Abstract
N-Linked glycosylation is an important determinant of protein structure and function. The interleukin-6 signal transducer glycoprotein 130 (gp130) is a common co-receptor for cytokines of the interleukin (IL)-6 family and is N-glycosylated at 9 of 11 potential sites. Whereas N-glycosylation of the extracellular domains D1–D3 of gp130 has been shown to be dispensable for binding of the gp130 ligand IL-6 and its cognate receptor in vitro, the role of the N-linked glycans on domains D4 and D6 is still unclear. We have mutated the asparagines of all nine functional N-glycosylation sites of gp130 to glutamine and systematically analyzed the consequences of deleted N-glycosylation (dNG) in both cellular gp130 and in a soluble gp130-IgG1-Fc fusion protein (sgp130Fc). Our results show that sgp130Fc-dNG is inherently unstable and degrades rapidly under conditions that do not harm wild-type sgp130Fc. Consistently, the bulk of cellular gp130-dNG is not transported to the plasma membrane but is degraded in the proteasome. However, the small quantities of gp130-dNG, which do reach the cell surface, are still able to activate the key gp130 signaling target signal transducer and activator of transcription-3 (STAT3) upon binding of the agonistic complex of IL-6 and soluble IL-6 receptor. In conclusion, N-linked glycosylation is required for the stability but not the signal-transducing function of gp130.
Keywords: Cytokines, Glycoproteins/Structure, Receptors/Cytokine, Signal Transduction/Jak-Stat, IL-6, STAT3, gp130
Introduction
N-Linked glycosylation is frequently but not always important for the folding, oligomerization, and stability of proteins (1). Some proteins need N-linked glycans as chaperone-like structures during protein synthesis to ensure correct folding by increasing their solubility and masking hydrophobic patches, but the N-glycans can then be dispensable for protein function (1). In contrast, N-linked glycosylation is essential for ligand binding and stability of diverse growth factor, cytokine, peptide, and pattern recognition receptors as well as adhesion molecules (2–8).
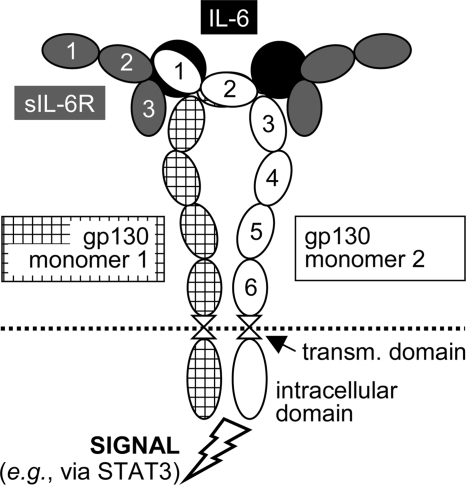
The interleukin (IL)2-6 cytokine family includes numerous members apart from IL-6, such as leukemia inhibitory factor (LIF), IL-11, or IL-27 (9, 10). Acting in complex with specific α-receptors, e.g. IL-6 receptor (IL-6R), and/or other β-receptors (e.g. LIF receptor), the transmembrane protein gp130 serves as a common β-receptor subunit and signal transducer of the IL-6 family (11). The pleiotropic cytokine IL-6 signals via a complex consisting of two gp130 molecules and either one or two molecules each of IL-6 and IL-6R (Fig. 1) (9, 12, 13). Only few cell types express IL-6R (e.g. hepatocytes and some leukocytes) (14), but virtually all cells in the body express gp130. A soluble form of the IL-6R (sIL-6R) is produced by protease shedding or alternative splicing and forms a soluble complex with IL-6. This agonistic IL-6·sIL-6R complex activates gp130 signaling in cells lacking membrane-bound IL-6R, a process termed trans-signaling (15, 16). Soluble gp130 (sgp130) molecules are produced by alternative splicing and are constitutively present in the plasma of healthy individuals at concentrations of 100–200 ng/ml (17). sgp130 selectively inhibits IL-6 trans-signaling (18), and sgp130Fc fusion proteins have considerable therapeutic potential in various chronic inflammatory disorders (19).
FIGURE 1.
Hexameric signaling complex of gp130, IL-6, and sIL-6R. In the tetrameric complex, only one molecule each of IL-6 and sIL-6R is present. 1–6, extracellular domains D1–D6 of gp130; transm., transmembrane.
gp130 belongs to the class of “tall cytokine receptors” that feature three fibronectin-type III-like domains between their ligand-binding domains and their transmembrane domain (20). In 2001, Moritz et al. (21) reported that of the 11 potential N-glycosylation sites, Asn21, Asn61, Asn109, Asn135, Asn205, Asn357, Asn361, Asn531, and Asn542 were glycosylated but Asn224 and Asn368 were not. Of these two sites, N-glycosylation on Asn224 is practically impossible, as Asn224 is followed by a proline and is also partially buried in the gp130 structure (21, 22). The crystal structure of the complete extracellular part of gp130 is still unknown, but the structure of the three ligand-binding domains D1–D3 or D2 + D3 has been solved in complex with viral IL-6 (23), LIF (24), or IL-6·sIL-6R (25). Two of these studies used gp130 fragments produced in insect cells in the presence of tunicamycin as a global N-glycosylation inhibitor (23, 24), whereas the third study employed a gp130 expression construct in which Asn21, Asn109, Asn135, Asn205, and Asn224 (but not Asn61) of gp130 were mutated to glutamine (25). Both tunicamycin and the Asn/Gln mutations resulted in reduced yield of the protein in insect cells but did not change its biochemical behavior or binding properties (23–25).
In contrast, the roles of O-glycans or the N-glycans on the fibronectin-type III-like domains D4 (Asn357 and Asn361) and D6 (Asn531 and Asn542) of gp130 have never been investigated. As the orientation of the domains D4–D6 is important for the positioning of the ligand-binding domains D1–D3 (26), N-glycosylation of domains D4 and D6 could be critical for ligand binding despite correct folding of domains D1–D3 in the absence of N-glycans.
Interestingly, a recent study with tunicamycin-treated murine neuroepithelial cells showed that gp130 expressed in these cells could be found on the cell surface but did not transduce LIF signals (27). These findings were analogous to a report on the α-subunit of the receptor for granulocyte-macrophage colony-stimulating factor (3). However, the authors conceded critical deficiencies of their model system; besides a strong nonspecific stress response induced by tunicamycin, which would interfere with any analyzed signaling process, they only managed to produce cells with both glycosylated and nonglycosylated gp130 species on the cell surface, which made any conclusions very difficult (27).
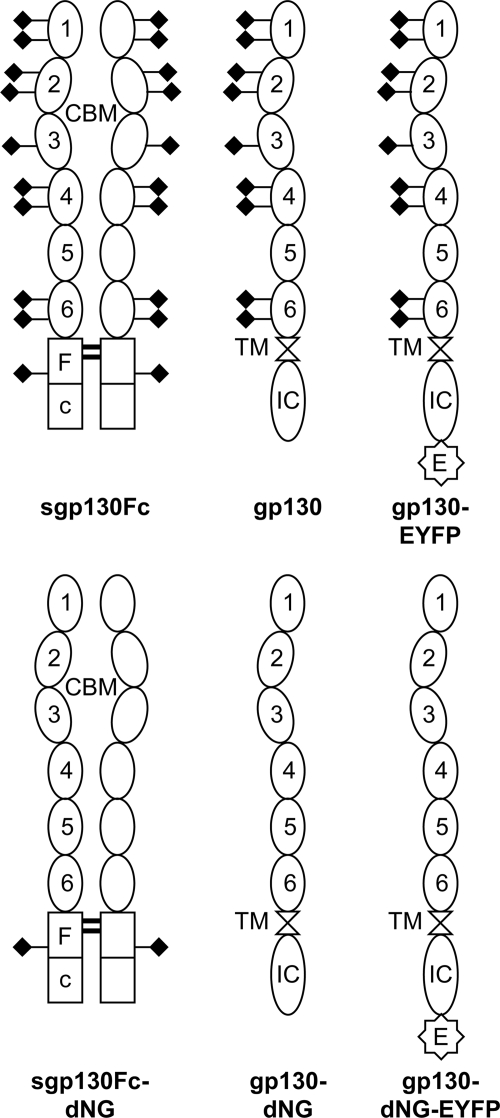
The aim of this study was therefore to clarify the importance of N-linked glycosylation for gp130. After assessing the contributions of O- and N-glycans to gp130 glycosylation, we have systematically analyzed the production, stability, and function of both wild-type gp130 and gp130 with deleted N-glycosylation (termed gp130-dNG) as well as of the analogous IgG1-Fc fusion proteins sgp130Fc and sgp130Fc-dNG (Fig. 2). Using transient and stable expression systems in CHO-K1 (hamster), HeLa (human), and BAF3 (murine) cells, we demonstrate that N-glycosylation of gp130 is essential for the stability and localization of both membrane-bound gp130 and the sgp130Fc fusion protein. Surprisingly, however, nonglycosylated gp130-dNG is still able to transduce an activation signal from the agonistic IL-6·sIL-6R complex to a key gp130 signaling target, the transcription factor STAT3. This has implications for the ligand binding mechanism of gp130 as well as for the production of sgp130Fc proteins for therapeutic purposes.
FIGURE 2.
Schematic representation of the engineered sgp130Fc and gp130 variants. The dNG variant sgp130Fc-dNG was constructed to study the influence of N-glycans on the production, stability, and biological activity of sgp130Fc fusion proteins. Overexpressed cellular gp130 with or without N-glycans (gp130-dNG) was used to analyze the influence of N-glycans on the surface expression and signaling function of gp130. EYFP-tagged variants of gp130 and gp130-dNG were used for the same purposes and, in addition, for microscopic visualization. The molecular weight of the sgp130Fc(−dNG) proteins is 2 × 93 kDa (without signal peptides). The molecular weight, without signal peptide, of the cellular gp130(-dNG) and gp130-(dNG)-EYFP proteins is 101 and 129 kDa, respectively. The N-glycans on one complete extracellular part of gp130 (D1–D6) add about 25 kDa to the apparent molecular weight of the protein. N-Glycans are symbolized by rhombi. 1–6, extracellular domains D1–D6 of gp130; CBM, cytokine-binding module (D2 + D3); E, C-terminal EYFP tag; Fc, IgG1-Fc; IC, intracellular domain; TM, transmembrane domain.
EXPERIMENTAL PROCEDURES
Constructs and Tools
A codon and structurally optimized version of human sgp130Fc has been described before (28, 29). An analogous codon-optimized variant sgp130Fc-dNG was synthesized by GENEART (Regensburg, Germany), in which all nine occupied N-glycosylation sites described by Moritz et al. (21) were mutated from Asn to Gln. The N-glycan on the IgG1-Fc (Asn297 of the IgG1 heavy chain) was left intact to retain the correct IgG1-Fc structure (30). Cloned full-length human gp130 (Gene ID IL6ST) with an enhanced yellow fluorescent protein (EYFP) tag (from pEYFP-N1; Clontech) was a kind gift from Dr. Stephanie Tenhumberg (Institute of Biochemistry, University of Kiel, Germany). The complete extracellular domains of codon-optimized gp130 and gp130-dNG from the synthesized sgp130Fc constructs were fused to the wild-type gp130 cDNA sequence coding for the transmembrane and intracellular domain of gp130 to ensure equal mRNA expression levels. Finally, the EYFP tag was deleted from the gp130(-dNG)-EYFP constructs to produce gp130 and gp130-dNG without a tag. All six variants, two of sgp130Fc and four of gp130 (Fig. 2), were produced using the high-yield Gateway expression vector pcDNA-DEST40 (Invitrogen).
Culture and Transient Transfection of CHO-K1 and HeLa Cells
CHO-K1 and HeLa cells were purchased from DSMZ (Braunschweig, Germany) and grown in Ham's F-12 (CHO-K1) or RPMI 1640 (HeLa) medium (both from PAA Laboratories, Cölbe, Germany) supplemented with 10% fetal bovine serum (FBS Superior; Biochrom, Berlin, Germany) at 37 °C with 5% CO2 in a water-saturated atmosphere. CHO-K1 and HeLa were transfected using Lipofectamine LTX (Invitrogen) according to the manufacturer's recommendations for these cells. For inhibition of proteasomal degradation in CHO-K1 cells, the inhibitor MG132 (benzyloxycarbonyl-Leu-Leu-Leu-al; Sigma) was added at 10 or 25 μm 2 h after transfection.
Production and Analysis of Cell Extracts
If not indicated otherwise, cells were harvested 24 h after transfection. Total RNA was extracted using the RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer's standard protocol. Reverse transcription-PCR was performed as described previously (31). The following two primer pairs were used in the same PCR: optimized sgp130Fc(-dNG), CCTGAGTTCACCTTCACCACC (forward) and TTGTGCACCTCCACGCC (reverse), amplicon, 239 bp (to avoid amplification of endogenous gp130 transcripts, the forward primer binds to gp130 and the reverse primer binds to Fc); neomycin resistance gene in pcDNA-DEST40, GATGCCTGCTTGCCGAATATC (forward) and CGCCAAGCTCTTCAGCAATATC (reverse), amplicon, 133 bp.
Denatured whole cell extracts were produced as described previously (31). N-Glycosylation was enzymatically removed from the denatured proteins in these extracts by peptide: N-glycosidase (PNGase) F treatment (New England Biolabs, Frankfurt/Main, Germany) according to the manufacturer's instructions. Secreted sgp130Fc was precipitated from cell culture supernatants by Protein A/G Plus-agarose beads (Santa Cruz Biotechnology, Heidelberg, Germany) using the manufacturer's standard protocol. Detergent-based total transmembrane protein extraction was performed with the ProteoExtract transmembrane protein extraction kit (Novagen/VWR, Darmstadt, Germany) according to the manufacturer's instructions. Selective isolation of plasma membrane proteins was achieved by surface biotinylation using the Pinpoint cell surface protein isolation kit (Pierce/Perbio/Thermo, Bonn, Germany). For BAF3 cell pools, we used a higher cell number (2.5 × 107) and a lower elution volume (100 μl) than recommended by the manufacturer.
sgp130Fc or sgp130Fc-dNG was detected by Western blotting as described previously (31) using anti-human IgG Fc (clone R10Z8E9; Chemicon/Millipore, Schwalbach, Germany) or anti-human gp130 (CD130) directed against the extracellular domain D4 of gp130 (clone B-P4; Diaclone, Besancon, France), both at 1:1,000 in TBS/Tween 20 (TBST) plus 5% blotting grade blocker (Bio-Rad) followed by anti-mouse IgG conjugated to horseradish peroxidase (HRP) (GE Healthcare) at 1:2,000 in 5% blocker/TBST. For normalization, β-actin was detected after stripping of the blot membranes (31) using anti-β-actin (clone AC-15; Sigma) at 1:10,000 in 4% blocker/TBST, followed by anti-mouse IgG-HRP as described. Ponceau S staining (Sigma) was used to confirm equal protein loading and transfer. Activation (phosphorylation) of STAT3 was measured by subsequent Western blot hybridizations (membranes were stripped in between as described previously (31)) with rabbit polyclonal anti-phospho-STAT3 (Tyr705) and mouse anti-STAT3 (clone 124H6) at 1:1,000 in 5% blocker/TBST, followed by anti-rabbit IgG-HRP at 1:2,000 in 5% blocker/TBST (all antibodies from Cell Signaling Technology, Frankfurt/Main, Germany) or anti-mouse IgG-HRP (GE Healthcare).
Production and Characterization of sgp130Fc-dNG
sgp130Fc and sgp130Fc-dNG were produced in stable clonal CHO-K1 production cell lines in Ham's F-12 medium supplemented with 5% IgG-stripped FBS (both from PAA Laboratories) in roller bottles (2,125 cm2; Greiner Bio-One, Frickenhausen, Germany) and purified by protein A affinity chromatography as described previously (18, 29). Protein integrity was analyzed by native PAGE and SDS-PAGE and subsequent silver staining (29). “Control-Fc” was constructed by fusing the IgG1-Fc contained in sgp130Fc and sgp130Fc-dNG to an Igκ signal peptide from pSecTag2 (Invitrogen). Control-Fc was produced in the same expression vector (pcDNA-DEST40) and production and purification system as the two sgp130Fc variants. Human IgG1 (for analysis of the shift in apparent molecular weight of the IgG1 heavy chain after deglycosylation by PNGase F) was purchased from Chemicon/Millipore.
Enzymatic analysis of N- and O-glycosylation of purified sgp130Fc was performed using the enzymatic protein deglycosylation kit from Sigma according to the manufacturer's instructions. The apparent molecular weight of sgp130Fc and the control protein fetuin were analyzed by SDS-PAGE with 7.5 and 10% gels, respectively, and subsequent silver staining (29).
Ligand binding capacity and biological activity of sgp130Fc and sgp130Fc-dNG were measured by enzyme-linked immunosorbent assay and BAF3/gp130 cell proliferation assay, respectively. For the enzyme-linked immunosorbent assay, Microlon 96-well enzyme-linked immunosorbent assay plates (Greiner Bio-One) were coated with 100 ng of sgp130Fc or sgp130Fc-dNG per well in 100 μl of cell culture grade Dulbecco's phosphate-buffered saline (PBS; PAA Laboratories) at 4 °C overnight. After blocking with 200 μl of 3% bovine serum albumin (BSA) (PAA Laboratories) in PBS (“BSA/PBS”) for 2 h at room temperature with gentle shaking (this applies for all following steps), plates were washed three times with “Tween/PBS,” i.e. PBS containing 0.05% Tween 20 (Merck), and dried by tapping on paper towels. A 1:2 BSA/PBS dilution series of human IL-6 (Invitrogen) plus half the amount of sIL-6R (Biochrom) ranging from 3.2 μg/ml IL-6 plus 1.6 μg/ml sIL-6R to 50 ng/ml IL-6 plus 25 ng/ml sIL-6R was added to triplicate columns per variant (100 μl/well) and incubated for 1 h at room temperature. After three washing steps with Tween/PBS as described above, bound sIL-6R was detected by mouse anti-IL-6R (clone M91; Beckman-Coulter, Krefeld, Germany) at 1:2,000 in BSA/PBS (100 μl/well) for 1 h at room temperature. Another three washing steps with Tween/PBS were followed by anti-mouse IgG-HRP (GE Healthcare) at 1:5,000 in BSA/PBS (100 μl/well) for 1 h at room temperature. Three washing steps with Tween/PBS and another three with distilled water preceded final exposure to 50 μl/well 3,3′,5,5′-tetramethylbenzidine substrate (Sigma). The HRP substrate reaction was stopped after ∼5 min by adding 50 μl/well 0.5 m sulfuric acid. A450 was measured in an Opsys MR plate reader equipped with Revelation QuickLink software (Dynex Technologies, Berlin, Germany).
BAF3 cells (murine pre-B-cells) stably transfected with human gp130 (BAF3/gp130 cells) have been described previously (18). Their proliferation upon stimulation with Hyper-IL-6, a covalent fusion protein of human IL-6 and sIL-6R (32), or IL-6·sIL-6R as well as the dose-dependent inhibition of this process by sgp130Fc (and lack of inhibition by sgp130Fc-dNG) were measured by the cell titer colorimetric assay (Promega) as described previously (29).
Microscopic Analysis of gp130(-dNG)-EYFP Localization in HeLa Cells
HeLa cells were seeded at 4 × 105 cells/ml in 6-well plates containing one sterile glass cover slide per well. The next day, cells were transfected with 1 μg/well pcDNA-DEST40_gp130-EYFP, pcDNA-DEST40_gp130-dNG- EYFP, or pcDNA-DEST40 (mock vector) and cultured for 24 h. Cells were fixed in 4% paraformaldehyde (Sigma) in PBS, and DNA was visualized by 4′,6-diamidino-2-phenylindole dihydrochloride staining as recommended by the manufacturer (Sigma). After washing, the cover slides with the attached cells were mounted onto glass slides, and EYFP fluorescence was examined using a Zeiss Axio Imager.Z1 apotome fluorescence microscope and the AxioVision Imaging software (Carl Zeiss MicroImaging, Göttingen, Germany).
Construction of Stably Transduced BAF3 Cell Pools Expressing gp130(-dNG) Variants
BAF3 cells were purchased from DSMZ. HEK-293 Phoenix-Eco cells were a kind gift from Dr. Ursula Klingmüller (DKFZ, Heidelberg, Germany) (33). Both cell lines were grown in Dulbecco's modified Eagle's medium high glucose culture medium (PAA Laboratories) supplemented with 10% FBS (Biochrom; see above). In addition, BAF3 medium was supplemented with 10 ng/ml murine IL-3 (Invitrogen) as the essential growth factor of these cells. BAF3 cells were retrovirally transduced with constructs based on the retroviral expression vector pMOWS (33); pMOWS_gp130, pMOWS_gp130-dNG, pMOWS_gp130-EYFP, and pMOWS_ gp130-dNG-EYFP as well as the control vector pMOWS-GFP (5 μg each) were transiently transfected into HEK-293 Phoenix-Eco cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The transfection efficiency was typically about 50%, as estimated by microscopic analysis of green fluorescent protein (GFP) expression 24 h after transfection (Axiovert 200 microscope; Carl Zeiss MicroImaging).
Supernatants containing retrovirus particles were produced as described (33). Subsequently, 250 μl of these supernatants were applied to 1 × 105 BAF3 cells (in 50 μl of medium) and supplemented with a final concentration of 8 μg/ml hexadimethrine bromide (Sigma). The cell suspension was mixed and then centrifuged at ∼300 × g for 2 h at room temperature. After medium exchange to standard medium supplemented with 10 ng/ml IL-3 and 1% penicillin/streptomycin (PAA Laboratories), cells were allowed to recover for 48 h in the incubator and were subsequently selected at the previously determined ideal dose of 0.1 μg/ml puromycin (Sigma) for 10 days.
Expression of gp130(-dNG) in total membrane protein or plasma membrane protein extracts (see above) was measured by Western blots using an anti-gp130 antibody directed against the intracellular domain of gp130 (sc-655; Santa Cruz Biotechnology) at 1:1,000 in 5% blocker/TBST (see above), followed by anti-rabbit IgG-HRP (Cell Signaling Technology) at 1:2,000 in 5% blocker/TBST. In addition, EYFP was detected either by microscopy (see above; >95% cells expressed EYFP after 10 days (data not shown)), by flow cytometry (see below), or by Western blots using an anti-EYFP/GFP antibody (clone JL-8; Clontech) at 1:1,000 in 5% blocker/TBST, followed by anti-mouse IgG-HRP (GE Healthcare) at 1:2,000 in 5% blocker/TBST.
Flow Cytometry Analysis of gp130(-dNG)-EYFP Expression in Transduced BAF3 Cell Pools
5 × 105 stably transduced BAF3 cells were washed once (centrifugation for 5 min, ∼500 × g, 4 °C) in flow cytometry buffer (PBS containing 1% BSA (PAA Laboratories) and 0.01% sodium azide (Sigma)) and were used either directly for flow cytometry analysis of the EYFP signal in the fluorescein isothiocyanate/GFP channel or were stained with antibodies raised against human gp130 (clone B-R3 from Abcam (Cambridge, UK) or clone B-P4 from Diaclone), followed by anti-mouse IgG-allophycocyanin (BD Biosciences), according to standard protocols of the manufacturers. Data were acquired from 10,000 gated events using a FACS-Canto analyzer (BD Biosciences).
Proliferation Analysis and Serum Starvation of Transduced BAF3 Cell Pools
BAF3 cell pools were washed twice in PBS to remove any traces of proliferation-inducing IL-3 from the culture medium (see above). In 96-well plates, 3 × 103 cells/well were seeded in 95 μl of growth medium without IL-3 and allowed to recover for 1 h in the incubator. Subsequently, cell pools received either 5 μl of growth medium (unstimulated control) or 5 μl of growth medium supplemented with either Hyper-IL-6 (final concentration 10 ng/ml) or IL-6 (Invitrogen) plus sIL-6R (Biochrom) with final concentrations of 100 and 50 ng/ml, respectively. The BAF3 proliferation assay was then performed as described previously (29). For measuring activation (Tyr705 phosphorylation) of STAT3 in Western blots (see above), cells were starved for 6 h in Dulbecco's modified Eagle's medium without FBS or IL-3. In some experiments, the proteasome inhibitor MG132 was added at 0.2–1 μm during this period. Subsequently, 1 × 106 cells were incubated in 1 ml of Dulbecco's modified Eagle's medium with or without 10 ng/ml Hyper-IL-6 in Eppendorf cups for 15 min (in some cases also for 5 min). After the stimulation period, cells were immediately harvested by centrifugation and lysed in denaturing lysis buffer (see above).
Data Presentation and Replication Rate
For all analyses, representative data of at least three independent experiments are shown. OD measurements were generally performed in triplicate, and most Western blot and flow cytometry measurements were performed in duplicate within one experiment. Statistical analyses were performed using the on-line software SISA (Daan G. Uitenbroek (1997), SISA Binomial). The normally distributed cell culture data were analyzed using the t test for independent samples.
RESULTS
Enzymatic Analysis of N- and O-Glycosylation of sgp130Fc- dNG
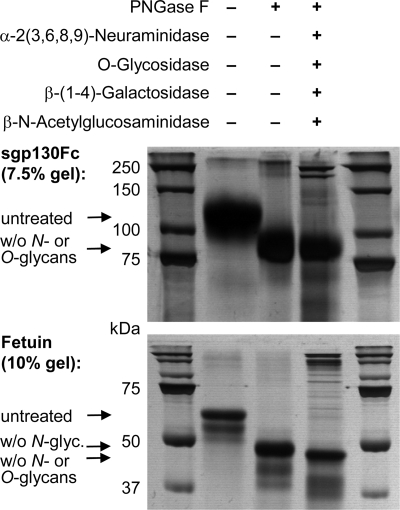
To assess the contributions of N- and O-glycans to gp130 glycosylation, sgp130Fc (Fig. 2) was incubated with PNGase F alone (to completely remove N-glycans) or with PNGase F in combination with a mixture of enzymes that together remove most O-glycans as follows: α-2(3,6,8,9)-neuraminidase, β-(1–4)-galactosidase, β-N-acetylglucosaminidase, and O-glycosidase. In contrast to the control protein fetuin, which is both N- and O-glycosylated, the apparent molecular weight of sgp130Fc was the same after treatment with PNGase F alone and after PNGase F plus the additional enzyme mix (Fig. 3). The slightly reduced width of the sgp130Fc band in the lane with the sample treated with all enzymes together might reflect a minimal amount of O-glycosylation (Fig. 3).
FIGURE 3.
Enzymatic assessment of N- and O-glycosylation of sgp130Fc. Both sgp130Fc (upper panel) and the N- and O-glycosylated control protein fetuin (lower panel) were treated with either PNGase F alone (to completely remove N-glycans) or with PNGase in combination with a mixture of enzymes, which together remove most O-glycans: α-2(3,6,8,9)-neuraminidase, β-(1–4)-galactosidase, β-N-acetylglucosaminidase, and O-glycosidase. In contrast to fetuin, the apparent molecular weight of sgp130Fc was not changed by O-deglycosylating enzymes. The stronger high molecular weight bands in experimental 3rd lane represent glycosidase proteins. w/o, without.
Characterization of sgp130Fc-dNG
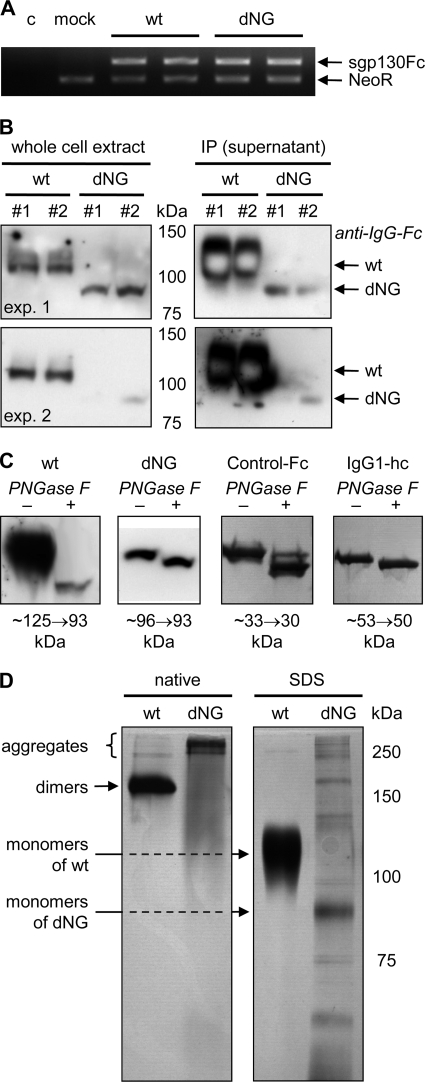
As a first approach to investigate the influence of N-glycosylation on gp130 stability, sgp130Fc-dNG (Fig. 2) was produced using the same protocol as established for sgp130Fc (18, 29). Whereas mRNA expression of wild-type sgp130Fc and sgp130Fc-dNG was similar (Fig. 4A), protein levels of sgp130Fc-dNG in whole cell extracts were strongly reduced compared with sgp130Fc (Fig. 4B, left panels). The difference between secreted sgp130Fc and sgp130Fc-dNG levels was even more extreme (Fig. 4B, right panels), indicating that almost no sgp130Fc-dNG was released from the cell. The most plausible explanation for these findings, which was investigated further with gp130(-dNG)(-EYFP) in the experiments presented below, was that the lack of N-glycans prevented sgp130Fc-dNG from passing the quality controls of the ER·Golgi complex and led to its degradation within the cell.
FIGURE 4.
Production and characterization of sgp130Fc-dNG in CHO-K1 cells. A, reverse transcription-PCR of sgp130Fc(-dNG) mRNA normalized by simultaneous amplification of cDNA from the neomycin resistance gene (NeoR) of the expression vector in the same PCR. sgp130Fc wild-type (wt) and sgp130Fc-dNG were amplified by the same primer pair. To avoid amplification of endogenous gp130 transcripts, the forward primer binds to gp130, and the reverse primer binds to Fc. The results show that sgp130Fc and sgp130Fc-dNG have the same mRNA expression levels. c, control; mock, only expression vector pcDNA-DEST40 (expressing NeoR). B, Western blots with anti-human IgG-Fc to detect wild-type (wt) sgp130Fc or sgp130Fc-dNG in denatured whole cell extracts (left panels) or in direct immunoprecipitations (IP) of the Fc part of the secreted sgp130Fc(-dNG) proteins from cell supernatants by protein A/G-agarose beads (right panels). Results from two (of five) experiments (exp. 1 and 2) with duplicate samples (#1 and #2) are shown to demonstrate the range of sgp130Fc-dNG protein levels. Whereas sgp130Fc-dNG protein can be readily detected in whole cell lysates, albeit at lower levels than wild-type sgp130Fc, almost no sgp130Fc-dNG is present in the cell supernatant (only visible with massive overexposure), indicating retention and degradation in the cell. C, silver-stained SDS-PAGE of wild-type (wt) sgp130Fc, sgp130Fc-dNG (dNG), control-Fc (i.e. IgG1-Fc alone), or IgG1 heavy chain (hc) after incubation without (−) or with (+) PNGase F to remove N-glycans. Shifts in apparent molecular weight are indicated below the panels. Gel strengths were 7.5% for sgp130Fc(−dNG) and 12% for control-Fc and IgG1 heavy chain. D, native and SDS-PAGE analysis of wild-type (wt) sgp130Fc and sgp130Fc-dNG preparations after elution from protein A. Molecular weight (MW) marker refers only to SDS-PAGE. The stronger band labeled monomers of dNG has the same apparent molecular weight as the single band detected in Western blots of whole cell extracts separated by SDS-PAGE under the same running conditions (B).
When denatured whole cell extracts were treated with PNGase F to remove N-glycans, sgp130Fc-dNG showed only a small reduction in apparent molecular weight. This was consistent with removal of the N-glycan from the IgG1-Fc, as evidenced by parallel analysis of the Fc of sgp130Fc and sgp130Fc-dNG alone, Control-Fc, and of the IgG1 heavy chain (Fig. 4C). These data confirmed the findings of Moritz et al. (21) and indicated that the gp130 part of sgp130Fc-dNG was indeed free of N-glycans.
The low secretion levels of sgp130Fc-dNG in transient transfection experiments were confirmed by analysis of stable CHO-K1 production cell lines. Although sgp130Fc is usually produced in the range of 10 mg/liter cell supernatant in roller bottles, the yield of sgp130Fc-dNG was <100 μg/liter, i.e. less than 1%. Native and SDS-PAGE analysis showed that sgp130Fc-dNG was fragmented and aggregated after the acidic elution from the protein A column (Fig. 4D). In contrast, sgp130Fc is stable under these conditions (18, 29) and shows only very little aggregation (Fig. 4D). Consistently, sgp130Fc-dNG preparations had no binding capacity for the IL-6·sIL-6R target complex in enzyme-linked immunosorbent assay experiments and no inhibitory biological activity in BAF3/gp130 cell proliferation assays (data not shown).
Localization and Degradation of gp130-dNG
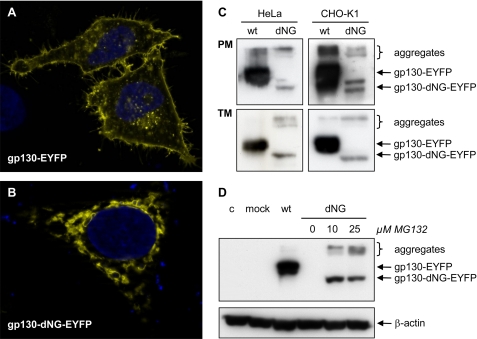
Fluorescence microscopy analyses of the localization of gp130-EYFP and gp130-dNG-EYFP in HeLa cells showed that, as expected, wild-type gp130-EYFP was mainly located in the plasma membrane (Fig. 5A). In contrast, gp130-dNG-EYFP largely remained within the ER·Golgi complex (Fig. 5B). This was confirmed by Western blot analyses of plasma membrane protein fractions of transiently transfected HeLa and CHO-K1 cells (Fig. 5C). The complete transmembrane protein fraction (including transmembrane proteins in the ER·Golgi complex) also showed significantly reduced levels of gp130-dNG(-EYFP) as compared with gp130(-EYFP), indicating strong and fast degradation (Fig. 5C).
FIGURE 5.
Localization and degradation of gp130-dNG. A and B, fluorescence micrographs of gp130(-dNG)-EYFP in HeLa cells. Whereas wild-type (wt) gp130-EYFP is localized on the cell surface (A), gp130-dNG-EYFP largely remains within the ER·Golgi complex (B). C, Western blots of plasma membrane proteins (PM) and of the complete transmembrane protein fraction (TM) from HeLa and CHO-K1 cells transiently transfected with gp130-EYFP or gp130-dNG-EYFP (detection by anti-EYFP). D, Western blots of denatured whole cell protein extracts from CHO-K1 cells transiently transfected with gp130-EYFP or gp130-dNG-EYFP (detection by anti-EYFP). Proteasome inhibition by MG132 significantly increased protein levels of gp130-dNG-EYFP (normalized by probing the stripped blot membrane with anti-β-actin). Lane 4 (dNG without MG132) does not show a signal due to short exposure time. c, control; mock, only expression vector pcDNA-DEST40. Controls for the experiments shown in C and D included Western blots with anti-gp130 (sc-655, directed against the intracellular domain of gp130) and confirmation of equal protein loading and transfer by Ponceau S staining (data not shown).
Consistently, proteasome inhibition by MG132 (benzyloxycarbonyl-Leu-Leu-Leu-al) significantly increased gp130-dNG(-EYFP) protein levels (Fig. 5D). A dose of 10 μm MG132 was sufficient to reach a maximum effect (Fig. 5D). In some experiments, 10 μm MG132 even increased gp130-dNG(-EYFP) expression to similar levels as observed with wild-type gp130(-EYFP) (data not shown).
Expression and Functionality of gp130-dNG in Transduced BAF3 Cell Pools
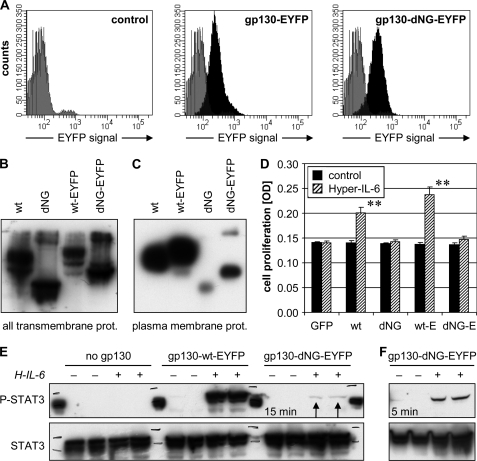
To avoid clone-specific artifacts in expression or signal transduction, stably transduced BAF3 cell pools were used for stimulation experiments and were thoroughly characterized. In contrast to transient expression in HeLa or CHO-K1 cells (Fig. 5, C and D), the BAF3 pools permanently produced gp130-dNG(-EYFP) to a similar if not higher extent as compared with its wild-type counterpart (Fig. 6, A and B). Flow cytometry showed very similar and homogeneous expression of gp130-EYFP and gp130-dNG-EYFP in the respective BAF3 pools (Fig. 6A). This was confirmed by microscopic counting of EYFP-positive cells (>95%; data not shown) and Western blot analyses of detergent extracts containing all transmembrane proteins, including those in the ER·Golgi complex (Fig. 6B).
FIGURE 6.
Characterization of gp130-dNG expression and function in stably transduced BAF3 cell pools. A, flow cytometry analysis of total gp130-EYFP and gp130-dNG-EYFP expression (fluorescein isothiocyanate/GFP channel; black) versus the nonspecific signal of untransduced BAF3 cells (gray). B and C, Western blot detection of wild-type (wt) gp130(-EYFP) and gp130-dNG(-EYFP) in complete transmembrane protein (prot.) fractions produced by detergent extraction (B) or plasma membrane protein fractions produced by surface biotinylation (C). In the experiments represented by B and C, gp130 was detected by an anti-gp130 (sc-655) directed against the intracellular domain of gp130. D, colorimetric cell proliferation assay of the BAF3 cell pools after stimulation with 10 ng/ml Hyper-IL-6 in the absence of the essential growth factor IL-3. E, EYFP tag; GFP, pool expressing green fluorescent protein but no gp130 (negative control); **, p < 0.01 versus unstimulated control. E, Western blot analysis of STAT3 activation (P, phosphorylation at Tyr705) in denatured whole cell protein extracts produced 15 min after stimulation with 10 ng/ml Hyper-IL-6 (H-IL-6) in the absence of IL-3 and after 6 h of serum starvation. BAF3 cells expressing no gp130 served as a negative control. After stripping, the blot membrane was probed with anti-STAT3 for normalization. Arrows mark weak phospho-STAT3 band in BAF3/gp130-dNG-EYFP extracts. F, STAT3 phosphorylation of BAF3/gp130-dNG-EYFP cells was determined as in E, but after a 5-min stimulation.
However, the amount of gp130-dNG(-EYFP), which finally reached the cell surface, was also extremely low in the BAF3 pools, as shown by Western blot analysis of plasma membrane protein fractions produced by surface biotinylation (Fig. 6C) and by flow cytometry analysis with antibodies directed against gp130 (data not shown). In contrast to the expression studies in CHO-K1 cells (Fig. 5D), increasing the transmembrane gp130-dNG levels by MG132 was no option in the BAF3 cell pools, as even short incubations (6 h) with very low concentrations (<1 μm) of MG132 (34) strongly reduced the STAT3 activation in response to Hyper-IL-6 and blocked cell proliferation (data not shown). Longer incubations with MG132 killed the BAF3 cells, even at 0.2 μm (data not shown).
When the BAF3 pools were stimulated with 10 ng/ml of the IL-6·sIL-6R fusion protein Hyper-IL-6 (or 100 ng/ml IL-6 plus 50 ng/ml sIL-6R; data not shown) in the absence of their normal essential growth factor IL-3, both the gp130- and the gp130-EYFP-transduced pools showed spontaneous proliferation (p < 0.01; Fig. 6D), whereas the pools expressing gp130-dNG or gp130-dNG-EYFP did not. In this experiment, the GFP-transduced BAF3 cell pool served as a negative control to exclude cytokine-independent proliferation as a result of viral transduction. Hyper-IL-6 was mainly used because it has been shown that Hyper-IL-6 bound to gp130 is less efficiently internalized, leading to longer lasting signaling activity of this protein on cells (35).
There were two plausible ways to explain this finding. (i) gp130 without N-glycans is inherently incapable of signal transduction. (ii) gp130-dNG(-EYFP) can transduce IL-6·sIL-6R signals, but its expression on the cell surface is too low to generate a sufficiently strong proliferation signal.
To address this question, we analyzed activation levels of the key gp130 target molecule, STAT3. We used the pools expressing the EYFP-tagged variants of gp130 and gp130-dNG for this purpose, as we consistently found a stronger proliferation response in the gp130-EYFP pool as compared with the pool expressing untagged gp130 (Fig. 6D). This could be due to the well known dimerization potential of EYFP (36, 37), which would further stabilize the gp130 dimers and thus increase gp130-dependent signaling. Fig. 6, B and C, shows that this observation did not result from a lower expression of untagged gp130.
Interestingly, the second explanation outlined above appears to be true, as BAF3/gp130-dNG-EYFP cells were indeed able to weakly phosphorylate STAT3 at Tyr705 upon stimulation with Hyper-IL-6 (Fig. 6, E and F). The intensity of the STAT3 phosphorylation (and thus activation) correlated with the amount of gp130-EYFP or gp130-dNG-EYFP expression on the BAF3 cell surface (Fig. 6, C and E), indicating that the few molecules of gp130-dNG(-EYFP), which finally do reach the cell surface, are capable of signal transduction. In contrast to wild-type gp130 (data not shown), activation of STAT3 in the BAF3/gp130-dNG-EYFP pool was more pronounced 5 min after stimulation than after the standard 15-min incubation (Fig. 6F). In summary, gp130 without N-glycans is functionally active and able to activate STAT3.
DISCUSSION
The first discovery of this study is that gp130 without N-glycans is inherently unstable, and most of it is degraded in the proteasome before it reaches the cell surface. Consistently, the production rate of the fusion protein sgp130Fc-dNG is less than 1% of a fully glycosylated sgp130Fc, and sgp130Fc-dNG is unstable during the affinity chromatography purification process involving acidic elution from protein A. Whereas sgp130Fc-dNG is too strongly damaged during its purification to be biologically active, the fraction of cellular gp130-dNG, which does reach the plasma membrane, is able to transduce the IL-6·sIL-6R signal, resulting in activation of STAT3.
These findings are in contrast to several studies reporting an essential role of N-glycosylation for ligand binding in diverse human receptors, e.g. epidermal growth factor receptor (2), the α-subunit of the receptor for human granulocyte-macrophage colony-stimulating factor (3), CXC chemokine receptor-4 (4), or formyl peptide receptor (5). Therefore, our data support the notion that N-glycans conferring stability to a receptor do not necessarily also play a role in ligand binding (1). The results of this study provide the first evidence that N-glycans on the ligand-binding domains D1–D3 of gp130 are indeed dispensable for ligand binding on living cells (23–25). The minimal surface expression (gp130-dNG) or secretion (sgp130Fc-dNG) due to proteosomal degradation is also in good agreement with the reduced yield of the gp130 ligand-binding domains D1–D3 or D2 + D3 in insect cells in the presence of tunicamycin or after mutational deletion of N-glycans (23–25).
Importantly, our data indicate that N-glycosylation of the domains D4 and D6 is not necessary for the formation of a functional gp130 signaling dimer. The three-dimensional orientation of the domains D4–D6 has been shown previously to be essential for signaling in response to IL-6·sIL-6R, even though they are not directly involved in ligand binding (26, 38).
Moreover, the identical apparent molecular weight differences of sgp130Fc-dNG, IgG1-Fc alone, or IgG1-Fc heavy chain before and after enzymatic removal of N-glycans by PNGase F indicate that the theoretical alternative N-glycosylation site in domain D4 (Asn368) is never used, even if all other N-glycosylation sites in gp130 are mutated (21). This is in agreement with a reduced likelihood of N-glycosylation at the motif Asn-Leu-Thr present at Asn368 (39). Finally, our enzymatic analyses of sgp130Fc suggest that O-glycosylation is not present or is minimal on gp130.
In summary, the results of this study show that N-glycosylation of gp130 is in principle dispensable for ligand binding both in vitro and in a cellular context, but it is essential for the structural stability and production yield of gp130 and gp130-based therapeutics, such as the fusion protein sgp130Fc.
Acknowledgments
We thank Tanja Kaacksteen and Melanie Schlapkohl for their expert technical assistance.
This work was supported in part by the German Excellence Initiative (Excellence Cluster “Inflammation at Interfaces”) and by Deutsche Forschungsgemeinschaft, Collaborative Research Center SFB415.
- IL
- interleukin
- LIF
- leukemia inhibitory factor
- IL-6R
- interleukin-6 receptor
- gp130
- glycoprotein 130
- sIL-6R
- soluble IL-6R
- dNG
- deleted N-glycosylation
- sgp130Fc
- soluble gp130-IgG1-Fc fusion protein
- STAT3
- signal transducer and activator of transcription-3
- EYFP
- enhanced yellow fluorescent protein
- FBS
- fetal bovine serum
- PNGase F
- peptide:N-glycosidase F
- HRP
- horseradish peroxidase
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- GFP
- green fluorescent protein
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Mitra N., Sinha S., Ramya T. N., Surolia A. (2006) Trends Biochem. Sci. 31, 156–163 [DOI] [PubMed] [Google Scholar]
- 2.Slieker L. J., Lane M. D. (1985) J. Biol. Chem. 260, 687–690 [PubMed] [Google Scholar]
- 3.Ding D. X., Vera J. C., Heaney M. L., Golde D. W. (1995) J. Biol. Chem. 270, 24580–24584 [DOI] [PubMed] [Google Scholar]
- 4.Zhou H., Tai H. H. (1999) Arch. Biochem. Biophys. 369, 267–276 [DOI] [PubMed] [Google Scholar]
- 5.Wenzel-Seifert K., Seifert R. (2003) Biochem. Biophys. Res. Commun. 301, 693–698 [DOI] [PubMed] [Google Scholar]
- 6.Meng J., Parroche P., Golenbock D. T., McKnight C. J. (2008) J. Biol. Chem. 283, 3376–3384 [DOI] [PubMed] [Google Scholar]
- 7.Isaji T., Sato Y., Fukuda T., Gu J. (2009) J. Biol. Chem. 284, 12207–12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato Y., Isaji T., Tajiri M., Yoshida-Yamamoto S., Yoshinaka T., Somehara T., Fukuda T., Wada Y., Gu J. (2009) J. Biol. Chem. 284, 11873–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishimoto T., Akira S., Narazaki M., Taga T. (1995) Blood 86, 1243–1254 [PubMed] [Google Scholar]
- 10.Hibi M., Nakajima K., Hirano T. (1996) J. Mol. Med. 74, 1–12 [DOI] [PubMed] [Google Scholar]
- 11.Hibi M., Murakami M., Saito M., Hirano T., Taga T., Kishimoto T. (1990) Cell 63, 1149–1157 [DOI] [PubMed] [Google Scholar]
- 12.Schroers A., Hecht O., Kallen K. J., Pachta M., Rose-John S., Grötzinger J. (2005) Protein Sci. 14, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow D., Ho J., Nguyen Pham T. L., Rose-John S., Garcia K. C. (2001) Biochemistry 40, 7593–7603 [DOI] [PubMed] [Google Scholar]
- 14.Rose-John S., Scheller J., Elson G., Jones S. A. (2006) J. Leukocyte Biol. 80, 227–236 [DOI] [PubMed] [Google Scholar]
- 15.Rose-John S., Heinrich P. C. (1994) Biochem. J. 300, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters M., Müller A. M., Rose-John S. (1998) Blood 92, 3495–3504 [PubMed] [Google Scholar]
- 17.Narazaki M., Yasukawa K., Saito T., Ohsugi Y., Fukui H., Koishihara Y., Yancopoulos G. D., Taga T., Kishimoto T. (1993) Blood 82, 1120–1126 [PubMed] [Google Scholar]
- 18.Jostock T., Müllberg J., Ozbek S., Atreya R., Blinn G., Voltz N., Fischer M., Neurath M. F., Rose-John S. (2001) Eur. J. Biochem. 268, 160–167 [DOI] [PubMed] [Google Scholar]
- 19.Rose-John S., Waetzig G. H., Scheller J., Grötzinger J., Seegert D. (2007) Expert Opin. Ther. Targets 11, 613–624 [DOI] [PubMed] [Google Scholar]
- 20.Sprang S. R., Bazan J. F. (1993) Curr. Opin. Struct. Biol. 3, 815–827 [Google Scholar]
- 21.Moritz R. L., Hall N. E., Connolly L. M., Simpson R. J. (2001) J. Biol. Chem. 276, 8244–8253 [DOI] [PubMed] [Google Scholar]
- 22.Bravo J., Staunton D., Heath J. K., Jones E. Y. (1998) EMBO J. 17, 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow D., He X., Snow A. L., Rose-John S., Garcia K. C. (2001) Science 291, 2150–2155 [DOI] [PubMed] [Google Scholar]
- 24.Boulanger M. J., Bankovich A. J., Kortemme T., Baker D., Garcia K. C. (2003) Mol. Cell 12, 577–589 [DOI] [PubMed] [Google Scholar]
- 25.Boulanger M. J., Chow D. C., Brevnova E. E., Garcia K. C. (2003) Science 300, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 26.Skiniotis G., Boulanger M. J., Garcia K. C., Walz T. (2005) Nat. Struct. Mol. Biol. 12, 545–551 [DOI] [PubMed] [Google Scholar]
- 27.Yanagisawa M., Yu R. K. (2009) Biochem. Biophys. Res. Commun. 386, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabe B., Chalaris A., May U., Waetzig G. H., Seegert D., Williams A. S., Jones S. A., Rose-John S., Scheller J. (2008) Blood 111, 1021–1028 [DOI] [PubMed] [Google Scholar]
- 29.Tenhumberg S., Waetzig G. H., Chalaris A., Rabe B., Seegert D., Scheller J., Rose-John S., Grötzinger J. (2008) J. Biol. Chem. 283, 27200–27207 [DOI] [PubMed] [Google Scholar]
- 30.Jefferis R., Lund J., Pound J. D. (1998) Immunol. Rev. 163, 59–76 [DOI] [PubMed] [Google Scholar]
- 31.Waetzig G. H., Seegert D., Rosenstiel P., Nikolaus S., Schreiber S. (2002) J. Immunol. 168, 5342–5351 [DOI] [PubMed] [Google Scholar]
- 32.Fischer M., Goldschmitt J., Peschel C., Brakenhoff J. P., Kallen K. J., Wollmer A., Grötzinger J., Rose-John S. (1997) Nat. Biotechnol. 15, 142–145 [DOI] [PubMed] [Google Scholar]
- 33.Ketteler R., Glaser S., Sandra O., Martens U. M., Klingmüller U. (2002) Gene Ther. 9, 477–487 [DOI] [PubMed] [Google Scholar]
- 34.Mu T. W., Ong D. S., Wang Y. J., Balch W. E., Yates J. R., 3rd, Segatori L., Kelly J. W. (2008) Cell 134, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters M., Blinn G., Solem F., Fischer M., Meyer zum Büschenfelde K. H., Rose-John S. (1998) J. Immunol. 161, 3575–3581 [PubMed] [Google Scholar]
- 36.Kukolka F., Niemeyer C. M. (2004) Org. Biomol. Chem. 2, 2203–2206 [DOI] [PubMed] [Google Scholar]
- 37.Jung G., Ma Y., Prall B. S., Fleming G. R. (2005) Chemphyschem. 6, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 38.Kurth I., Horsten U., Pflanz S., Timmermann A., Küster A., Dahmen H., Tacken I., Heinrich P. C., Müller-Newen G. (2000) J. Immunol. 164, 273–282 [DOI] [PubMed] [Google Scholar]
- 39.Shakin-Eshleman S. H., Spitalnik S. L., Kasturi L. (1996) J. Biol. Chem. 271, 6363–6366 [DOI] [PubMed] [Google Scholar]