Abstract
GATA5 is a member of the zinc finger transcription factor GATA family (GATA1–6) that plays a wide variety of roles in embryonic and adult development. Experiments in multiple model systems have emphasized the importance of the GATA family members 4–6 in the development of the endoderm and mesoderm. Yet despite overlapping expression patterns, there is little evidence of an important role for GATA5 in mammalian cardiac development. We have generated a new Gata5 mutant allele lacking exons 2 and 3 that encodes both zinc finger domains (Gata5tm2Eem), and we show that although Gata5−/− mice are viable, Gata4+/−5−/− mutants die at mid-gestation and exhibit profound cardiovascular defects, including abnormalities of cardiomyocyte proliferation and cardiac chamber maturation. These results demonstrate functional redundancy between Gata4 and Gata5 during cardiac development and implicate Gata5 as a candidate modifier gene for congenital heart disease.
Keywords: Development Differentiation/Organ, DNA/Transcription, Genetics/Mouse, Tissue/Organ Systems/Heart, Transcription/Development, GATA
Introduction
All six members of the GATA family (GATA1–6) of zinc finger transcription factors play important roles in cell fate decision, differentiation, and morphogenesis (1). Members of this family recognize the GATA motif, which is present in the promoters of many genes. These factors have been divided into two subfamilies, Gata1/2/3 and Gata4/5/6, based on their expression patterns and amino acid sequence homology (2). Gata1, Gata2, and Gata3 are preferentially expressed in hematopoietic cells and regulate proliferation and differentiation during hematopoiesis (3). Gata4, Gata5, and Gata6 are predominantly expressed during embryonic heart development, in addition to other sites. The DNA binding domains of GATA4, GATA5, and GATA6 are ∼85% similar at the protein level. During early cardiac development, Gata4 expression is confined to the cardiac crescent, and later the transcripts are detected throughout the myocardium and endocardium (4, 5). Gata4-deficient mice die between E8.54 and E10.5 because of defects in ventral morphogenesis, including a failure of the cardiac mesoderm to form a linear heart tube (6, 7). A frameshift mutation in human GATA4 (E359del) is linked to cardiac septal defects (8). Like Gata4, Gata6 is expressed in the precardiac mesoderm and later in myocardial cells. Gata6 is also expressed in vascular smooth muscle (9). Gata6 null mice die prior to cardiac development because of defects in the visceral endoderm function and extraembryonic development (10, 11).
The temporal and spatial expression pattern of Gata5 suggests its involvement in tissue-specific transcriptional regulation during embryonic development (12). Gata5 is expressed in the precardiac crescent between E7 and E8 (12). By E9.5 in the heart, Gata5 is expressed at high levels in the atria with lower levels observed in the ventricle and outflow tract. By E12.5, expression is primarily restricted to endocardial cells of the atria, and by E16.5, Gata5 transcripts are no longer detected in cardiac tissues (12). In chick, GATA5 is transcribed in the cardiac crescent prior to formation of the primordial heart tube (13). In Xenopus, gata5 is expressed in both cardiac mesoderm and hepatogenic endoderm. Down-regulation of gata5 using two nonoverlapping translation-blocking and splice site-blocking morpholino causes severe reduction in the number of heart and liver precursors at the time of or shortly after their specification (14). In zebrafish, gata5 is encoded by the faust locus, and mutants reveal that gata5 is required for the production of normal numbers of developing myocardial precursors (15, 16). In addition, gata5 transcriptionally regulates several cardiac genes important in cardiac development, including nkx2.5 (14).
In a previous study, Molkentin et al. (17) reported that disruption of the mouse Gata5 gene had no effect on cardiac development or morphology. Homozygous Gata5 mutants (Gata5tm1Eno) are viable and fertile, with female mice harboring a genitourinary abnormality. Considering the prominent and essential role that Gata5 appeared to play in the development of the heart in other model systems, this was a surprising result (14, 18). Indeed, subsequent analysis of potential Gata5 isoforms expressed in disparate model systems suggests that the Gata5tm1Eno allele might be hypomorphic. For example, the chick harbors a novel GATA5 isoform that contains a DNA binding domain composed of a single zinc finger that can bind to DNA and promote transcription (19). This is a common feature in other GATA family members with mouse Gata1, human and mouse Gata2, human and mouse Gata3, and human and mouse Gata6 genes all possessing two promoters and two initiation codons (20–24). Analysis of the previously targeted Gata5tm1Eno allele suggests that a partially functional transcript analogous to this isoform could be produced. Thus, we sought to generate a novel Gata5 allele to assess the complete loss of Gata5 in mammals.
Here, we report the creation and characterization of a new Gata5 mutant allele lacking both zinc finger domains (Gata5tm2Eem, hereafter designated as Gata5−/−), and we demonstrate striking functional redundancy between Gata4 and Gata5 during cardiac development. Murine embryos are exquisitely sensitive to the loss of a single Gata5 allele in the setting of Gata4 heterozygosity resulting in profound cardiac defects, in part, due to the cooperative regulation of cardiomyocyte proliferation.
EXPERIMENTAL PROCEDURES
Targeting of the Mouse Gata5 Locus and Generation of Gata4/5 Compound Mutants
A Gata5 targeting vector was constructed in pPNT containing pGK-Neo and pGK-TK cassettes for positive and negative selection, respectively (25). The 5′ arm was a 1.9-kb fragment of Gata5 genomic DNA that includes the majority of exon1, and 20 bp of exon2 (first zinc finger) was cloned upstream of lacZ. The 3′ arm was an 8-kb genomic fragment beginning immediately after exon 3 (second zinc finger) (Fig. 1A). The targeting vector was linearized with SacII and electroporated into R1 ES cells (26). After 24 h, neomycin-resistant colonies were selected in 250 μg/ml G418 and 1.5 m ganciclovir for 7 days. Genomic DNA from resistant ES cell clones was isolated and analyzed by Southern blot after KpnI digestion (Fig. 1B). Hybridization of the probe to KpnI-digested DNA yielded a 29.8-kb wild-type band and a 6.2-kb mutant band. Seven of 200 individual ES cell colonies contained the 6.2-kb mutant band for a targeting efficiency of 3.5%. Positive clones were microinjected into E3.5 mouse C57BL/6 blastocysts and subsequently implanted into pseudo-pregnant CD1 females using standard techniques. Male chimeras were mated with C57BL/6 females to obtain agouti germ line Gata5 heterozygotes. Gata5 heterozygotes were mated to generate Gata5 mutant mice (Gata5tm2Eem). Male Gata5−/− were mated with female Gata4+/− mice (7) to generate Gata4+/−5+/− compound mutants. Further mating between male Gata5−/− with female Gata4+/−5+/− compound mutants generated Gata4+/−5−/− mutant mice.
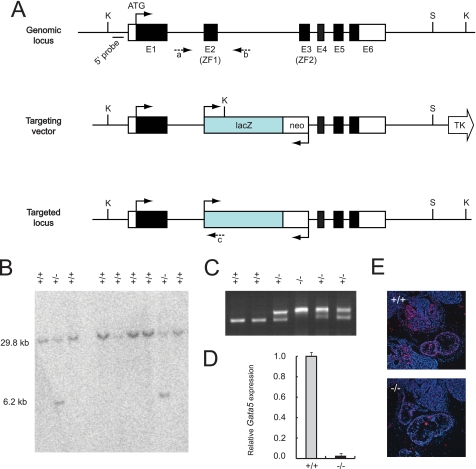
FIGURE 1.
Generation and confirmation of Gata5 mutant mice. A, Gata5 null mice were generated by standard gene-targeting techniques using homologous recombination in murine R1 ES cells. Exons 2 and 3 were replaced with a LacZ-pGK-Neo cassette. K, KpnI; S, SalI; TK, thymidine kinase; c, primer c as described under “Experimental Procedures.” B, Southern blot with a 5′ probe identified multiple positive ES clones that were used to generate chimeras. These were backcrossed with C57/Bl6 mice to identify germ line transmission. C, intercrossing Gata5 heterozygous germ line mice gave rise to viable and fertile Gata5 nulls identified by PCR with wild-type primers “a and b” and mutant primers “a–c”. D, RT-qPCR for Gata5 transcripts revealed no accumulation indicating that the new allele was a complete null. E, null allele was further confirmed by in situ hybridization at E10.5 revealing a complete absence of Gata5 transcript in all tissues.
PCR Genotyping
Agouti offspring from Gata5+/− intercrosses were genotyped by Southern blot or PCR analysis (Fig. 1, B and C). Genomic DNA was analyzed by Southern blot as described above. The following primers (listed 5′ to 3′) were used for PCR genotyping analysis: primer a, GGCATCTACCTTAGAGTGTGG; primer b, GCTCCCATCTTTAATCCATCCT; and primer c, TAGGTTACGTTGGTGTAGATGGG. Primers a and b flank exon2 (first zinc finger) in introns 1 and 2, respectively, and amplify a 500-bp fragment from the wild-type Gata5 gene. Primer c is a reverse primer in lacZ and together with primer a amplifies a 750-bp fragment from mutant Gata5 mice. Gata4+/−5−/− mutants from Gata5−/− crosses with Gata4+/−5+/− were genotyped by PCR and Southern blot, as described previously and above (10).
Histology and in Situ Hybridization
Histology was performed as described previously (27, 28). Briefly, E12.5 and E14.5 mouse embryos were harvested from timed matings, fixed in 4% paraformaldehyde at 4 °C overnight, dehydrated through graded ethanol (30, 50, 70, 95, and 100%), embedded in paraffin, sectioned at 8 μm, and stained with hematoxylin and eosin according to standard protocol (12). Radioactive in situ hybridization was performed on paraformaldehyde-fixed, paraffin-embedded heart sections according to standard protocols (29). A 601-nucleotide in situ probe was generated containing the 3′-untranslated region of the mouse Gata5 gene. The following primers (listed 5′ to 3′) were used to generate this fragment: forward, AGCCCCCAGGCTGGTCTCAG; reverse, CAGAAGTCTGCGATGATGGG.
Quantitative Real Time PCR and Microarray
RT-qPCR was performed as described previously (30). Briefly, total RNA was isolated from dissected mouse hearts at E12.5 and E14.5 using TRIzol (Invitrogen). RNA was reverse-transcribed using random hexamers and the SuperScript First Strand Synthesis kit (Invitrogen). Gene expression was measured by quantitative RT-PCR (ABI PRISM 7900) using SYBR Green (Applied Biosystems) or ABI Taqman chemistry (Gata4, Mm03053570_s1; Gata5, Mm03053574_s1; and Gata6, Mm00802636_m1). Signals were normalized to corresponding glyceraldehyde-3-phosphate dehydrogenase controls, and the ratios expressed as fold changes when compared with wild-type controls. PCR conditions and primer set sequences are available upon request. Microarray analysis was performed on RNA isolated from E10.5 Gata5+/−(n = 3), Gata5−/− (n = 3), and Gata4+/−5−/− (n = 3) hearts using Affymetrix GeneChip mouse 430 2.0 microarray. Differences in gene expression levels were considered significant at p < 0.05, and raw data were deposited in GEO.
ChIP Assay
Chromatin was isolated from E12.5 murine hearts using a commercially available kit (Upstate Biotechnology, catalogue no. 17-295). In brief, the entire heart was excised from the embryo (including systemic and pulmonary venous attachments, atria, ventricles, and proximal great vessels), manually minced, and fixed in 1% formaldehyde. Chromatin was sheared by sonication to an average length of 200–500 bp, immunoprecipitated with either a GATA5- or GATA4-specific antibody (Santa Cruz Biotechnology, M-20, C-20, respectively) or with an IgG negative control. Reverse, cross-linked immunoprecipitated chromatin was then subjected to PCR analysis using primers listed as follows (gene/GATA-binding site position with respect to transcription start site/product size (bp))/forward PCR primer sequence/reverse PCR primer sequence: Cdkn1a/−195/93, ACACTTCCTCTTCCTTCCTGGGTAGGGCTGATAGAAAGGAAATCCAAGGG; Cdkn1b/−1158/133, ACGACAGGCCTGGCTATTCTGTTTACCACCACCACGGTATTACCAACA; Cdkn1c/−636/80, CTCTACACCCAGGCCTAAGTATCTGTCTAACTCCGTGATAGAGGGTCTTAGGG; and Cdkn2c/−1460, −1431/98, GGAAGCTATGATAGGAGATTGGCACCCAAGCAAGACACCATTAATTTCCAG.
Proliferation and TUNEL Analyses
Cardiomyocyte proliferation was evaluated by immunohistochemistry on WT, Gata5 mutant, and Gata4/5 compound mutant heart sections by Ki67 (1:50), phosphohistone H3 (1:2000, Cell Signaling, Beverly, MA), and MF20 (1:25, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City). DAPI (Vector Laboratories, Burlingame, VT) was used to stain nuclei. ImageJ software was used to count Ki67 and phosphohistone H3-positive myocyte nuclei and the total number of myocytes in 6–8 different sections of 3–4 independent heart samples. Apoptosis was assayed by TUNEL assay using the manufacturer's instructions (Invitrogen).
Statistics
All measurement data are expressed as mean ± S.D. The statistical significance of differences between groups was analyzed by Student's t test. Differences were considered significant at a p value <0.05.
RESULTS
Gata5tm2EemNull Mice Are Viable, Fertile, and Demonstrate Normal Cardiac Morphology
To elucidate the function of GATA5 in cardiac development, we generated Gata5 null mice by standard gene-targeting techniques using homologous recombination in murine ES cells (25). A new Gata5 allele (Gata5tm2Eem) was created with a targeting construct that substituted both zinc finger domains (exons 2 and 3) with a pGK-Neo cassette (Fig. 1A). Southern blot and PCR analysis of 200 individual ES clones confirmed appropriate integration of the targeting construct in seven clones, representing a 3.5% targeting efficiency (Fig. 1, B and C). Quantitative RT-PCR and in situ hybridization analysis confirmed that the targeted modification of the Gata5 locus resulted in a functionally null allele (Fig. 1, D and E). Mice heterozygous for the mutant Gata5 allele appeared normal and fertile. Offspring from Gata5+/− intercrosses resulted in an expected Mendelian ratio of 18 (26%) Gata5+/+, 36 (52%) Gata5+/−, and 15 (22%) Gata5−/− (p = 0.822). Gata5 null males were phenotypically normal, whereas females exhibited genitourinary abnormalities identical to those exhibited by the Gata5tm1Eno allele (17). Gata5−/− females showed a reduction in the distance between the vagina and anus identical to that seen in the Gata5tm1Eno allele with reduced fertility. Histological analysis of WT and Gata5 mutant (n = 6) hearts from embryonic and adult stages revealed no obvious cardiac defects (Fig. 3 and data not shown). Echocardiographic analysis of three-month-old Gata5 null mice revealed normal cardiac function when compared with WT littermates (data not shown).
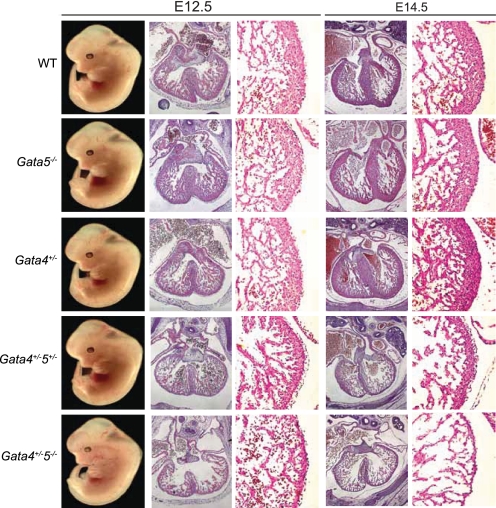
FIGURE 3.
Ventricular phenotype of Gata4/5 compound mutant mice. Compared with WT embryos, Gata5−/− and Gata4+/− (1st to 3rd rows, respectively) mice appeared grossly normal, whereas Gata4+/−5−/− (5th row) compound mutants all died mid-gestation with no survivors past E14.5. There was progressive ventricular wall thinning, and the compact myocardium was hypoplastic with increasingly fine, abnormal trabecular structures with loss of each additional Gata5 allele in the setting of Gata4 heterozygosity (rows 3rd to 5th rows).
Compound Gata4+/−5−/− Mutant Mice Are Embryonic Lethal and Display Profound Cardiac Defects
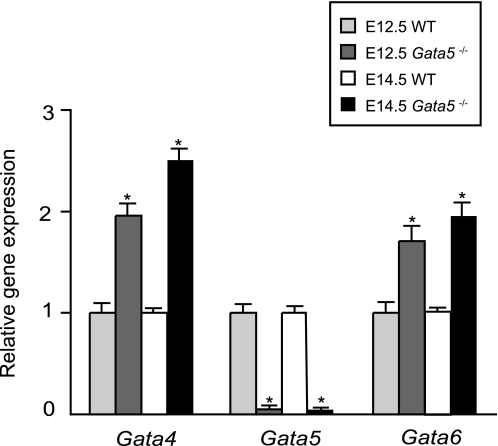
Gata4, Gata5, and Gata6 have partially overlapping expression patterns and are transcribed in both extra-embryonic and embryonic mesoderm early in development (9, 13, 31). Transcript abundance for Gata4, Gata5, and Gata6 was analyzed by RT-qPCR in Gata5 null mice and wild-type littermates at E12.5 and E14.5 (Fig. 2). We noted a greater than 2-fold increased expression of Gata4 and Gata6 in Gata5 null hearts suggesting the possibility of functional compensation.
FIGURE 2.
RT-qPCR of Gata4, Gata5, and Gata6 in Gata5 null embryos. Gata4 and Gata6 expression are significantly unregulated in whole heart extracts of Gata5 null mice at two time points in midgestation, E12.5 and E14.5 (n = 3 per time point). *, p < 0.05.
To test the hypothesis that Gata4 might compensate for Gata5 function in Gata5 mutant hearts, we generated Gata4/5 compound mutants. Mice heterozygous for Gata4 were viable and demonstrated no obvious cardiovascular phenotypes, consistent with previous reports (6, 7). Gata4/5 compound heterozygotes (Gata4+/−5+/−) were also viable and fertile. At E12.5, gross morphology of Gata4+/−5+/− hearts is indistinguishable compared with WT controls. However, histological analysis of Gata4+/−5+/− hearts demonstrated a 43% reduction in ventricular compact zone thickness as early as E12.5 (Fig. 3). By E14.5, differences in myocardial thickness between WT and Gata4+/−5+/− hearts were also prominent with a 32% reduction in compact zone thickness (Fig. 3).
To determine whether complete loss of GATA5 increases the severity of cardiac phenotype, we generated Gata4+/−5−/− compound mutant mice. No Gata4+/−5−/− progeny was identified in newborn litters, suggesting that absence of a single Gata4 allele in the presence of Gata5 null is critical for embryonic development (Table 1). At E12.5, Gata4+/−5−/− mutant embryos were present at a Mendelian ration but harbored significant cardiac defects (Figs. 3 and 4). First, the ventricular, compact myocardium was hypoplastic (E12.5, 61% reduction in thickness with respect to WT; E14.5, 84% reduction in thickness with respect to WT) with a fine, abnormal trabecular structure (Fig. 3). Second, there was hypoplasia of the endocardial cushions by E12.5 leading to complete common atrioventricular canal (Fig. 4). This was occasionally observed as an unbalanced canal leading to a functionally univentricular heart (Fig. 4J). This abnormality was associated with a primum atrial septal defect as well as a large inlet-type ventricular septal defect modeling complete common atrioventricular canal. Third, the conotruncal cushions harbored defects in both decreased size and location leading to multiple abnormalities of ventriculoarterial positioning (Fig. 4, A–F). By E14.5, most of the Gata4+/−5−/− embryos exhibited systemic hemorrhage and body wall edema suggestive of heart failure (data not shown).
TABLE 1.
Viability of Gata45 compound mutant mice at four developmental time points
Numbers of embryos under Gata genotype portion of table refer to observed (and expected) pups.
| Age | Total no. |
Gata genotype |
p value | |||
|---|---|---|---|---|---|---|
| 4+/+ 5+/− | 4+/− 5+/− | 4+/+ 5−/− | 4 +/− 5−/− | |||
| >8 weeks | 60 | 23 (15) | 26 (15) | 11 (15) | 0 (15) | p < 0.0001 |
| E14.5 | 16 | 4 (4) | 4 (4) | 3 (4) | 5 (4) | p = 0.97 |
| E12.5 | 91 | 24 (23) | 21 (23) | 24 (23) | 22 (23) | p = 0.26 |
| E10.5 | 79 | 18 (20) | 27 (20) | 16 (20) | 18 (20) | p = 0.30 |
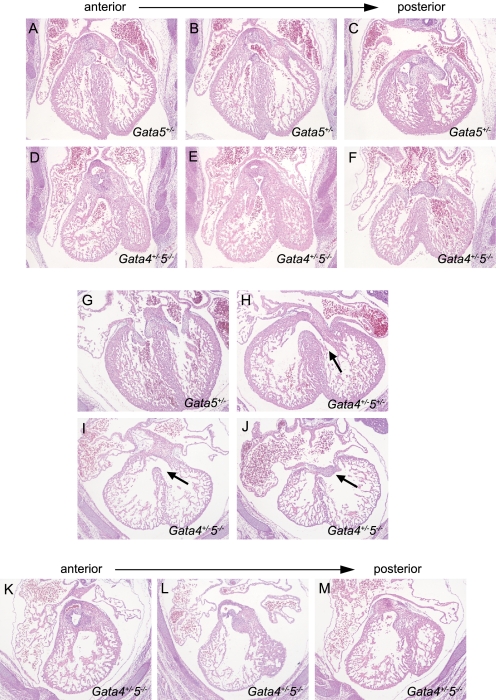
FIGURE 4.
Conotruncal and endocardial cushion phenotype of E14.5. Gata4/5 compound mutant mice. Compound Gata4/5 mutant mice harbored multiple defects in distinct anatomic compartments. Gata5+/− mice appeared normal with no identifiable intracardiac defects (A–C and G). Gata4+/−5+/− double heterozygotes were viable but demonstrated conoventricular septal defects with incomplete penetrance (H, arrow). Gata4+/−5−/− compound mutants demonstrated moderately hypoplastic and anteriorly malaligned left ventricular outflow tracts (D–F). Similarly, ventriculoarterial positioning of the right ventricular outflow tract was more posteriorly positioned than normal (K–M). There was hypoplasia of the endocardial cushions leading to complete common atrioventricular canal defects. This was associated with a primum atrial septal defect as well as a large ventricular septal defect modeling human complete common atrioventricular canal (I and J).
Myocardial Proliferation and Apoptosis in Compound Gata4+/−5−/− Mutant Hearts
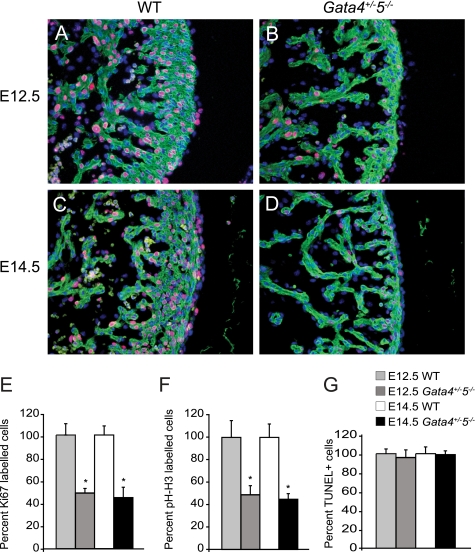
GATA transcription factors have been previously implicated in regulating proliferation and apoptosis. GATA4 has been shown to regulate myocardial proliferation and survival, whereas GATA6 inhibits cell cycle progression in vascular smooth muscle cells (32, 33). Ki67 was used to assess cell proliferation rates in WT and Gata4+/−5−/− mutant hearts. Gata4+/−5−/− mutant hearts showed 51% (at E12.5) and 55% (at E14.5) reduction in Ki67 staining throughout the ventricular myocardium when compared with WT controls (Fig. 5, A–E). There was also a trend toward decreased proliferation in the endocardial and epicardial compartments, although this observation did not reach significance to p < 0.05. Similar results were observed when proliferation was analyzed by phosphohistone H3 (Fig. 5F), yet there was no increase in apoptosis as evaluated by TUNEL staining (Fig. 5G). By contrast, proliferation in skeletal muscle was unchanged in the Gata4+/−5−/− mutant embryos (data not shown). These experiments suggest that the two alleles of Gata4 and Gata5 are crucial for cardiomyocyte proliferation during embryonic development.
FIGURE 5.
Cellular proliferation and apoptosis in Gata4/5 compound mutant hearts. Cardiac myocytes in Gata4+/−5−/− compound mutant embryos (B and D) demonstrate a 51% (at E12.5) and 55% (at E14.5) reduced staining with either Ki67 or phosphohistone H3 (red) compared with WT embryos (A, C, and summarized in E), MF20 (green), and 4′,6-diamidino-2-phenylindole (blue). Similar decreases in proliferation in Gata4+/−5−/− compound mutants results were observed by immunohistochemistry analysis with phosphohistone H3 (pH-H3, F). There was no change in apoptosis with respect to WT embryos in Gata4+/−5−/− compound mutants as assessed by TUNEL stain (G). *, p < 0.05.
Altered Expression of Cell Cycle Genes in Compound Gata4+/−5−/− Mutant Hearts
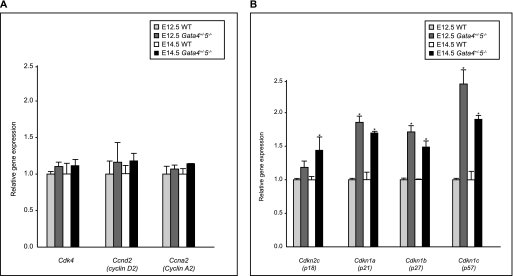
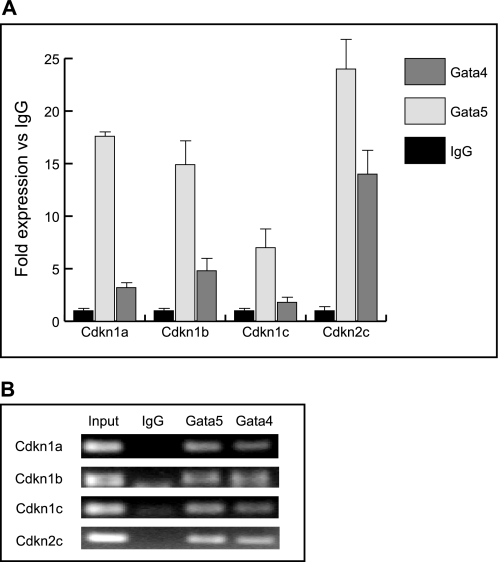
Cardiomyocytes proliferate during embryogenesis but have a severely decreased capacity soon after birth. Cell cycle progression is tightly controlled by both positive (i.e. cyclins and cyclin-dependent kinases) and negative regulators (i.e. Cdks-cyclin-dependent kinase inhibitors) (33–35). Recent experiments demonstrated that GATA4 is required for cyclin D2, Cdk4, and cyclin A2 expression in the ventricular myocardium. Furthermore, cyclin D2 and Cdk4 are direct targets of GATA4 (32). To determine whether the ventricular myocardial hypoplasia observed in Gata4+/−5−/− mutant hearts was due to altered expression of cell cycle regulators, we analyzed the abundance of cyclin D2, cyclin A2, Cdk4, Cdkn2c (p18INC4c), Cdkn1a (p21WAF/CIP1), Cdkn1b (p27Kip1), and Cdkn1c (p57Kip2) transcripts by quantitative RT-PCR on E12.5 and E14.5 heart samples from WT and Gata4+/−5−/− mutant embryos. Gene expression levels of cyclin D2, cyclin A2, and Cdk4 were unchanged in Gata4+/−5−/− mutant hearts compare with WT controls (Fig. 6A). In contrast, expression levels of the cell cycle inhibitors Cdkn2c (p18INC4c), Cdkn1a (p21WAF/CIP1), Cdkn1b (p27Kip1), and Cdkn1c (p57Kip2) were significantly increased in Gata4+/−5−/− mutant heart samples compared with WT controls (Fig. 6B). We next investigated if up-regulation of Cdkn2c (p18INC4c), Cdkn1a (p21WAF/CIP1), Cdkn1b (p27Kip1), or Cdkn1c (p57Kip2) in Gata4+/−5−/− mutants were reflected by direct binding by GATA4 or GATA5 in wild-type hearts. Bioinformatic analysis of 2 kb upstream of the transcription start site of four CDKI promoters revealed some potential conserved GATA factor-binding sites. We performed ChIP assay on WT E12.5 heart extracts and revealed that GATA4 and GATA5 both are recruited to cyclin-dependent kinase I promoters and thus likely directly regulate their gene expression in vivo (Fig. 7). Taken together, our quantitative expression analyses and ChIP results suggest that reduction in the myocardial cell proliferation of Gata4+/−5−/− mutant hearts is due to de-repression and early activation of cell cycle inhibitors.
FIGURE 6.
RT-qPCR of candidate cell cycle targets. A, RT-qPCR of candidate cyclin targets in cardiac extracts from E12.5 and E14.5 gene-targeted murine embryos. At two developmental time points, E12.5 and E14.5, Cdk4, Ccnd2, and Ccna2 expression show no statistically significant differences to wild-type levels. B, at two developmental time points, E12.5 and E14.5, levels of Cdkn1a, and Cdkn1b, and Cdkn1c transcripts are all increased in compound mutants relative to WT levels at E12.5. For Cdkn2c, RNA levels are increased at E14.5 (*, p < 0.05, n = 3).
FIGURE 7.
ChIP assay of GATA factor binding to cell cycle targets. ChIP assays demonstrate that GATA4 and GATA5 were associated with the mouse Cdkn1a, Cdkn1b, Cdkn1c, and Cdkn12c promoters. Chromatin DNA from “input” or individual transcription factor pulldown by IgG, GATA5, or GATA4 antibodies was amplified around the promoter region of Cdkn1a, Cdkn1b, Cdkn1c, and Cdkn2c containing the WGATAR motif(s). The littermate product is reported in fold IgG input (A) and by agarose gel electrophoresis (B).
DISCUSSION
The formation of vertebrate heart is a multistep process comprising patterning, cell differentiation, and morphogenesis (36). Transcription factors and their combinatorial activity have been shown to govern many of the underlying molecular pathways. GATA factors (GATA4, GATA5, and GATA6) are essential regulators of mesodermal and endodermal organ development (1). GATA transcription factors are among the first to be expressed in the developing heart and regulate different aspects of cardiac development. In rodents, GATA4 and GATA6 are critical for activation of the cardiomyocyte gene expression program (37). In contrast to GATA4 and GATA6, a role for GATA5 in vertebrate cardiomyocyte gene expression regulation and development has not been demonstrated. Using a new null allele of Gata5 (Gata5tm2Eem), we now show that Gata5 acts cooperatively with Gata4 to regulate cardiomyocyte proliferation and development. Our data demonstrate that in concert with Gata4, Gata5 negatively regulates expression levels of the cell cycle inhibitors Cdkn2c (p18INC4c), Cdkn1a (p21WAF/CIP1), Cdkn1b (p27Kip1), and Cdkn1c (p57Kip2). In addition, we show that loss of a single allele of Gata5 in Gata4 heterozygotes leads to thinning of the ventricular myocardium, an intermediate cardiac phenotype compared with the profound defects seen in Gata4+/−5−/− mutant embryos. GATA5 now appears to act in concert with GATA4 consistent with their overlapping expression pattern during early cardiac development. Together, our data reveal a previously unappreciated role for GATA5 in cardiac proliferation and development.
GATA Factors and Cell Cycle Regulation during Mouse Cardiac Development
To ensure proper progression through the cell cycle, a series of orchestrated checkpoints modulate cyclin-dependent kinase (Cdk) complexes. Their activities are regulated in both a positive and negative manner (38). In vivo and in vitro data suggest that loss or down-regulation of CDKI increases the proliferative capacity of neonatal cardiac myocytes (39, 40). GATA family of transcription factors has been previously implicated in cell cycle progression. GATA1 inhibits expression of Cdk6 and cyclin D2 (Ccdn2) and induces the cyclin-dependent kinase inhibitors Cdkn2c (p18INC4c) and Cdkn1b (p27Kip1) promoting proliferation arrest during erythroid maturation (34, 41, 42). GATA2 inhibits the growth of hematopoietic stem/progenitor cells by regulating expression levels of Cdkn2a (p21WAF1) and Cdkn1b (p27Kip1) (35, 43). Overexpression of Gata6 inhibits cell cycle progression of vascular smooth muscle cells by regulating expression of Cdkn2a p21WAF1 (33). Finally, GATA4 directly regulates expression levels of Ccdn2 (cyclin D) and Cdk4 controlling myocardial cell proliferation in the anterior heart field (32).
In this study, we demonstrate that haploinsufficiency of both Gata4 and Gata5 result in cardiac defects characterized by thinning of the ventricular myocardial wall. In addition, loss of a second Gata5 allele leads to profound cardiac defects including atrial septal defects, ventricular septal defects, ventricular hypoplasia, and malpositioning/hypoplasia of conotruncal and atrioventricular cushions associated with an increased expression of the cell cycle inhibitors Cdkn2c (p18INC4c), Cdkn1a (p21WAF/CIP1), Cdkn1b (p27Kip1), and Cdkn1c (p57Kip2). These data implicate GATA5 as a novel negative regulator of cyclin-dependent kinase I in the cardiac myocyte that helps balance the combined, important roles of GATA4–6 in cardiomyocyte proliferation and differentiation.
Functional Redundancy between GATA4 and GATA5 in the Heart
The transcription factors, GATA4–6, are ∼89% identical in their zinc finger domain, recognize identical DNA sequence motifs, and are all expressed during early cardiac development (2). Gata4 null mice that survive gastrulation form differentiated myocardium with increased Gata6 expression suggesting compensation by Gata6 for loss of Gata4 (6, 7). Although Gata6 null mice undergo growth arrest during gastrulation due to an essential role of this factor in extra-embryonic endoderm development, ES cells deficient for Gata6 are able to differentiate into cardiac myocytes in vitro and contribute to myocardium in chimeric embryos (37). As reported previously, Gata5 null (Gata5tm1Eno) did not exhibit any overt cardiac defects (17). In contrast to Gata4 null mice, Gata5tm1Eno null mice do not exhibit up-regulation of Gata4 or Gata6 expression. Despite intense scrutiny, we did not uncover any cardiac defects in our newly generated Gata5 null (Gata5tm2Eem) mice. However, we did identify increased expression of Gata4 and Gata6, indicating a significant difference between the two alleles and suggesting that in the Gata5tm1Eem mutant, Gata4 and/or Gata6 might compensate for the loss of Gata5. One possibility is that combined GATA4 and GATA5 function are predominantly mediated by the total level of GATA4, whereas in the absence of GATA4, GATA5 takes on a prominent role. Similarly, because GATA6 is also unregulated in Gata5 null mice, albeit to a lesser degree than GATA4, there may be a combined compensatory up-regulation of both GATA4 and GATA6 messenger RNA. Another possibility is that GATA factor redundancy may be mediated by differential activity of the protein products. For example, Nemer and Nemer (44) suggest a specific interaction between GATA4 and GATA6 to regulate target genes, and a similar interaction might exist between GATA4/GATA5 or GATA5/GATA6.
Functional redundancy between GATA4 and GATA6 in the murine heart has been addressed by generating compound heterozygous Gata4/6 mice. Compound heterozygous Gata4+/−6+/− embryos die during embryonic development due to profound vascular and cardiac defects, including outflow tract defects, thin myocardium, and ventricular septal defects, whereas a complete loss of both Gata4 and Gata6 results in a loss of cardiac specification (37, 45). Thus, there is ample evidence for functional redundancy between Gata4 and Gata6 in mammalian cardiac development.
Despite these important findings, functional redundancy between GATA4 and GATA5 is less understood. In a previous study, made an effort to determine whether GATA4 or GATA6 substitute GATA5 functions by generating compound Gata4+/−5−/− and Gata5−/−6+/− mutants using the Gata5tm1Eno allele (17, 45). Compound mutant hearts appeared to be identical to Gata5 null (Gata5tm1Eno) mice. This might have been due to residual expression of a truncated GATA5 protein (from an alternative promoter) in Gata5tm1Eno null mice. Alternatively, there might be strain differences between the analyses of the Gata5tm1Eno and Gata5tm2Eem alleles. In this study, we clearly demonstrate that compound heterozygous mice for Gata4/5 (Gata4+/−5+/−) survive and exhibit thinning of the ventricular myocardium. However, further loss of the second allele of Gata5 in compound mutant Gata4+/−5−/− mice leads to embryonic lethality. No viable Gata4+/−5−/− animals were ever observed after E14.5, indicating that this Gata4/5 genotypic combination results in embryonic lethality with complete penetrance by mid-gestation. Thus, our data support the notion that Gata5 plays an important role in vertebrate cardiac development, in part, by acting cooperatively with Gata4.
Acknowledgments
We are grateful to Michael Parmacek, Alvin Chin, the Jonathan Epstein lab members, and Thomas L. Spray for helpful discussions.
This work was supported by grants from the Pliezowicz Family Foundation (to P. J. G.) and National Institutes of Health Grants R01-HL064632 (to E. E. M.) and R01-HL071546 (to J. A. E.).
- E
- embryonic day
- ChIP
- chromatin immunoprecipitation
- TUNEL
- terminal dUTP nick-end labeling
- WT
- wild type
- RT-qPCR
- reverse transcription-quantitative PCR
- ES
- embryonic stem.
REFERENCES
- 1.Peterkin T., Gibson A., Loose M., Patient R. (2005) Semin. Cell Dev. Biol. 16, 83–94 [DOI] [PubMed] [Google Scholar]
- 2.Molkentin J. D. (2000) J. Biol. Chem. 275, 38949–38952 [DOI] [PubMed] [Google Scholar]
- 3.Orkin S. H. (1992) Blood 80, 575–581 [PubMed] [Google Scholar]
- 4.Grépin C., Dagnino L., Robitaille L., Haberstroh L., Antakly T., Nemer M. (1994) Mol. Cell. Biol. 14, 3115–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heikinheimo M., Scandrett J. M., Wilson D. B. (1994) Dev. Biol. 164, 361–373 [DOI] [PubMed] [Google Scholar]
- 6.Molkentin J. D., Lin Q., Duncan S. A., Olson E. N. (1997) Genes Dev. 11, 1061–1072 [DOI] [PubMed] [Google Scholar]
- 7.Kuo C. T., Morrisey E. E., Anandappa R., Sigrist K., Lu M. M., Parmacek M. S., Soudais C., Leiden J. M. (1997) Genes Dev. 11, 1048–1060 [DOI] [PubMed] [Google Scholar]
- 8.Garg V., Kathiriya I. S., Barnes R., Schluterman M. K., King I. N., Butler C. A., Rothrock C. R., Eapen R. S., Hirayama-Yamada K., Joo K., Matsuoka R., Cohen J. C., Srivastava D. (2003) Nature 424, 443–447 [DOI] [PubMed] [Google Scholar]
- 9.Morrisey E. E., Ip H. S., Lu M. M., Parmacek M. S. (1996) Dev. Biol. 177, 309–322 [DOI] [PubMed] [Google Scholar]
- 10.Morrisey E. E., Tang Z., Sigrist K., Lu M. M., Jiang F., Ip H. S., Parmacek M. S. (1998) Genes Dev. 12, 3579–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koutsourakis M., Langeveld A., Patient R., Beddington R., Grosveld F. (1999) Development 126, 723–732 [PubMed] [Google Scholar]
- 12.Morrisey E. E., Ip H. S., Tang Z., Lu M. M., Parmacek M. S. (1997) Dev. Biol. 183, 21–36 [DOI] [PubMed] [Google Scholar]
- 13.Laverriere A. C., MacNeill C., Mueller C., Poelmann R. E., Burch J. B., Evans T. (1994) J. Biol. Chem. 269, 23177–23184 [PubMed] [Google Scholar]
- 14.Reiter J. F., Alexander J., Rodaway A., Yelon D., Patient R., Holder N., Stainier D. Y. (1999) Genes Dev. 13, 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J. N., Haffter P., Odenthal J., Vogelsang E., Brand M., van Eeden F. J., Furutani-Seiki M., Granato M., Hammerschmidt M., Heisenberg C. P., Jiang Y. J., Kane D. A., Kelsh R. N., Mullins M. C., Nüsslein-Volhard C. (1996) Development 123, 293–302 [DOI] [PubMed] [Google Scholar]
- 16.Stainier D. Y., Fouquet B., Chen J. N., Warren K. S., Weinstein B. M., Meiler S. E., Mohideen M. A., Neuhauss S. C., Solnica-Krezel L., Schier A. F., Zwartkruis F., Stemple D. L., Malicki J., Driever W., Fishman M. C. (1996) Development 123, 285–292 [DOI] [PubMed] [Google Scholar]
- 17.Molkentin J. D., Tymitz K. M., Richardson J. A., Olson E. N. (2000) Mol. Cell. Biol. 20, 5256–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haworth K. E., Kotecha S., Mohun T. J., Latinkic B. V. (2008) BMC Dev. Biol. 8, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacNeill C., Ayres B., Laverriere A. C., Burch J. B. (1997) J. Biol. Chem. 272, 8396–8401 [DOI] [PubMed] [Google Scholar]
- 20.Ito E., Toki T., Ishihara H., Ohtani H., Gu L., Yokoyama M., Engel J. D., Yamamoto M. (1993) Nature 362, 466–468 [DOI] [PubMed] [Google Scholar]
- 21.Minegishi N., Ohta J., Suwabe N., Nakauchi H., Ishihara H., Hayashi N., Yamamoto M. (1998) J. Biol. Chem. 273, 3625–3634 [DOI] [PubMed] [Google Scholar]
- 22.Pan X., Minegishi N., Harigae H., Yamagiwa H., Minegishi M., Akine Y., Yamamoto M. (2000) J. Biochem. 127, 105–112 [DOI] [PubMed] [Google Scholar]
- 23.Asnagli H., Afkarian M., Murphy K. M. (2002) J. Immunol. 168, 4268–4271 [DOI] [PubMed] [Google Scholar]
- 24.Brewer A., Gove C., Davies A., McNulty C., Barrow D., Koutsourakis M., Farzaneh F., Pizzey J., Bomford A., Patient R. (1999) J. Biol. Chem. 274, 38004–38016 [DOI] [PubMed] [Google Scholar]
- 25.Tybulewicz V. L., Crawford C. E., Jackson P. K., Bronson R. T., Mulligan R. C. (1991) Cell 65, 1153–1163 [DOI] [PubMed] [Google Scholar]
- 26.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh M. K., Petry M., Haenig B., Lescher B., Leitges M., Kispert A. (2005) Mech. Dev. 122, 131–144 [DOI] [PubMed] [Google Scholar]
- 28.Singh M. K., Christoffels V. M., Dias J. M., Trowe M. O., Petry M., Schuster-Gossler K., Bürger A., Ericson J., Kispert A. (2005) Development 132, 2697–2707 [DOI] [PubMed] [Google Scholar]
- 29.Wawersik S., Epstein J. A. (2000) Methods Mol. Biol. 137, 87–96 [DOI] [PubMed] [Google Scholar]
- 30.Trivedi C. M., Lu M. M., Wang Q., Epstein J. A. (2008) J. Biol. Chem. 283, 26484–26489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arceci R. J., King A. A., Simon M. C., Orkin S. H., Wilson D. B. (1993) Mol. Cell. Biol. 13, 2235–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rojas A., Kong S. W., Agarwal P., Gilliss B., Pu W. T., Black B. L. (2008) Mol. Cell. Biol. 28, 5420–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlman H., Suzuki E., Simonson M., Smith R. C., Walsh K. (1998) J. Biol. Chem. 273, 13713–13718 [DOI] [PubMed] [Google Scholar]
- 34.Rylski M., Welch J. J., Chen Y. Y., Letting D. L., Diehl J. A., Chodosh L. A., Blobel G. A., Weiss M. J. (2003) Mol. Cell. Biol. 23, 5031–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ezoe S., Matsumura I., Nakata S., Gale K., Ishihara K., Minegishi N., Machii T., Kitamura T., Yamamoto M., Enver T., Kanakura Y. (2002) Blood 100, 3512–3520 [DOI] [PubMed] [Google Scholar]
- 36.Bruneau B. G. (2008) Nature 451, 943–948 [DOI] [PubMed] [Google Scholar]
- 37.Zhao R., Watt A. J., Battle M. A., Li J., Bondow B. J., Duncan S. A. (2008) Dev. Biol. 317, 614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sánchez I., Dynlacht B. D. (2005) Semin. Cell Dev. Biol. 16, 311–321 [DOI] [PubMed] [Google Scholar]
- 39.Bicknell K. A., Coxon C. H., Brooks G. (2007) J. Mol. Cell. Cardiol. 42, 706–721 [DOI] [PubMed] [Google Scholar]
- 40.Pasumarthi K. B., Field L. J. (2002) Circ. Res. 90, 1044–1054 [DOI] [PubMed] [Google Scholar]
- 41.Dubart A., Roméo P. H., Vainchenker W., Dumenil D. (1996) Blood 87, 3711–3721 [PubMed] [Google Scholar]
- 42.Muntean A. G., Pang L., Poncz M., Dowdy S. F., Blobel G. A., Crispino J. D. (2007) Blood 109, 5199–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tipping A. J., Pina C., Castor A., Hong D., Rodrigues N. P., Lazzari L., May G. E., Jacobsen S. E., Enver T. (2009) Blood 113, 2661–2672 [DOI] [PubMed] [Google Scholar]
- 44.Nemer G., Nemer M. (2003) Dev. Biol. 254, 131–148 [DOI] [PubMed] [Google Scholar]
- 45.Xin M., Davis C. A., Molkentin J. D., Lien C. L., Duncan S. A., Richardson J. A., Olson E. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11189–11194 [DOI] [PMC free article] [PubMed] [Google Scholar]