Abstract
Members of the conserved 14-3-3 protein family spontaneously self-assemble as homo- and heterodimers via conserved sequences in the first four (αA-αD) of the nine helices that comprise them. Dimeric 14-3-3s bind conserved motifs in diverse protein targets involved in multiple essential cellular processes including signaling, intracellular trafficking, cell cycle regulation, and modulation of enzymatic activities. However, recent mostly in vitro evidence has emerged, suggesting functional and regulatory roles for monomeric 14-3-3s. We capitalized on the simplicity of the 14-3-3 family in Drosophila to investigate in vivo 14-3-3ζ monomer properties and functionality. We report that dimerization is essential for the stability and function of 14-3-3ζ in neurons. Moreover, we reveal the contribution of conserved amino acids in helices A and D to homo- and heterodimerization and their functional consequences on the viability of animals devoid of endogenous 14-3-3ζ. Finally, we present evidence suggesting endogenous homeostatic adjustment of the levels of the second family member in Drosophila, D14-3-3ϵ, to transgenic monomeric and dimerization-competent 14-3-3ζ.
Keywords: Cell/Neuron, Cell/Intracellular Processing, Chaperones, Genetics/Drosophila, Protein/Conformation, Tissue/Organ Systems/Brain, 14-3-3
Introduction
14-3-3s are a ubiquitous family of highly conserved polypeptides, present in all eukaryotes, with the number of isoforms varying between species, from a single protein in Giardia lamblia to nine in mammals and over fifteen in plants (1). 14-3-3s bind phosphorylated serines or phosphorylated threonines primarily in the conserved motifs RSXpSXP or RXXXpSXP (pS is phosphoserine) of over 200 protein targets (2–6). Binding to phosphoserines and threonines is a functional molecular hallmark of these proteins and may alter the subcellular localization, phosphorylation status, and enzymatic activity (6–9) of client proteins. 14-3-3s targets are of cardinal importance in diverse signaling cascades, metabolism, cell cycle regulation, apoptosis, and protein trafficking (6, 10, 11). Involvement of 14-3-3s in such essential processes, which underlie cell fate determination, function, and carcinogenesis in a broad range of organisms and cell types, demonstrates the importance of understanding their functional properties and regulation.
In addition to phospho-Ser/Thr binding, dimerization is another elemental functional property of 14-3-3s. Each monomer consists of nine antiparallel α-helices (αA to aI) organized in two domains (12–14). The N-terminal domain consists of αA-αD and mediates dimer interactions, where αA and αB of one subunit interact with αC and αD of the dimerization partner (12, 15). The C-terminal helices (αE-αI) form an amphipathic target binding groove. Although each monomer contains a ligand binding site and can associate with targets independently (16–18), 14-3-3s self-assemble spontaneously into homo- and heterodimers (1, 12). Importantly, the U-shaped groove formed by homo- and heterodimers can interact with two motifs on a single, or different client proteins. This is thought to promote interactions between distinct 14-3-3 targets (9, 19, 20), or to alter the conformation and activity of a single client (18, 21–23). Therefore, the precise composition of each dimer is highly significant functionally, because it probably dictates the range of its possible clients. This was nicely demonstrated by the obligatory 14-3-3β/14-3-3ϵ heterodimer for aldosterone regulation of a kidney epithelial sodium channel (24). Such requirements may also be reflected in the intrinsic dimerization properties of 14-3-3s, illustrated by 14-3-3σ selectively forming homodimers and 14-3-3ϵ preferentially heterodimers, in the extremes of the range of possibilities.
In vitro and in cultured cells, certain mutant dimerization-impaired 14-3-3s have been shown to bind clients (25, 26), often with similar affinities as their dimeric counterparts, but with some exceptions, they appear unable to support normal target activity (21, 25, 27, 28). In addition, phosphorylation of Ser58 on vertebrate 14-3-3ζ renders it unable to dimerize in transfected cultured cells (29), suggesting that monomerization may regulate some 14-3-3 functions. Similarly, a monomeric Drosophila 14-3-3ζ has been reported to interact with and inhibit the activity of the calcium-dependent potassium channel Slowpole (dSlo), equally well with the wild-type protein, but in a heterologous system (Zhou et al. (34)). Although provocative, these results have not to date been examined in the context of an intact animal and more specifically in a tissue where 14-3-3s are found normally. Given the role of these proteins in multiple vital processes and the potential regulatory role of monomerization, it is essential to evaluate these conclusions in vivo and in a native experimental cellular environment expected to contain natural regulators of 14-3-3 homeostasis and function.
We used Drosophila melanogaster to study 14-3-3 dimerization and whether monomers exist stably and function in vivo or are transient, possibly regulatory intermediate species because it offers distinct advantages for this study. It has only two well characterized genes representing the two 14-3-3 conservation groups, leonardo (leo) encoding three D14-3-3ζ (LEO) isoforms and D14-3-3ϵ, encoding the 14-3-3ϵ ortholog (30–32). Secondly, null and hypomorphic mutants of both genes are available (30, 33). Thus, unlike the situation in cultured cells, the effects of dimerization mutant 14-3-3s can be studied largely without potential interference from resident wild-type proteins. Third, the transgenic D14-3-3ζ proteins utilized were studied largely in the fly central nervous system (CNS)2 system, a tissue where they are abundant in wild-type animals (32), and are expected to harbor resident mechanisms to regulate their levels and activity (30). Finally, with the observation described herein that these isoforms homodimerize and heterodimerize with each other, Drosophila constitutes a simple, but highly representative system to functionally dissect 14-3-3s in vivo. We show that under conditions of homeostasis, dimerization is necessary for LEO protein stability and function.
EXPERIMENTAL PROCEDURES
Drosophila Culture and Strains
D. melanogaster strains were cultured in standard wheat-flour-sugar food supplemented with soy flour and CaCl2, at 21–23 °C (30). Wild-type (WW) cDNAs and ones encoding the single (WM and MW) mutations and the combined double mutant (MM) described previously (34) were a kind gift from Yi Zhou and Irwin B. Levitan. These cDNAs were subcloned into the pUAST vector, and the resulting constructs were used to generate leoFLAG transformants followed by genetic background normalization to that of the Cantonised w1188 resident in the laboratory (35). Determination of transgene chromosomal localization and introduction in leoP1188/CyO or leoP2335/CyO mutant background was achieved with standard genetic crosses. The lethal leoP1188 and leoP2335 alleles have been described previously (32, 33). Because the leo gene resides on the second chromosome, transgenic lines bearing leoFLAG on the X or third chromosomes were used to introduce the transgenes in leoP1188/CyO or leoP2335/CyO mutant backgrounds for ease of genetic manipulations.
The Elav-Gal4, Actin-Gal4, and Tub-Gal80ts have been described previously (–31), whereas Elav-Gal4; leoP1188/CyO and Elav-Gal4; Tub-Gal80ts were generated by standard crosses. Negative controls for all experiments were heterozygotes of each Gal4 driver obtained by crossing driver homozygotes with w1188. Transgene expression under Elav-Gal4; Tub-Gal80ts was induced specifically in adult flies by incubation at 30 °C for 24 h. Temporal regulation of UAS controlled transgenes under the ubiquitously expressed temperature sensitive Gal80ts has been described before (37). Briefly, at 18 °C the ubiquitously expressed under the tubulin promoter Gal80ts competes effectively with Gal4 for binding to the UAS and blocks transgene transcription. In contrast, Gal80ts becomes inactive and allows Gal4-driven transcription after incubation at temperatures of 29–31 °C, thus enabling temporal control of transgene expression.
To assess the ability of leoFLAG transgenes to rescue the embryonic lethality of leoP1188or leoP2335 homozygotes, males bearing leoFLAG in leoP1188/ CyO or leoP2335/ CyO background were crossed en masse with Elav-Gal4; leoP1188/CyO females. The number of non CyO progeny denoting leoP1188, leoP2335 homozygotes or leoP1188/leoP2335 trans-heterozygotes, and the total number of progeny from such a cross were determined. If these mutant animals were fully viable, they would amount to one third of total progeny because homozygotes for the CyO chromosome die as embryos. Therefore, rescue from lethality was calculated as the percentage of the expected one third of total progeny, which was comprised by non CyO flies. For the leoFLAGMW transgene residing on the X chromosome, only females were counted. Data were averaged from at least three independent crosses.
Western Blotting
Four fly heads were homogenized in 40 μl of lysis buffer (50 mm Tris pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, containing protease and phosphatase inhibitor mixture, Sigma). Equivalence between samples in total protein was confirmed using Quant-iT (Molecular Probes). Laemmli buffer was added, samples were boiled for 5 min at 92 °C, and centrifuged for 5 min at 14,000 × g. Proteins were separated by SDS gel electrophoresis (1.5 h, 200 V) and transferred to polyvinylidene difluoride membrane (1 h, 100 V). Total protein equivalent to half a head/lane was loaded, unless stated otherwise, and each sample was run on duplicate gels. Membranes were probed with rabbit anti-LEO pAb (32) at 1:20,000, mouse anti-FLAG M2 mAb (Sigma) at 1:600, chicken anti-D14-3-3ϵ pAb (30) at 1:5,000 or mouse anti-syntaxin mAb (8C3, DSHB) at 1:5,000; followed by appropriate secondary horseradish peroxidase-conjugate Ab (1:5,000; Jackson ImmunoResearch). Bands were visualized with chemiluminesence (Pierce). The results from three independent experiments were quantified densitometrically using ImageQuant 5.0 (Molecular Dynamics) and analyzed statistically.
Cross-linking
100 heads were lysed in 250 μl of ice-cold buffer (20 mm sodium phosphate, pH 7.4, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride with phosphatase and protease inhibitor mixture). Head lysates were centrifuged for 15 min at 14,000 × g at 4 °C to remove lipids and carbohydrates and protein amounts were measured using the Quant-iT (Molecular Probes). Approximately 4 mg of protein from each sample in a final volume of 200 μl were cross-linked by incubation with 1.5 mm BS3 (Pierce), for 2 h on ice. The reaction was stopped by the addition of lysis buffer. Samples were subjected to a pull-down assay with anti-FLAG beads, as described below, and analyzed by Western blotting.
Pull-down Assay
100 heads were lysed in 250 μl of ice-cold lysis buffer (50 mm Tris pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, including protease, and phosphatase inhibitor mixture). Lipids and carbohydrates were removed by centrifugation and equal amounts of protein from each sample subjected to a pull-down assay using anti-FLAG M2 Affinity Gel (Sigma) according to the manufacturer's instructions. Briefly, lysates were incubated with 50 μl of anti-FLAG beads for 4 h, at 4 °C, with rotation. Beads were collected by centrifugation, washed twice with lysis buffer and three times with wash buffer (25 mm Tris, pH 7.5, 250 mm LiCl, 1% Triton X-100, protease, and phosphatase inhibitor mixture). Pulled-down material was eluted by boiling for 10 min in 40 μl of Laemmli buffer, run on a SDS gel and blotted as described above.
Reverse Transcription and Real-time PCR
20 heads were lysed in TRIzol (Invitrogen), and RNA was extracted according to the manufacturer's instructions. 1 μg of mRNA was reverse transcribed with random primers using ImProm-I reverse transcriptase (Promega). The resulting cDNA was diluted 1:50 and 4 μl were used per PCR reaction. Reactions were performed using the MiniOpticon System for Real Time PCR (Bio-Rad), with Platinum SYBR Green qPCR Supermix UDG (Invitrogen). leoFLAG transcripts were amplified with leoforward (GCAGCCCACACATCCAATCAG) and FLAG reverse primers (TCATCATCATCCTTATAATCG). Act5C was also amplified to control for relative amounts of RNA between samples, using act5C forward and reverse primers (30). For each sample, leoFLAG and act5C were assayed in separate wells and in triplicate. Reactions were monitored with the MJ Opticon Monitor Analysis software (v3.1), and data analyzed with the relative quantification method previously described by Pfaffl (2001) (40). Results presented are the average of three runs from two independent reverse transcription reactions.
Statistical Analysis
Statistical analysis was performed using Student's paired t-tests.
RESULTS
For consistency, we have kept the nomenclature of mutations in 14-3-3ζ used to generate the transgenes for our study as originally described by Zhou et al. (34), where M denotes the mutated site and W the wild-type sequence. The first mutations, Leu15-Ala-Glu to Gln15-Gln-Arg, in leoFLAGMW are located in helix A, and the Arg88-Val-Glu to Asn88-Val-Gln in leoFLAGWM mutations are located in helix D (supplemental Fig. S1). The leoFLAGMM transgene carries both mutations. Because endogenous LEO is highly enriched in the CNS, we targeted expression of transgenes to this tissue using the Elav-Gal4 driver (38) and determined the levels of the resultant proteins in adult head lysates.
Dimerization Is Essential for LEO Stability
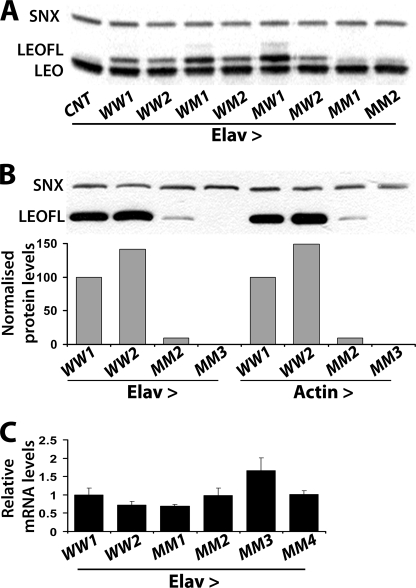
Initially we determined the abundance of the putative monomeric transgenic proteins to address the possibility that impaired dimerization might render them unstable as we have previously reported for subunits of other multimeric complexes (41). The FLAG-tagged transgenic proteins were easily discernable from endogenous LEO because of their larger size (Fig. 1A, compare lanes 1 and 2) and appeared highly expressed in the Drosophila CNS. In fact, one each of the two independent lines for leoFLAGWM and leoFLAGMW accumulated 2 and 2.5 times higher than the wild-type transgenic protein respectively (quantification not shown). Surprisingly however given previous reports (34), doubly mutant transgenic protein was not detectable for the lines shown in Fig. 1 and additional independent leoFLAGMM lines (supplemental Fig. 2A). The leoFLAGMM insert in the transgene was then re-cloned from the transgenic flies, sequenced in its entirety and additional mutations, such as spurious stop codons were not uncovered. Therefore, to determine whether the doubly mutant protein is produced but is unstable, we blotted higher amounts of extracts from animals expressing the transgenes under Elav. Furthermore, to investigate whether the protein is unstable only in neurons, we expressed the same transgenes ubiquitously under the Actin-Gal4 driver. To increase the resolution of the assay, we probed for the presence of the transgenic proteins with the anti-FLAG mAb. The LEOFLAGMM protein in line MM2 is present both ubiquitously and specifically in neurons albeit at least 10-fold lower than its wild-type transgenic counterparts (Fig. 1B). Transgenic protein was not detected from the MM3 line under these conditions. Therefore, the levels of doubly mutant transgenic proteins are low irrespective of the tissue where they are found, suggesting that they may be intrinsically unstable.
FIGURE 1.
The doubly mutant LEOFLAGMM protein is not abundant relative to its single mutant or wild-type counterparts in vivo. A, Western blot of head lysates from animals expressing the indicated transgenes under Elav-Gal4-driven. WW, WM, MW, and MM represent the respective LEOFLAG proteins yielded by independent transgenes as denoted by the numbers 1, 2. Elav-Gal4 heterozygotes with w1118 were used as the negative control (CNT). LEO and the transgenic LEOFLAG proteins were revealed with the anti-LEO pAb, whereas anti-Syntaxin (SNX) was used to control for the amount loaded per lane. B, similar Western blot of head lysates from animals expressing the indicated wild-type (WW) and double mutant (MM) proteins from independent transgenes under Elav or Actin-Gal4 probed with the anti-FLAG mAb. A quantification of the protein levels from three such independent blots is shown below. LEOFLAG/SNX levels are expressed as a percentage of levels obtained in the arbitrarily selected as control WW1 line. C, leoFLAG mRNA levels normalized to those of act5C measured by Q-RT-PCR. leoFLAG/act5C ratios are presented as fold change relative to those obtained in the WW1 transgenic line. Error bars show the S.D. from three independent experiments.
Although unlikely, because similar results were obtained with multiple independent lines, the marginal levels of LEOFLAGMM could be a consequence of impaired transcription because of transgene genomic location (positional) effects. To resolve this, we performed quantitative PCR with transgene-specific primers on RNA from adult brains expressing pan-neuronally leoFLAGMM and two wild-type transgenes. The results clearly demonstrate that all leoFLAGMM transgenes were expressed equally with, or higher than their wild-type counterparts (Fig. 1C). Collectively, these results indicate that unlike the reports from the heterologous system (34), the combined Gln15-Gln-Arg and Asn88-Val-Gln mutations result in a dramatic post-transcriptional reduction in the steady state LEOFLAG levels in vivo.
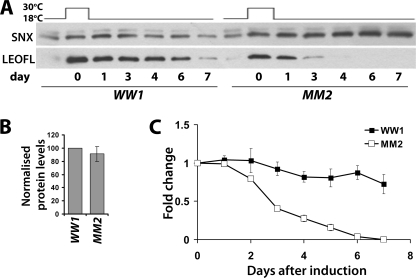
To determine whether the low levels of LEOFLAGMM reflect instability of the mutant protein rather than decreased protein synthesis, we controlled its expression both spatially and temporally using the Gal80ts system (Elav-Gal4; Gal80ts, see “Experimental Procedures”). The leoFLAGWW and leoFLAGMM transgenes were kept inactive by culturing at 18 °C throughout development. Pan-neuronal transgene transcription was induced for 24 h under the Elav-Gal4 driver by shifting adult flies to 30 °C. Further transcription was blocked by returning to the non-permissive temperature (18 °C), and the levels of transgenic proteins accumulated during the permissive 24 h were monitored over the next several days (Fig. 2). Immediately following transgene induction, the relative level of the doubly mutant LEOFLAGMM was not statistically different from that of the wild-type LEOFLAGWW control (Fig. 2, A and B). However, the doubly mutant LEOFLAGMM protein declined rapidly to nearly undetectable levels by day 4, whereas the wild-type transgenic protein was easily detectable 7 days post-induction (Fig. 2A). Quantification of multiple independent such experiments demonstrated that LEOFLAGMM was less that 50% of control 3 days post-induction and declined below detection under these conditions within 6 days (Fig. 2B). In contrast, the level of control LEOFLAGWW remained robust even beyond the seventh day that was systematically monitored, in agreement with the reported long perdurance of LEO in vivo (33). These observations were confirmed with the independent lines WW2 and MM1 (data not shown). The results, in congruence with the quantitative PCR data, demonstrate that neither transcription nor translation of the doubly mutant LEOFLAGMM are impaired. Rather, its low steady state levels reflect its instability. Therefore, as in cultured cells after transfection, the levels of the monomeric LEO protein were similar to controls immediately after conditions of elevated transcription (repression of Gal80ts), but the protein is unstable under steady state conditions. Because the mutant protein is thought unable to dimerize, these results suggest that dimerization is essential for LEO stability.
FIGURE 2.
LEOFLAGMM is an unstable protein in vivo. A, leoFLAGWW and leoFLAGMM were conditionally expressed specifically in the adult nervous system under Elav-Gal4; Gal80ts by 24 h of incubation at 30 °C. Head lysates were collected prior to induction (blank), after 24 h of induction (0), and after transferring the flies to 18 °C at the time points indicated. A representative Western blot of these samples challenged with anti-FLAG mAb blot to monitor the transgenic proteins is shown, with syntaxin (SNX) serving as a loading control. B, mean LEOFLAG/SNX levels ± S.E. directly after induction (day 0), presented as a percentage of that ratio in the WW1 line, quantified over three independent experiments run in duplicate. C, degradation of the wild-type and double mutant transgenic LEOFLAG presented as the respective ratio with SNX for each indicated time point as an average fold change relative to the ratio at day 0 and plotted as a function of time. Error bars are as in B.
Differential LEO Dimerization by Interface Mutations
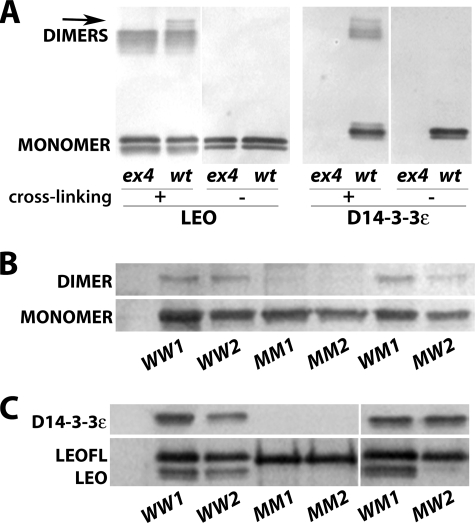
Although assumed (30, 42), it had not been demonstrated that Drosophila 14-3-3s homo- and heterodimerize in vivo, especially in the CNS. To establish this, we cross-linked proteins in head lysates from wild-type animals and D14-3-3ϵex4 mutant homozygotes lacking D14-3-3ϵ (30). The anti-LEO antibody detected both monomers and dimers in extracts from wild-type and D14-3-3ϵex4 animals (Fig. 3A). Importantly, the dimers detected with the LEO antibody in extracts from D14-3-3ϵex4 animals can only be LEO homodimers (Fig. 3A, first lane). As expected, the anti-D14-3-3ϵ antibody detected dimers only in wild-type animals, but interestingly both antibodies detected a band of very slow mobility in extracts from wild-type animals, absent from D14-3-3ϵex4 mutant lysates (arrow in Fig. 3A, lanes 2 and 6). Because the latter harbor only LEO homodimers, these complexes pointed out by the arrows must represent LEO/D14-3-3ϵ heterodimers detected in vivo for the first time.
FIGURE 3.
The wild-type and single mutant LEOFLAG proteins dimerize, but the double mutant LEOFLAGMM is monomeric. A, cross-linked proteins from head extracts of either wild type-w1118 (wt) or D14-3-3ϵex4 (ex4) homozygous mutant flies were Western blotted with anti-LEO or anti-D14-3-3ϵ pAbs. B, cross-linked LEOFLAG complexes from head extracts of flies expressing the indicated leoFLAG under Elav-Gal4 were harvested with anti-FLAG beads and detected by anti-FLAG mAb Western blot. C, LEOFLAG proteins under Elav-Gal4 were immunoprecipitated from head lysates with anti-FLAG mAb, and co-immunoprecipitated LEO and D14-3-3ϵ were detected with their respective antibodies by Western blotting.
Provided that Drosophila 14-3-3s homo- and heterodimerize, we tested the hypothesis that impaired dimerization is detrimental to LEOFLAGMM stability in vivo. Hence, we determined the dimerization profile of wild-type and mutant LEOFLAG proteins in the adult CNS. Proteins in the lysates were cross-linked, pulled-down with anti-FLAG beads, run on a Western blot, and probed with the anti-FLAG mAb (Fig. 3B). Loading was adjusted such that approximately equivalent LEOFLAG amounts from the different transgenic lines were assayed. Slow migrating bands corresponding to the position expected of 14-3-3 dimers were detectable in head lysates containing LEOFLAGWW from two independent lines, LEOFLAGWM and to a lesser degree LEOFLAGMW. However, such bands were absent from the two independent LEOFLAGMM-containing lysates (Fig. 3B). To confirm the cross-linking data independently and to probe whether the transgenic proteins dimerize with endogenous 14-3-3s, FLAG-containing proteins were immunoprecipitated from head lysates and probed for endogenous LEO and D14-3-3ϵ (Fig. 3C). As for the native proteins (Fig. 3A), LEOFLAGWW and LEOFLAGWM formed homodimers with endogenous LEO and heterodimers with D14-3-3ϵ (Fig. 3C). In contrast, LEOFLAGMM did not dimerize with either of the endogenous 14-3-3s, in congruence with the cross-linking data (Fig. 3C). Surprisingly, the LEOFLAGMW single mutant protein appeared unable to dimerize with endogenous LEO, but was clearly able to form heterodimers with D14-3-3ϵ (Fig. 3C). These results indicate that interactions involving Leu15-Ala-Glu in helix αA are required for homodimerization, but seem dispensable for heterodimer formation.
Dimerization Is Necessary for LEO Function
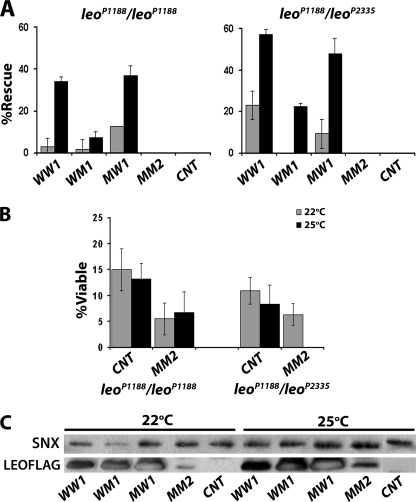
We capitalized on the distinct dimerization properties of the four transgenic LEO proteins to investigate whether they are functional in vivo. We employed a previously established assay (30, 43), to test the ability of leoFLAG transgenes to reverse the homozygous lethality of leo mutant alleles. Because survival of an embryo lacking endogenous LEO depends on proper operation of a number of essential molecular processes involving the transgenic protein, this assay constitutes a broad test of 14-3-3 functionality. LEO is essential for embryonic development and for the function of the nervous system during hatching into larvae (32, 44, 45), but apparently largely dispensable for viability in later stages (33). Thus, we tested for improvement of the 10–15% (background) viability exhibited by homozygotes or heteroallelics for the transposon insertion alleles leoP1188 and leoP2335 (32, 33) under the conditions we employed (Fig. 4B). The highest expressing transgenic line for each LEOFLAG protein was crossed into these mutant strains and driven by Elav-Gal4. Pan-neuronal expression of leoFLAGWW at 25 °C increased the number of leoP1188 homozygotes and leoP1188/leoP2335 heteroallelics surviving to adulthood to 30% above the background for the former and nearly 60% for the latter (Fig. 4A). The different rescue levels for leoP1188 homozygotes and leoP1188/leoP2335 heteroallelics reflect the relative strength of the mutations as described previously (33). Lowering the culture temperature to 22 °C reduced the effect to near elimination for leoP1188 homozygotes, demonstrating that it specifically depends on transgenic protein dosage, because Gal4-driven expression is elevated at higher temperatures (36, 46). This is also clearly demonstrated by monitoring LEOFLAG abundance in the heads of leoP1188 homozygotes, which should contain only transgenic LEO if any at all (Fig. 4C). Therefore improvements in mutant homozygote survival reflect levels and activity of the transgenic proteins.
FIGURE 4.
Differential rescue of leo mutant lethality by transgenic LEOFLAG proteins. A, indicated leoFLAG transgenes were expressed in the nervous system under Elav-Gal4 in leoP1188/leoP1188 homozygous or leoP1188/leoP2335 heteroallelic animals, and the number of resultant adult flies of each genotype was determined. Rescue was calculated as the % of the expected number of homozygous or heteroallelic flies carrying the particular leoFLAG transgene if these flies were fully viable. CNT indicates rescue when no transgene is expressed. Experiments (n = 2–7) were performed at 22 and 25 °C (gray and black bars, respectively). B, % viable leoP1188/leoP1188 homozygotes or leoP1188/leoP2335 heteroallelic animals obtained without or with the Elav-Gal4 driver. Viability was calculated as the % of the expected number of homozygous or heteroallelic flies if these flies were fully viable. Experiments were performed at 22 and 25 °C (gray and black bars, respectively). n = 2–7. C, level of each transgenic protein used for the rescue experiments in A was assessed at 22 and 25 °C in head lysates from rescued leoP1188/leoP1188 homozygotes and no transgene control (CNT) escapers by Western blotting of head lysates challenged with anti-FLAG mAb and anti-SNX as a loading control.
In contrast to the wild-type transgenic protein and despite its significant accumulation (Fig. 4C), LEOFLAGWM yielded marginal rescue of both tester mutant strains (Fig. 4A). Thus, although not deficient in dimerization (Fig. 3, B and C), LEOFLAGWM appears functionally compromised, at least with respect to processes required for embryonic hatching. Similar results were obtained with another leoFLAGWM line (not shown). It appears, therefore, that dimerization is necessary but not sufficient for LEOFLAGWM functionality. Interestingly, the homodimerization-defective LEOFLAGMW supported survival of both mutants substantially more than LEOFLAGWM and nearly as efficiently as the wild type (Fig. 4A). The levels of both mutant proteins appeared equivalent (Fig. 4C), hence this cannot account for the difference. Because LEOFLAGMW heterodimerized selectively with D14-3-3ϵ (Fig. 3C), results strongly suggest that such heterodimers can support and may in fact be sufficient for survival to adulthood.
Under the steady state conditions of these genetic experiments, accumulation of the doubly mutant LEOFLAGMM was drastically reduced (Figs. 1B and 4C), and the protein did not form homo- or heterodimers (Fig. 3, B and C). It is not surprising then that this protein did not improve survival of the tester lethal homozygotes and heteroallelics. In fact, normalization of the results for the background viability of leoP1188 homozygotes and leoP1188/leoP2335 heteroallelics, suggested that pan-neuronal expression of leoFLAGMM actually decreased survival of the tester animals, especially at 25 °C (Fig. 4B). Given the low steady state levels of this protein, the results suggest that it may act as a dominant negative or be toxic for processes mediating survival to adulthood. These effects may be a consequence of competition with endogenous dimeric LEO for target proteins, impairing their function or rendering them inactive. Additional support for this interpretation was provided by overexpression of LEOFLAGMM in wild-type flies (supplemental Fig. S2), where its ubiquitous accumulation under the Actin-Gal4 reduced survival by nearly 50% compared with accumulation of the wild-type LEOFLAG. Milder effects were observed upon accumulation of the monomeric protein specifically in the CNS.
14-3-3 Homeostatic Responses to Dimerization Mutant LEO
Previous results have indicated that the overall level of 14-3-3s in the CNS is regulated in a homeostatic manner, such that LEO is up-regulated in 14-3-3ϵ-null embryos (30), while drastic LEO reduction in the adult CNS yields an increase in D14-3-3ϵ.3 This model predicts that an overall increase in LEO due to expression of leoFLAG transgenes may lead to changes in endogenous D14-3-3ϵ levels. One plausible hypothesis to explain this phenomenon posits that LEO/D14-3-3ϵ heterodimer formation is essential for this response. To investigate this possibility and gain insights on the mechanism of this putative homeostatic response, we determined the levels of endogenous D14-3-3ϵ in animals rescued from lethality and thus containing solely LEOFLAG transgenic proteins.
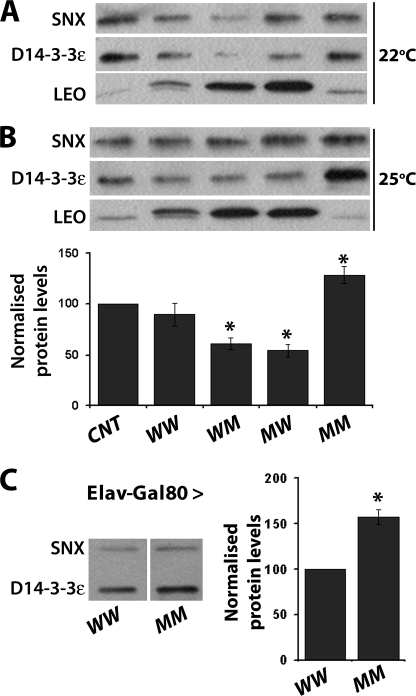
Western blots from single heads of leo1188 homozygous escapers grown either at 22 °C or 25 °C revealed residual levels of endogenous LEO (Fig. 5, A and B), which is a likely explanation of their survival to adulthood. Compared with its level in these escapers, D14-3-3ϵ was somewhat reduced in homozygotes rescued from lethality by the wild-type LEOFLAGWW (Fig. 5B). In contrast, either at 22 °C or 25 °C, the steady state level of D14-3-3ϵ was significantly reduced as the level of LEOFLAGWM and LEOFLAGMW proteins increased (Fig. 5B). These results raise two points. First, D14-3-3ϵ levels are decreased in response to LEOFLAG elevation. Because the leoFLAGWM and leoFLAGMW transgenes utilized appear to be expressed higher than the respective wild-type transgenic protein (Fig. 1A), D14-3-3ϵ levels decline proportionally to the levels of these dimerization competent LEO. These data provide independent confirmation of the 14-3-3 homeostasis notion, but in contrast to LEO elevation upon D14-3-3ϵ loss (30), here it is the latter that adjusts its levels in response to LEO elevation. This is significant, because rescue of leo homozygotes could in principle be a consequence of compensatory D14-3-3ϵ elevation to depletion of endogenous LEO. However, because D14-3-3ϵ levels actually decline, rescue from lethality is a consequence of transgenic protein accumulation at least in the case of single mutants and wild-type LEOFLAG.
FIGURE 5.
Distinct homeostatic responses of D14-3-3ϵ levels to LEOFLAG accumulation. A, assessment of endogenous and transgenic LEO and endogenous D14-3-3ϵ in head lysates from leoP1188/leoP1188 homozygotes rescued from lethality by the indicated transgenes under Elav-Gal4 and control escapers (CNT) raised at 22 °C. SNX served as the loading control. B, same as A, except the flies were raised at 25 °C. In the plot below, the results of three independent such blots run in duplicate are quantified. The mean ± S.E. of D14-3-3ϵ relative to SNX levels are presented as a percentage of the normalized D14-3-3ϵ/SNX level in CNT flies. Significant differences from the levels in CNT are indicated by the * at p < 0.05. C, LEOFLAGWW and LEOFLAGMM were induced in adult wild-type flies with Elav-Gal4; Gal80ts driver by a 24-h induction at 30 °C, and the levels of D14-3-3ϵ and SNX were assessed in head lysates by Western blots (panel on the left) and quantified as described above; n = 3 experiments run in duplicate.
In contrast, a large increase in the steady state levels of D14-3-3ϵ was revealed in the few leo1188 homozygotes recovered expressing the dimerization deficient LEOFLAGMM at 25 °C (Fig. 5B). Although not quantified, this effect was also observed in animals obtained at 22 °C. Because these flies contain traces of functional endogenous LEO and the transgenic protein cannot heterodimerize, the data suggest that D14-3-3ϵ elevation is likely a compensatory response to the presence of dimerization and functionally deficient LEO. Alternatively, D14-3-3ϵ elevation may be a consequence of functional LEO loss as an attempt to increase overall 14-3-3 levels above a threshold requisite for successful development and hatching (30, 44). To differentiate between these two possibilities, we expressed the dimerization deficient LEOFLAGMM in wild-type animals. Doubly mutant and wild-type transgenes were expressed for 24 h in adult animals under Elav-Gal4; Gal80ts, conditions that lead to equivalent accumulation of the transgenic proteins as shown above (Fig. 2A). D14-3-3ϵ was significantly elevated in the CNS of animals harboring the dimerization defective LEOFLAGMM, in contrast to ones accumulating the wild-type transgenic protein (Fig. 5C). This suggests that the relatively acute elevation of D14-3-3ϵ is a direct response to LEOFLAGMM, likely because of its inability to dimerize and function. Conversely, the data are also consistent with the notion that LEOFLAGMM acts as a dominant negative protein by competing for target binding with the endogenous dimeric proteins and elevation of D14-3-3ϵ is a cellular homeostatic response to neutralize this effect. This is consistent with the apparent dominant effect of LEOFLAGMM on the viability of leoP1188 homozygotes and leoP1188/leoP2335 heteroallelics (Fig. 4A) and wild-type animals (supplemental Fig. 2B).
DISCUSSION
There are three LEO isoforms (LEOI-LEOIII) in Drosophila, which differ in 5–10 amino acids, in the ligand binding helices αF, αG and αH encoded by alternatively spliced exons 6 (43). The work presented here and that published previously (34), utilized LEOIII. However, because the sequences of all three isoforms are invariant in helices αA-αE, the results described herein and in previous studies on LEO dimerization should be applicable to all LEO isoforms.
We have used such transgenic constructs to investigate the ability of 14-3-3ζ to function as a steady state monomer in vivo. Collectively, the evidence presented here strongly suggests that dimerization is necessary for LEO stability and functionality. An important advancement from our results is that they support the notion that in vivo, dimerization is essential for LEO stability in the Drosophila CNS. An additional significant difference with the cell culture data of Zhou et al. (34) is that both singly mutated proteins can dimerize. Importantly, however LEOFLAGMW appears to heterodimerize exclusively with endogenous D14-3-3ϵ and not with LEO, indicating a role for Leu15-Ala-Glu in selection of dimerization partners.
Analysis of the dimerization properties of LEO single mutants in αA and αD sequences essential for dimerization yielded some surprising results. The crystal structure of the mammalian 14-3-3ζ (13) predicts that highly conserved residues Leu15-Ala-Glu in αA at the dimer interface are essential for dimerization. Indeed, mutation of Leu15-Ala-Glu to Gln15-Gln-Arg yielded LEO proteins unable to homodimerize either with the transgenic or the endogenous protein in agreement with previously published data (34). This is also congruent with the proposed essential role for Ala16 mutated here to Gln in maintaining the 14-3-3ζ homodimer interface (12). Surprisingly however, our results clearly establish that in the Drosophila CNS LEOFLAGMW is not monomeric as reported (34), but rather it selectively heterodimerizes with D14-3-3ϵ (Fig. 3C). This deviation from the study of Zhou et al. (34) reporting this protein unable to dimerize, could be explained considering that these transfected vertebrate cells must also contain 14-3-3ϵ and other proteins from this family, likely engaged in such interactions. However, such heterodimers were not probed for and could also be obscured as emphasis was placed solely on interactions among the abundant transfected Drosophila LEO variants.
The LEOFLAGMW/D14-3-3ϵ heterodimers are functional, at least with respect to supporting vital functions as they yielded rescue of leoP1188 homozygotes nearly as well as heterodimers with the wild-type protein (Fig. 4A). This indicates that LEO/D14-3-3ϵ heterodimers may be normally used for processes requisite for viability. Furthermore, the LEOFLAGMW transgenic protein may be used as a tool for in vivo differentiation of the suggested (30) processes requiring homodimers from those where heterodimers are functional.
The stability of LEOFLAGMW/D14-3-3ϵ heterodimers may be mediated by the reported salt bridges between them (15). In addition to polar and hydrophobic residues in helices αA-αD mediating dimerization, 14-3-3ϵ homodimers can form only a single stabilizing salt bridge involving Arg21 and Glu90 (using Drosophila numbering, see supplemental Fig. S1). However, upon heterodimerization with 14-3-3ζ, two more salt bridges can be formed providing additional stability to 14-3-3ϵ heterodimers (12, 15). However, Glu90 has been mutated into Gln in LEOFLAGMW, and similar to the human 14-3-3ζ, LEO contains Glu84 and the conserved Glu87. These residues might be used in the case of the LEOFLAGMW to selectively form salt bridges with Arg21 in D14-3-3ϵ. Although these interactions have to be confirmed by crystal structures or mutagenesis, additional salt bridges can be formed between the conserved Glu5 of D14-3-3ϵ with the invariant Lys81 of LEO and Asp23 of LEO with Met84 which is not present in LEOIII, but characterizes all 14-3-3ϵ species (supplemental Fig. S1).
In contrast, mutating the second site in αD from Arg88-Val-Glu to Asn88-Val-Gln did not prevent homodimerization, at least with endogenous wild-type LEO (Fig. 3C), in agreement with Zhou et al. (34), or heterodimerization with D14-3-3ϵ. Therefore, the particular mutations in helix D alone do not appear to affect LEO homo- or heterodimerization properties. However these dimers were not fully functional, at least with respect to processes essential for viability (Fig. 4A). Therefore, although stable enough to be detectable by our methods, such dimers may not assume correct conformation to properly engage clients. Alternatively, though unlikely, the mutated amino acids in αD in addition to mediating dimerization are important for target binding or coordination. It is possible that in such mutant/wild-type dimers, only the wild-type LEO or D14-3-3ϵ engages clients properly, similar to the proposed action of dominant negative R59A/R63A mutations of vertebrate and Drosophila 14-3-3ζ (34, 47, 48). This is consistent with the interpretation that though essential, dimerization does not always suffice for proper functionality.
Our results also demonstrate that LEO carrying both mutant sites on αA and αD simultaneously, is unstable relative to wild-type protein. Nevertheless, in Zhou et al. (34) and other studies (25, 26, 49), monomeric species appear abundant enough to permit functional analyses. However, these results were obtained by transfecting cultured cells with constructs encoding the monomeric proteins (25, 26, 34, 49). Expression from transfected constructs is relatively high and acute, approximating our own experiments with the Gal80ts system (Fig. 2). Under these conditions, the monomeric protein accumulated to levels identical with those of dimerization competent proteins in our system as well. Therefore, we propose that the monomeric proteins are not intrinsically unstable, but rather are subject to regulated degradation occurring at least twice as fast as that of their dimeric counterparts (Fig. 2). This interpretation is also consistent with the observed low steady state levels of the monomeric protein when chronically expressed under Elav-Gal4. Based on the above, we predict that levels of monomeric 14-3-3s should decline over time even in transfected cells in contrast to their dimeric counterparts.
Although monomeric LEO was able to regulate dSlo activity in a heterologous system (34), in most cases monomeric 14-3-3s seem to bind clients but are unable to support their activity (25, 26, 28, 29, 48, 49). Thus, endogenous 14-3-3 that fails to dimerize may be degraded to prevent unproductive target binding. It is then possible that stimulus-induced dimer dissociation and consequent degradation may serve to terminate 14-3-3 binding and modulate client activity. Interestingly, Ser58 in helix αC of in the dimer interface of vertebrate 14-3-3ζ becomes phosphorylated in mouse fibroblasts by the sphingosine-dependent kinase SDK1 (50). This phosphorylation suffices to disrupt dimer formation apparently in an inducible manner, as it requires sphingolipids (29). In support of this, a phosphomimic mutant 14-3-3ζ (S58E) was shown unable to dimerize (39), but the stabilities of this endogenous inducible monomer, or the phosphomimic mutant were not examined, hence it is unknown whether they degraded over time as we hypothesize. However, these data demonstrate that inducible monomerization and monomer degradation could be utilized as a mechanism to functionally regulate 14-3-3s with significant implications on the multiple cellular activities requiring these proteins. LEO contains an equivalent serine (Ser60), but it is currently unknown whether it is utilized for monomerization in vivo. It is also possible that regulated monomerization occurs in a tissue- and temporal-specific manner, and the resultant monomers are used for specific purposes as that of dSlo activity regulation (34). If extant, such monomers are likely to be rare, perhaps because of their proposed instability, as we have not detected them despite a systematic search.4
Finally the results presented herein provide independent confirmatory evidence for our proposal that overall 14-3-3 levels are subject to homeostatic regulation. Furthermore, they strongly suggest that heterodimerization of LEO isoforms with D14-3-3ϵ is likely part of the homeostatic mechanism. Transgenic LEOFLAG proteins able to heterodimerize with the endogenous D14-3-3ϵ did not alter its levels. However, acute elevation of the dimerization defective mutant protein LEOFLAGMM resulted in a significant D14-3-3ϵ elevation (Fig. 5). This is consistent with the notion that cells devoid of endogenous LEO do not perceive the dimerization defective protein as a functional 14-3-3ζ and elevate D14-3-3ϵ in compensation. Significantly, this in vivo analysis of LEO dimerization was performed in the CNS where 14-3-3 proteins are abundant in Drosophila and vertebrates (42). The genetic versatility and power of the Drosophila system provides a general experimental platform for such in vivo functional analyses of 14-3-3 properties in different tissues of the fly, aimed at addressing the specificity of their interactions with client proteins.
Supplementary Material
Acknowledgments
We thank Drs. I. Levitan and Y. Zhou for cDNA clones, T. Tzortzopoulos for generation of transgenic strains, and the Developmental Studies Hybridoma Bank (University of Iowa) for antibodies. We also thank K. Papanikolopoulou for constructive comments on the manuscript.
This work was supported in part by Hellenic General Secretariat for Research and Technology PENED Grant 01EΔ207 (to S. G.) and European Union Marie Curie TOK Grant 003141 (to G. M. and E. M. C. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
F. Leptourgidou and E. M. C. Skoulakis, unpublished data.
S. Grammenoudi and E. M. C. Skoulakis, unpublished observations.
- CNS
- central nervous system
- SNX
- syntaxin
- mAb
- monoclonal antibody.
REFERENCES
- 1.Aitken A., Baxter H., Dubois T., Clokie S., Mackie S., Mitchell K., Peden A., Zemlickova E. (2002) Biochem. Soc. Trans. 30, 351–360 [DOI] [PubMed] [Google Scholar]
- 2.Angrand P. O., Segura I., Völkel P., Ghidelli S., Terry R., Brajenovic M., Vintersten K., Klein R., Superti-Furga G., Drewes G., Kuster B., Bouwmeester T., Acker-Palmer A. (2006) Mol. Cell. Proteomics 5, 2211–2227 [DOI] [PubMed] [Google Scholar]
- 3.Jin J., Smith F. D., Stark C., Wells C. D., Fawcett J. P., Kulkarni S., Metalnikov P., O'Donnell P., Taylor P., Taylor L., Zougman A., Woodgett J. R., Langeberg L. K., Scott J. D., Pawson T. (2004) Curr. Biol. 14, 1436–1450 [DOI] [PubMed] [Google Scholar]
- 4.Meek S. E., Lane W. S., Piwnica-Worms H. (2004) J. Biol. Chem. 279, 32046–32054 [DOI] [PubMed] [Google Scholar]
- 5.Pozuelo Rubio M., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., Mackintosh C. (2004) Biochem. J. 379, 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridges D., Moorehead G. B. (2005) Sci. STKE 2005, re10. [DOI] [PubMed] [Google Scholar]
- 7.Obsil T., Ghirlando R., Klein D. C., Ganguly S., Dyda F. (2001) Cell 105, 257–267 [DOI] [PubMed] [Google Scholar]
- 8.Shikano S., Coblitz B., Wu M., Li M. (2006) Trends Cell Biol. 16, 370–375 [DOI] [PubMed] [Google Scholar]
- 9.Tzivion G., Avruch J. (2002) J. Biol. Chem. 277, 3061–3064 [DOI] [PubMed] [Google Scholar]
- 10.Dougherty M. K., Morrison D. K. (2004) J. Cell Sci. 117, 1875–1884 [DOI] [PubMed] [Google Scholar]
- 11.van Heudsen G. P. (2005) IUBMB Life 57, 623–629 [DOI] [PubMed] [Google Scholar]
- 12.Gardino A. K., Smerdon S. J., Yaffe M. B. (2006) Semin. Cancer Biol. 16, 173–182 [DOI] [PubMed] [Google Scholar]
- 13.Liu D., Bienkowska J., Petosa C., Collier R. J., Fu H., Liddington R. (1995) Nature 376, 191–194 [DOI] [PubMed] [Google Scholar]
- 14.Xiao B., Smerdon S. J., Jones D. H., Dodson G. G., Soneji Y., Aitken A., Gamblin S. J. (1995) Nature 376, 188–191 [DOI] [PubMed] [Google Scholar]
- 15.Yang X., Lee W. H., Sobott F., Papagrigoriou E., Robinson C. V., Grossmann J. G., Sundström M., Doyle D. A., Elkins J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petosa C., Masters S. C., Bankston L. A., Pohl J., Wang B., Fu H., Lidington R. C. (1998) J. Biol. Chem. 273, 16305–16310 [DOI] [PubMed] [Google Scholar]
- 17.Rittinger K., Budman J., Xu J., Volinia S., Cantley L. C., Smerdon S. J., Gamblin S. J., Yaffe M. B. (1999) Mol. Cell 4, 153–166 [DOI] [PubMed] [Google Scholar]
- 18.Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 19.Garnett M. J., Rana S., Paterson H., Barford D., Marais R. (2005) Mol. Cell 20, 963–969 [DOI] [PubMed] [Google Scholar]
- 20.Yaffe M. B. (2002) FEBS Lett. 513, 53–57 [DOI] [PubMed] [Google Scholar]
- 21.Gu Y. M., Jin Y. H., Choi J. K., Baek K. H., Yeo C. Y., Lee K. Y. (2006) FEBS Lett. 580, 305–310 [DOI] [PubMed] [Google Scholar]
- 22.Hekman M., Wiese S., Metz R., Albert S., Troppmair J., Nickel J., Sendtner M., Rapp U. R. (2004) J. Biol. Chem. 279, 14074–14086 [DOI] [PubMed] [Google Scholar]
- 23.Tzivion G., Luo Z. J., Avruch J. (2000) J. Biol. Chem. 275, 29772–29778 [DOI] [PubMed] [Google Scholar]
- 24.Liang X., Butterworth M. B., Peters K. W., Walker W. H., Frizzell R. A. (2008) J. Biol. Chem. 283, 27418–27425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichimura T., Ito M., Itagaki C., Takahashi M., Horigome T., Omata S., Ohno S., Isobe T. (1997) FEBS Lett. 413, 273–276 [DOI] [PubMed] [Google Scholar]
- 26.Shen Y. H., Godlewski J., Bronisz A., Zhu J., Comb M. J., Avruch J., Tzivion G. (2003) Mol. Biol. Cell 14, 4721–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Z. J., Zhang X. F., Rapp U., Avruch J. (1995) J. Biol. Chem. 270, 23681–23687 [DOI] [PubMed] [Google Scholar]
- 28.Tzivion G., Luo Z., Avruch J. (1998) Nature 394, 88–92 [DOI] [PubMed] [Google Scholar]
- 29.Woodcock J. M., Murphy J., Stomski F. C., Berndt M. C., Lopez A. F. (2003) J. Biol. Chem. 278, 36323–36327 [DOI] [PubMed] [Google Scholar]
- 30.Acevedo S. F., Tsigkari K. K., Grammenoudi S., Skoulakis E. M. (2007) Genetics 177, 239–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang H. C., Rubin G. M. (1997) Genes Dev. 11, 1132–1139 [DOI] [PubMed] [Google Scholar]
- 32.Skoulakis E. M., Davis R. L. (1996) Neuron 17, 931–944 [DOI] [PubMed] [Google Scholar]
- 33.Philip N., Acevedo S. F., Skoulakis E. M. (2001) J. Neurosci. 21, 8417–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y., Reddy S., Murrey H., Fei H., Levitan I. B. (2003) J. Biol. Chem. 278, 10073–10080 [DOI] [PubMed] [Google Scholar]
- 35.Acevedo S. F., Froudarakis E. I., Tsiorva A. A., Skoulakis E. M. (2007) Mol. Cell Neurosci. 34, 378–389 [DOI] [PubMed] [Google Scholar]
- 36.Duffy J. B. (2002) Genesis 34, 1–15 [DOI] [PubMed] [Google Scholar]
- 37.McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L. (2003) Science 302, 1765–1768 [DOI] [PubMed] [Google Scholar]
- 38.Lin D. M., Goodman C. S. (1994) Neuron 13, 507–523 [DOI] [PubMed] [Google Scholar]
- 39.Powell D. W., Rane M. J., Joughin B. A., Kalmukova R., Hong J. H., Tidor B., Dean W. L., Pierce W. M., Klein J. B., Yaffe M. B., McLeish K. R. (2003) Mol. Cell. Biol. 23, 5376–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl M. W., Hageleit M. (2001) Biotechnol. Lett. 23, 275–282 [Google Scholar]
- 41.Missirlis F., Kosmidis S., Brody T., Mavrakis M., Holmberg S., Odenwald W. F., Skoulakis E. M., Rouault T. A. (2007) Genetics 177, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skoulakis E. M., Davis R. L. (1998) Mol. Neurobiol. 16, 269–284 [DOI] [PubMed] [Google Scholar]
- 43.Messaritou G., Leptourgidou F., Franco M., Skoulakis E. M. (2009) FEBS Lett. 582, 2934–2938 [DOI] [PubMed] [Google Scholar]
- 44.Broadie K., Rushton E., Skoulakis E. M., Davis R. L. (1997) Neuron 19, 391–402 [DOI] [PubMed] [Google Scholar]
- 45.Li W., Skoulakis E. M., Davis R. L., Perrimon N. (1997) Development 124, 4163–4171 [DOI] [PubMed] [Google Scholar]
- 46.Grammenoudi S., Kosmidis S., Skoulakis E. M. (2006) FEBS Lett. 580, 4602–4606 [DOI] [PubMed] [Google Scholar]
- 47.Kagan A., Melman Y. F., Krumerman A., McDonald T. V. (2002) EMBO J. 21, 1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S., Xing H., Muslin A. J. (1999) J. Biol. Chem. 274, 24865–24872 [DOI] [PubMed] [Google Scholar]
- 49.Gu M., Du X. (1998) J. Biol. Chem. 273, 33465–33471 [DOI] [PubMed] [Google Scholar]
- 50.Megidish T., Cooper J., Zhang L., Fu H., Hakomori S. (1998) J. Biol. Chem. 273, 21834–21845 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.