCR signaling machinery from signal initiation to signal termination. CD3-mediated activation of protein kinase C leads to phospholipase C-β-3 phosphorylation, which in turn inhibits the activation of Gqα (62). However, it is also possible that the signaling pathways of both the TCR and CXCR4 use the same Gqα subunit. Therefore, stimulation of CD4+ T cells with anti-CD3 antibody prior to the gp120 chemotaxis assay transiently desensitizes CXCR4, rendering the cell unresponsive to the agonistic effects of the gp120MN-CXCR4 interaction. Pretreating resting CD4+ T cells with B Tat enhanced gp120MN-induced F-actin polymerization and Erk1/2 phosphorylation while also increasing the chemotactic response. However, conditioning the cells with C Tat resulted in a decrease of gp120MN-mediated events. This difference is possibly due to C Tat binding CXCR4 and acting as a CXCR4 antagonist (16). B Tat also binds CXCR4 with the same Kd as C Tat, but the augmentation of surface CXCR4 offsets any antagonistic events that may occur.
In addition to up-regulating the surface expression of CXCR4, B Tat enhanced gp120MN-induced chemotaxis signaling events, suggesting that B Tat may increase the susceptibility of resting CD4+ T cells to infection by X4 HIV-1. The best characterized latently infected cells are resting CD4+ T cells that are infected while in an activated state prior to returning to a resting state (63). However, HIV-1 can be isolated from CD45RA+ naïve CD4+ T cells in HIV-1-infected organ cultures and from HIV-1-infected individuals (64, 65). Moreover, resting CD4+ T cells can be latently infected in vivo without the requirement for a secondary stimulus (66). Taken together, this suggests that an activation step is not required for either HIV-1 entry or for HIV-1 integration. We show that B Tat increases the efficiency of X4 but not R5 viral entry into resting CD4+ T cells. In contrast, C Tat slightly inhibits X4 HIV-1IIIB infection. However, in activated CD4+ T cells, both Tat proteins inhibit X4-mediated but not R5-mediated viral infection/entry of cells. Therefore, B Tat may play a dual role—promoting the infection of resting CD4+ T cells while inhibiting the infection of activated cells, whereas C Tat inhibits infection of both resting and activated CD4+ T cells.
In a recent study of HIV-1-infected individuals, CXCR4-dependent CD4+ T cell migration was greater in subsets with increased CXCR4 expression (naïve and central memory) than in subsets with normal CXCR4 levels (effector memory and effector) (67). This finding is consistent with the present study and suggests that an increase of CD4+ T cells with increased CXCR4 expression in vivo may signal an enhanced response to HIV-1 particles. This increased capacity of X4 HIV-1 to infect resting CD4+ T cells may contribute to the depletion of this cell population observed during advanced HIV-1 infection and affect HIV-1 pathogenesis.
It is well established that R5 viruses are efficiently transmitted at the time of infection irrespective of clade, whereas R5X4 and X4 variants emerge in ∼40% of late stage clade B HIV-1 infection and is associated with more rapid disease progression and clinical deterioration of AIDS patients (20). Importantly, the R5 to X4 shift is both a consequence of low CD4+ T cell counts, promoting naïve CD4+ T cell division, and a cause of rapid CD4+ T cell decline, because X4 virus targets the naïve cell pool (24, 26). In contrast, usage of CXCR4 by HIV-1 clade C is uncommon (28, 29). It is possible that C Tat is less pathogenic, because monocytes treated with B Tat produce significantly more TNF and CCL2 than those treated with C Tat (16). TNF induces the down-regulation of CCR5 expression on monocytes and CD4+ T cells (68), whereas CCL2 induces increased surface expression of CXCR4 on CD4+ T cells (42). This may create a favorable environment for the expansion of CXCR4-using HIV-1 variants.
The quantity of Tat detected in the plasma of HIV-1-infected individuals (60) corresponds to the concentrations of Tat used in this study that significantly up-regulate CXCR4 surface expression on resting CD4+ T cells, suggesting that effective Tat concentrations might be reached in vivo. Moreover, higher concentrations than those present in the plasma may be found in lymphoid tissue, where productively infected cells are most frequent and where Tat may act locally after secretion from HIV-1-infected cells (60). In our experiments, we took care to work with reduced Tat, because oxidation of B Tat reduces its full in vitro activity (35, 69), which we observed when oxidation abrogated the ability of B Tat to up-regulate CXCR4 (data not shown). However, despite our precautions, we still used synthetic Tat proteins, and Tat found secreted in vivo may be modified and/or possess different activities.
A hallmark of HIV-1 infection is the progressive loss of CD4+ T cells via both apoptosis and activation-induced cell death (44, 55, 56). Studies have suggested that both Tat and gp120 may contribute to this depletion (57, 58). However, we were unable to detect an apoptotic effect of either Tat protein or gp120 on CD4+ T cells in the absence of any activating or other apoptotic stimuli (data not shown). However, when we serum-deprived the CD4+ T cells or stimulated them with anti-CD3, we observed that the Tat proteins induced comparable levels of apoptosis in accordance with previously published data (data not shown and Refs. 15 and 58).
Genetic variations in CCR2 play a role in HIV-1 pathogenesis. The CCR2-V64I allele also has a significant influence on the rate of HIV-1 disease progression in adults, with the presence of CCR2-V64I associated with a 2–3-year delay in progression to AIDS and death, although it has no impact upon HIV-1 transmission where CCR5 using viruses are dominant (70). Interestingly, although peripheral blood mononuclear cells isolated from CCR2-V64I heterologous donors have normal levels of both CCR5 and CCR2, and the calcium flux mediated by CCL2 through CCR2-V64I was unaffected, they have reduced surface expression of CXCR4 (71).
In summary, B Tat but not C Tat up-regulates CXCR4 expression on resting CD4+ T cells through a CCR2b-dependent mechanism that augments the ability of these cells to be chemoattracted to X4 gp120 and increases their permissiveness to X4 HIV-1 entry. Thus, B Tat has the capacity to render a large population of lymphocytes more susceptible to X4 HIV-1 late in the course of infection.
Acknowledgments
We thank Carol Mundy (Pediatrics, University of California, San Diego) for assistance in obtaining peripheral blood mononuclear cells. We also thank Jennifer D. Watkins, Didier Esquieu, and Daniel Lafitte (INSERM U911, Université de la Méditerranée, Marseille, France) for the synthesis and purification of the Tat proteins and Dennis Young (Flow Cytometry Core Facility, University of California, San Diego) for assistance in fluorescence-activated cell sorter analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant AI068632.
G. R. Campbell and S. A. Spector, unpublished observations.
- HIV-1
- human immunodeficiency virus, type 1
- APC
- allophycocyanin
- CCL
- chemokine (C-C motif) ligand
- CCR
- chemokine (C-C motif) receptor
- CXCL
- chemokine (CXC motif) ligand
- CXCR
- chemokine (CXC motif) receptor
- CRF
- circulating recombinant form
- Erk
- extracellular signal-regulated kinase
- F-actin
- filamentous actin
- FITC
- fluorescein isothiocyanate
- IL
- interleukin
- LTR
- long terminal repeat
- MAPK
- mitogen-activated protein kinase
- DPBS
- Dulbecco's phosphate-buffered saline
- PE
- phycoerythrin
- PerCP
- peridin-chlorophyll protein
- PHA
- phytohemagglutinin
- PTX
- pertussis toxin
- TCR
- T cell receptor
- SAPK
- stress-activated protein kinase
- JNK
- Jun amino-terminal kinase
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Hemelaar J., Gouws E., Ghys P. D., Osmanov S. (2006) AIDS 20, W13–W23 [DOI] [PubMed] [Google Scholar]
- 2.Renjifo B., Gilbert P., Chaplin B., Msamanga G., Mwakagile D., Fawzi W., Essex M.the Tanzanian Vitamin and HIV Study Group (2004) AIDS 18, 1629–1636 [DOI] [PubMed] [Google Scholar]
- 3.Bhoopat L., Rithaporn T. S., Khunamornpong S., Bhoopat T., Taylor C. R., Thorner P. S. (2006) Mod. Pathol. 19, 255–263 [DOI] [PubMed] [Google Scholar]
- 4.Vasan A., Renjifo B., Hertzmark E., Chaplin B., Msamanga G., Essex M., Fawzi W., Hunter D. (2006) Clin. Infect. Dis. 42, 843–852 [DOI] [PubMed] [Google Scholar]
- 5.Kaleebu P., Nankya I. L., Yirrell D. L., Shafer L. A., Kyosiimire-Lugemwa J., Lule D. B., Morgan D., Beddows S., Weber J., Whitworth J. A. (2007) J. Acquir. Immune Defic. Syndr. 45, 28–33 [DOI] [PubMed] [Google Scholar]
- 6.Coetzer M., Cilliers T., Ping L. H., Swanstrom R., Morris L. (2006) Virology 356, 95–105 [DOI] [PubMed] [Google Scholar]
- 7.Kumar M., Jain S. K., Pasha S. T., Chattopadhaya D., Lal S., Rai A. (2006) AIDS Res. Hum. Retroviruses 22, 1206–1219 [DOI] [PubMed] [Google Scholar]
- 8.Walker P. R., Ketunuti M., Choge I. A., Meyers T., Gray G., Holmes E. C., Morris L. (2007) AIDS Res. Hum. Retroviruses 23, 204–215 [DOI] [PubMed] [Google Scholar]
- 9.Pacyniak E., Gomez M. L., Gomez L. M., Mulcahy E. R., Jackson M., Hout D. R., Wisdom B. J., Stephens E. B. (2005) AIDS Res. Hum. Retroviruses 21, 379–394 [DOI] [PubMed] [Google Scholar]
- 10.Bell C. M., Connell B. J., Capovilla A., Venter W. D., Stevens W. S., Papathanasopoulos M. A. (2007) AIDS Res. Hum. Retroviruses 23, 322–330 [DOI] [PubMed] [Google Scholar]
- 11.Péloponèse J. M., Jr., Collette Y., Grégoire C., Bailly C., Campèse D., Meurs E. F., Olive D., Loret E. P. (1999) J. Biol. Chem. 274, 11473–11478 [DOI] [PubMed] [Google Scholar]
- 12.Ranga U., Shankarappa R., Siddappa N. B., Ramakrishna L., Nagendran R., Mahalingam M., Mahadevan A., Jayasuryan N., Satishchandra P., Shankar S. K., Prasad V. R. (2004) J. Virol. 78, 2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roof P., Ricci M., Genin P., Montano M. A., Essex M., Wainberg M. A., Gatignol A., Hiscott J. (2002) Virology 296, 77–83 [DOI] [PubMed] [Google Scholar]
- 14.Campbell G. R., Pasquier E., Watkins J., Bourgarel-Rey V., Peyrot V., Esquieu D., Barbier P., de Mareuil J., Braguer D., Kaleebu P., Yirrell D. L., Loret E. P. (2004) J. Biol. Chem. 279, 48197–48204 [DOI] [PubMed] [Google Scholar]
- 15.Campbell G. R., Watkins J. D., Esquieu D., Pasquier E., Loret E. P., Spector S. A. (2005) J. Biol. Chem. 280, 38376–38382 [DOI] [PubMed] [Google Scholar]
- 16.Campbell G. R., Watkins J. D., Singh K. K., Loret E. P., Spector S. A. (2007) J. Virol. 81, 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell G. R., Senkaali D., Watkins J. D., Esquieu D., Opi S., Yirrell D. L., Kaleebu P., Loret E. P. (2007) Vaccine 25, 8441–8447 [DOI] [PubMed] [Google Scholar]
- 18.de Mareuil J., Carre M., Barbier P., Campbell G. R., Lancelot S., Opi S., Esquieu D., Watkins J. D., Prevot C., Braguer D., Peyrot V., Loret E. P. (2005) Retrovirology 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranjbar S., Rajsbaum R., Goldfeld A. E. (2006) J. Immunol. 176, 4182–4190 [DOI] [PubMed] [Google Scholar]
- 20.van Opijnen T., Jeeninga R. E., Boerlijst M. C., Pollakis G. P., Zetterberg V., Salminen M., Berkhout B. (2004) J. Virol. 78, 3675–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger E. A. (1997) AIDS 11, S3–S16 [PubMed] [Google Scholar]
- 22.Philpott S. M. (2003) Curr. HIV Res. 1, 217–227 [DOI] [PubMed] [Google Scholar]
- 23.Koot M., van't Wout A. B., Kootstra N. A., de Goede R. E., Tersmette M., Schuitemaker H. (1996) J. Infect. Dis. 173, 349–354 [DOI] [PubMed] [Google Scholar]
- 24.Blaak H., van't Wout A. B., Brouwer M., Hooibrink B., Hovenkamp E., Schuitemaker H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Correa R., Muñoz-Fernández M. A. (2001) AIDS 15, 1959–1963 [DOI] [PubMed] [Google Scholar]
- 26.Davenport M. P., Zaunders J. J., Hazenberg M. D., Schuitemaker H., van Rij R. P. (2002) Trends Microbiol. 10, 275–278 [DOI] [PubMed] [Google Scholar]
- 27.Björndal A., Deng H., Jansson M., Fiore J. R., Colognesi C., Karlsson A., Albert J., Scarlatti G., Littman D. R., Fenyö E. M. (1997) J. Virol. 71, 7478–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cecilia D., Kulkarni S. S., Tripathy S. P., Gangakhedkar R. R., Paranjape R. S., Gadkari D. A. (2000) Virology 271, 253–258 [DOI] [PubMed] [Google Scholar]
- 29.Ndung'u T., Sepako E., McLane M. F., Chand F., Bedi K., Gaseitsiwe S., Doualla-Bell F., Peter T., Thior I., Moyo S. M., Gilbert P. B., Novitsky V. A., Essex M. (2006) Virology 347, 247–260 [DOI] [PubMed] [Google Scholar]
- 30.Sotsios Y., Ward S. G. (2000) Immunol. Rev. 177, 217–235 [DOI] [PubMed] [Google Scholar]
- 31.Huang L., Bosch I., Hofmann W., Sodroski J., Pardee A. B. (1998) J. Virol. 72, 8952–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Secchiero P., Zella D., Capitani S., Gallo R. C., Zauli G. (1999) J. Immunol. 162, 2427–2431 [PubMed] [Google Scholar]
- 33.Jeang K. T., Xiao H., Rich E. A. (1999) J. Biol. Chem. 274, 28837–28840 [DOI] [PubMed] [Google Scholar]
- 34.Huigen M. C., Kamp W., Nottet H. S. (2004) Eur. J. Clin. Invest. 34, 57–66 [DOI] [PubMed] [Google Scholar]
- 35.Campbell G. R., Loret E. P. (2009) Retrovirology 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albini A., Benelli R., Giunciuglio D., Cai T., Mariani G., Ferrini S., Noonan D. M. (1998) J. Biol. Chem. 273, 15895–15900 [DOI] [PubMed] [Google Scholar]
- 37.Barany G., Merrifield R. B. (1980) in The Peptide: Analysis, Synthesis, Biology (Gross E., Meinhofer J. eds) Vol. 2, pp. 1–284, Academic Press, New York [Google Scholar]
- 38.Barré-Sinoussi F., Chermann J. C., Rey F., Nugeyre M. T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vézinet-Brun F., Rouzioux C., Rozenbaum W., Montagnier L. (1983) Science 220, 868–871 [DOI] [PubMed] [Google Scholar]
- 39.Wain-Hobson S., Vartanian J. P., Henry M., Chenciner N., Cheynier R., Delassus S., Martins L. P., Sala M., Nugeyre M. T., Guétard D. (1991) Science 252, 961–965 [DOI] [PubMed] [Google Scholar]
- 40.Lole K. S., Bollinger R. C., Paranjape R. S., Gadkari D., Kulkarni S. S., Novak N. G., Ingersoll R., Sheppard H. W., Ray S. C. (1999) J. Virol. 73, 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Japour A. J., Mayers D. L., Johnson V. A., Kuritzkes D. R., Beckett L. A., Arduino J. M., Lane J., Black R. J., Reichelderfer P. S., D'Aquila R. T. (1993) Antimicrob. Agents Chemother. 37, 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell G. R., Spector S. A. (2008) J. Biol. Chem. 283, 30745–30753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balabanian K., Harriague J., Décrion C., Lagane B., Shorte S., Baleux F., Virelizier J. L., Arenzana-Seisdedos F., Chakrabarti L. A. (2004) J. Immunol. 173, 7150–7160 [DOI] [PubMed] [Google Scholar]
- 44.Patrussi L., Ulivieri C., Lucherini O. M., Paccani S. R., Gamberucci A., Lanfrancone L., Pelicci P. G., Baldari C. T. (2007) Blood 110, 1730–1738 [DOI] [PubMed] [Google Scholar]
- 45.Opi S., Péloponèse J. M., Jr., Esquieu D., Campbell G., de Mareuil J., Walburger A., Solomiac M., Grégoire C., Bouveret E., Yirrell D. L., Loret E. P. (2002) J. Biol. Chem. 277, 35915–35919 [DOI] [PubMed] [Google Scholar]
- 46.Opi S., Péloponèse J. M., Jr., Esquieu D., Watkins J., Campbell G., De Mareuil J., Jeang K. T., Yirrell D. L., Kaleebu P., Loret E. P. (2004) Vaccine 22, 3105–3111 [DOI] [PubMed] [Google Scholar]
- 47.Mirzadegan T., Diehl F., Ebi B., Bhakta S., Polsky I., McCarley D., Mulkins M., Weatherhead G. S., Lapierre J. M., Dankwardt J., Morgans D., Jr., Wilhelm R., Jarnagin K. (2000) J. Biol. Chem. 275, 25562–25571 [DOI] [PubMed] [Google Scholar]
- 48.Pelchen-Matthews A., Armes J. E., Marsh M. (1989) EMBO J. 8, 3641–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Y., Zhang L., Goodwin J. S., Wang Z., Liu B., Zhang J., Fan G. H. (2008) Exp. Cell Res. 314, 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babcock G. J., Mirzabekov T., Wojtowicz W., Sodroski J. (2001) J. Biol. Chem. 276, 38433–38440 [DOI] [PubMed] [Google Scholar]
- 51.Riol-Blanco L., Sánchez-Sánchez N., Torres A., Tejedor A., Narumiya S., Corbí A. L., Sánchez-Mateos P., Rodríguez-Fernández J. L. (2005) J. Immunol. 174, 4070–4080 [DOI] [PubMed] [Google Scholar]
- 52.Kumar A., Humphreys T. D., Kremer K. N., Bramati P. S., Bradfield L., Edgar C. E., Hedin K. E. (2006) Immunity 25, 213–224 [DOI] [PubMed] [Google Scholar]
- 53.Wu L., Gerard N. P., Wyatt R., Choe H., Parolin C., Ruffing N., Borsetti A., Cardoso A. A., Desjardin E., Newman W., Gerard C., Sodroski J. (1996) Nature 384, 179–183 [DOI] [PubMed] [Google Scholar]
- 54.Curnock A. P., Sotsios Y., Wright K. L., Ward S. G. (2003) J. Immunol. 170, 4021–4030 [DOI] [PubMed] [Google Scholar]
- 55.Finkel T. H., Tudor-Williams G., Banda N. K., Cotton M. F., Curiel T., Monks C., Baba T. W., Ruprecht R. M., Kupfer A. (1995) Nat. Med. 1, 129–134 [DOI] [PubMed] [Google Scholar]
- 56.Noraz N., Gozlan J., Corbeil J., Brunner T., Spector S. A. (1997) AIDS 11, 1671–1680 [DOI] [PubMed] [Google Scholar]
- 57.Régulier E. G., Reiss K., Khalili K., Amini S., Zagury J. F., Katsikis P. D., Rappaport J. (2004) Int. Rev. Immunol. 23, 25–59 [DOI] [PubMed] [Google Scholar]
- 58.Westendorp M. O., Frank R., Ochsenbauer C., Stricker K., Dhein J., Walczak H., Debatin K. M., Krammer P. H. (1995) Nature 375, 497–500 [DOI] [PubMed] [Google Scholar]
- 59.Ghezzi S., Noonan D. M., Aluigi M. G., Vallanti G., Cota M., Benelli R., Morini M., Reeves J. D., Vicenzi E., Poli G., Albini A. (2000) Biochem. Biophys. Res. Commun. 270, 992–996 [DOI] [PubMed] [Google Scholar]
- 60.Xiao H., Neuveut C., Tiffany H. L., Benkirane M., Rich E. A., Murphy P. M., Jeang K. T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11466–11471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peacock J. W., Jirik F. R. (1999) J. Immunol. 162, 215–223 [PubMed] [Google Scholar]
- 62.Stanners J., Kabouridis P. S., McGuire K. L., Tsoukas C. D. (1995) J. Biol. Chem. 270, 30635–30642 [DOI] [PubMed] [Google Scholar]
- 63.Han Y., Wind-Rotolo M., Yang H. C., Siliciano J. D., Siliciano R. F. (2007) Nat. Rev. Microbiol. 5, 95–106 [DOI] [PubMed] [Google Scholar]
- 64.Eckstein D. A., Penn M. L., Korin Y. D., Scripture-Adams D. D., Zack J. A., Kreisberg J. F., Roederer M., Sherman M. P., Chin P. S., Goldsmith M. A. (2001) Immunity 15, 671–682 [DOI] [PubMed] [Google Scholar]
- 65.Kinter A. L., Umscheid C. A., Arthos J., Cicala C., Lin Y., Jackson R., Donoghue E., Ehler L., Adelsberger J., Rabin R. L., Fauci A. S. (2003) J. Immunol. 170, 2449–2455 [DOI] [PubMed] [Google Scholar]
- 66.Agosto L. M., Yu J. J., Dai J., Kaletsky R., Monie D., O'Doherty U. (2007) Virology 368, 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Patrigeon S., Vingert B., Lambotte O., Viard J. P., Delfraissy J. F., Thèze J., Chakrabarti L. A. (2009) AIDS 23, 1197–1207 [DOI] [PubMed] [Google Scholar]
- 68.Lane B. R., Markovitz D. M., Woodford N. L., Rochford R., Strieter R. M., Coffey M. J. (1999) J. Immunol. 163, 3653–3661 [PubMed] [Google Scholar]
- 69.Fanales-Belasio E., Moretti S., Nappi F., Barillari G., Micheletti F., Cafaro A., Ensoli B. (2002) J. Immunol. 168, 197–206 [DOI] [PubMed] [Google Scholar]
- 70.Smith M. W., Dean M., Carrington M., Winkler C., Huttley G. A., Lomb D. A., Goedert J. J., O'Brien T. R., Jacobson L. P., Kaslow R., Buchbinder S., Vittinghoff E., Vlahov D., Hoots K., Hilgartner M. W., O'Brien S. J. (1997) Science 277, 959–965 [DOI] [PubMed] [Google Scholar]
- 71.Lee B., Doranz B. J., Rana S., Yi Y., Mellado M., Frade J. M., Martinez-A C., O'Brien S. J., Dean M., Collman R. G., Doms R. W. (1998) J. Virol. 72, 7450–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]