Abstract
Cyclophilin B (CyPB) induces migration and adhesion of T lymphocytes via a mechanism that requires interaction with 3-O-sulfated heparan sulfate (HS). HS biosynthesis is a complex process with many sulfotransferases involved. N-Deacetylases/N-sulfotransferases are responsible for N-sulfation, which is essential for subsequent modification steps, whereas 3-O-sulfotransferases (3-OSTs) catalyze the least abundant modification. These enzymes are represented by several isoforms, which differ in term of distribution pattern, suggesting their involvement in making tissue-specific HS. To elucidate how the specificity of CyPB binding is determined, we explored the relationships between the expression of these sulfotransferases and the generation of HS motifs with CyPB-binding properties. We demonstrated that high N-sulfate density and the presence of 2-O- and 3-O-sulfates determine binding of CyPB, as evidenced by competitive experiments with heparin derivatives, soluble HS, and anti-HS antibodies. We then showed that target cells, i.e. CD4+ lymphocyte subsets, monocytes/macrophages, and related cell lines, specifically expressed high levels of NDST2 and 3-OST3 isoforms. Silencing the expression of NDST1, NDST2, 2-OST, and 3-OST3 by RNA interference efficiently decreased binding and activity of CyPB, thus confirming their involvement in the biosynthesis of binding sequences for CyPB. Moreover, we demonstrated that NDST1 was able to partially sulfate exogenous substrate in the absence of NDST2 but not vice versa, suggesting that both isoenzymes do not have redundant activities but do have rather complementary activities in making N-sulfated sequences with CyPB-binding properties. Altogether, these results suggest a regulatory mechanism in which cell type-specific expression of certain HS sulfotransferases determines the specific binding of CyPB to target cells.
Keywords: Carbohydrate/Binding Protein, Carbohydrate/Biosynthesis, Carbohydrate/Function, Enzymes/Sulfotransferase, Extracellular Matrix/Heparan Sulfate, Protein/Binding/Heparin, N-Deacetylase/N-Sulfotransferase, Cyclophilin
Introduction
Initially identified as cyclosporin A-binding proteins, cyclophilins are peptidyl-prolyl cis-trans isomerases involved in various biological processes, including protein folding, mitochondrial functions, apoptosis, and regulation of trafficking and signaling (1, 2). Besides the repertoire of intracellular functions in which they have been implicated, secreted cyclophilins A and B (CyPB)2 were reported to be mediators of inflammation and innate immunity. They trigger chemotaxis of neutrophils, T lymphocytes, and monocytes/macrophages by way of interactions with CD147 and cell surface heparan sulfate (HS) (3–7). CyPB also induces integrin-mediated adhesion of CD4+ T lymphocytes and monocytes/macrophages to fibronectin, by a mechanism that requires interaction with the HS moieties of syndecan-1 and association of CD147 with CD98, the latter being an activator of β1 integrins (4, 8, 9).
HS consists of alternating N-acetyl/N-sulfate glucosamine (GlcNAc/GlcNS) and GlcUA/IdoUA residues clustered in a series of domains of relatively high IdoUA content and sulfate density (NS domains), bound by short transition zones with intermediate sulfation patterns and separated by N-acetylated domains (NA domains). HS is involved in a plethora of biological processes, which relies on its ability to selectively interact with various proteins. Heparin, which is modified predominantly to IdoUA2S-GlcNS6S disaccharides, has been widely used as a structural surrogate of the NS domains of HS. In particular, characterization of heparin oligosaccharides with high affinity to distinct proteins has led to the identification of specialized HS sequences with precisely located N- and O-sulfate groups (10, 11).
The structural distinctions in HS motifs are derived from enzymatic modifications of the nascent polymer composed of alternating GlcUA and GlcNAc units. The non-sulfated precursor is first subject to partial N-deacetylation/N-sulfation of GlcNAc residues, which leads to the occurrence of consecutively N-sulfated regions, regions that escape modifications and remain N-acetylated, and regions of alternating N-acetylated and N-sulfated disaccharide units. Sometimes, the N-deacetylation/N-sulfation reaction is incomplete and gives rise to GlcNH2. The further modifications include C5-epimerization of some GlcUA into IdoUA, 2-O-sulfation of IdoUA, and 6-O-sulfation of GlcN units. Rarely, O-sulfation also occurs at position 3 of GlcNS and GlcNH2 units. These modifications involve HS biosynthetic enzymes, including N-deacetylases/N-sulfotransferases (NDSTs), C5-epimerase, and 2-O, 3-O, and 6-O-sulfotransferases (2-OST, 3-OSTs, and 6-OSTs) (11–13). In the general scheme of HS biosynthesis, GlcNAc N-deacetylation and N-sulfation by NDSTs create the prerequisite substrate needed for the next enzymatic modifications. Four NDSTs have been cloned and characterized, NDST1 and NDST2 being widely expressed in all the tissues analyzed (14, 15). Although both isoenzymes exhibit similar activity in vitro, accumulating data have suggested that they are probably not biologically redundant. Lack of NDST1 affects HS structure in all tissues tested, with a dramatic reduction in N- and O-sulfation of the polysaccharide. In contrast, mice lacking NDST2 develop and reproduce normally. No significant alteration in tissue HS structure was found, except in mast cells where heparin biosynthesis is severely disturbed. These studies suggest that NDST1 is required for initiation of N-sulfation of the nascent precursor, whereas NDST2 may fill in or extend the sections of N-sulfated residues in heparin and highly sulfated HS species (16–19). 3-OSTs catalyze the least abundant modification in HS. Seven 3-OST isoforms have been recognized in humans, with 3-OST1, 3-OST3 (3A and 3B), and 3-OST5 being the most widely expressed in various tissues and cell types. Interestingly, these isoenzymes exhibit fine differences in substrate specificity, suggesting their involvement in making tissue-specific HS with different biological functions. Although 3-OST1 is known to exclusively generate an HS-binding site for antithrombin III, 3-OST3 isoforms transfer sulfate groups to the 3-OH position of GlcNH2 and/or GlcNS adjacent to the IdoUA2S residue, thus providing an entry receptor for herpesvirus type I. In contrast, 3-OST5 exhibits broad substrate specificity and generates products with affinity to herpesvirus type I and antithrombin III (20–25).
Our preliminary works have illustrated the importance of a 3-O-sulfated N-unsubstituted GlcN (GlcNH23S) residue in high affinity CyPB binding to heparin and cell surface HS expressed on peripheral blood CD4+ T lymphocytes and Jurkat T cells (26). In addition to the requirement of this structural modification, we also found that interaction of CyPB with HS/heparin was likely to be dependent on the density of N-sulfates. Moreover, the protein displayed comparable high affinities for both heparin and membrane-associated HS on T cells, indicating that the preferred binding site of CyPB on responsive cells is probably located in a highly N-sulfated sequence (6, 26, 27). Several lines of evidence indicate that the sulfation pattern of HS may be determined by cell type-specific expression of certain isoforms of HS sulfotransferases and by their relative expression as a response to different cellular conditions. In this study, we explored the relationships between the expression of HS sulfotransferases and their ability to generate HS motifs with CyPB-binding and -activating properties. Following the demonstration that N-, 2-O-, and 3-O-sulfations of HS sequences were critically important to support the efficient binding of CyPB, we showed that activated/memory CD4+ T lymphocytes, monocytes/macrophages, Jurkat and THP-1 cells, which are target cells of CyPB, specifically expressed high levels of mRNA encoding NDST2 and 3-OST3 isoforms. To know whether the binding of CyPB may be regulated at the biosynthetic stage of HS modifications, we used RNA interference to specifically down-regulate the expression of HS sulfotransferases. The ability of cell surface HS to bind to CyPB was dramatically reduced by silencing the expression of NDST1, NDST2, 2-OST, and 3-OST3. Moreover, we found that NDST1 and NDST2 have no redundant activities but rather have complementary activities in making N-sulfated HS motifs with CyPB-binding properties. Altogether, these results suggest a regulatory mechanism in which the expression of some HS sulfotransferases determines the specificity of binding of CyPB to target cells.
EXPERIMENTAL PROCEDURES
Materials
Recombinant human CyPB and human milk lactoferrin were produced and purified as described previously (28, 29). Porcine mucosal heparin, bovine kidney HS, heparinases I (EC 4.2.2.7), II (no EC number assigned), and III (EC 4.2.2.8), and DNase I (EC 3.1.21.1) were purchased from Sigma. Porcine intestine mucosal HS was obtained from Celsius Laboratories (Cincinnati, OH). Rabbit antibodies to ERK1/2, which recognize p44/p42 MAPK regardless of their phosphorylation status, and mouse antibodies to phosphorylated forms of p44/p42 MAPK were purchased from Sigma and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Horseradish peroxidase-conjugated anti-IgG antibodies were from Amersham Biosciences. The expression level of cell surface glycosaminoglycans was analyzed by using phage display-derived vesicular stomatitis virus (VSV)-tagged single-chain antibodies, which have been described previously for their ability to specifically recognize different glycosaminoglycan epitopes. The antibodies were as follows: MPB49 (irrelevant) and AO4B08, HS4C3, HS4E4, RB4Ea12 (specific for HS epitopes), and IO3H10 (specific for chondroitin sulfate) (30–35). The characteristics of these antibodies are presented in supplemental Table S1. Mouse anti-VSV antibody (P5D4) and fluorescein-conjugated anti-mouse IgG were from Sigma. Reagents for electrophoresis and cell culture were from Bio-Rad and Lonza BioWhittaker (Basel, Switzerland), respectively. All other chemicals, except where otherwise mentioned, were purchased from Sigma.
Cells
Human lymphoblastic Jurkat T cells (clone E6-1, ATCC TIB-152, Manassas, VA) and promonocytic leukemia THP-1 cells (88081201, ECACC, Porton Down, Salisbury, UK) were routinely cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal calf serum, 2 mm l-glutamine, 10 mm gentamycin. To induce responsiveness to CyPB, THP-1 cells were differentiated for 72 h with 50 nm 1,25-dihydroxyvitamin D3, as described previously (9). Human epithelium-like cervical tumor cell line HeLa (ATCC CCL-2) and breast tumor cell lines MCF7 (ATCC HTB-22), MDA-MB-231 (ATCC HTB-26), and T-47D (ATCC HTB-133) were grown in Dulbecco's modified Eagle's medium, containing 10% (v/v) fetal calf serum, 2 mm l-glutamine, and 1% (w/v) penicillin/streptomycin. Cell lines were cultured at 37 °C in a humidified atmosphere with 5% CO2. Human citrated venous blood samples were obtained from the local blood transfusion center (Etablissement de Transfusion Sanguine, Lille, France). Isolation of peripheral blood T lymphocytes and monocytes was conducted by selection with magnetic beads coupled to appropriate antibodies, according to the instructions of the manufacturer (BD Biosciences). Stimulation of T lymphocytes was obtained by incubating CD4+CD45RA+ T cells in the presence of precoated anti-CD3 antibody (5 μg/ml) (Immunotech, Marseille, France) in complete RPMI 1640 medium for 16 h. Monocytes were activated with lipopolysaccharide (10 ng/ml) (Sigma) for 16 h in complete RPMI 1640 medium. Macrophages were obtained by incubating freshly isolated monocytes (106 cells/ml) in complete RPMI 1640 medium supplemented with 10 ng/ml macrophage colony-stimulating factor (AbCys, Paris, France) for 5 days. The purity of cell populations was assessed by flow cytofluorimetry and found to be >95%.
Preparation of HS and Heparin Derivatives
Partially N-desulfated heparin was produced as described previously (36). Briefly, pyridinium salt of heparin was treated with dimethyl sulfoxide/water (95:5, v/v) at 20 °C for 60 min. The sample was then dialyzed extensively and freeze-dried. Fully N-desulfated heparin was a gift of M. Lyon (Department of Medical Oncology, University of Manchester, Christie Hospital, Manchester, UK). The degrees of N-desulfation were calculated from the increase in N-acetylated disaccharides obtained after N-reacetylation and digestion with heparinases (36). Disaccharide analysis revealed 25 and 92% reduction in N-sulfation for partially and fully N-desulfated heparins, respectively, with no significant loss of 2-O and 6-O-sulfates. Information regarding the density of GlcNS residues was obtained by deamination of HS or oligosaccharides with nitrous acid at pH 1.5 (37). Freshly prepared HNO2 reagent was added to the dry oligosaccharide sample and incubated for 15 min at room temperature. Reaction was stopped by the addition of saturated Na2CO3 solution. The supernatant was immediately desalted on a Sephadex G-10 column (Amersham Biosciences) and dried under vacuum.
Fluorescent derivatization of oligosaccharides was performed by reductive amination as described previously (27). Briefly, dried samples (5–50 nmol) were dissolved in 5 μl of acetic acid/water (3:17, v/v) containing 0.2 m of 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) (Molecular Probes, Leiden, Netherlands) and 5 μl of dimethyl sulfoxide containing 1 m sodium cyanoborohydride (Fluka, Buchs, Switzerland). The mixture was incubated at 37 °C for 16 h, desalted on a 10 ml-Sephadex G-15 column, and thereafter dried under vacuum.
Real Time PCR
Total RNA was isolated from 3 × 106 cells using a NucleoSpin RNA II kit, according to the instructions of the manufacturer (Macherey-Nagel, Hoerdt, France). Reverse transcription was performed from 2 μg of total RNA with an oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). PCR amplifications were performed using a Mx4000 Multiplex Quantitative PCR system (Stratagene, La Jolla, CA). Synthetic primers for NDST1, NDST2, 2-OST, 3-OST1, 3-OST3, and 3-OST5 were designed by using Primer Premier 5.0 (Biosoft International, Palo Alto, CA), according to the published cDNA sequences. The primer set for 3-OST3 detects the same sequence in both isoenzymes 3-OST3A and 3-OST3B, which have the identical catalytic domain. The transcript of hypoxanthine-guanine phosphoribosyltransferase was used as a control to normalize the expression of our genes of interest. The primer sets are presented in supplemental Table S2. The sequence of each amplified product was confirmed by sequencing (Genoscreen, Lille, France). Each reaction consisted of 25 μl containing 2 μl of diluted cDNA sample (1:5), 12.5 μl of Brilliant SYBR Green quantitative PCR master mix (two times) (Stratagene), 1 μl of forward primer (7.5 μm for 2-OST and 3-OST3; 15 μm for NDST1, NDST2, and 3-OST5; and 22.5 μm for 3-OST1 and hypoxanthine-guanine phosphoribosyltransferase), 1 μl of reverse primer (7.5 μm for 2-OST; 15 μm for NDST1, NDST2, 3-OST1, 3-OST5, and hypoxanthine-guanine phosphoribosyltransferase; and 22.5 μm for 3-OST3) (Eurogentec, Seraing, Belgium), and 8.5 μl of water. Each PCR also included a reverse transcription negative control and a non-template negative control to check for the absence of genomic DNA and primer dimers, respectively. The conditions of PCR were as follows: 1 cycle of denaturation at 95 °C for 10 min, followed by 40 cycles of 30 s at 95 °C, 1 min at specific temperature of annealing (60 °C for NDST1, NDST2, 2-OST, and 3-OST5; 67 °C for 3-OST1; 68 °C for 3-OST3; and 51 °C for hypoxanthine-guanine phosphoribosyltransferase) and 30 s at 72 °C. The fluorescence data were measured at the end of each cycle. A melting curve (55–95 °C at 1 °C interval) was constructed for each primer pair to check for the presence of one gene-specific peak. The amplification efficiency of each primer pair was performed on serial dilutions of cDNA. Triplicate PCRs were prepared for each sample. The point at which the PCR product is first detected above a fixed threshold, termed cycle threshold (Ct), was determined for each sample, and the average Ct of triplicate samples was used for further analysis. The relative quantification of transcripts was calculated as described previously (38).
RNA Interference
Synthetic small interfering RNA (siRNA) duplexes with symmetric 3′-deoxythymidine overhangs (Eurogentec) were used to carry out RNA interference. A set of three distinct synthetic siRNA duplexes for the same target sequences was designed and tested for their efficiency to down-regulate the expression of mRNA encoding NDST1, NDST2, 2-OST, and 3-OST3. siRNAs giving at least 75% of gene inhibition were retained for next experiments (supplemental Table S3). The oligonucleotide sequences were subjected to a BLAST search analysis, and no significant identity to other sequences could be detected. A synthetic siRNA duplex (siGFP) targeting green fluorescent protein mRNA was used as an irrelevant control. Negative control siRNAs, in which two nucleotides have been changed from the target sequence, were used to demonstrate the specificity of silencing. Jurkat T cells were transiently transfected using the nucleofection technology according to Amaxa Biosystems protocol (Amaxa, Cologne, Germany). Briefly, cells were resuspended in 100 μl of Cell Line Nucleofector Solution V, and the cell suspension were nucleofected with 4 μg of siRNA using the program V-001. After nucleofection, cells were transferred into prewarmed complete maintenance medium and were cultured as described before. To monitor the transfection efficiency, a fluorescein-tagged siRNA duplex was transfected in parallel, and the transfection rate was evaluated by flow cytofluorimetry and found to be >85%. We also used rescuing reagents to demonstrate the specificity of the siRNA-mediated silencing of each target sequence. For expression of sequences refractory to siNDST1 and siNDST2, we decided to use plasmids expressing mRNA of mouse NDST1 (pBud-NDST1) and NDST2 (pcDNA-NDST2). These plasmids have been provided by L. Kjellén (Uppsala University) and are presented in Ref. 39. Transient cell transfection with these plasmids allowed the expression of enzymes for which the catalytic activity is closely related to the one of human NDST1 and NDST2. However, the mRNA sequences encoding mouse enzymes were distinct enough from human target sequences to make the expression plasmids refractory to silencing. Primers for verification of mouse NDST1 and NDST2 expression have been designed in Ref. 39 and were used here to check for the expression of mRNA encoding both enzymes in transfected Jurkat T cells. To make 2-OST and 3-OST3 resistant to related siRNA, we used pcDNA-2-OST and pcDNA-3-OST3B plasmids as templates for site-directed mutagenesis, according to the QuickChange XL site-directed mutagenesis protocol (Stratagene). Both plasmids have been produced at home from human full-length cDNA, according to Refs. 21 and 40. pcDNA-2OSTREF and pcDNA-3OST3BREF were generated by introducing six silent mutations without changing the amino acid sequences of human 2-OST and 3-OST3B, respectively. The sense primers for the mutagenesis were as follows: 5′-G TCA TTG CAA GAT CAG GTG CGG TTC GTT AAA AAC ATT ACT TCC TGG AAA GAG ATG-3′ and 5′-G GGC CTC AAG AGG ATC ATC ACC GAT AAA CAT TTT TAT TTC AAC AAG ACC AAG GGC-3′, for 2-OST and 3-OST3B, respectively (letters in boldface are silently mutated nucleotides and underlined are the target sequences of siRNA). Point mutations were checked by full DNA sequencing. For rescue experiments, cells were first treated with siRNA as described above and cultured for 24 h. Thereafter, cells were transfected with 5 μg of plasmids expressing sequences refractory to siRNA. To avoid dramatic change in HS maturation or toxic side effect related to a decrease in the availability of sulfate donor, cells were used 24 h post-transfection to limit the expression of exogenous sulfotransferases. As expected, mortality was not increased in cells transfected with refractory plasmids by comparison with mock-transfected cells. Moreover, binding of anti-HS antibodies was increased by less than 20% in cells transfected with refractory plasmids, thus indicating that a 24-h treatment had only a moderate effect on the overall HS modifications.
Carbohydrate Electrophoresis and Mobility Shift Assay
ANTS-labeled dodecasaccharides (0.6 nmol) and CyPB (0.1 nmol) were mixed in 20 μl of electrophoresis binding buffer, containing 20 mm Tris-HCl, pH 7.9, 400 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, and incubated in the absence or presence of heparin derivatives or soluble HS (20 μg) for 30 min at room temperature. Following addition of 5 μl of 60% (v/v) glycerol, the samples were subjected to electrophoresis in a 10% (w/v) native polyacrylamide gel. Electrophoresis was carried at 100 V for 1–2 h at 4 °C in a Mini-PROTEAN 3 system (Bio-Rad). The electrophoretic buffer used was 40 mm Tris/acetic acid, 1 mm EDTA, pH 7.8. A mixture of bromphenol blue and phenol red was used as electrophoresis markers. At the end of the electrophoresis, images were acquired with the Gel Doc 2000 system from Bio-Rad, equipped with a 365-nm UV transilluminator. Analysis was performed with supplied software Quantity One.
In Vitro N-Sulfation Assays
Jurkat T cells (25 × 106/ml) were lysed in solubilization buffer containing 50 mm MES, 10 mm MnCl2, 10 mm MgCl2, 5 mm CaCl2, 1% Triton X-100 (v/v), pH 7.0, supplemented with protease inhibitor mixture (Roche Applied Science) and DNase I for 16 h at 4 °C. Following centrifugation at 10,000 × g for 15 min, anionic molecules were removed by chromatography on DEAE-Sepharose, and the effluent was used as the source of endogenous enzymes. Desulfated heparin has been commonly used as a sulfate acceptor to quantify N-sulfotransferase activity in vitro (41). We therefore applied a similar experimental procedure, with the following modifications. Heparin-derived oligosaccharides (dp14) were fully O- and N-desulfated by treatment with 10% (v/v) methanol in dimethyl sulfoxide at 100 °C for 2 h and thereafter N-reacetylated as described previously (36). Thereafter, oligosaccharides were labeled at the reducing end with ANTS and used as sulfate acceptor. The sulfation reaction was performed by incubating cell lysate (100 μg of solubilized proteins) with 5 μg of acceptor and 10 μm of 3′-phosphoadenosyl-5′-phosphosulfate in a total volume of 50 μl of solubilization buffer, for 3 h at 37 °C. The reaction was stopped by the addition of 200 μl of a solution of 10% (v/v) trichloroacetic acid containing 1 m NaCl. After centrifugation and passage through a Sephadex G-15 column equilibrated with 50 mm Tris-HCl, pH 7.5, 0.15 m NaCl, the fractions containing oligosaccharides was loaded on a 1-ml DEAE-Sepharose column. After washing with the same buffer containing 0.4 m NaCl, N-sulfated oligosaccharides were eluted in the presence of 1.5 m NaCl, desalted, dried under vacuum, and subjected to a deaminative cleavage with HNO2 at pH 1.5. The cleavage products were fractionated on Bio-Gel P-6 (Bio-Rad) column equilibrated in 0.1 m NH4HCO3 (20 × 0.8 cm). Fractions of 500 μl were collected and analyzed for fluorescence by using a TriStar LB 941 multimode microplate reader (Berthold, Bad Wildbad, Germany).
Flow Cytofluorimetry Analysis
The expression level of cell surface glycosaminoglycans was determined by incubating cells (5 × 105 cells per sample) with VSV-tagged single-chain antibodies in Dulbecco's phosphate-buffered saline containing 0.5% BSA (w/v) (DPBS/BSA) for 1 h at 4 °C. After washing, mouse anti-VSV antibody (1:1000) was added for another 1-h incubation, and cells were stained with fluorescein-conjugated anti-mouse IgG (1:64). The following dilutions were retained: HS4E4, 1:50; HS4C3, 1:150; AO4B08, 1:100; RB4Ea12, 1:100. Nonspecific binding was determined individually by using the isotype control antibody MPB49 at the same dilution. Increasing the amount of antibodies did not modify the variations in fluorescence mean values, thus validating the experimental procedure. Moreover, pretreatment of cells with DPBS containing 1 m NaCl did not modify the binding of antibodies, ruling out the possibility that HS epitopes could be masked by extracellular ligands. The expression of cell markers on peripheral blood T lymphocytes and monocytes was determined by incubating cells in DPBS/BSA with the appropriate monoclonal anti-marker antibodies or the respective isotype-matched control IgG (all from BD Biosciences). After washing, cells were labeled with fluorescein-conjugated anti-mouse IgG. In all experiments, cells were washed twice and immediately used for analysis. Data were monitored on a flow cytofluorimeter (FACSCalibur, BD Biosciences) and analyzed with CellQuest software (Mountain View, CA). Results are expressed as variations of fluorescence mean values (ΔFMV %).
Cell Binding Assays
Interaction between immobilized ligands (CyPB or lactoferrin) and cell surface HS was determined in 96-well microtiter plates (Nunc-Polylabo, Strasbourg, France), as described in Ref. 42. Briefly, cells in DPBS/BSA (at the concentrations of 8 × 106 per ml for T cells, monocyte/macrophages, Jurkat and THP-1 cells, and 5 × 106 per ml for epithelia cells) were allowed to adhere to immobilized ligand (1 μg of CyPB or 4 μg of lactoferrin per well, respectively) for 45 min at 20 °C. After washing with phosphate-buffered saline, adherent cells were fixed with 3% (w/v) formaldehyde, pH 7.8, stained with 1% (w/v) methylene blue in 100 mm borate buffer, pH 8.2, and lysed with 0.1 m HCl. The absorbance, which is proportional to the number of adhered cells, was measured at 590 nm with a microplate Bio-Rad reader model 550. Cell adhesion was estimated by using standard curves in which absorbance was related to cell numbers. In control experiments, cells were either pretreated for 1 h at 37 °C with heparinases (0.2 unit/106 cells), prior adhesion assays, or incubated in the presence of soluble ligands (100 μg/ml of CyPB or 400 μg/ml of lactoferrin) to determine nonspecific binding. For competitive binding assays, cells were allowed to adhere to immobilized ligands in the presence of increasing concentrations of heparin derivatives, soluble HS, or anti-HS antibodies. Statistical significance was determined using the Student's t test, and p values < 0.05 were considered as significant.
Analysis of p44/p42 MAPK Activation
Activation of p44/p42 MAPK was analyzed essentially as described in Ref. 26. Jurkat T cells were cultured overnight in the presence of 1% (v/v) fetal calf serum and stimulated with 50 nm CyPB or 250 nm lactoferrin at 37 °C. At various times, cells (106 per sample) were washed in cold phosphate-buffered saline and lysed for 3 h at 4 °C in 75 μl of lysis buffer (20 mm phosphate, pH 7.4, 350 mm NaCl, 10 mm KCl, 1 mm EDTA, 1% (v/v) Triton X-100, 20% (v/v) glycerol) supplemented with 1 mm sodium orthovanadate, 10 mm sodium fluoride, and protease inhibitor mixture. The lysates were clarified by centrifugation and used for immunoblotting. Proteins were separated on 10% (w/v) SDS-PAGE and transferred onto nitrocellulose membrane (Schleicher & Schüll). The blots were blocked for 1 h at room temperature in 20 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1% (v/v) Tween 20, 3% (w/v) BSA. Membrane was probed for 2 h with primary antibodies in Tris buffer supplemented with 1% (w/v) BSA. After washing, immunoreactive proteins were visualized with horseradish peroxidase-conjugated secondary antibodies by using a chemiluminescence detection kit (Amersham Biosciences).
RESULTS
CyPB Binding to HS/Heparin Is Dependent on N-Sulfate Density
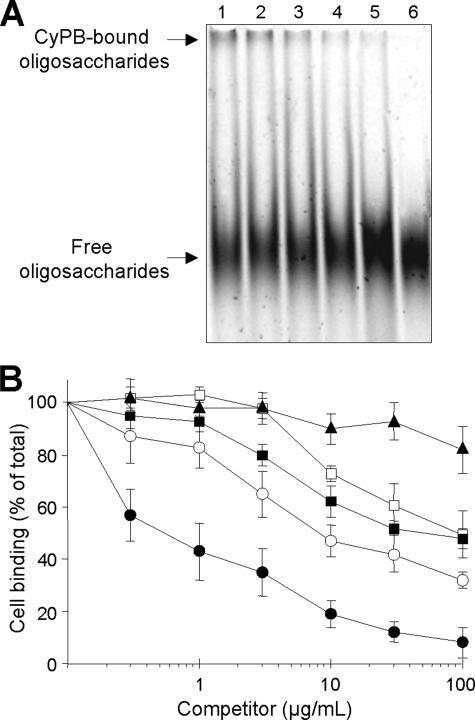
We recently used electrophoretic mobility shift assay with fluorescent oligosaccharides to demonstrate that an octasaccharide is the minimal length unit required for efficient binding of CyPB to heparin and cell surface HS (27). In a first set of experiments, we used the same approach to investigate the importance of N-sulfate density in CyPB binding. To this end, we analyzed the capability of chemically N-desulfated heparin derivatives and soluble HS to compete for the binding of CyPB to a heparin-derived dp12 (Fig. 1A). As reported previously, interaction of CyPB with fluorescent oligosaccharides led to formation of a complex that did not migrate and therefore could be visualized at the top of the gel. As expected, a 10-fold excess (w/w) of unmodified heparin was efficient to get a complete inhibition of CyPB binding to ANTS-labeled dp12. In contrast, fully N-desulfated heparin was not able to dissociate the complex between CyPB and ANTS-labeled dp12, showing that N-desulfated heparin did not inhibit the interactions between CyPB and fluorescent oligosaccharides (less than 10% of inhibition). The formation of an apparent stable complex could be visualized in the presence of partially N-desulfated heparin. The relative intensity of the complex was reduced by ∼35%, indicating that partial removal of N-sulfate groups was sufficient to dramatically alter the ability of heparin to inhibit the interactions between CyPB and fluorescent oligosaccharides. The intensity of the complexes was reduced by almost 70 and 55% in the presence of HS from bovine kidney and porcine mucosae, respectively. This indicated that soluble HS were able to inhibit the interaction between CyPB and fluorescent oligosaccharides, although to a lesser extent than unmodified heparin. However, they were more efficient competitors than partially N-desulfated heparin. In our hands, partial N-desulfation of heparin resulted in a loss of 25% N-sulfate density (71.9% versus 95.2% for unmodified heparin). Moreover, treatment with HNO2 at pH 4 generated fragments of homogeneous size, demonstrating that partial chemical N-desulfation was random (data not shown). In contrast, the overall N-sulfate densities of bovine kidney and porcine mucosal HS, in which GlcNS are clustered in NS domains, are 48.3 and 42.2%, respectively. This indicates that CyPB binding is dependent on N-sulfate density but also on the structural arrangement of GlcNS in HS sequences.
FIGURE 1.
Competition of CyPB binding to heparin-derived oligosaccharides and cell surface HS. A, ANTS-labeled dp12 (0.6 nmol) and CyPB (0.1 nmol) were mixed in the absence (control) or presence of heparin derivatives or soluble HS (20 μg). After a 30-min incubation, samples were subjected to electrophoretic mobility shift assay. Lane 1, control; lane 2, fully N-desulfated heparin; lane 3, partially N-desulfated heparin; lane 4, porcine mucosal HS; lane 5, bovine kidney HS; lane 6, unmodified heparin. At the end of the electrophoresis, the profile of migration of ANTS-labeled dp12 was imaged after exposure to UV transilluminator for 0.60 s. Representative gel of three separate experiments is shown. B, inhibition of the interaction of CyPB with cell surface HS was analyzed by measuring the binding of Jurkat T cells to immobilized CyPB (1 μg/well) in the absence (control) or presence of increasing concentrations of heparin derivatives or soluble HS as follows: unmodified heparin (●), partially N-desulfated heparin (■), fully N-desulfated heparin (▴), bovine kidney HS (○), and porcine mucosal HS (□). Cell binding was related to the number of initially added cells (0.8 × 106 per well) remaining fixed to the adhesive substrate. Maximal binding in the absence of competitor was 0.52 ± 0.06 × 106 cells per well. Results are expressed as percentages of this maximal value. Points are means ± S.D. of triplicates from at least three separate experiments.
To validate these results, we compared the ability of chemically N-modified heparins and soluble HS to inhibit the binding of CyPB to Jurkat T cells (Fig. 1B). Unmodified heparin was used as a control to ensure that soluble heparin was effective to give a complete inhibition of CyPB binding to cell surface. The 50% inhibitory concentration (IC50) of unmodified heparin was approximately 0.5 μg/ml, although the IC50 of partially N-desulfated heparin was 100 times higher. Completely N-desulfated heparin was devoid of any inhibitory properties. As expected, we found that bovine kidney and porcine mucosal HS were also poor competitors, with IC50 around 10 and 100 μg/ml, respectively.
CyPB Binding to Cell Surface HS Is Dependent on the Interaction with Specific N-Sulfated Motifs
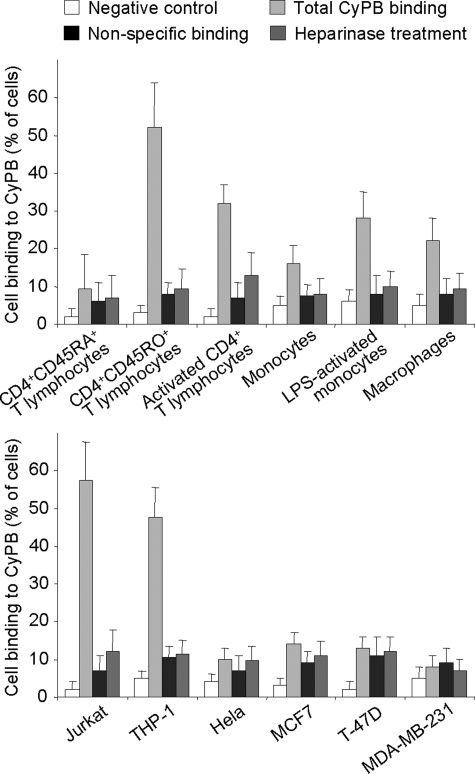
In previous studies, we analyzed the interactions between CyPB and cell surface HS by using a method in which T lymphocytes were allowed to adhere to immobilized ligand (26, 42). We therefore used the same approach to compare the binding of CyPB to various subsets of T lymphocytes and monocytes/macrophages. As shown in Fig. 2, no specific interaction could be observed between CyPB and CD4+CD45RA+ T cells. In contrast, CD4+CD45RO+ T cells and activated CD4+ T lymphocytes strongly interacted with the immobilized ligand, thus indicating that CyPB binding is dependent on the stage of activation and/or maturation of CD4+ T cells. Freshly isolated monocytes and monocyte-derived macrophages also specifically interacted with immobilized CyPB, although to a lesser extent than T lymphocytes did. However, the binding was significantly increased following lipopolysaccharide stimulation of monocytes, indicating that the expression of binding sites for CyPB could be modulated by inflammatory stimuli. We then demonstrated that treatment with heparinases strongly reduced the binding of activated/memory CD4+ T cells and monocytes/macrophages to immobilized CyPB, thus confirming the involvement of cell surface HS in the interactions. We reproduced the experiments with various cells lines. As already reported (9, 42), THP-1 and Jurkat T cells specifically interacted with CyPB. Interaction was efficiently reduced following heparinase treatment, thus confirming that both cell lines are appropriate cellular models to analyze the interaction between CyPB and cell surface HS. Epithelia cell lines express high levels of membrane-associated HS, which have been reported to support interactions with various extracellular ligands (43, 44). However, we found here that HeLa, MCF7, T-47D, and MDA-MB-231 epithelia cells were unable to specifically interact with immobilized CyPB. These last results suggest that the expression of HS with specialized binding sequences is required to support the specific binding of CyPB to target cells.
FIGURE 2.
CyPB binding to primary T lymphocytes subsets, monocytes/macrophages, and cell lines. To analyze the interaction between CyPB and cell surface HS, cells were allowed to adhere to immobilized ligand (1 μg per well) for 45 min at 20 °C. After washing, adherent cells were fixed with 3% (w/v) formaldehyde, stained with 1% (w/v) methylene blue, and lysed with 0.1 m HCl. In control experiments, cells were either allowed to adhere to plastic in the absence of CyPB (negative control), pretreated for 1 h at 37 °C with heparinases (0.2 units/106 cells) prior to adhesion assays, or incubated in the presence of 100 μg/ml soluble CyPB to determine nonspecific binding. Cell adhesion was estimated by using standard curves in which absorbance was related to cell numbers. Results are presented as percentages of initially added cells (8 × 106 per ml for T cell subsets, monocytes/macrophages, Jurkat and THP-1 cells, and 5 × 106 per ml for epithelia cells lines), which remained fixed to the adhesive substrate. Each bar of histograms represents mean ± S.D. of three independent experiments. Statistical significance was determined using the Student's t test, and p values <0.05 were considered as significant.
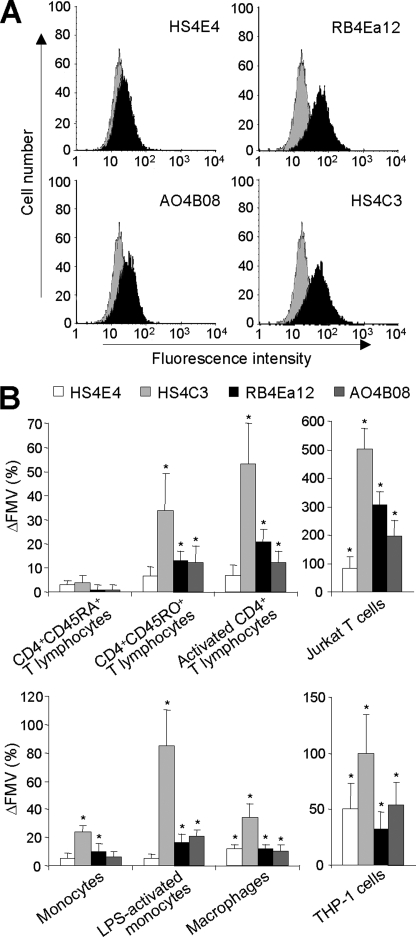
To gain insights into the structural features of HS expressed at the surface of activated/memory CD4+ T cells and monocytes/macrophages, flow cytometric analysis was performed with antibodies recognizing specific sulfated HS epitopes. The reactivity of anti-HS antibodies was first determined with Jurkat T cells (Fig. 3A). HS4C3 and AO4B08, which react with highly sulfated HS/heparin epitopes, efficiently stained the cell surface, even though the epitope recognized by AO4B08 appeared less represented. RB4Ea12, which reacts with a less sulfated epitope, was as efficient as HS4C3 to stain the cell surface of Jurkat T cells, whereas HS4E4, which recognizes distinct HS epitope with low O-sulfation, was the less reactive anti-HS antibody. We reproduced the experiments with CD4+ T lymphocytes and monocytes/macrophages (Fig. 3B). CD4+ CD45RA+ T cells were not significantly immunostained with anti-HS antibodies, indicating that HS was absent or barely expressed on the cell surface of naive T cells. In contrast, a significant staining could be observed on CD4+CD45RO+ and activated CD4+ T cells. Experiments were conducted with peripheral blood T lymphocytes isolated from at least three different healthy donors. Although some inter-individual variations in the fluorescence intensity have been observed, the anti-HS antibody binding profiles were similar. These data indicate that the expression level of HS on CD4+ T lymphocytes was dependent on the stage of activation and/or maturation. Fluorescence values were low compared with Jurkat T cells, indicating that HS was less expressed at the surface of primary T cells. However, the profiles of cell immunostaining resembled each other, suggesting that activated/memory CD4+ T lymphocytes and Jurkat T cells expressed cell surface HS with similar structural features. The reactivity of antibodies with primary monocytes was low (HS4C3 and RB4Ea12) or not significant (HS4E4 and AO4B08). Cell immunostaining with HS4C3, RB4Ea12, and AO4B08 was increased after stimulation with lipopolysaccharide, suggesting that monocyte activation induced the expression of cell surface HS with specific epitopes. Following monocyte differentiation into macrophages, a low but significant staining was observed with anti-HS antibodies, demonstrating that in vitro differentiation of monocytes did not dramatically increase the level of expression of cell surface HS. Finally, we found that anti-HS antibodies markedly reacted with THP-1 cells. Although the values of fluorescence were high compared with primary monocytes/macrophages, the profiles of cell immunostaining resembled each other, indicating that these cell types express cell surface HS with similar structural features.
FIGURE 3.
Detection of cell surface HS on T lymphocytes and monocytes/macrophages. Cells were immunostained with VSV-tagged antibodies to HS epitopes or isotype control. After incubation with mouse anti-VSV and fluorescein-conjugated anti-mouse antibodies, fluorescence was detected by flow cytofluorimetry. A, reactivity of anti-HS antibodies with Jurkat T cells. The black histogram represents staining with anti-HS antibodies, and the gray histogram represents the negative control. Data are representative of three separate experiments. B, detection of cell surface HS on CD4+ T lymphocyte subsets, monocytes/macrophages, Jurkat T cells, and THP-1 cells. Data are expressed as variation of fluorescence mean value (ΔFMV %) and correspond to means ± S.D. from at least three separate experiments obtained with peripheral blood cells from different donors. *, significant difference compared with isotype control (p < 0.05).
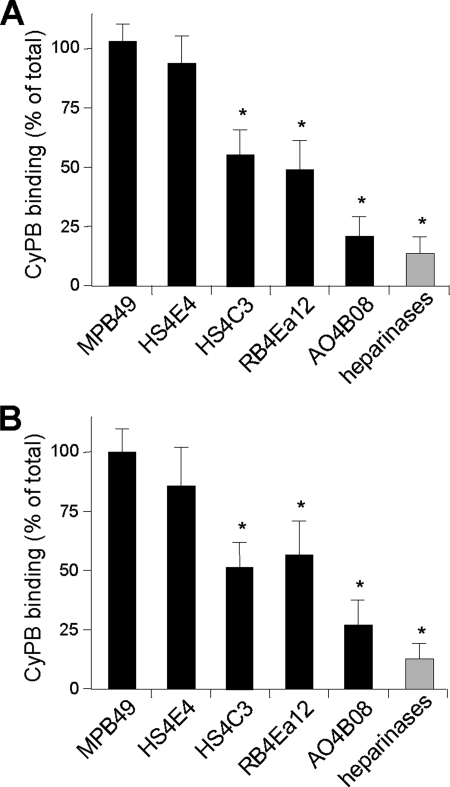
Taking into account the reactivity of anti-HS antibodies (supplemental Table S1), these results indicate that membrane-associated HS on activated/memory CD4+ T lymphocytes, Jurkat T cells, monocytes/macrophages, and THP-1 cells contained highly sulfated sequences. We then analyzed the ability of anti-HS antibodies to inhibit the binding of CyPB to both THP-1and Jurkat cells (Fig. 4). By comparison with the irrelevant antibody, HS4E4 did not inhibit the interaction between CyPB and responsive cells, indicating that the HS sequences recognized by both the antibody and CyPB did not overlap. In contrast, HS4C3, RB4Ea12, and AO4B08 efficiently reduced the interaction between CyPB and cell surface HS, confirming that CyPB interacted with highly modified HS motifs. HS4C3 was described to react with HS epitopes containing 3-O-sulfated groups, thus confirming the involvement of this modification on the interaction with CyPB. Surprisingly, AO4B08 was more efficient than HS4C3 and RB4Ea12 to inhibit the interaction, and also the epitope recognized by this antibody appeared less represented. Given that this antibody recognizes highly sulfated epitopes containing internal 2-O-sulfated IdoUA residue, these results indicate that HS sequences containing these structural modifications are also critically involved in the efficient interaction with CyPB.
FIGURE 4.
Effect of anti-HS antibodies on the interaction of CyPB with cell surface HS. Jurkat T cells (A) and THP-1 cells (B) were preincubated in the presence of anti-HS antibodies or isotype control and allowed to adhere to immobilized CyPB (1 μg/well). Cell binding was related to the number of initially added cells (0.8 × 106 per well) remaining fixed to the adhesive substrate. Heparinase-treated cells were used as a control to estimate the participation of HS in the interaction. Maximal binding obtained in the absence of antibody was estimated at 0.48 ± 0.08 × 106 and 0.33 ± 0.05 × 106 cells per well for Jurkat and THP-1, respectively. Results are normalized to these control values, which were set at 100%. Each bar of the histograms represents the mean ± S.D. of triplicates from three separate experiments. *, significant difference compared with isotype control (p < 0.05).
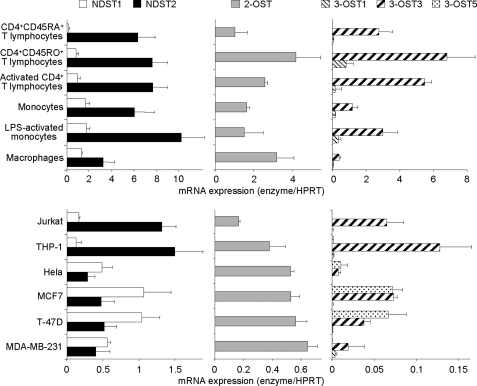
T Lymphocytes and Monocytes/Macrophages Specifically Express High Levels of NDST2 and 3-OST3 Isoforms
In the following experiments, we analyzed whether structural features of the high affinity CyPB-binding sites could be related to cell type-specific expression of certain HS sulfotransferases. Although 2-OST is represented by a single isoform, four NDST and seven 3-OST have been cloned and characterized. Among these isoenzymes, NDST1, NDST2, 3-OST1, 3-OST3 (3A and 3B), and 3-OST5 are widely expressed in various tissues, whereas the expression of other isoforms is mainly restricted to embryonic and/or brain tissues. We then decided to analyze the respective levels of expression of mRNAs encoding NDST1, NDST2, 2-OST, 3-OST1, 3-OST3, and 3-OST5 by real time reverse transcription-PCR. As shown in Fig. 5, NDST1 was barely detected in naive T lymphocytes by comparison with memory and activated CD4+ T cells, whereas no notable variation in the expression of NDST2 could be distinguished between T cell subsets. Conversely, the expression of NDST2 in monocytes was modified following cell activation or maturation, although the same treatment had no significant effect on the level of mRNA encoding NDST1. These results suggest that the expression of both isoenzymes could be regulated by distinct mechanisms in lymphocyte and monocyte subsets. Nevertheless, a common feature of these cells is the relative expression of mRNAs encoding both isoenzymes. Indeed, the level of transcripts encoding NDST1 remained low in all the subsets of CD4+ T lymphocytes and monocytes/macrophages by comparison with the high expression of NDST2. The level of expression of NDST1 and NDST2 in Jurkat T cells and THP-1 cells was lower than in primary cells, which could be due, however, to the transformed phenotype of cell lines. Most importantly, the relative expression of both isoenzymes was quite similar to those observed in CD4+ T lymphocytes and monocytes/macrophages. We then analyzed the expression of mRNAs encoding these enzymes in HeLa, MCF7, T-47D, and MDA-MB-231 cell lines (Fig. 5). In contrast to Jurkat and THP-1, the transcript encoding NDST1 was the main NDST mRNA expressed in epithelia cells, thus suggesting that high expression of NDST2 relative to NDST1 may be a common feature of cells from lymphoid and myeloid lineages.
FIGURE 5.
Expression of mRNAs encoding HS sulfotransferases. Total RNA was extracted from primary CD4+ T lymphocyte subsets, monocytes/macrophages, and various cell lines (Jurkat, THP-1, HeLa, MCF7, T-47D, and MDA-MB-231). Following reverse transcription, variations in the levels of expression of NDST1, NDST2, 2-OST, 3-OST1, 3-OST3, and 3-OST5 transcripts were quantified by real time PCR, as described under “Experimental Procedures.” Relative transcript abundance was normalized to endogenous control hypoxanthine-guanine phosphoribosyltransferase (HPRT) mRNA. Data are means ± S.D. from triplicates and are representative of at least three experiments performed independently.
CD4+ T lymphocytes and monocytes/macrophages express mRNA encoding 2-OST. As above, some variations could be observed between subsets of T cells and monocytes/macrophages, indicating that the expression of 2-OST was also modulated by cell activation and/or maturation. Moreover, the expression of mRNA encoding 2-OST was low in Jurkat and THP-1 cells by comparison with primary cells, thus supporting the idea that the transformed phenotype of established cell lines decreases the expression of HS biosynthetic enzymes. As expected, epithelial cell lines also express mRNA encoding 2-OST. However, no notable difference could be observed with Jurkat and THP-1 cells, as the levels of expression remained in the same range of values. Finally, we quantified the levels of expression of mRNAs encoding 3-OST1, 3-OST3 (including 3-OST3A and -3B), and 3-OST5. As expected, we found that CD4+ T lymphocytes and monocytes/macrophages specifically expressed high levels of 3-OST3. 3-OST1 was barely detected, although no notable expression of 3-OST5 could be observed. The profiles of expression of 3-OST isoforms were quite similar in THP-1 and Jurkat, indicating that high expression of 3-OST3 relative to other 3-OST isoforms may also be a common feature of cells from lymphoid and myeloid lineages. In support of this idea, we found that the expressions of these isoenzymes were quite different in epithelial cell lines. Indeed, 3-OST3 (and other isoforms) were poorly expressed in HeLa and MDA-MB-231 cells, whereas mRNAs encoding 3-OST3 and 3-OST5 were both detected at a similar level in MCF7 and T-47D cells lines. On the assumption that cell type-specific expression of HS sulfotransferases may regulate the structure of HS species, our results raise the intriguing possibility that synthesis of HS with CyPB-binding properties could be determined by the relative expression of certain isoforms of some sulfotransferases.
Silencing the Expression of HS Sulfotransferases Efficiently Alters the Pattern of Sulfation of Cell Surface HS
To gain evidence regarding the relationships between the expression of sulfotransferases in CyPB-responsive cells and structural features in HS sequences, we developed a model based on RNA interference in Jurkat T cells. We checked that treatment of cells with either irrelevant siRNA (siGFP) or negative control siRNAs, in which two nucleotides have been changed from the target sequence, did not induce any decrease in the expression of target mRNAs. The specific siRNAs were thereafter tested for their ability to suppress mRNAs encoding each enzyme. Cell treatment with siRNA targeting NDST1 (siNDST1) resulted in a significant down-regulation of the corresponding mRNA. Inhibition was found to reach 80% at day 1, and the inhibitory effect was maintained for 3 days. A similar decrease in the expression of mRNA encoding NDST2, 2-OST, and 3-OST3 was observed in Jurkat T cells treated with corresponding specific siRNA (supplemental Fig. S1A). Moreover, we checked for the absence of any cross-reactivity. As expected, the expression of mRNA encoding NDST2, 2-OST, and 3-OST3 was not significantly altered by siNDST1, nor was the expression of mRNA encoding NDST1, 2-OST, and 3-OST3 by siNDST2, demonstrating the specificity of both siRNAs used in this assay. In the same way, si2-OST and si3-OST3 significantly reduced the expression of target mRNA, without affecting the expression of others (supplemental Fig. S1B). Notably, we also checked that the transcripts of other HS biosynthetic enzymes were expressed at the same level in control and siRNA-treated cells (data not shown), thus indicating that the expected changes in the structural and functional properties of HS would only be due to silencing of target sequences encoding sulfotransferase isoforms.
We then analyzed whether silencing the expression of sulfotransferases led to a consequent decrease in HS sulfation (supplemental Fig. S2). By comparison with cells transfected with irrelevant siRNA or negative control siRNA, silencing the expression of NDSTs resulted in a dramatic loss of the reactivity of HS4C3, AO4B08, and RB4Ea12, which all recognize N-sulfated epitopes. Moreover, specific siRNAs did not reduce cell immunostaining with anti-proteoglycan antibodies, confirming that the loss of reactivity of anti-HS antibodies was due to a decrease in N-sulfation of HS moieties rather than inhibition of cell surface expression of proteoglycans (data not shown). We also analyzed the binding of IO3H10 (1:150), which reacts with an epitope in chondroitin sulfate (32), to check whether down-regulation of NDSTs did not affect sulfation of other cell surface glycosaminoglycans. Surprisingly, we found that silencing the expression of NDST1, and to a lesser extent of NDST2, increased the binding of IO3H10 to Jurkat T cells. These findings suggest that the loss of HS sulfation had probably made available the amount of sulfate donor to modify other membrane-associated glycosaminoglycans. To further demonstrate the specificity of siRNA-mediated silencing of NDST isoforms, we performed rescue experiments with plasmids expressing NDST sequences refractory to siRNA. As expected, transfection with plasmid encoding either NDST1 or NDST2 partially restored the binding of anti-HS antibodies, indicating that the loss of activity of endogenous NDST isoforms has been compensated by the expression of exogenous NDST isoforms refractory to siRNA. Silencing the expression of 2-OST and 3-OST3 had less effect on the reactivity of anti-HS antibodies. Cell immunostaining by HS4E4 and RB4Ea12 was not significantly modified, confirming that 2-O- and 3-O-sulfation are not required to support binding of these antibodies. Although the binding of AO4B08 was reported to require the presence of internal IdoUA2S residue, the reactivity of this antibody was weakly altered by silencing the expression of 2-OST. However, we found that the expression of mRNA encoding 2-OST was reduced by ∼90% in transfected cells. Therefore, residual 2-O-sulfation of HS could be sufficient to support the binding of AO4B08. Finally, silencing the expression of 2-OST and 3-OST3 significantly reduced the binding of HS4C3. Among other HS modifications, the reactivity of this antibody is dependent on 2-O and 3-O sulfation, thus indicating that siRNAs had efficiently reduced the expression of target sequences. To confirm these results, rescue experiments were performed with plasmids expressing 2-OST and 3-OST3B sequences refractory to siRNA. As above, transfection of cells with refractory plasmids restored the binding of anti-HS antibodies, confirming the specificity of corresponding siRNA. Altogether, these data demonstrate that specific silencing of mRNAs encoding NDST1, NDST2, 2-OST, or 3-OST3 has led to significant changes in the pattern of HS sulfation, thus confirming that these HS sulfotransferases were enzymatically active in Jurkat T cells.
Silencing the Expression of NDST1 or NDST2 Differently Alters N-Sulfation
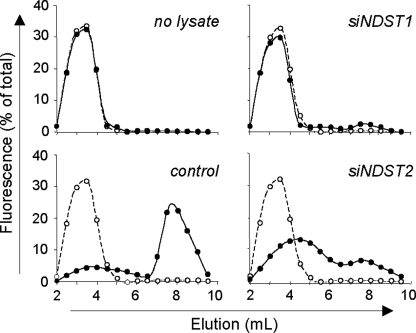
Although the reactivity of anti-HS antibodies was informative to analyze siRNA-induced alterations in HS sulfation, it was not discriminant enough to know whether NDST1 and NDST2 have redundant or distinct activities. To establish how silencing the expression of these isoenzymes modified N-sulfate density of HS, we therefore used an in vitro approach, in which Jurkat T cell lysates were used as enzyme sources and ANTS-labeled desulfated/N-reacetylated dp14 as substrates. The oligosaccharides were first incubated with cell lysates and thereafter treated with HNO2, pH 1.5, which specifically target GlcNS residues (37). We could expect that in vitro modified dp14, which contain newly synthesized GlcNS residues, were sensitive to attack by low pH nitrous acid and generated degradation products smaller than parent oligosaccharides. The products of cleavage were subsequently separated by gel filtration, and the ANTS moiety served as a tag to differentiate small oligosaccharides derived at the reducing end from the parent compounds (Fig. 6). In the absence of incubation with cell lysate, treatment by nitrous acid did not modify the elution of fluorescent oligosaccharides, confirming that this substrate did not contain detectable N-sulfate groups. In contrast, incubation with cell lysate obtained from control cells and subsequent treatment with HNO2 resulted in the production of small fluorescent species. The degradation products were mainly eluted as di- and tetrasaccharide standards, indicating that cell lysate contained active NDSTs that have efficiently modified fluorescently tagged oligosaccharides. As expected, down-regulation of the expression of NDST1 dramatically reduced the capability of cell lysate to catalyze N-sulfation of oligosaccharides. A small fraction (∼10%) of fluorescent oligosaccharides was sensitive to nitrous acid degradation, indicating that only partial N-sulfation had occurred. Given that the expression of NDST2 was not altered in these cells, the fluorescent species that migrated as small oligosaccharides could result from N-sulfation by this isoenzyme. Nevertheless, the lack of efficient N-sulfation suggested that NDST2 had a weak activity in the absence of NDST1. On the other hand, we found that the expression of NDST1 was reduced by 80% in transfected cells. Therefore, partial N-sulfation could also be the consequence of a remaining pool of NDST1. We reproduced the same experiment with cell lysate in which the expression of NDST2 was down-regulated. Most of oligosaccharides were degraded by HNO2 treatment, indicating that N-sulfation of fluorescent substrate occurred in the absence of NDST2. However, the degradation products were eluted earlier than di- and tetrasaccharides, suggesting that the fluorescent oligosaccharides had a lower density of N-sulfate groups and were consequently less sensitive to HNO2 cleavage. We demonstrated that NDST1 expression was not altered in cells transfected with siNDST2. Therefore, NDST1 was probably active in the absence of NDST2 and could initiate N-sulfation of the fluorescent substrate. However, the loss of NDST2 probably led to a decrease in the addition of N-sulfate groups in the fluorescent substrate. These data indicate that NDST1 and NDST2 do not have redundant activity but rather have complementary activity, probably acting sequentially to fully modify HS sequences.
FIGURE 6.
Contribution of NDST1 and NDST2 to N-sulfotransferase activity in Jurkat T cells. N-sulfotransferase activity was analyzed in siRNA-transfected cells by using an in vitro N-sulfation assay, in which cell lysates were used as enzyme sources to modify fluorescently tagged desulfated dp14. Jurkat T cells (25 × 106/ml) were lysed 48 h post-transfection. The sulfation reaction was performed by incubating cell lysate (100 μg of solubilized proteins) with 5 μg of substrate and 10 μm 3′-phosphoadenosyl-5′-phosphosulfate. Newly modified N-sulfated oligosaccharides were purified and subjected to a deaminative cleavage with HNO2 at pH 1.5, as described under “Experimental Procedures.” The products of deaminative cleavage (●) and parent oligosaccharides (○) were fractionated on Bio-Gel P-6 column (20 × 0.8 cm), and fractions of 500 μl were analyzed for fluorescence. Total fluorescence intensity, relative to N-sulfotransferase activity in cell lysates, was set at 100%. Data are representative of three independent experiments.
Specific Binding and Activity of CyPB Are Controlled by the Expression of HS Sulfotransferases
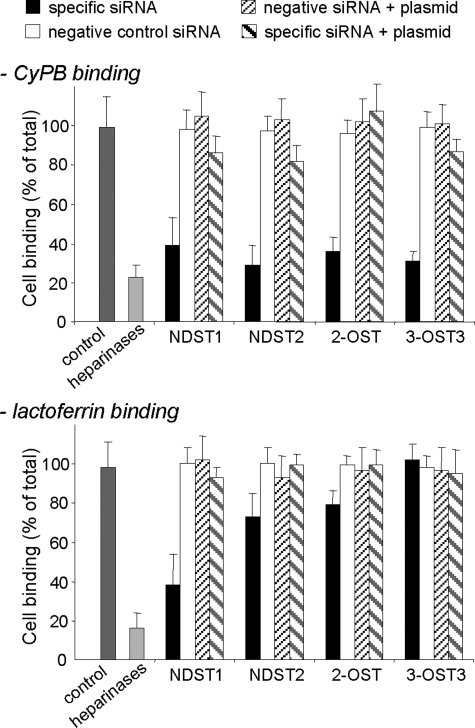
To study the biological consequence of silencing sulfotransferase expression, we analyzed the interaction between CyPB and cell surface HS in siRNA-treated cells (Fig. 7). Lactoferrin has been demonstrated to interact with leukocytes and epithelial cells, mainly as a consequence of interaction with N-sulfate groups in NS domains of HS (43, 44). We therefore used this protein as a model of HS-binding ligand in the following experiments. We first checked that binding of Jurkat T cells to immobilized CyPB or lactoferrin was potently inhibited (>75% of initial binding) by heparinase treatment, thus confirming that the method was also appropriate to analyze interactions between lactoferrin and cell surface HS. Knockdown of NDST1 mRNA reduced by 60% the binding of T cells to immobilized CyPB and lactoferrin, indicating that N-sulfation of GlcN residues by this isoenzyme was of critical importance to support the interactions. Moreover, these data further suggest that NDST2 was unable to compensate for the lack of NDST1 and to initiate HS maturation. We reproduced the same experiment with siRNA specific for NDST2. Interestingly, we found that binding of transfected T cells to CyPB was dramatically reduced (70% of initial binding). In contrast, down-regulation of the expression of NDST2 was less inhibitory for lactoferrin, because binding of Jurkat T cells was only reduced by ∼30%. Taking into account that NDST1 was fully active to initiate N-sulfation in siNDST2-treated cells, these data demonstrate that NDST2 critically contributes to the synthesis of the HS motifs that specifically interact with CyPB. We then analyzed the effects of siRNA specific for 2-OST and 3-OST3 isoforms. Silencing the expression of 2-OST and 3-OST3 strongly reduced the binding of treated cells to immobilized CyPB (65 and 70% of inhibition, respectively), confirming that 2-O- and 3-O-sulfate groups were required to support the binding of the ligand. In contrast, cell binding to lactoferrin was poorly altered by the decrease of 2-O-sulfation (∼20% of inhibition), and down-regulation of the expression of 3-OST3 had no inhibitory effect on the interaction between this ligand and cell surface HS. In control experiments, we checked that the inhibitory effect produced by siRNA treatment could be significantly rescued by exogenous expression of refractory sequences, thus confirming the specificity of siRNA-mediated silencing.
FIGURE 7.
Effect of siRNA targeting HS sulfotransferases on the binding of Jurkat T cells to immobilized ligands. Involvement of NDST1, NDST2, 2-OST, and 3-OST3 in the generation of HS motifs with binding properties was analyzed by measuring the interaction of siRNA-treated Jurkat T cells with immobilized CyPB (1 μg/well) or lactoferrin (4 μg/well), 48 h post-transfection. In control experiments, cells were either treated with irrelevant siGFP or negative control siRNA. In rescue experiments, cells were first treated with specific siRNA or corresponding negative control siRNA for 24 h. Thereafter, cells were transfected with plasmids expressing sequences refractory to specific siRNA and used 24 h later for binding experiments. Cell binding was related to the number of initially added cells (0.8 × 106 per well) remaining fixed to the adhesive substrate. The binding of siGFP-treated cells (control) was estimated at 0.45 ± 0.06 × 106 and at 0.31 ± 0.05 × 106 cells per well for CyPB and lactoferrin, respectively. Results are presented as percentages of these maximal values. Heparinase-treated cells were used as a control to estimate the participation of HS in the interaction. Each bar of histograms represents the mean ± S.D. of three independent experiments.
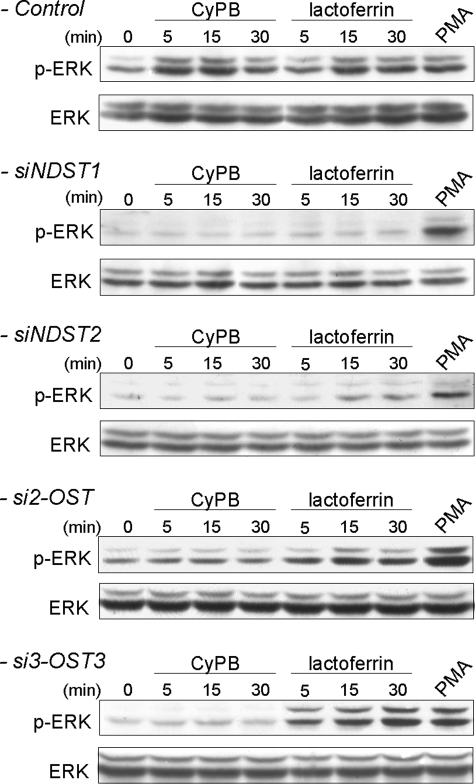
The main intracellular event following CyPB or lactoferrin binding to T lymphocytes is the activation of the p44/p42 MAPK pathway (8, 26, 45). When analyzing the phosphorylated status of ERK1/2, we found that transfection of Jurkat T cells with siNDST1 dramatically reduced the ability of CyPB and lactoferrin to activate p44/p42 MAPK. Inhibition was only due to down-regulation of NDST1 expression, because phorbol myristate acetate, which is a strong activator of the p44/p42 MAPK pathway, efficiently induced the phosphorylation of ERK1/2 in transfected cells (Fig. 8). Although down-regulation of the expression of NDST2 slightly affected cell surface binding of lactoferrin, the activation of p44/p42 MAPK was reduced in both treated cells. However, the inhibitory effect was not as dramatic as that observed with CyPB. Indeed, the ability of CyPB to activate p44/p42 MAPK pathway was abolished in siNDST2-treated cells, indicating that silencing the expression of NDST2 has rendered the cells unresponsive to the inflammatory factor. Previous data have shown that NDST2 was not required to initiate N-sulfation but would rather contribute to increase the density of N-sulfate groups. Consistent with these findings, our results suggest that N-sulfation by NDST1 is sufficient to support the binding of lactoferrin and partial activation of the signaling pathway. In contrast, the contribution of both NDST1 and NDST2 to high N-sulfation is absolutely required for generating HS motifs with CyPB-binding and -activating properties. As expected, down-regulation of 2-OST and 3-OST3 strongly inhibited the ability of CyPB to activate p44/p42 MAPK pathway, thus confirming the involvement of 2-O- and 3-O-sulfate groups in the binding of the protein. In contrast, the response induced by lactoferrin was only slightly reduced in si2-OST2-treated cells and remained unmodified in si3-OST3-treated cells, thus confirming the binding data (Fig. 8). Altogether, these results indicate that specific binding of CyPB to responsive cells and the consequent activation of the signaling pathway are dependent on the expression of highly modified HS.
FIGURE 8.
Effect of siRNA targeting HS sulfotransferases on CyPB- and lactoferrin-induced activation of p44/p42 MAPK. The contribution of NDST1, NDST2, 2-OST, and 3-OST3 in the generation of HS motifs with functional properties was analyzed by measuring the activation of p44/p42 MAPK in siRNA-transfected cells, 48 h post-transfection. Jurkat T cells were stimulated in the presence of 50 nm CyPB or 250 nm lactoferrin for various times, and the phosphorylation of ERK1/2 (p-ERK) was analyzed by Western blot. Parallel immunoblotting with anti-total ERK1/2 confirmed equal loading of samples. Phorbol myristate acetate (PMA) was used as a positive control for activation of p44/p42 MAPK in siRNA-transfected cells. Representative results from three independent experiments are shown.
DISCUSSION
Our previous studies (6, 26, 27) demonstrated that efficient binding of CyPB to heparin and cell surface HS expressed on T cells was dependent on the interaction with a highly sulfated HS sequence, in which a critical modification was the presence of a 3-O-sulfated GlcNH2 residue. Although GlcNH2 residues have been reported to be preferentially located in NA domains and in transition zones between NA and NS domains, 3-O-sulfated GlcNH2-containing sequences have been characterized within heparin and the highly sulfated NS domain of some HS species (21, 23, 46). These data prompted us to propose that the high affinity binding site of CyPB may be located in specialized NS domains of HS expressed at the membrane of target cells. This study further supports this hypothesis. We found that CyPB binding to partially N-desulfated heparin, which contained high levels of GlcNH2 compared with unmodified heparin, was not improved but rather was inhibited. This clearly demonstrates that GlcNH2 residues are required but not sufficient to support interaction with CyPB. Moreover, these results indicate that N-sulfation rather than 6-O- and/or 2-O-sulfations was important to support CyPB binding, because chemical N-desulfation of heparin did not significantly reduce the rate of 6-O- and 2-O-sulfate groups. Soluble HS inhibited the binding of CyPB to fluorescent oligosaccharides to a lesser extent than heparin did. However, they were better competitors than partially N-desulfated heparin, indicating that structural arrangement of sulfate groups in NS domains was also important for optimal binding of CyPB. Competitive binding experiments with anti-HS antibodies allowed us to further characterize the interaction of CyPB with cell surface HS. We demonstrated that HS4C3 reduced CyPB binding to responsive cells. HS4C3 was demonstrated to selectively identify 3-O-sulfated sequences (30, 33, 34), thus confirming the involvement of 3-O-sulfated GlcN residues in the interaction. Importantly, the epitope recognized by HS4C3 was reported to colocalize with the HS motifs that interact with the envelope glycoprotein gD of HSV-1 (35). Given that the disaccharide unit IdoUA2S-GlcNH23S±6S contributes to the specific binding of HSV-1 gD to heparin and cell surface HS of target cells (23), our findings strongly suggest that the HS sequences that react with CyPB and envelope protein of HSV-1 share common structural features. We also report that the binding reactions of CyPB to heparin and cell surface HS from T lymphocytes are quite similar (6, 26, 27). Interestingly, AO4B08 was more efficient than HS4C3 and RB4Ea12 to inhibit the interaction between CyPB and target cells. AO4B08 recognizes highly N- and O-sulfated HS motifs containing internal IdoUA2S residues (31, 33), thus suggesting that this modification was also important for the binding of CyPB. In contrast to AO4B08, HS4E4 recognizes low sulfated HS motifs but does not bind to highly sulfated structures such as some found in the heparin and NS domains of HS (31, 33), explaining why it was unable to inhibit the interaction between CyPB and responsive cells. Altogether, these data support the idea that CyPB interacts with a heparin-like sequence expressed at the surface of target cells.
The sulfation pattern varies considerably among HS species, which has been related to cell type-specific expression of certain isoforms of HS sulfotransferases and their relative expression depending on cellular conditions. Given that the density of N-sulfates and the presence of 2-O- and 3-O-sulfates appeared critical to support CyPB binding, we have then analyzed the expression of related HS sulfotransferases in various cell types. We found that target cells of CyPB, i.e. memory/activated T lymphocytes, monocytes/macrophages, and related established cell lines specifically expressed high levels of mRNA encoding NDST2 and 3-OST3. Although cell activation or maturation differently affects the expression of each isoenzyme in T cells and monocytes/macrophages, the level of transcript encoding NDST2 remains higher than that of NDST1 in all cases. These data are in correspondence with previous studies reporting a higher level of NDST2 relative to NDST1 in mast cells (16), lymphoid tissues, e.g. thymus and spleen, and whole blood leukocytes (14, 47), thus indicating that high expression of NDST2 may be a common feature of cells from the hematopoietic lineages. Although epithelial cells produce high level of HS by comparison with T lymphocytes and monocytes/macrophages, they are devoid of any high affinity binding sites for CyPB. Interestingly, we found that epithelial cell lines express a lower level of NDST2 relative to NDST1, thus arguing for a role of this isoenzyme in making HS sequence with CyPB binding properties. Surprisingly, we found that naive CD4+ T cells expressed high levels of NDST2 and 3-OST3, although they were devoid of any binding activity for CyPB. However, we demonstrated that these cells were not immunostained by anti-HS antibodies, indicating that they did not expressed cell surface HS. These findings are in agreement with previously published data reporting that naive T cells did not express any membrane-associated HS proteoglycans. For example, Forster-Horvàth et al. (48) reported that CD44v3 is expressed intracellularly by naive T cells and rapidly exposed at the cell surface following cell stimulation. Likewise, we and others have shown that CD4+ T lymphocytes and monocytes/macrophages express various HS proteoglycans, e.g. syndecans and CD44v3, for which the mRNA level and cell surface expression are increased following cell activation and differentiation (8, 49, 50). In accordance with these data, we speculate that HS proteoglycans are rapidly exposed to the cell surface upon stimulation of naive T cells. Thereafter, cell differentiation may be accompanied by an increase in HS proteoglycan expression, leading to their constitutive association to the cell membrane. Further investigations are in progress to validate this model and to elucidate its relevance to CyPB functions.
Because high expression of NDST2 has been reported to correlate with high N-sulfate density in heparin and HS (51, 52), we checked the possibility that biosynthesis of HS motifs with CyPB-binding properties could be NDST2-dependent. As expected, down-regulation of the expression of this isoenzyme inhibited the binding of CyPB and consequent activation of p44/p42 MAPK in Jurkat T cells. However, silencing the expression of NDST1 also reduced the binding and activity of CyPB, indicating that both NDSTs are required to synthesize HS motifs with CyPB-binding and -activating properties. Obviously, several aspects regarding the regulation of expression of NDST isoforms and the relative contribution of each isoenzyme to the synthesis of HS remain unclear. Overexpression of NDST1 and NDST2 in HEK293 cells was reported to result in an increased N-sulfation of HS produced by these cells, with no parallel increase in O-sulfation and C5-epimerization, thus arguing for a direct relationship between NDST activity and N-sulfate density. Moreover, cells stably transfected with NDST2 synthesized HS with higher N-sulfate density by comparison with NDST1-transfected cells, suggesting that the N-sulfation pattern may be isoform-dependent (51). Systemic inactivation of NDST1 expression affects HS structure in all tissues tested. Animals succumb perinatally, thus supporting the idea that NDST1 plays a crucial role in HS maturation. In contrast, mice carrying a disruption of the gene encoding NDST2 develop and reproduce normally. They have no notable alteration in HS biosynthesis except in mast cells, in which decreased N-sulfation of heparin results in defective granule formation (16–18). By using genetically modified mice, Ledin et al. (19) demonstrated that liver HS from wild-type and knock-out animals all had the same structure despite significantly reduced NDST enzyme activity in NDST1+/−/NDST2−/− embryos. Moreover, the presence of active NDST2 did not appear to affect HS structure as long as NDST1 was also present. They concluded from their experiments that NDST1 might be preferentially incorporated within a complex formed by the association of different enzymes of HS biosynthesis, although NDST2 would be frozen out. In the absence of NDST1, NDST2 could occupy the NDST position in the HS biosynthesis complex and thus take place on N-sulfation of HS. Another intriguing hypothesis may be that NDST1 is required for partial N-sulfation of the nascent precursor and consequent initiation of other modifications, whereas NDST2 could fill in or extend the sections of N-sulfated residues to create highly sulfated domains in some HS species. Previous results have indeed reported that HS is a better substrate for recombinant NDST isoforms than bacterial polysaccharide K5, a surrogate of unsulfated HS precursor, thus suggesting that NDSTs can act on a previously modified substrate (53). By using the Cre-loxP system of gene inactivation, Wang et al. (54) demonstrated that NDST1 and NDST2 contribute to N-sulfation of HS at a ratio of approximately 3:1 in endothelial cells. In this model, NDST1 was able to compensate for the absence of NDST2 but not vice versa. Our results are in agreement with these latest studies. We demonstrated that down-regulation of the expression of NDST1 dramatically reduced NDST enzymatic activity by comparison with control, indicating that NDST2 was unable to compensate for the absence of NDST1. In contrast, N-sulfation still occurred in the absence of NDST2. However, a significant decrease in the N-sulfate density was observed, suggesting that NDST1 was not sufficient to fully compensate for the absence of NDST2. According to these results, we proposed a model in which both NDSTs have not redundant but rather complementary enzymatic activities. This would be possible if NDST1 and NDST2 localized to different Golgi compartments. Actually, NDST1 was reported to interact with the HS polymerase EXT2, thus suggesting that it takes part in the first modifications of the HS precursor (55). In cells where NDST2 expression is high by comparison with NDST1, NDST2 may act in a late compartment and increase the level of N-sulfation in sequences that have been previously modified by enzymes in a more proximal part of the Golgi network.
We demonstrated in previous works that the binding of CyPB was dependent on the interaction with a HS sequence, in which a key modification was the presence of a 3-O-sulfated GlcNH2 (26). As expected, we demonstrated here that silencing the expression of 3-OST3 dramatically reduced the binding of CyPB to target cells and consequently the activation of the signaling pathway, thus confirming previous results. In addition, we found that down-regulation of 2-OST also reduced CyPB binding and consequent activation of p44/p42 MAPK in responsive cells. These results suggested a direct role for 2-O-sulfate groups in the interaction between CyPB and cell surface HS. This is consistent with the demonstration that AO4B08, which recognizes internal IdoUA2S residues, was highly efficient at reducing the binding of CyPB to target cells. On the other hand, we found that silencing the expression of 2-OST significantly reduced the binding of HS4C3, indicating that alteration of the activity of 2-OST had reduced the subsequent reaction of HS 3-O-sulfation. Actually, 3-OST3 isoforms have been reported to transfer sulfate groups to the 3-OH position of GlcNH2 and/or GlcNS residues, which are adjacent to IdoUA2S residues (21, 23, 25). Therefore, 2-O-sulfation of IdoUA residues is probably a limiting step for the unique substrate specificity of 3-OST3, suggesting that both 2-OST and 3-OST3 also have complementary activities to generate the HS-binding motif of CyPB.
An overall conclusion from our studies is that cell type-specific expression of certain isoforms of sulfotransferases may determine the specificity of extracellular ligand binding. Considering that a number of inflammatory and immunoregulatory mediators interact with cell surface HS to target subsets of T lymphocytes and monocytes/macrophages (56), this could play an important role in inflammatory and immune responses. Moreover, a growing interest has focused on the use of recombinant enzymes to synthesize HS-derived molecules with therapeutic application (57). In this context, a better understanding of the tight regulation of HS biosynthesis will be helpful for the selection of recombinant isoenzymes that will take part in the synthesis of sulfated oligosaccharides with expected activity in the treatment of some inflammatory disorders.
Supplementary Material
Acknowledgments
We are grateful to Christophe Vanpouille (National Institutes of Health, Bethesda) for helpful discussions, Malcolm Lyon (University of Manchester, United Kingdom) for providing fully N-desulfated heparin, and Lena Kjellén (University of Uppsala, Sweden) for the generous gift of plasmids expressing NDST1 and NDST2.
This work was supported by the CNRS and the Université des Sciences et Technologies de Lille, France.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I–III and Figs. S1–S2.
- CyPB
- cyclophilin B
- ANTS
- 8-aminonaphthalene-1,3,6-trisulfonic acid
- dp
- degree of polymerization
- DPBS
- Dulbecco's phosphate-buffered saline
- ERK
- extracellular signal-regulated kinase
- FCS
- fetal calf serum
- GlcNH2
- N-unsubstituted d-glucosamine
- GlcNS
- N-sulfated d-glucosamine
- HS
- heparan sulfate
- MAPK
- mitogen-activated protein kinase
- NDST
- N-deacetylase/N-sulfotransferase
- OST
- O-sulfotransferase
- siRNA
- small interfering RNA
- VSV
- vesicular stomatitis virus
- BSA
- bovine serum albumin
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Handschumacher R. E., Harding M. W., Rice J., Drugge R. J., Speicher D. W. (1984) Science 226, 544–547 [DOI] [PubMed] [Google Scholar]
- 2.Barik S. (2006) Cell. Mol. Life Sci. 63, 2889–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yurchenko V., O'Connor M., Dai W. W., Guo H, Toole B., Sherry B., Bukrinsky M. (2001) Biochem. Biophys. Res. Commun. 288, 786–788 [DOI] [PubMed] [Google Scholar]
- 4.Allain F., Vanpouille C., Carpentier M., Slomianny M. C., Durieux S., Spik G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2714–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yurchenko V., Constant S., Bukrinsky M. (2006) Immunology 117, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denys A., Allain F., Carpentier M., Spik G. (1998) Biochem. J. 336, 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arora K., Gwinn W. M., Bower M. A., Watson A., Okwumabua I., MacDonald H. R., Bukrinsky M. I., Constant S. L. (2005) J. Immunol. 175, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pakula R., Melchior A., Denys A., Vanpouille C., Mazurier J., Allain F. (2007) Glycobiology 17, 492–503 [DOI] [PubMed] [Google Scholar]
- 9.Melchior A, Denys A., Deligny A., Mazurier J., Allain F. (2008) Exp. Cell Res. 314, 616–628 [DOI] [PubMed] [Google Scholar]
- 10.Capila I., Linhardt R. J. (2002) Angew. Chem. Int. Ed. Engl. 41, 391–412 [DOI] [PubMed] [Google Scholar]
- 11.Ori A., Wilkinson M. C., Fernig D. G. (2008) Front. Biosci. 13, 4309–4338 [DOI] [PubMed] [Google Scholar]
- 12.Turnbull J., Powell A., Guimond S. (2001) Trends Cell Biol. 11, 75–82 [DOI] [PubMed] [Google Scholar]
- 13.Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 14.Aikawa J., Esko J. D. (1999) J. Biol. Chem. 274, 2690–2695 [DOI] [PubMed] [Google Scholar]
- 15.Aikawa J., Grobe K., Tsujimoto M., Esko J. D. (2001) J. Biol. Chem. 276, 5876–5882 [DOI] [PubMed] [Google Scholar]
- 16.Forsberg E., Pejler G., Ringvall M., Lunderius C., Tomasini-Johansson B., Kusche-Gullberg M., Eriksson I., Ledin J., Hellman L., Kjellén L. (1999) Nature 400, 773–776 [DOI] [PubMed] [Google Scholar]
- 17.Ringvall M., Ledin J., Holmborn K., van Kuppevelt T., Ellin F., Eriksson I., Olofsson A. M., Kjellen L., Forsberg E. (2000) J. Biol. Chem. 275, 25926–25930 [DOI] [PubMed] [Google Scholar]
- 18.Ledin J., Staatz W., Li J. P., Götte M., Selleck S., Kjellén L., Spillmann D. (2004) J. Biol. Chem. 279, 42732–42741 [DOI] [PubMed] [Google Scholar]
- 19.Ledin J., Ringvall M., Thuveson M., Eriksson I., Wilén M., Kusche-Gullberg M., Forsberg E., Kjellén L. (2006) J. Biol. Chem. 281, 35727–35734 [DOI] [PubMed] [Google Scholar]
- 20.Shworak N. W., Liu J., Fritze L. M., Schwartz J. J., Zhang L., Logeart D., Rosenberg R. D. (1997) J. Biol. Chem. 272, 28008–28019 [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Shriver Z., Blaiklock P., Yoshida K., Sasisekharan R., Rosenberg R. D. (1999) J. Biol. Chem. 274, 38155–38162 [DOI] [PubMed] [Google Scholar]
- 22.Shukla D., Liu J., Blaiklock P., Shworak N. W., Bai X., Esko J. D., Cohen G. H., Eisenberg R. J., Rosenberg R. D., Spear P. G. (1999) Cell 99, 13–22 [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Shriver Z., Pope R. M., Thorp S. C., Duncan M. B., Copeland R. J., Raska C. S., Yoshida K., Eisenberg R. J., Cohen G., Linhardt R. J., Sasisekharan R. (2002) J. Biol. Chem. 277, 33456–33467 [DOI] [PubMed] [Google Scholar]
- 24.Xu D., Tiwari V., Xia G., Clement C., Shukla D., Liu J. (2005) Biochem. J. 385, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mochizuki H., Yoshida K., Shibata Y., Kimata K. (2008) J. Biol. Chem. 283, 31237–31245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanpouille C., Deligny A., Delehedde M., Denys A., Melchior A., Liénard X., Lyon M., Mazurier J., Fernig D. G., Allain F. (2007) J. Biol. Chem. 282, 24416–24429 [DOI] [PubMed] [Google Scholar]
- 27.Vanpouille C., Denys A., Carpentier M., Pakula R., Mazurier J., Allain F. (2004) Biochem. J. 382, 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spik G., Strecker G., Fournet B., Bouquelet S., Montreuil J., Dorland L., van Halbeek H., Vliegenthart J. F. (1982) Eur. J. Biochem. 121, 413–419 [DOI] [PubMed] [Google Scholar]
- 29.Spik G., Haendler B., Delmas O., Mariller C., Chamoux M., Maes P., Tartar A., Montreuil J., Stedman K., Kocher H. P., Keller R., Hiestand P. C., Movva N. R. (1991) J. Biol. Chem. 266, 10735–10738 [PubMed] [Google Scholar]
- 30.van Kuppevelt T. H., Dennissen M. A., van Venrooij W. J., Hoet R. M., Veerkamp J. H. (1998) J. Biol. Chem. 273, 12960–12966 [DOI] [PubMed] [Google Scholar]
- 31.Dennissen M. A., Jenniskens G. J., Pieffers M., Versteeg E. M., Petitou M., Veerkamp J. H., van Kuppevelt T. H. (2002) J. Biol. Chem. 277, 10982–10986 [DOI] [PubMed] [Google Scholar]
- 32.Smetsers T. F., van de Westerlo E. M., ten Dam G. B., Overes I. M., Schalkwijk J., van Muijen G. N, van Kuppevelt T. H. (2004) J. Invest. Dermatol. 122, 707–716 [DOI] [PubMed] [Google Scholar]
- 33.Smits N. C., Lensen J. F., Wijnhoven T. J., Ten Dam G. B., Jenniskens G. J., van Kuppevelt T. H. (2006) Methods Enzymol. 416, 61–87 [DOI] [PubMed] [Google Scholar]
- 34.Ten Dam G. B., Kurup S., van de Westerlo E. M., Versteeg E. M., Lindahl U., Spillmann D., van Kuppevelt T. H. (2006) J. Biol. Chem. 281, 4654–4662 [DOI] [PubMed] [Google Scholar]
- 35.Tiwari V., ten Dam G. B., Yue B. Y., van Kuppevelt T. H., Shukla D. (2007) FEBS Lett. 581, 4468–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Z., Lyon M., Gallagher J. T. (2005) J. Biol. Chem. 280, 15742–15748 [DOI] [PubMed] [Google Scholar]
- 37.Shively J. E., Conrad H. E. (1976) Biochemistry 15, 3932–3942 [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlsson P., Presto J., Spillmann D., Lindahl U., Kjellén L. (2008) J. Biol. Chem. 283, 20008–20014 [DOI] [PubMed] [Google Scholar]
- 40.Rong J., Habuchi H., Kimata K., Lindahl U., Kusche-Gullberg M. (2000) Biochem. J. 346, 463–468 [PMC free article] [PubMed] [Google Scholar]
- 41.Bame K. J., Reddy R. V., Esko J. D. (1991) J. Biol. Chem. 266, 12461–12468 [PubMed] [Google Scholar]
- 42.Carpentier M., Allain F., Slomianny M. C., Durieux S., Vanpouille C., Haendler B., Spik G. (2002) Biochemistry 41, 5222–5229 [DOI] [PubMed] [Google Scholar]
- 43.Pejler G. (1996) Biochem. J. 320, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Damiens E., El Yazidi I., Mazurier J., Elass-Rochard E., Duthille I., Spik G., Boilly-Marer Y. (1998) Eur. J. Cell Biol. 77, 344–351 [DOI] [PubMed] [Google Scholar]
- 45.Dhennin-Duthille I., Masson M., Damiens E., Fillebeen C., Spik G., Mazurier J. (2000) J. Cell. Biochem. 79, 583–593 [PubMed] [Google Scholar]
- 46.Westling C., Lindahl U. (2002) J. Biol. Chem. 277, 49247–49255 [DOI] [PubMed] [Google Scholar]
- 47.Pallerla S. R., Lawrence R., Lewejohann L., Pan Y., Fischer T., Schlomann U., Zhang X., Esko J. D., Grobe K. (2008) J. Biol. Chem. 283, 16885–16894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forster-Horváth C., Bocsi J., Rásó E., Orbán T. I., Olah E., Tímár J., Ladányi A. (2001) Eur. J. Immunol. 31, 600–608 [DOI] [PubMed] [Google Scholar]
- 49.Jones K. S., Petrow-Sadowski C., Bertolette D. C., Huang Y., Ruscetti F. W. (2005) J. Virol. 79, 12692–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wegrowski Y., Milard A. L., Kotlarz G., Toulmonde E., Maquart F. X., Bernard J. (2006) Clin. Exp. Immunol. 144, 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pikas D. S., Eriksson I., Kjellén L. (2000) Biochemistry 39, 4552–4558 [DOI] [PubMed] [Google Scholar]
- 52.Kusche-Gullberg M., Eriksson I., Pikas D. S., Kjellén L. (1998) J. Biol. Chem. 273, 11902–11907 [DOI] [PubMed] [Google Scholar]
- 53.van den Born J., Pikas D. S., Pisa B. J., Eriksson I., Kjellen L., Berden J. H. (2003) Glycobiology 13, 1–10 [DOI] [PubMed] [Google Scholar]
- 54.Wang L., Fuster M., Sriramarao P., Esko J. D. (2005) Nat. Immunol. 6, 902–910 [DOI] [PubMed] [Google Scholar]
- 55.Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor K. R., Gallo R. L. (2006) FASEB J. 1, 9–22 [DOI] [PubMed] [Google Scholar]
- 57.Lindahl U. (2007) Thromb. Haemostasis 98, 109–115 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.