Abstract
Sterol 14α-demethylase (14DM, the CYP51 family of cytochrome P450) is an essential enzyme in sterol biosynthesis in eukaryotes. It serves as a major drug target for fungal diseases and can potentially become a target for treatment of human infections with protozoa. Here we present 1.9 Å resolution crystal structures of 14DM from the protozoan pathogen Trypanosoma brucei, ligand-free and complexed with a strong chemically selected inhibitor N-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadi-azol-2-yl)benzamide that we previously found to produce potent antiparasitic effects in Trypanosomatidae. This is the first structure of a eukaryotic microsomal 14DM that acts on sterol biosynthesis, and it differs profoundly from that of the water-soluble CYP51 family member from Mycobacterium tuberculosis, both in organization of the active site cavity and in the substrate access channel location. Inhibitor binding does not cause large scale conformational rearrangements, yet induces unanticipated local alterations in the active site, including formation of a hydrogen bond network that connects, via the inhibitor amide group fragment, two remote functionally essential protein segments and alters the heme environment. The inhibitor binding mode provides a possible explanation for both its functionally irreversible effect on the enzyme activity and its selectivity toward the 14DM from human pathogens versus the human 14DM ortholog. The structures shed new light on 14DM functional conservation and open an excellent opportunity for directed design of novel antiparasitic drugs.
Keywords: Cytochromes/Cytochrome P450, Enzymes/Inhibitors, Lipid/Sterol, Methods/X-ray Crystallography, Organisms/Trypanosome, Protein/Structure, Sterol 14 Alpha-Demethylase (CYP51), Sterol Biosynthesis
Introduction
Human infections by Trypanosomatidae affect millions of people, yet current treatment remains unsatisfactory (accessible through the World Health Organization Overview of Diseases site). It relies on a handful of drugs that are nonspecific, highly toxic, and have severe side effects and low efficacy (1, 2). Sequencing of protozoan genomes has evoked a broad search for new, parasite-specific drug targets (3–6). Trypanosomal sterol 14α-demethylase (14DM)2 is a highly promising potential drug target because inhibitors of its fungal ortholog (imidazole and triazole derivatives) have been successfully used for many years for treatment of human infections with fungi (7, 8). These cytochrome P450 (CYP) enzymes are found in all biological kingdoms, and regardless of their low amino acid sequence identity, ranging from 22 to 27% across phylogeny, have been joined into one (CYP51) family because of their strict functional conservation (9). From bacteria to mammals, they all are known to catalyze the same, three-step stereospecific reaction of the oxidative removal of the 14α-methyl group from Δ8-sterol precursors formed upon sterol biosynthesis (supplemental Fig. S1). In eukaryotes, this 14DM-dependent reaction is required for production of sterol components for eukaryotic membrane biogenesis, which is the basis for 14DM being an effective drug target. A complete sterol biosynthetic pathway including the CYP51 gene is present in all sequenced Trypanosomatidae genomes.
Recently, randomly selected 14DM inhibitors have been found efficient against human infections with Trypanosoma cruzi (Chagas disease) (2, 10). However, the search for such drugs has remained empirical, focusing on cellular-based inhibition (blind screening), their influence on enzyme catalysis not being estimated. Using broad screening of a large number of chemical structures for binding with the purified trypanosomal 14DMs followed by direct testing of enzyme inhibition in the in vitro reconstituted 14DM reaction and evaluation of cellular effects of the best inhibitors in Trypanosomatidae versus human leukemia cell lines (a control for cellular toxicity), we have identified several novel highly promising scaffolds including non-azole structures (11, 12), N-1-(2,4-dichlorophenyl)- 2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide (VNI) being the best among them. Contrary to all pyridine or pyrimidine derivatives (12) and the majority of azoles, which act as reversible, competitive 14DM inhibitors, the effect of the VNI scaffold on trypanosomal 14DMs is functionally irreversible. At 1/1 molar ratio, the inhibitors completely inactivate the enzymes and cannot be displaced from the active site by substrate. Moreover, opposite to many antifungal azoles, which are highly toxic (e.g. posaconazole that has been reported to enter clinical trials as a drug candidate for Chagas disease (2)), VNI has low cytotoxicity. At 1 μm concentration, it kills more than 99% of T. cruzi amastigotes, the intracellular form that is prevalent at the chronic currently incurable stage of infection, by stopping sterol production in the parasite at the formation of the 14α-methylated precursors (11). Electron microscopy clearly shows the resulting damage of the cellular membranes (13).
In this study, we have determined the x-ray structure of 14DM from Trypanosoma brucei brucei (Tbb), which is the first structure reported for a microsomal 14DM essential for sterol biosynthesis. Comparison of the ligand-free structure with that of the enzyme complexed with VNI provides the molecular basis for the profound inhibitory potency of this scaffold on trypanosomal 14DMs and demonstrates how the binding mode of a chemically selected compound can direct the way for design of novel 14DM inhibitors. Particularly important, being a strong inhibitor of 14DMs from infectious microbes such as T. brucei and T. cruzi and Candida albicans, VNI does not inhibit human 14DM. Consequently, structure of the Tbb14DM-VNI complex reveals chemical details needed for design of potent 14DM inhibitors for at least some human pathogens, which would have a much weaker effect on the host enzyme, although both catalyze the same reaction.
EXPERIMENTAL PROCEDURES
Protein Design and Crystallization
The full-length Tbb14DM expression construct (14) was modified by replacing its N-terminal transmembrane domain upstream of Pro-32 with the MAKKTSSKGKL N-terminal sequence from CYP2C3 (15). The protein was expressed in Escherichia coli (HMS174) and purified in two chromatographic steps including nickel-nitrilotriacetic acid and Q-Sepharose columns as described previously for full-length Tbb14DM (14) except that Triton X-100 was replaced by 20 mm n-tetradecyl-β-d-maltoside (Anatrace). The modified Tbb14DM has a spectrophotometric index A417/A278 1.8 (supplemental Fig. S2a), it retains the expected enzymatic activity, including strict preference for the C4-monomethylated substrate (obtusifoliol). Ligand-free and VNI-bound (molar ratio inhibitor: enzyme = 1.2:1) Tbb14DM were crystallized under the same conditions. Crystals were obtained by hanging-drop vapor diffusion at 25 °C, with a 240 μm P450 solution in 20 mm potassium phosphate buffer, pH 7.3, containing 200 mm NaCl, 0.1 mm EDTA, 10% glycerol, and 0.02 mm n-tetradecyl-β-d-maltoside against a well solution containing 50 mm potassium phosphate buffer, pH 7.3, and 20% polyethylene glycol 3500 (supplemental Fig. S2c).
Data Collection and Structure Determination
Crystals were soaked briefly in a cryo-buffer (well solution with 40% glycerol) and flash-cooled in liquid nitrogen. Data were collected at Life Sciences Collaborative Access Team (LS-CAT), Advanced Photon Source, Argonne National Laboratory, beamlines 21ID-F (native) and 21ID-D (iron-single anomalous dispersion), and processed with the HKL-2000 software package (16). The ligand-free structure was solved by combination of iron-single anomalous dispersion (2.6 Å) and molecular replacement (1.9 Å) methods. The positions of four iron atoms were determined in SOLVE (17) and used to identify the correct molecular replacement solutions. A number of CYP coordinates from the Protein Data Bank (PDB) were tested in PHASER (CCP4 suite (18)), and the solution for four molecules in the asymmetric unit with the iron coordinates that were symmetry-related to the four iron positions from SOLVE was found using as a search ensemble the poly(A) model of CYP120 (2VE4). Restrained refinement using the Hendrickson Lattman coefficient option in REFMAC5 of the CCP4 suite followed by solvent flattening by RESOLVE (19) resulted in an interpretable map. Model building and refinement were performed with O (20) and REFMAC5, respectively. The resulting structure of ligand-free Tbb14DM was used as a search ensemble for the VNI-bound structure. Four independent monomers were located in the asymmetric unit yielding a solution with an Rfactor of 0.31 and log-likelihood gain of 8,632. Model building and refinement were performed with COOT (21) and REFMAC5. Supplemental Table S1 summarizes the diffraction and refinement data statistics. The coordinates and structure factors of ligand-free and VNI-bound Tb 14DM have been deposited at the RCSB Protein Data Bank under ID codes 3G1Q and 3GW9, respectively.
Structure Analysis
Active site volume was determined in VOIDOO (22) using rolling probe algorithm (probe radius 1.4 Å). Cavities were specified using the coordinates of the C13 atom of VNI for ligand-bound molecules and an equivalent point in the cavity space for ligand-free molecules. Molecular volume was calculated in MSMS 2.6.1 (from the MGLTools web site). Structure-based sequence alignments and structure superposition were done using LSQKAB of the CCP4 suite. Figures were prepared with PyMOL and SETOR.
Immunoanalysis of 14DM Expression in Bloodstream Tbb
Bloodstream forms of Tbb (AnTat 1.3A clone of the EATRO1125 strain) were isolated by DE-52 column purification of infected rodent blood (23). Procyclic forms from the AnTat 1.1 strain were cultivated at 27 °C in SDM79 medium (24) supplemented with 15% fetal calf serum. Rat and mouse antibodies were obtained after subcutaneous immunizations with recombinant Tbb14DM according to standard protocols. For Western blotting, each protein sample was solubilized for 10 min at 96 °C in a non-reducing SDS mixture (2% SDS, 10% glycerol, 0.005% bromphenol blue, and 80 mm Tris-HCl, pH 6.8) and subjected to SDS-PAGE following transfer to a nitrocellulose membrane (Hybond-C Extra, Amersham Biosciences), which was then blocked with 5% (w/v) nonfat milk powder in 20 mm Tris-HCl, pH 7.4, 150 mm NaCl and incubated for 2 h at 20 °C with a 1:1000 dilution of mouse anti-Tbb14DM antiserum in 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.05% (w/v) Tween 20, and 1% nonfat milk. Peroxidase-conjugated goat anti-mouse antibodies (GE Healthcare) were diluted in the same buffer and used as indicated by the manufacturer. Bound antibody was detected by chemiluminescence (GE Healthcare). For immunoprecipitation, frozen pellets of 109 AnTat 1.3 A trypanosomes, isolated from rats and purified on a DE-52 column, were resuspended for 1 h at 4 °C in 1 ml of lysis buffer (137 mm NaCl, 10% (v/v) glycerol, 1% Triton X-100, 20 mm Tris-HCl (pH 8), and EDTA-free protease inhibitor mixture (Roche Applied Science)). The suspension was centrifuged at 15,000 × g, and the supernatant was incubated overnight with 10 μl of rat anti-Tbb14DM antiserum and 50 μl of GammaBind G Sepharose® (GE Healthcare). Unbound material was removed by repeated washings in phosphate-buffered saline buffer. Resin was then dissolved in the non-reducing SDS mixture (see above).
Treatment of Tbb-infected Mice with 14DM Inhibitors
Mouse care and experimental procedures were performed under approval from the Institutional Animal Care and Use Committee. Groups of five mice (BALB/c, 10 weeks old) were injected intraperitoneally with 104 bloodstream forms of Tbb (strain 427). Oral treatment with 14DM inhibitors dissolved in 5% Arabic gum was initiated 2 h later, and subsequent oral doses were given every 12 h. Control mice received only the vehicle. Parasitemia was measured in tail-vein blood, and Student's t test was used to compare values between the groups.
RESULTS
Ligand-free 14DM
The Tbb14DM structure has the characteristic P450 fold (supplemental Fig. S3) with 12 major helices (A to L), 10 shorter helices (′ and ″) between them, and 12 β-strands assembled into four β-bundles. In the multiple sequence alignment of CYP51 from different phyla (supplemental Fig. S4), the location and length of these secondary structural elements in Tbb14DM match rather well to those in the water-soluble CYP51 family member from Mycobacterium tuberculosis (Mt), the only CYP51 structure previously determined (25). The main differences include: shifting of the L′ helix toward the C terminus and two additional 310 helices (B″ and G′). The G′-helix is found in most microsomal P450s, whereas the B″-310 helical turn is unique to Tbb14DM, and based on the sequence alignment, it will be characteristic for all 14DMs from Trypanosomatidae.
Regardless of this similarity at the secondary structure level, spatial arrangement of the structural elements in the bacterial and eukaryotic structures display profound differences (Fig. 1a). Only the most conserved P450 structural regions superimpose well (C-terminal part of helix I, helices L, K″, J, E, D, and β-bundle 3; root mean square deviation for the Cα atom positions in these elements is <0.5 Å). These regions are responsible for maintenance of the heme environment and overall P450 fold. In the upper part of the molecule, which forms the substrate-binding pocket, the deviation is drastic. In Tbb14DM, the N-terminal half of the I-helix is much less bent outward, and its medium part is also positioned closer to the heme (left inset). Helix G, especially its C-terminal portion, and helix F are located above and closer to the structural core, deviating about 9 and 6 Å, respectively, from the corresponding helices in MtCYP51. The B′-helix (CYP51 B′ signature (9)) is ∼3 Å nearer the heme plane. Most remarkably, the B′C loop in Tbb14DM is closed (right inset), the phyla-specific (animals/fungi/protists) tyrosine (Tyr-116 in Tbb14DM) forming a hydrogen bond with the heme propionate (2.7 Å to the heme versus 20.3 Å for the corresponding Phe-89 in MtCYP51).
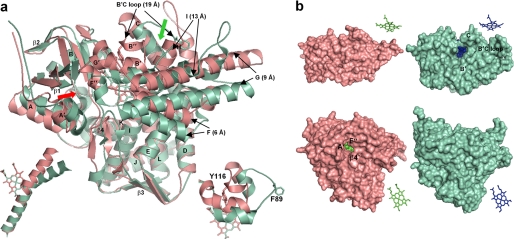
FIGURE 1.
Ligand-free Tbb14DM (salmon). a, superposition with MtCYP51 (1e9x) (green), ribbon diagram. The heme, Tyr-116 (Tbb), and Phe-89 (Mt) are shown in stick representation. The most divergent active site-defining regions are indicated with black arrows, and distances between the equivalent Cα atoms are provided in parentheses. The locations of the putative substrate entrance in Tbb and Mt structures are shown with red and green arrows, respectively. The structure of ligand-free MtCYP51 (1h5z) is identical to 1e9x except for its larger opening to the upper surface due to missing density for helix C. Left inset, I-helix in Tbb and Mt CYP51, distal view. Right inset, BC loop, upper view. b, surface representations of Tbb and Mt CYP51 from the upper and distal sides; the corresponding heme orientation is shown on the right. Inside the proteins, the heme is presented as spheres. In Tbb14DM, the channel extends about 20 Å from the distal surface to the iron, forming an angle of about 50° to the heme plane. In MTCYP51, the funnel-like opening runs from above perpendicular to the heme plane.
As a result, in contrast to MtCYP51, there is no opening in the upper surface of the Tbb14DM molecule, and the heme is completely buried by an additional helical structure (B″) and not visible from above (Fig. 1b). Instead, a narrow channel (∼6 by 9 Å) runs through the left part of the distal surface, with a glimpse of the heme seen between the F″ helix, β4-hairpin and helix A′. Although involvement of the A′-helix in substrate binding has not been observed in other P450 structures, such a channel location is more likely for eukaryotic 14DMs because this part of the molecule in microsomal P450s is predicted to be immersed into the endoplasmic reticulum membrane (26–28), allowing highly lipophilic sterol substrates to enter the enzyme active site directly from the lipid bilayer and offering no access to cytosolic (foreign) metabolites.
The active site cavity in front of the heme plane is surrounded by helix B′ from above, helix I from the right, β-bundle 1 from the left, K-β1–4 loop from the bottom, and helix F/β4-hairpin from the front surface (also see supplemental Fig. S3). Long hydrophobic side chains (predominantly phenylalanines, leucines, and methionines) are spread inside the cavity so that its calculated solvent-accessible volume varies among the four molecules in the asymmetric unit (variation range 450–740 Å3), the average being ∼600 Å3, which is more than four times smaller than the value determined for MtCYP51 (2,600 Å3 (25)).
In the absence of substrate, the binding cavity contains water. One water molecule occupies the sixth coordination site of the heme iron. Interestingly, this water also forms a unique direct contact with the I-helix, not seen on other P450 structures, and thus is involved in a hydrogen bond network with the CYP51 I-helix signature residues (Fig. 2). Via the Ala-291 carbonyl (4.3 Å from the iron), the network connects the side chain hydroxyl of Thr-295, two other water molecules in the I-helix groove, and the Phe-290 carbonyl. It further continues to His-294, which in turn forms a salt bridge with Glu-205 (C terminus of helix F). This network is likely to strengthen the water-iron coordination, which explains why 14DMs are always found in a hexacoordinated (low spin) form (9) and experience only partial low-to-high spin transition upon substrate binding. It is possible that the water is not expelled by substrate in 14DMs but only slightly shifts, as observed for substrate-bound cholesterol 24-hydroxylase (CYP46A1) (27), remaining in the iron proximity. Protons derived from this water molecule can be required for O2 activation prior to substrate monooxygenations, the observed hydrogen-bond network being indicative of an I helix-based proton relay system (29, 30) in 14DMs, especially because synchronized sequential delivery of six catalytic protons for three rounds of oxygen cleavage is necessary for their three-step stereospecific reaction (supplemental Fig. S1). This structural information supports catalytic significance of the CYP51 I-helix signature residues previously demonstrated by site-directed mutagenesis (9).
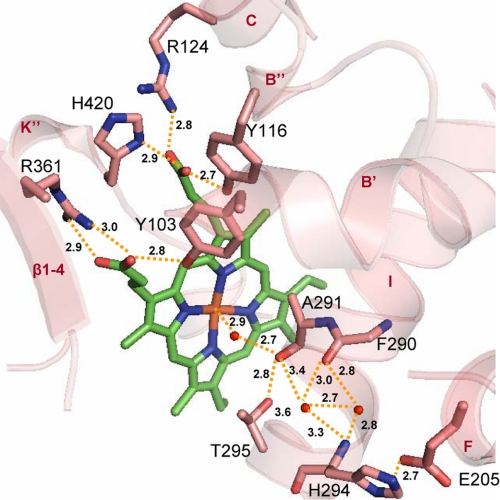
FIGURE 2.
Heme environment in ligand-free Tbb14DM. Heme and surrounding residues are shown in stick representation, H-bonds are indicated with yellow dashes, and the distances (Å) are marked. Upper region, residues interacting with the heme propionates. The A-ring forms H-bonds with Tyr-103 (helix B′) and Arg-361 (β1–4), and the D-ring is supported by Tyr-116 (helix B″), Arg-124 (helix C), and His-420 (helix K″). Lower region, potential I-helix-based proton delivery route. The water molecules included into the H-bond network are presented as small red spheres. The I-helix groove is formed by protrusion of the Ala-291 carbonyl toward the heme and probably modulated by flexible Gly-292. As a result, two helical H-bonds (Phe-290–His-294 and Ala-291–Thr-295) are missing, the peptide carbonyl of Phe-290 being connected through the waters. Ala-291, Gly-292, His-294, and Thr-295 (“the conserved threonine” in the majority of CYP families) are part of the CYP51 I-helix signature (–aGqHTS–). His-294 is the residue specific for the 14DM; the other P450s usually have Asp/Glu in this position. Glu-205 (helix F), which forms a salt bridge with His-294, is also conserved in all 14DM.
On the proximal side of the heme plane, the fifth coordination site of the iron is occupied by Cys-422. Additional support to the heme is provided by five residues forming hydrogen-bonding interactions with the porphyrin ring propionates (Fig. 2). Tyr-103, Arg-361, and His-420 are invariant in 14DMs from all phyla. Arg-124 is conserved in all Trypanosomatidae and aligns with Lys-97 in MtCYP51, which also interacts with the heme. Finally, Tyr-116 is conserved among protozoan, fungal, and animal 14DM orthologs. In plants, the same role is likely played by the conserved Tyr, which is positioned four residues to the C terminus from Tbb14DM Tyr-116 (Tyr-124, potato sequence numbering), but in MtCYP51, the corresponding hydrogen bond is absent (see also Fig. 1a, left inset, Phe-89). Hydrogen-bonding interactions with the peripheral heme substituents are known to be crucial for P450 heme stabilization, for maintaining the iron redox potential, and for controlling enzymatic activity (29, 31). Lack of heme support from the protein moiety may be the cause of MtCYP51 rapid inactivation upon reduction and one of the reasons for the 1–2 orders of magnitude lower in vitro catalytic activity when compared with eukaryotic 14DMs (14), whereas larger active site volume is likely to explain weak inhibition of MtCYP51 by azole derivatives.
VNI-bound 14DM
Binding of VNI (Fig. 3a) does not cause large scale conformational rearrangements in Tbb14DM. The root mean square deviation for all Cα atoms between the VNI-bound and ligand-free structures is ∼0.75 Å (only slightly higher than the deviations observed within the four molecules in the asymmetric unit for each structure (supplemental Table S1)), which may be indicative of the general 14DM structural rigidity. The minor alteration in the backbone can be described as slight expansion of several binding site-defining segments (Fig. 3b) 0.5–1 Å away from the protein core, as if the enzyme inhales, sparing additional space for the bulky inhibitor. The total volume of the molecule, however, remains unchanged (∼61,000 Å3), perhaps because of the subtle backward movement and slight increase in the helical content on the opposite (proximal) surface (supplemental Fig. S5). As helices A′ and F″ shift away from each other, the access channel entrance opens ∼2 Å wider so that the two outer rings of the VNI δ-arm within the channel can be clearly seen from the distal side. A hint of an additional opening appears nearby due to the I-helix shift and altered position of the His-458 side chain (Fig. 3c). Although small, this additional channel might have relevance because it is bordered by the conserved His-294–Glu-205 H-bond pair (discussed above as potentially involved in the surface-exposed part of the proton delivery route).
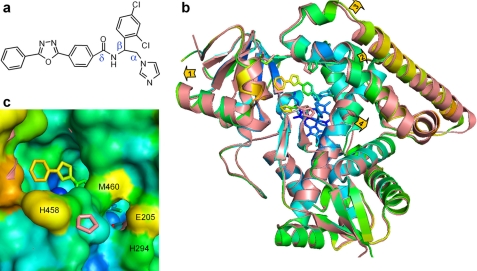
FIGURE 3.
VNI-bound Tbb14DM. a, the chemical structure of VNI. b, superposition of the ligand-free (salmon) and VNI-bound (colored by B-factor) Tbb14DM structures, distal view. The B-factor ranges from blue minimum to red maximum within 17–68 Å2. The heme, VNI, and three residues surrounding the entrance (Ile-45, Pro-210, and His-458) are presented as stick models. Four structural segments where the backbone in all four molecules moves >0.5 Å away from the catalytic core relative to the ligand-free structure are marked with arrows showing the directions of coordinate shifts. 1, N terminus (residues 32–65); 2, BC segment (95–114); 3, FG segment (185–232); and 4, middle part of helix I (288–292). Superimposition of four ligand-free and four VNI-bound molecules can be seen in supplemental Fig. S5. c, enlarged view of the access channel in surface representation; orientation is the same as in b. Some of the heme atoms are seen as blue spheres. The salmon ribbon of the ligand-free structure is partially visible where the shift in the backbone is larger than 1 Å (helices A, F″, and I). The second channel is bordered by helices I (His-294), F (Glu-205), and β4 hairpin (Met-460).
Inside the protein globule, the average active site volume increases to 770 Å3 (about 28%, variation range 650–880 Å3). In all four molecules, the electron density map unambiguously shows the single orientation of the inhibitor (supplemental Fig. S7). The imidazole nitrogen coordinates to the heme iron, and the right side of the ring forms van der Waals contacts with the I-helix (this unusual I-helix proximity to the heme can be one possible reason for the elevated 14DM susceptibility to azole inhibitors). The β-phenyl ring lies above the imidazole ring and is tightly packed against the heme, helices I, B′, and B′C loop; the three-ring δ-arm is positioned within the channel. At the distance of 5 Å, VNI is surrounded by 25 residues, 15 of them being located within van der Waals contacts (4 Å) (Fig. 4a). Except for Phe-105 (Ile-105 in T. cruzi 14DM, which is the substrate preference-defining residue (32)) and Phe-214 (alanine in T. brucei gambiense), these VNI-contacting amino acids are conserved among all 14DMs from Trypanosomatidae. Some of the side chains are shifted away from the inhibitor in comparison with the ligand-free structure, including Ala-291. The distance from the iron to the I-helix increases by 1.3 Å, giving an impression that the helix straightens. The water molecules leave the I-helical groove and form a new H-bonding network, now connecting Ala-291, Thr-295, and the inhibitor amide carbonyl. The only residue that moves (∼1.9 Å) closer to VNI is Tyr-103 (Fig. 4b). Its hydrogen bond with the porphyrin ring is lost and, instead, it now forms a new H-bond with the amide nitrogen of VNI. Interestingly, substitution of the corresponding Tyr (conserved across the CYP51 family) to Phe in Mt and human 14DM completely inactivates the enzymes (33), which implies that loss of this heme support observed in the Tbb14DM structure upon the inhibitor binding should also play an important role in the enzyme inactivation. Most likely, as a result of all these multiple contacts with the protein, the VNI molecule is pushed closer toward the heme iron, the length of the coordination bond N–Fe (∼2.0 Å) being shorter than that observed for other P450-azole complexes such as MtCYP51 (25), and this might indicate a stronger interaction. Concomitantly, on the proximal side, Cys-422 is found farther from the iron atom (2.4 Å), suggesting that the strength of thiolate coordination might be decreasing (31, 34) (the quality of the electron density for this region at 2.5 σ is shown in supplemental Fig. S7b). The altered heme iron environment in the 14DM-VNI complex is experimentally expressed by affecting the ability of Tbb14DM to bind carbon monoxide (the rate of CO complex formation is ∼70-fold slower, see supplemental Fig. S2b). Hence, based on the structural information, we believe that the profound inhibitory effect of VNI can be a consequence of several events including hand-in-glove fit induced by minor rearrangements required to increase the enzyme active site volume, multiple hydrophobic interactions with the side chains inside the cavity, the H-bond network around the amide group of VNI, and weakened heme support from the protein moiety (Tyr-103 and perhaps Cys-422), possibly resulting in stronger coordination of the basic imidazole nitrogen to the heme iron.
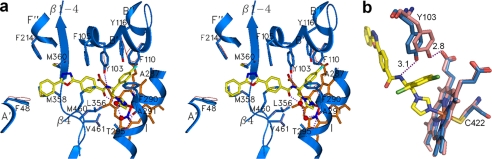
FIGURE 4.
Active site of Tbb14DM with bound VNI. a, secondary structure and stick representations of the 15 residues (blue) within 4 Å from VNI (yellow) shown in stereo view. Location of the corresponding side chains in the ligand-free structure, if different, can be seen as pink lines. The hydrogen bonds around the VNI amide group fragment are indicated as purple dashes. b, altered location of Tyr-103 in ligand-free (salmon) and VNI-bound (blue) Tbb14DM.
The mode of VNI interaction with the protein is in good agreement with our experimental data. Thus, the distant aromatic ring in the δ-arm of VNI forms the smallest number of contacts with the protein (Fig. 4a) and has the highest flexibility (the B-factor increases along the VNI molecule from 21 to 51 Å2 (Fig. 3b)). This explains our finding (11) that analogous compounds with significant variations in the δ-arm composition retain their inhibitory effect on the enzyme and trypanosomal cells, whereas even minor alterations in the β-phenyl ring substituents are often crucial for the inhibitory potency, and offers the real possibility for further directed modifications of this lead structure aiming to fulfill the requirements of high cellular permeability, optimal life time in human blood, and preserve low toxicity.
DISCUSSION
The highly hydrophobic, membrane-bound eukaryotic 14DMs have remained recalcitrant to crystallization, and the only CYP51 structure previously determined is that of the water-soluble CYP51 family member from Mt. Although this P450 possesses in vitro-14DM activity, its biological role is unknown; there is no sterol biosynthetic pathway in Mycobacteria genomes, and the CYP51 gene knock-out does not affect Mt growth (35). The latter point is particularly meaningful because inhibition of CYP51 in other microbes leads to their inability to multiply. Although similarity between Tbb14DM and MtCYP51 at the secondary structure level predicts a common evolutionary origin for bacterial and eukaryotic CYP51s and provides an additional, secondary structure signature for 14DMs across phylogeny (13 of 33 amino acid residues conserved in more than 150 known CYP51 sequences are glycines and prolines at the beginning or at the end of the helices and strands), profound differences in organization of their substrate-binding cavities might indicate that in Mycobacteria, the CYP51 gene has acquired another function. Such a possibility is supported by the ability of MtCYP51 to bind estriol (36), a sterol that does not interact with eukaryotic 14DMs. Structural differences between Tbb14DM and MtCYP51 explain both low efficacy of MtCYP51-based homology modeling in predicting potencies for the inhibitors aimed to target 14DMs from eukaryotic pathogens and limited applicability of the MtCYP51 structure in understanding structure-function relations in the CYP51 family (33, 36–39). Based on the MtCYP51 structure, large scale conformational changes were expected for 14DM catalysis and inhibition (25, 33). Using fluorescence resonance energy transfer analysis, they were confirmed to occur upon the MtCYP51-lanosterol complex formation (40), but we were unable to detect them in human or trypanosomal 14DMs.3
Being a step in the essential eukaryotic metabolic pathway, the 14DM reaction must remain conserved across phylogeny, and the enzymes preserve their strict substrate specificity and retain the three-step monooxygenation mechanism at amino acid identities as low as 22%. We surmise that opposite to high plasticity as the basis for catalytic promiscuity of xenobiotic-metabolizing P450s (41), it must be the structural rigidity of the substrate-binding cavity that maintains the 14DM functional conservation. Minimal conformational changes in the Tbb14DM structure upon VNI binding support this hypothesis and have allowed us to predict that the Tbb14DM structure will serve as a good model for other eukaryotic 14DM orthologs.4
As seen in supplemental Fig. S8a, VNI is also a highly potent inhibitor for 14DM from C. albicans, but it does not inhibit the human enzyme. The Tbb14DM structure provides an opportunity to understand this selectivity. Although there are four variations in the VNI-binding residues between trypanosomal and human 14DMs, three of them are unlikely to cause the observed VNI resistance. Two substitutions (A287G and M358I) are also found in C. albicans, which, nevertheless, is strongly inhibited by VNI. Leu-356 in Tbb14DM is positioned 3.6 Å from the VNI imidazole ring. If the corresponding isoleucine in the human enzyme were to prevent imidazole coordination to the heme iron, this would abolish the P450 spectral response to VNI, which does not take place (supplemental Fig. S8b). VNI binds to human 14DM with the apparent Kd being close to that of ketoconazole (0.11 μm), which we found to be a strong inhibitor of human 14DM (supplemental Fig. S9a). Similar lack of inhibition at high apparent binding affinity to Tbb and T. cruzi 14DMs was previously observed for the VNI scaffold derivatives that contain the phenyl ring at the α-position (11). We find that the α-phenyl azoles, even those showing apparent Kd values <0.1 μm, act as “short term inhibitors” and can be easily replaced in the enzyme active site by substrate (an example is shown in supplemental Fig. S9b).
The VNI binding mode suggests that the observed resistance in human 14DM is most likely connected with its Ile-488, which aligns with Val-461 in Tbb14DM. In the VNI-Tbb14DM structure, Val-461 is close (3.5 Å) to the water molecule that mediates the H-bond network between the VNI amide carbonyl and helix I. The bulkier side chain of isoleucine in human 14DM (supplemental Fig. S9c) may interfere with this water molecule, thus eliminating the VNI-I helix interaction. If in human 14DM the H-bonding between the VNI carbonyl and helix I is not formed, as a result of altered position of the inhibitor, it might also not be formed between the VNI NH and Tyr-103, so this residue will retain its crucial coordination to the porphyrin ring, which in turn may preserve higher affinity of the heme environment to the substrate. Thus, the amide group fragment, unique for the VNI scaffold, must be responsible for the inhibitor selectivity to 14DMs from pathogenic microbes, and the VNI-bound 14DM structure can serve as a basis for directed design of novel pathogen-specific drugs of low toxicity that would not affect the host counterpart.
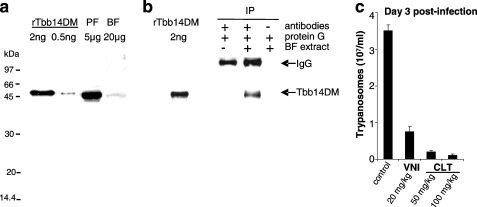
Among Trypanosomatidae, where endogenously produced ergosterol-like sterols are essential (2, 4, 5), T. brucei appears to be the only exception. These extracellular parasites, spending the majority of their mammalian life stage in the bloodstream, utilize a great part of their genome capacity to evade the host immunosystem, whereas the metabolic repertoire of the parasite is highly restricted (42). Using the plasma as a source of nutrients, the bloodstream trypanosomes have also developed the ability to build their membranes from host cholesterol (43). Our data, however, clearly show that VNI and its close derivatives produce significant dose-dependent antiparasitic effects in Tbb cells (11). The efficiency of cellular responses (ED50 1–5 μm) is comparable in bloodstream and procyclic (insect) forms, low general cytotoxicity implying that the observed effect is likely to be 14DM-specific. This is in line with the finding that Tbb cells deficient in the heme delivery system demonstrate a slow growing phenotype and increased susceptibility to host macrophages (20). Because trypanosome genomes are highly regulated at all stages of their life cycle (43), in this study, we analyzed 14DM expression in Tbb and found that, although at a level significantly lower than in the procyclic forms (where production of endogenous sterols is required), the gene is expressed in the bloodstream forms of the parasite (Fig. 5, a and b). Moreover, our in vivo studies demonstrate that 14DM inhibitors are able to slow down, in a dose-dependent manner, parasitemia in Tbb-infected mice (Fig. 5c). This supports the notion (44–46) that sterol derivatives produced by protists must have functions beyond membrane biogenesis. Similar to sterols in animals and plants, they can serve as precursors for biologically active molecules essential for the growth and development processes (45), and therefore, potent 14DM inhibitors might be helpful in combination with immunostimulants with traditional anti-sleeping sickness drugs to decrease their effective dosage and thus ease the burden of toxicity, adverse side effects, and resistance. Because azoles are generally known to cross the blood-brain barrier (47), they might be advantageous in the chronic stage of the disease when the parasite enters the central nervous system because in the cerebrospinal fluid, the source of cholesterol becomes hundreds of times more limited (48). More detailed studies are underway, and we believe that identification of regulatory sterols in T. brucei will not only advance knowledge of protist biology but may uncover novel parasite-specific drug targets as well.
FIGURE 5.
14DM in T. brucei. a and b, immunodetection of 14DM in Tbb procyclic (PF) and bloodstream forms (BF). a, whole cell extracts of procyclic and mice-isolated bloodstream form trypanosomes were probed by Western blotting with mouse anti-Tbb14DM antiserum. Recombinant (r) Tbb14DM served as positive control. b, mice-isolated bloodstream form cell extracts (10 mg) were immunoprecipitated with rat anti-Tbb14DM antiserum. The immunoprecipitated material (IP) was analyzed by Western blotting after non-reducing electrophoresis with mouse polyclonal anti-Tbb14DM antiserum. Controls were performed in the absence of either cell extracts or rat antiserum. Tbb14DM served as positive control. c, suppression of Tbb infection in mice by oral administration of VNI (20 mg/kg) or clotrimazole (CLT, 50 and 100 mg/kg). Clotrimazole was selected among several commercially available antifungal azoles by testing their inhibitory potency in reconstituted Tbb14DM reaction in vitro due to the limited amount of VNI. The differences observed between the mean of treated and mock-treated mice are statistically significant as assessed by Student's t test (p < 0.01). Error bars indicate S.E.
Supplementary Material
Acknowledgments
Use of the Advanced Photon Source and the LS-CAT Sector 21 was supported by Grant DE-AC02-06CH11357 and 085P1000817. Support was also provided by the Structural Genomics Consortium, Toronto, registered charity number 1097737. We thank Novartis Research Institute, Vienna, Austria, for VNI, Dr. U. K. Singha (Meharry Medical College, Nashville, TN) for help with in vivo studies, and Dr. D. C. Lamb (University of Swansea, Wales, UK) for discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants GM 067871 (to M. R. W. and G. I. L.), P30ES00267 (to M. R. W.), AI 080580 (to F. V.), GM 081146 (to M. C.), and WF D-1276 (to W. D. N.). The work was also supported by American Heart Association Grant 0535121N (to G. I. L.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3G1Q and 3GW9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9 and supplemental Table 1.
G. I. Lepesheva and M. R. Waterman, unpublished results.
While this manuscript was in preparation, using Tbb14DM (3G1Q) as a search model, the human 14DM structure was solved (PDB accession number 3I3K (H. W. P.)), and we found the mammalian and protozoan orthologs to be very similar (the root mean square deviation for the Cα atoms for the Tbb and human 14DM structures being only 1.7 Å).
- 14DM
- sterol 14α-demethylase (CYP51)
- CYP
- cytochrome P450
- Tbb
- T. brucei brucei
- Mt
- M. tuberculosis
- VNI
- N-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide.
REFERENCES
- 1.Barrett M. P., Burchmore R. J., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., Krishna S. (2003) Lancet 362, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 2.Croft S. L., Barrett M. P., Urbina J. A. (2005) Trends Parasitol. 21, 508–512 [DOI] [PubMed] [Google Scholar]
- 3.Myler P. J. (2008) Adv. Exp. Med. Biol. 625, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soeiro M. N., de Castro S. L. (2009) Expert. Opin. Ther. Targets 13, 105–121 [DOI] [PubMed] [Google Scholar]
- 5.Lüscher A., de Koning H. P., Mäser P. (2007) Curr. Pharm. Des. 13, 555–567 [DOI] [PubMed] [Google Scholar]
- 6.Cruz A. K., de Toledo J. S., Falade M., Terrão M. C., Kamchonwongpaisan S., Kyle D. E., Uthaipibull C. (2009) Curr. Drug Targets 10, 178–192 [DOI] [PubMed] [Google Scholar]
- 7.Como J. A., Dismukes W. E. (1994) N. Engl. J. Med. 330, 263–272 [DOI] [PubMed] [Google Scholar]
- 8.Thompson G. R., 3rd, Cadena J., Patterson T. F. (2009) Clin. Chest. Med. 30, 203–215 [DOI] [PubMed] [Google Scholar]
- 9.Lepesheva G. I., Waterman M. R. (2007) Biochim. Biophys. Acta 1770, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckner F., Yokoyama K., Lockman J., Aikenhead K., Ohkanda J., Sadilek M., Sebti S., Van Voorhis W., Hamilton A., Gelb M. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15149–15153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepesheva G. I., Ott R. D., Hargrove T. Y., Kleshchenko Y. Y., Schuster I., Nes W. D., Hill G. C., Villalta F., Waterman M. R. (2007) Chem. Biol. 14, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konkle M. E., Hargrove T. Y., Kleshchenko Y. Y., von Kries J. P., Ridenour W., Uddin M. J., Caprioli R. M., Marnett L. J., Nes W. D., Villalta F., Waterman M. R., Lepesheva G. I. (2009) J. Med. Chem. 52, 2846–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepesheva G. I., Hargrove T. Y., Kleshchenko Y., Nes W. D., Villalta F., Waterman M. R. (2008) Lipids 43, 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepesheva G. I., Nes W. D., Zhou W., Hill G. C., Waterman M. R. (2004) Biochemistry 43, 10789–10799 [DOI] [PubMed] [Google Scholar]
- 15.von Wachenfeldt C., Richardson T. H., Cosme J., Johnson E. F. (1997) Arch. Biochem. Biophys. 339, 107–114 [DOI] [PubMed] [Google Scholar]
- 16.Otninowski Z., Minor W. (1995) HKL Manual, Yale University, New Haven, CT [Google Scholar]
- 17.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 19.Terwilliger T. C. (2000) Acta Crystallogr. D 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 21.Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22.Kleywegt G. J., Jones T. A. (1994) Acta Crystallogr. D 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 23.Vanhollebeke B., De Muylder G., Nielsen M. J., Pays A., Tebabi P., Dieu M., Raes M., Moestrup S. K., Pays E. (2008) Science 320, 677–681 [DOI] [PubMed] [Google Scholar]
- 24.Brun R., Jenni L. (1977) Acta Trop. 34, 21–33 [PubMed] [Google Scholar]
- 25.Podust L. M., Poulos T. L., Waterman M. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh D., Griswold J., Erman M., Pangborn W. (2009) Nature 457, 219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mast N., White M. A., Bjorkhem I., Johnson E. F., Stout C. D., Pikuleva I. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9546–9551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams P. A., Cosme J., Sridhar V., Johnson E. F., McRee D. E. (2000) Mol. Cell 5, 121–131 [DOI] [PubMed] [Google Scholar]
- 29.Hasemann C. A., Kurumbail R. G., Boddupalli S. S., Peterson J. A., Deisenhofer J. (1995) Structure 3, 41–62 [DOI] [PubMed] [Google Scholar]
- 30.Poulos T. L. (2007) Drug Metab. Rev. 39, 557–566 [DOI] [PubMed] [Google Scholar]
- 31.Guallar V., Baik M. H., Lippard S. J., Friesner R. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6998–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lepesheva G. I., Zaitseva N. G., Nes W. D., Zhou W., Arase M., Liu J., Hill G. C., Waterman M. R. (2006) J. Biol. Chem. 281, 3577–3585 [DOI] [PubMed] [Google Scholar]
- 33.Lepesheva G. I., Virus C., Waterman M. R. (2003) Biochemistry 42, 9091–9101 [DOI] [PubMed] [Google Scholar]
- 34.Schlichting I., Berendzen J., Chu K., Stock A. M., Maves S. A., Benson D. E., Sweet R. M., Ringe D., Petsko G. A., Sligar S. G. (2000) Science 287, 1615–1622 [DOI] [PubMed] [Google Scholar]
- 35.McLean K. J., Dunford A. J., Sabri M., Neeli R., Girvan H. M., Balding P. R., Leys D., Seward H. E., Marshall K. R., Munro A. W. (2006) Biochem. Soc. Trans. 34, 1178–1182 [DOI] [PubMed] [Google Scholar]
- 36.Podust L. M., Yermalitskaya L. V., Lepesheva G. I., Podust V. N., Dalmasso E. A., Waterman M. R. (2004) Structure 12, 1937–1945 [DOI] [PubMed] [Google Scholar]
- 37.Lepesheva G. I., Podust L. M., Bellamine A., Waterman M. R. (2001) J. Biol. Chem. 276, 28413–28420 [DOI] [PubMed] [Google Scholar]
- 38.Bellamine A., Lepesheva G. I., Waterman M. R. (2004) J. Lipid Res. 45, 2000–2007 [DOI] [PubMed] [Google Scholar]
- 39.Matsuura K., Yoshioka S., Tosha T., Hori H., Ishimori K., Kitagawa T., Morishima I., Kagawa N., Waterman M. R. (2005) J. Biol. Chem. 280, 9088–9096 [DOI] [PubMed] [Google Scholar]
- 40.Lepesheva G. I., Seliskar M., Knutson C. G., Stourman N. V., Rozman D., Waterman M. R. (2007) Arch. Biochem. Biophys. 464, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekroos M., Sjögren T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Böhme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E., El-Sayed N. M. (2005) Science 309, 416–42216020726 [Google Scholar]
- 43.Coppens I., Courtoy P. J. (2000) Annu. Rev. Microbiol. 54, 129–156 [DOI] [PubMed] [Google Scholar]
- 44.Vanden Bossche H., Koymans L., Moereels H. (1995) Pharmacol. Ther. 67, 79–100 [DOI] [PubMed] [Google Scholar]
- 45.Roberts C. W., McLeod R., Rice D. W., Ginger M., Chance M. L., Goad L. J. (2003) Mol. Biochem. Parasitol. 126, 129–142 [DOI] [PubMed] [Google Scholar]
- 46.Zhou W., Cross G. A., Nes W. D. (2007) J. Lipid Res. 48, 665–673 [DOI] [PubMed] [Google Scholar]
- 47.Goodpasture H. C., Hershberger R. E., Barnett A. M., Peterie J. D. (1985) Arch. Intern. Med. 145, 879–880 [PubMed] [Google Scholar]
- 48.Leoni V., Lütjohann D., Masterman T. (2005) J. Lipid Res. 46, 191–195 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.