Abstract
Integrin αIIbβ3 affinity regulation by talin binding to the cytoplasmic tail of β3 is a generally accepted model for explaining activation of this integrin in Chinese hamster ovary cells and human platelets. Most of the evidence for this model comes from the use of multivalent ligands. This raises the possibility that the activation being measured is that of increased clustering of the integrin rather than affinity. Using a newly developed assay that probes integrins on the surface of cells with only monovalent ligands prior to fixation, I do not find increases in affinity of αIIbβ3 integrins by talin head fragments in Chinese hamster ovary cells, nor do I observe affinity increases in human platelets stimulated with thrombin. Binding to a multivalent ligand does increase in both of these cases. This assay does report affinity increases induced by either Mn2+, a cytoplasmic domain mutant (D723R) in the cytoplasmic domain of β3, or preincubation with a peptide ligand. These results reconcile the previously observed differences between talin effects on integrin activation in Drosophila and vertebrate systems and suggest new models for talin regulation of integrin activity in human platelets.
Keywords: Blood Coagulation, Cell/Adhesion, Extracellular Matrix/Integrin, Protein/Adhesion, Protein/Ligand Binding, Receptors/Extracellular Matrix, Receptors/Structure-Function, Regulation Allosteric
Introduction
Integrins are adhesive heterodimeric transmembrane proteins that bind to extracellular matrix ligands or to cell surface proteins on adjacent cells. The cytoplasmic tails of the integrins are linked directly, or via adaptors, to numerous cytoskeletal and signaling proteins and transmit signals from the outside of the cell to the inside. The adhesive properties of integrins are dynamically regulated as these receptors shift between different conformations upon binding to extracellular ligands or cytoplasmic proteins. Thus, integrins are present in high or low affinity states on the surface of cells depending on the cellular environment. Regulation of integrin activation is critical in controlling cell adhesion, migration, and extracellular matrix assembly. This regulation is therefore important in normal development, hemostasis, inflammation, angiogenesis, tumor cell metastasis, and immune responses (1–4).
Talin is one of the most intensively studied cytoplasmic activators of integrin activity (5, 6). The N-terminal globular head region of talin contains a FERM2 (band four-point-one, ezrin, radixin, moesin homology) domain that has the ability to bind to β3 integrin cytoplasmic tails, and this results in the activation of αIIbβ3 integrins. The ability of talin to interact with integrins is itself regulated as the FERM domain is autoinhibited by binding to its C-terminal tail in an intra- or intermolecular manner (2, 3, 7, 8). Autoinhibition of talin can be removed by the Rap1 effector Rap1-GTP-interacting adaptor molecule in response to extracellular cues (9). Experimentally, the autoinhibition can be removed by expressing the talin head (or FERM) domain in the absence of the inhibitory tail. Overexpression of the talin head or FERM domain activates the β3 and β1 integrins, and inhibition of talin expression reduces integrin activity in mammalian cell culture (5, 10).
Intriguingly, recent experiments have not been able to detect an effect of talin FERM domain expression or reduction in talin expression on the activation state of the Drosophila PS2 integrins. This lead to speculation of a fundamental difference between Drosophila talin-PS2 integrin interactions and those observed for talin-αIIbβ3 in vertebrates (11). However, the methodologies used to measure integrin affinity for ligand in these two studies were fundamentally different. In this report, I find that when identical binding assays are conducted for αIIbβ3 integrins, as were used for the Drosophila PS2 integrins, talin has no effect on the affinity of αIIbβ3 for monovalent ligand. Thus, I find no difference between the Drosophila talin-PS2 integrin interactions and those seen for vertebrate talin-αIIbβ3 integrin.
I do find that the talin FERM domain increases the ability of αIIbβ3 to bind to multivalent ligands, and this appears to be the source of discrepancy between the binding assays. Numerous studies have shown that a clustering mechanism activates β3 integrins. In platelets, avidity is an important component of αIIbβ3 binding to ligand as PAC-1 IgM showed 60-fold greater binding capacity as compared with the PAC-1 Fab even in the presence of secondary antibodies (12). Ligand binding, talin head expression, and agonist-induced activation in CHO cells all result in integrin clustering (13–15).
A clear understanding of αIIbβ3 activity regulation that distinguishes affinity from clustering effects is important as it may impact the development of therapeutic agents designed to modulate integrin activity to treat pathologies involving inflammation, thrombosis, angiogenesis, and tumor progression. If integrin activation or integrin activity is a general term that encompasses clustering and/or affinity changes, then clearly talin activates αIIbβ3 integrins in CHO cells. However, integrin activation is commonly interpreted to mean an increase in the affinity of an individual integrin heterodimer for ligand prior to encountering ligand (3). It is this latter definition of integrin regulation by talin that is found in at least one cell biology textbook that states “Thus when talin binds to the β chain it undoes the intracellular α-β linkage, allowing the two legs of the integrin molecule to spring apart. This drives the extracellular portion of the integrin into its extended, active conformation.” (16) My results in CHO cells and additional experiments in human platelets contradict this view of integrin affinity regulation by talin and point instead to the role of talin in integrin clustering.
EXPERIMENTAL PROCEDURES
Antibodies
PAC-1 IgM (17) was purchased from BD Biosciences (catalog number 340535). PAC-1 Fab was expressed as a His-tagged fusion protein expressed by cultured Drosophila cells (18) and prepared as described for TWOW-1 (19). R-Phycoerythrin-labeled or unlabeled HIP8 (Pharmingen catalog numbers 555467 and 555465) was used to quantify αIIbβ3 levels in CHO cells or platelets. This antibody gave similar results in CHO cells and platelets as the conformation-insensitive monoclonal antibody SSA6 (a generous gift from Sandy Shattil). The secondary antibody used to detect PAC-1 and unlabeled HIP8 was R-phycoerythrin-conjugated goat anti-mouse IgG (Invitrogen catalog number P852). Alexa Fluor 568 goat anti-mouse IgG (Invitrogen catalog number A11031) was used for microscopy of PAC-1 bound to CHO cells.
Cell Culture
CHO cells stably transformed to express human integrin αIIbβ3 (A5 cells) or constitutively active αIIbβ3(D723R) have been described (20, 21). CHO cells were grown in Dulbecco's modified Eagle's medium (Mediatech catalog number 15-013-CV) supplemented with essential amino acids (Invitrogen catalog number 11140), penicillin-streptomycin-glutamine (Invitrogen catalog number 10378-016), and 10% fetal bovine serum (FBS; Sigma catalog number F6178). For PAC-1 binding experiments, cells were thawed and used within three passages. Outdated platelets were obtained from the University of Arizona blood bank and were used within 2 days.
Transfections
Transfections were done using Lipofectamine 2000 reagent (Invitrogen) as recommended by the supplier. Cells were seeded at 2.5 × 105 cells/ml in 60-mm tissue culture plates (4.5 ml/plate). On the following day, the medium was replaced with the same volume of medium containing FBS but lacking antibiotics. 30 μl of Lipofectamine-2000 was mixed with 500 μl of serum-free medium and allowed to incubate for 5 min and then mixed with 500 μl of serum-free medium containing 6 μg of plasmid DNA. After 20 min, this was added directly to the cells. On the following day, the transformed cells were rinsed with phosphate-buffered saline (PBS) (0.8 mm KH2PO4, 5.6 mm Na2HPO4, 154 mm NaCl, pH 7.4) and trypsinized (Mediatech trypsin EDTA catalog number 25-052-Cl) for 2–3 min. Trypsinization was stopped with the addition of medium containing FBS. Following one wash, the cells were diluted in 10 ml of fresh medium containing FBS and antibiotics in a 100-mm tissue culture plate. Cells were used in binding experiments 48 h after transfection. Plasmids used for transfections were either a GFP-murine talin head F2-F3 domain (amino acids 206–405) chimera or an empty vector expressing only GFP (10, 22).
PAC-1 Binding
CHO cells expressing integrins were rinsed with PBS and trypsinized (Mediatech Cellgro trypsin EDTA catalog number 25-052-Cl) for 2–3 min. Trypsinization was stopped with the addition of medium containing FBS. Cells were centrifuged and washed with medium and then with PBS. 5 × 105 cells were then resuspended in 30 μl of Tyrode's buffer (12.1 mm NaHCO3, 5 mm HEPES, 137 mm NaCl, 2.6 mm KCl, and 5.6 mm glucose) containing 1 mg/ml bovine serum albumin (BSA) and 1.66 mm CaCl2 and MgCl2; 1.66 mm CaCl2, MgCl2, and MnCl2; or 8.3 mm EDTA. 20 μl of PAC-1 IgM or PAC-1 Fab was added, yielding the final concentrations of 1 mm Ca2+, 1 mm Mg2+, 1 mm Mn2+, and 5 mm EDTA. PAC-1 IgM binding was performed using a standard protocol (23). PAC-1 IgM was incubated with cells for 30 min at room temperature. Cells were washed by adding 1.5 ml of Tyrode's buffer with appropriate divalent cations or EDTA followed by centrifugation. Cells were then resuspended in 50 μl of R-phycoerythrin-conjugated secondary antibody (10 μg/ml) in Tyrode's buffer containing 1 mg/ml BSA. After a 25-min incubation on ice, cells were diluted with ice-cold PBS and analyzed immediately by flow cytometry. PAC-1 Fab binding was done basically as described (19) with slight modifications of the buffer so that it was identical to that used for PAC-1 IgM binding. A similar protocol has been used to measure the affinity of LFA-1 and its ligand ICAM-1 (24). Cells were incubated as above with PAC-1 Fab for 10 min followed by the addition of 50 μl of 4% formaldehyde in Tyrode's buffer to fix bound PAC-1 to the cells. Following a 5-min fixation, cells were diluted by the addition of 1.5 ml of Tyrode's buffer. Cells were collected by centrifugation and resuspended in 50 μl of R-phycoerythrin-conjugated secondary antibody (10 μg/ml) in Tyrode's buffer containing 1 mg/ml BSA. After a 25-min incubation, on ice, cells were collected by centrifugation and resuspended in 0.5 ml of PBS containing 2% formaldehyde. To determine αIIbβ3 integrin expression levels, 5 × 105 cells were also incubated for 30 min with 50 μl of R-phycoerythrin-labeled HIP8 (diluted 1:1 with PBS). All centrifugations for CHO cells were done for 2 min at 1,000 × g.
For each experiment, phycoerythrin fluorescence levels for 1,000–5,000 strongly GFP-positive cells were analyzed by flow cytometry. To determine integrin-dependent binding, I subtracted nonspecific binding: the amount observed when divalent cations had been removed (by EDTA). Integrin-dependent mean fluorescence intensity (MFI) of PAC-1 binding was divided by MFI of HIP8 binding, in the strongly GFP-positive cells, thereby allowing adjustment for differences in expression levels between samples, which were 25% or less. Significant differences in binding are given as p values using Student's t test.
Platelet Activation and PAC-1 Binding
For thrombin activation, platelets were collected by centrifugation and resuspended at 2.9 × 108 cells/ml in Tyrode's buffer containing 1 mm MgCl2, 1 mg/ml BSA. Thrombin (Sigma T7513) from a 100 units/ml frozen stock was added to a final concentration of 0.5 units/ml to activate the platelets. Platelets were incubated for 30 min at room temperature with intermittent vigorous pipetting to reduce aggregation. 35 μl of cells (1 × 107) was added to tubes containing 5 μl of Tyrode's buffer with 1 mg/ml BSA and 10 mm CaCl2, 3 mm MgCl2; 10 mm CaCl2, 3 mm MgCl2, 10 mm MnCl2; or 50 mm EDTA. Control platelets were treated identically but without the addition of thrombin. The vigorous pipetting was necessary for only the thrombin-activated platelets to reduce their aggregation. This treatment did not inadvertently activate the platelets in the absence of thrombin as determined by microscopic and flow cytometry analysis or the levels of surface fibrinogen (supplemental Fig. S2).
For RGD activation of integrins, platelets were collected by centrifugation and resuspended at 2 × 108 cells/ml in Tyrode's buffer containing 1 mg/ml BSA, 1 mm MgCl2, 1 mm CaCl2. GRGDSP, GRGESP (AnaSpec, Inc. catalog numbers 22945 and 22949), or no peptide was added to a final concentration of 1 mm and incubated for 5 min at room temperature. Cells were then fixed by the addition of an equal volume of Tyrode's buffer containing 1 mm MgCl2, 1 mm CaCl2, 4% formaldehyde. After 5 min of fixation, the fix was diluted by the addition of 30 volumes of Tyrode's buffer (containing 1 mm MgCl2, 1 mm CaCl2 or 5 mm EDTA). 1.5 ml of cells (5 × 106 cells) was centrifuged and resuspended in Tyrode's buffer containing 1 mg/ml BSA and 1.25 mm CaCl2, 1.25 mm MgCl2; 1.25 mm CaCl2, 1.25 mm MgCl2, 1.25 mm MnCl2; or 6.25 mm EDTA.
For PAC-1 binding to thrombin-activated platelets (or their controls), 10 μl of PAC-1 (IgM or Fab) was added to the above tubes, and they were incubated for 10 min at room temperature. Final concentrations of divalent cations were all 1 mm, and EDTA was 5 mm. Bound PAC-1 was fixed to the platelets by the addition of 50 μl of Tyrode's buffer containing 4% formaldehyde. Following 5 min of fixation at room temperature, the fix was diluted by the addition of 1.5 ml of Tyrode's buffer. Fixed platelets were collected by centrifugation and resuspended in 50 μl of R-phycoerythrin-conjugated secondary antibody (10 μg/ml) in Tyrode's buffer containing 1 mg/ml BSA. After a 25-min incubation on ice, cells were collected by centrifugation and resuspended in 0.5 ml of PBS containing 2% formaldehyde.
10,000 platelets were analyzed by flow cytometry for each experiment. As for CHO cells, nonspecific binding (in the presence of EDTA) was subtracted, and PAC-1 values were divided by total integrin expression (as assessed by HIP8 staining) to adjust for any expression differences or increased values due to aggregating platelets. For the platelet experiments, unlabeled HIP8 binding was done identically to PAC-1 binding in the presence Ca2+ and Mg2+: the same times, fixations, and detection with the same secondary antibody. HIP8 concentration in these experiments was 2 μg/ml. As for CHO cell binding experiments, tests for significant differences in binding are given as p values using Student's t test.
Immunofluorescence
CHO cells expressing GFP-Talin F2-F3 were processed as in the PAC-1 IgM binding assay through removal of unbound PAC-1. Bound PAC-1 was then fixed on the cells with formaldehyde for 5 min. Following washing to remove the formaldehyde, cells were allowed to attach to glass slides and fixed to the slides with formaldehyde, and then PAC-1 was detected using Alexa Fluor 568 goat anti-mouse IgG secondary antibody (2 μg/ml in Tyrode's buffer containing 1 mg/ml BSA). Following washing, cells were mounted in VECTASHIELD and examined on a DeltaVision microscope using a ×100, 1.4 NA Olympus UPlanSApo objective. The z-series was deconvolved and quick-projected using SoftWoRx (Applied Precision). All exposure times and processing were held constant (with the exception of the number of optical slices). The National Institutes of Health ImageJ software was used to determine particle sizes of fluorescent clusters. Particle sizes for unclustered PAC-1 IgM antibodies were determined by imaging PAC-1 fixed to glass slides in the absence of cells. The threshold intensity for positive pixels was set at ∼2× background (minimum pixel intensity) and was held constant for all images analyzed. Particle (cluster) areas and intensities are given as the mean ± S.E.
RESULTS
Monovalent Fixation Assay Reports on αIIbβ3 Affinity
Talin head domain or its F2-F3 subdomain has been demonstrated to increase PAC-1 binding to αIIbβ3 expressed in CHO cells (5, 22). The assays used to demonstrate integrin αIIbβ3 binding affinity regulation utilized the multivalent IgM ligand mimetic antibody, PAC-1. This antibody contains the integrin binding motif Arg-Tyr-Asp (RYD) in each of its 10 complementary determining region 3 areas (H-CDR3s) (25). Alternatively, the binding assays used monovalent ligands, such as PAC-1 Fab (with only one integrin binding motif). In both assays, binding capacity was assessed by the addition of labeled polyclonal secondary antibodies that bind to the PAC-1. Binding levels were then measured by flow cytometry without fixation (6, 12, 18, 23). One potential artifact in the monovalent binding assay is that PAC-1 Fab and secondary antibodies were present at the same time during binding, and this likely produced a clustered multivalent PAC-1 ligand. Using a modified assay that I call the monovalent fixation assay, which does test integrin affinity for monovalent ligands (19), we were unable to detect any effect of talin on Drosophila PS2 integrin affinity for ligand (11). I therefore set out to test whether the monovalent fixation assay detects an increase in αIIbβ3 affinity by Talin F2-F3 in CHO cells.
In the monovalent fixation assay, PAC-1 Fab binding to cells was achieved by incubating the cells with PAC-1 Fab for 10 min. Bound monovalent PAC-1 was then formaldehyde-fixed to the cells. Unbound PAC-1 was washed away, and the bound PAC-1 was detected using a labeled secondary antibody and flow cytometry. Staining for total integrin levels was used to adjust for expression levels that may vary between experiments or cell lines. Using this assay, PAC-1 Fab bound in a dose-dependent manner to CHO cells expressing αIIbβ3 integrins (Fig. 1). To determine whether the monovalent fixation assay reports on previously described activators of integrin affinity, I also determined binding curves for cells expressing an integrin containing a β cytoplasmic domain-activating mutation, αIIbβ3(D723R) (21). Also, the divalent cation Mn2+ is a well known integrin activator (26–28), and I asked whether its presence in the binding assay increased monovalent ligand binding to wild type αIIbβ3 and αIIbβ3(D723R) integrins expressed in CHO cells. αIIbβ3(D723R) confers a significant increase in affinity in the absence and in the presence of Mn2+ (Fig. 1). The ability of Mn2+ to activate the integrins is much greater than the cytoplasmic domain mutation and is additive with it. This suggests that neither the mutation nor Mn2+ alone fully activates the integrins (Figs. 1 and 3). This is entirely consistent with observations on the PS2 integrins in Drosophila cells (19). Importantly, these results demonstrate the validity of the monovalent fixation assay for reporting on the affinity state of αIIbβ3 integrins.
FIGURE 1.
PAC-1 Fab binding to CHO cells expressing αIIbβ3 and αIIbβ3(D723R). PAC-1 Fab binding levels were determined for cells expressing wild type αIIbβ3 (solid lines) or αIIbβ3(D723R) (dashed lines) in the absence (solid squares) or presence of Mn2+(open circles). In this and subsequent figures, binding is expressed as a ratio of specific PAC-1 immunofluorescence (PAC-1 MFI) over total integrin detected by the αIIbβ3 antibody HIP8 (HIP8 MFI). Specific PAC-1 binding is the total PAC-1 immunofluorescence minus that seen in the presence of EDTA. Values shown are the mean ± S.E. (error bars) from 3 independent experiments.
FIGURE 3.
β3(D723R) increases αIIbβ3 binding to PAC-1 IgM and Fab. Binding of PAC-1 IgM (10 μg/ml) was followed by washes and analysis of live unfixed cells. Binding of PAC-1 Fab (20 μg/ml) was done with a fixation step prior to washing. Values are as in Fig. 1 but are the mean ± S.E. (error bars) for 4 and 5 experiments for the IgM and Fab samples, respectively. The difference between PAC-1 Fab binding to the cell lines expressing wild type (WT) β3 and β3 (D723R) (D>R) is significant (p = 0.011).
αIIbβ3 Binding to Multivalent but Not Monovalent Ligand Is Regulated by Talin in CHO Cells
To determine the contributions of affinity and clustering on integrin regulation by talin, I measured multivalent ligand binding using a standard PAC-1 IgM binding assay and measured monovalent ligand binding using the monovalent fixation binding assay. In both cases, CHO cells expressing human αIIbβ3 were transiently transfected to express either the GFP-tagged activating talin head fragment (GFP-Talin F2-F3) or GFP alone. Similar to previous reports (10, 22), I found that cells expressing GFP-Talin F2-F3 bind more multivalent PAC-1 IgM than cells expressing GFP (Fig. 2A). Surprisingly, I found no difference between cells expressing the GFP-Talin F2-F3 or GFP in their ability to bind PAC-1 Fab (Fig. 2A). The levels of PAC-1 Fab used in this experiment (20 μg/ml) result in binding to ∼15% of the available integrins (Fig. 1) and were chosen to give similar binding levels as compared with PAC-1 IgM (used at a typical concentration of 10 μg/ml). Preliminary experiments at higher concentrations of PAC-1 Fab (up to 300 μg/ml) also found binding to be unaffected by the presence of GFP-Talin F2-F3 (supplemental Fig. S1). Taken together, my results suggest that talin head expression increases the clustering and avidity but not the affinity of αIIbβ3 for PAC-1.
FIGURE 2.
Talin increases αIIbβ3 binding to multivalent PAC-1 IgM but not monovalent PAC-1 Fab. A, binding of PAC-1 IgM (10 μg/ml) to CHO cells expressing αIIbβ3 is increased when the cells also express GFP-talin head F2-F3 domain (TH-GFP) as compared with cells expressing GFP alone (p = 0.0001). Binding of PAC-1 Fab (20 μg/ml) was not increased (p = 0.79). B, activation of αIIbβ3 by Mn2+ resulted in increased binding of both forms of PAC-1 (p = 0.0001 for both). Binding of PAC-1 IgM was followed by washes and analysis of live unfixed cells, whereas binding of PAC-1 Fab was done with a fixation step prior to washing. Values are as in Fig. 1 but are the mean ± S.E. (error bars) from 6 experiments.
To begin to determine the relative contributions of affinity and clustering on integrin regulation by the artificial activators Mn2+ and the β3(D723R)-activating mutation, I compared their abilities to increase binding to multivalent and monovalent PAC-1. I found that Mn2+ strongly increases binding to both ligands (Fig. 2B), and therefore, the increase in binding could largely be explained by increased affinity. αIIbβ3(D723R) bound significantly more PAC-1 Fab than wild type αIIbβ3, and this increase was much more pronounced for the multivalent ligand PAC-1 IgM (Fig. 3). This suggests that the β3(D723R) mutation increases both affinity and the clustering of αIIbβ3.
PAC-1 IgM Binding to αIIbβ3 Is Clustered
To determine whether the PAC-1 IgM bound to cells is clustered, I examined PAC-1 IgM on the surface of cells by epifluorescence microscopy (Fig. 4A). Bound PAC-1 IgM appears to be clustered. As a control for fluorescent particles resulting from PAC-1 IgM binding to secondary antibodies in the absence of a cellular context, I examined PAC-1 IgM attached to glass (Fig. 4B). The fluorescence on slides was due to PAC-1 IgM as examination of slides not first incubated with PAC-1 IgM showed a reduction of 99.6% in particle number (not shown).
FIGURE 4.
PAC-1 IgM binding to CHO cells is clustered. PAC-1 IgM bound to CHO cells expressing αIIbβ3 and GFP-talin head F2-F3 domain was fixed, labeled with fluorescent secondary antibodies, and visualized by epifluorescence microscopy. A, maximum intensity projection image of a cell showing clustered PAC-1 IgM on the cell surface. PAC-1 IgM fixed to a glass slide and detected with secondary antibodies show significantly less clustering (B).
The apparent clustered binding was confirmed by quantitative analysis of particle size using the National Institutes of Health ImageJ software. Fluorescent particles from four fields (total of 800 particles) of PAC-1 IgM on glass found none greater in area than 0.2 μm2 and only 3 ± 0.6% greater than 0.1 μm2. For PAC-1 bound to cells, 18% of the particles (clusters) examined on 11 cells (total of 1,059 particles) were greater in area than 0.2 μm2. This population of clusters was responsible for an average of 55 ± 6% (range 24–82%) of the total fluorescent area and 59 ± 7% (range 29–89%) of the fluorescence intensity in particles on the cell surface. Thus, PAC-1 IgM bound to CHO cells expressing αIIbβ3 and GFP-Talin F2-F3 was clustered in the typical affinity assay.
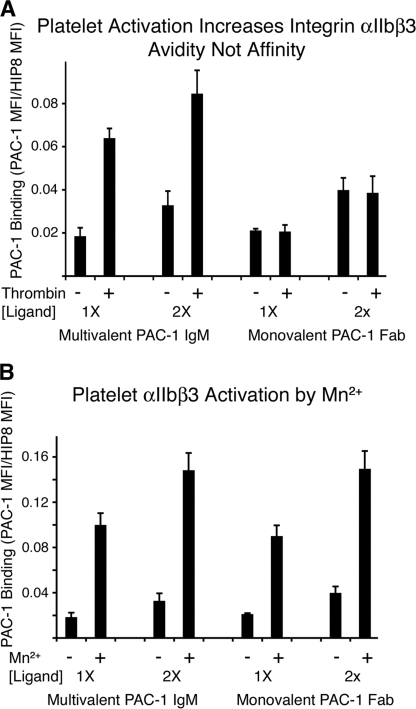
αIIbβ3 Binding to Multivalent but Not Monovalent Ligand Is Increased by Thrombin Activation of Human Platelets
To ask whether my findings in CHO cells are relevant to what actually occurs in human platelets, where regulation of αIIbβ3 binding to ligands is biologically relevant to the process of hemostasis, thrombosis, and inflammation, I examined αIIbβ3 binding to monovalent and multivalent ligands in thrombin-activated human platelets. To probe contributions of affinity and clustering on the increase in αIIbβ3 activity that occurs upon platelet activation, thrombin-activated platelets were compared with resting (non-activated) platelets for their binding to either PAC-1 IgM or PAC-1 Fab at two different ligand concentrations for each. Binding to PAC-1 (IgM or Fab) was followed directly by fixation, and then unbound PAC-1 was removed by washing. Bound PAC-1 was then detected by labeled secondary antibody and flow cytometry. Surprisingly, thrombin activation increased multivalent PAC-1 IgM binding but not monovalent PAC-1 Fab binding (Fig. 5A). Again, Mn2+ was able to increase the affinity of the integrin as measured by monovalent PAC-1 binding (Fig. 5B). Thus, my results indicate that in platelets, as in CHO cells, αIIbβ3 regulation is at the level of their ability to bind multivalent but not monovalent ligands.
FIGURE 5.
Thrombin activation of human platelets increases the binding of multivalent PAC-1 IgM but not monovalent PAC-1 Fab. A, binding of the ligands PAC-1 IgM and PAC-1 Fab, at two different concentrations each, was stopped by the addition of fixative after 10 min and then detected by secondary antibodies. B, activation of αIIbβ3 by Mn2+ increases binding of both forms of PAC-1. PAC-1 IgM 1× and 2× concentrations were 10 and 20 μg/ml. PAC-1 Fab 1× and 2× concentrations were 37.5 and 75 μg/ml. Values are as in Fig. 1 but are the mean ± S.E. (error bars) from 3 experiments.
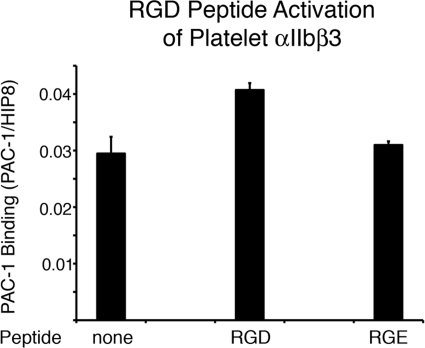
The binding of integrins to their ligands increases their affinity for ligands, in vitro, and results in changes in conformation as detected by ligand-induced binding site antibodies (29–31). Therefore, I next tested whether the affinity of αIIbβ3 for monovalent PAC-1 could be modulated by prior ligand binding. Platelets were incubated with no ligand, a GRGDSP peptide ligand (RGD is a recognized integrin binding motif), or a GRGESP control peptide and then briefly fixed with formaldehyde. The platelets were then washed to remove the fixative as well as unbound peptides. Those bound peptides, possessing only one reactive amine group at the N terminus, are unlikely to be fixed to the integrin and are also removed during washing. The fixed integrins on the surface of the platelets were tested for ligand binding in the monovalent fixation assay. I found that binding of monovalent PAC-1 was increased by the GRGDSP peptide but not by the GRGESP peptide (Fig. 6). Thus, I confirmed that ligand binding increases integrin affinity.
FIGURE 6.
RGD peptide binding increases αIIbβ3 binding to PAC-1 Fab. Platelets were incubated without peptide or with 1 mm GRGDSP (RGD) or GRGESP (RGE) peptide, fixed, and then used in a standard binding assay for PAC-1 Fab. Preincubation with the RGD peptide resulted in increased PAC-1 Fab binding. Binding assays were done using 50 μg/ml PAC-1 Fab, and the values given are the mean ± S.E. (error bars) from 4 experiments. Differences between RGD and no peptide or RGD and RGE are significant (p = 0.013 and p = 0.0004, respectively).
DISCUSSION
Talin has been demonstrated to be a key regulator of integrin adhesive activity both in whole organisms and in cell culture models (5, 6, 32–35). Regulating integrin activity can occur by either changing the affinity of individual integrins for their ligands and/or clustering of integrins, thereby altering their ability to bind multivalent ligands.
αIIbβ3 Affinity Versus Clustering
My goal has been to distinguish between changes in integrin-ligand affinity versus avidity (clustering plus affinity) on the surface of living cells. Monovalent ligands, like PAC-1 Fab, in solution with cells, can probe the affinity of individual integrins for individual ligands. Simply clustering the integrins without changing their individual affinities should not affect their ability to bind a monovalent ligand. Multivalent ligands, like PAC-1 IgM, are able to detect clustering of integrins even in the absence of affinity changes. Each individual multivalent ligand can make multiple interactions with multiple integrins when these receptors are in close proximity (clustered), and this results in increased binding. When the integrins are not in close proximity (unclustered), multiple interactions are not possible, and reduced binding results. The binding of multivalent ligands will also be altered by changes in integrin affinity, and initial binding of the multivalent ligand can promote clustering of the integrins. The combination of affinity and clustering is often referred to as avidity. By using both types of assays, monovalent and multivalent, I can distinguish between affinity and avidity regulation of integrins.
Using a newly developed binding assay for monovalent ligands that exposes integrins solely to the ligand in its monovalent state, we have reported on affinity increases in the Drosophila PS2 integrin due to the presence of Mn2+, integrin cytoplasmic and extracellular point mutations, and deletion of a βPS integrin plexin-semaphorin-integrin domain (19). Here I show that this same assay reports αIIbβ3 integrin affinity modulation by Mn2+, a β3 cytoplasmic domain mutation, and ligand-induced activation. All of these results validate the ability of the monovalent fixation assay to detect affinity differences in Drosophila and vertebrate integrins.
In contrast to previously reported results, I do not find increased binding of monovalent ligand (PAC-1 Fab) to αIIbβ3 integrins in the presence of high levels of talin head domains in CHO cells. The same cells did show increased binding to the same ligand presented in a multivalent state (PAC-1 IgM) in the traditional binding assay. Using the monovalent fixation assay, I also do not find increased affinity of αIIbβ3 on human platelets activated with thrombin. Despite being able to detect numerous activating conditions, I have considered that there might be an artifact of this assay, such as the presence of formaldehyde during the binding phase, that renders it unable to detect integrin activity differences resulting from platelet activation. To test this, I used the multivalent PAC-1 IgM in the exact same assay, including the presence of formaldehyde during the binding phase. This ligand did demonstrate an activation-dependent increase in binding. Thus, the monovalent fixation assay does not do away the ability to detect integrin activity increases upon activation; only changing the valency of the ligand does that. Therefore, I propose that talin increases integrin activity, both in CHO cell culture model assays and in human platelets, by increasing their clustering but not, directly, their affinity.
Differences between the Monovalent Fixation Assay and Those Previously Used
The monovalent fixation assay differs markedly from the ones currently used (monovalent plus multivalent secondary) to detect monovalent PAC-1 ligand binding to αIIbβ3, and I suggest that this difference is the cause of my different results in the cases of talin and thrombin activation of αIIbβ3 integrins in CHO cells and platelets. In the commonly used monovalent plus multivalent secondary assay, PAC-1 Fab fragment is incubated with cells for 15–30 min at room temperature, and then polyclonal secondary antibodies are added, and the incubation continues for an additional 20–30 min on ice. This is typically followed by a wash step and analysis by flow cytometry of the live cells. I have a number of concerns regarding this procedure. Most importantly, as soon as the secondary antibodies are added to the binding reaction, the PAC-1 Fab ceases to be a monovalent ligand. It is then capable of reporting on clustering as well as affinity changes. Although multivalent, the exact nature of the newly created ligand will depend on the exact concentration of the PAC-1 Fab and the polyclonal secondary antibodies and where the secondary antibodies bind to the PAC-1 Fab. Therefore, it is not expected that this ligand will behave exactly the same as the PAC-1 IgM in all binding assays.
That multivalency is a component of the standard monovalent ligand secondary assay is illustrated by the fact that in all the reports based on this assay, PAC-1 Fab (and secondary antibody) remains bound even following a wash and/or dilution of live cells prior to flow cytometry that removes most or all unbound PAC-1 Fab. If unbound and bound PAC-1 Fab were truly in equilibrium with the integrins, then washes bringing the unbound PAC-1 concentration essentially to 0 should have resulted in dissociation of the bound ligand from the integrins. That it did not demonstrates that the reactions were not at equilibrium and therefore were not measuring the affinity of the integrins at that time. In preliminary experiments with Drosophila cells, I have found that washing the cells after binding of a monovalent ligand to PS2 integrins (without adding secondary antibodies) resulted in dissociation of the ligand (data not shown). Therefore, the monovalent ligand must be fixed to the integrins prior to washing.
In the case of activated platelets, additional complications suggest that previous binding assays were not a reliable measure of the affinity of integrins prior to ligand binding. On the surface of activated platelets, the binding of ligands to αIIbβ3 is a multiphasic, energy-dependent process where ligand binding becomes irreversible over 15–30 min (36–38). Time-dependent irreversibility has also been seen in CHO cell experiments measuring αIIbβ3 binding to fibrinogen or PAC-1 (18). As the commonly used assays involve binding times of 15–30 min at room temperature followed by a 30-min incubation with secondary antibodies on ice, time-dependent changes make it difficult to determine what the affinity of the integrin is prior to ligand binding at a time when talin is proposed to increase integrin affinity. I suggest that a binding time of 10 min followed directly by fixation gives a more accurate measure of the effects of talin on early integrin-ligand interactions.
The prediction of low affinity interactions between PAC-1 and αIIbβ3 integrins that have not been activated (presumably of low affinity and not clustered) is seen in the monovalent fixation assay because the bound ligand is fixed to the integrins prior to washing (Fig. 1). In the common monovalent plus multivalent secondary or multivalent IgM assays, almost no binding is detected. This is exactly what would be expected for assays that involve a wash that removes unbound ligand in the absence of integrin clustering, as in CHO cells expressing only αIIbβ3 integrins or non-activated platelets. Even dimerizing the low affinity integrins resulted in their binding to the PAC-1 IgM and fibrinogen (39). This increase in binding was seen even though the integrins did not show an increased exposure to multiple conformation reporting antibodies, demonstrating that low affinity binding to soluble ligands does occur. This assay did not find binding to PAC-1 Fab under dimerizing conditions, suggesting that integrin affinity had not been increased. I would suggest that the PAC-1 Fab-secondary assay used in this study gave different results than PAC-1 IgM due to the sensitivity of clusters of only two integrins, linked by a disulfide bond, to the nature of the multivalent ligand formed by the PAC-1 Fab and secondary antibodies.
Clustering of integrins in cultured cells and platelets when binding to PAC-1 IgM and other multivalent ligands has been repeatedly observed. In addition to my observations of PAC-1 IgM binding to CHO cells, integrin αIIbβ3 redistribution into macroclusters as a result of agonist-induced activation of talin has been demonstrated in CHO cells that express high levels of talin and PKCα (15). Additionally, β-galactosidase complementation and bioluminescence resonance energy transfer assays demonstrated that αIIbβ3 is clustered in CHO cells binding to either bivalent antibodies or bivalent fibrinogen (13). In spreading B16F1 cells, talin head expression induced integrin αVβ3 activation and macroclusters (14). Finally, in platelets, there is extensive evidence, by both confocal and electron microscopy, for the clustering of αIIbβ3 bound to PAC-1 IgM, fibrinogen, and peptides in thrombin- and ADP-activated human platelets (37, 40, 41). Thus, in suggesting that the binding assays that rely on multivalent ligands introduce a clustering component, I am proposing something that is perhaps underappreciated but not unobserved.
Re-evaluation of Differences between the Effects of Talin on αIIbβ3 and Drosophila Integrins
My results explain the apparent differences observed between talin effects on Drosophila PS2 and human αIIbβ3 integrins. The binding assays done on the Drosophila integrins were performed with the monovalent fixation protocol, whereas the experiments probing αIIbβ3 binding were done with the monovalent plus multivalent secondary protocol (11). My data show that human αIIbβ3 gives the same result as was reported for Drosophila PS2 integrins when the experiments are conducted in the same manner. Talin head expression does not result in increased PAC-1 Fab binding in the monovalent fixation assay (Fig. 2).
Models for Integrin Activation by Talin
The data presented here call into question simple models proposing that the mechanism of action of talin is to directly increase integrin affinity for ligands. My data are consistent with a model where the direct role of talin is one of regulating the clustering of αIIbβ3 integrins (Fig. 7A). Clustering may also be accompanied by changes in integrin conformation, but my data would suggest that the integrin is still in a low affinity state. Clustering increases the integrin αIIbβ3 avidity for multivalent ligands such as the widely used PAC-1 (either IgM or Fab with secondary antibodies) or fibrinogen. Once these ligands bind to the clustered low affinity integrins, they are predicted to stabilize the high affinity state of the integrins. As the ligand is retained in the vicinity of the clustered integrins, due to the talin-induced clustering, it binds to many of the integrins in the cluster and stabilizes the high affinity state that triggers sustained inside-out signaling. Once the integrins are clustered and converted to high affinity states, the multivalent ligand is essentially irreversibly bound. In this model, it is the ligand binding that directly induces the high affinity state of the αIIbβ3 integrin. Talin facilitates this indirectly by increasing the clustering of the integrins prior to ligand binding. Talin does not convert the conformation of the integrin heterodimers to a high affinity state prior to ligand binding. The model would predict that in the absence of integrin clustering, individual integrins do interact with ligands, such as PAC-1 IgM, and become activated to a high affinity state (Fig. 7B). However, dissociation of the ligand can and does occur because the integrins and ligand are in true equilibrium, and the integrin reverts to a low affinity state. The level of inside-out signaling resulting from these transient interactions is expected to be low and not result in dramatic cellular responses. A slightly more complicated model takes into consideration the observation that ligand binding to αIIbβ3 integrins has been shown to promote their clustering (13). It may be that it is ligand binding to the low affinity integrin that triggers clustering in CHO cells and platelets. Talin, in this model, would be required for executing the clustering that again would result in large increases in multivalent, but not monovalent, ligand binding.
FIGURE 7.
Model for αIIbβ3 activity regulation by talin. A, upper diagram, integrins (αIIb, green; β3 blue) are present on the surface of CHO cells or platelets in a non-clustered, bent, low affinity state. Talin (fuchsia) in the cytoplasm is inhibited from binding to integrins due to intra- or intermolecular interactions between its head and tail domains. Unbound multivalent ligands are shown as black lines with triangles for binding motifs. Middle diagrams, upon activation of platelets, the inhibition of talin is released, and it binds to the cytoplasmic domains of the β3 subunit resulting in clustered integrins. This clustering may leave the integrins in the bent state or a new conformation, but in either case, the integrin remains in a low affinity state (represented by vertical blue rectangles). The increase in clustering results in multiple low affinity interactions between ligand and integrins. Lower diagram, binding of ligand to the integrins triggers a change in integrin head domains so that they are ultimately in a high affinity state (represented by diagonal blue rectangles). B, in the absence of clustering, integrins interact with ligands and promote a change in affinity of individual integrins. Upon dissociation, they return to their low affinity state.
In addition to being different in valency, PAC-1 IgM (or PAC-1 Fab clustered with secondary antibodies) and PAC-1 Fab are different in size, and this could contribute to differences in binding to different integrin conformations. Although some models propose ligand binding to integrins in an extended conformation, results consistent with regulated binding to integrin αVβ3 in the bent conformation, without conversion to the extended form, have also been observed (42). The ligand binding head domain of αIIbβ3 in the bent conformation might be accessible to the smaller Fab probe but not to the larger IgM or (or PAC-1 Fab clustered with secondary antibodies). If this is the case, the monovalent fixation assay specifically probes the affinity state of the integrin head domain in either the bent or the extended conformation. IgM and other large multivalent ligands are sensitive to three factors: the affinity state of the head domain, the bent state of the integrin, and clustering.
My work does not address the mechanisms of how ligand binding stabilizes or induces the high affinity state of integrins. One model, with support from structural data, of how this may occur proposes that binding of ligand to the MIDAS domain of β3 results in rearrangements in MIDAS contacts that are transmitted through changes in interactions between helixes α1 and α7 in the β integrin I-domain. Helix α7 connects directly to the hybrid domain, and ligand binding facilitates an outward of the hybrid domain from the I-domain leading to separation of the stalks and cytoplasmic tails of the α and β subunits. Separation of the α and β cytoplasmic domains then leads to cytoplasmic signaling and clustering (31).
Although the use of monovalent ligands is important experimentally, it is likely that most extracellular or cell surface integrin ligands in the organism are multivalent. Therefore, regulation of the clustering of low affinity integrins followed by the subsequent multivalent ligand-induced activation of integrin affinity is a parsimonious strategy to convert integrins on the cell surface to a clustered high affinity state.
Acknowledgments
I thank Hisashi Kato and Teresa Helsten for cell lines, antibodies, protocols, and advice. I thank Paula Campbell and Barb Carolus of the Arizona Research Laboratories Cytometry Service for much assistance and the University Medical Center Blood Bank for outdated platelets. Greg Rogers and Joyce Schroeder provided useful comments on the manuscript. I thank Candida Morris, who assisted with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM42474 (to D. Bower and T. B.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
This work is dedicated to the memory of my close colleague and friend, Danny Brower, whose untimely death happened just prior to the beginning of the work reported here. I thank him for helpful discussions regarding this project and for securing the funding that made this work possible.
- FERM
- band four-point-one, ezrin, radixin, moesin homology
- PS
- position-specific
- PBS
- phosphate-buffered saline
- CHO
- Chinese hamster ovary
- FBS
- fetal bovine serum
- BSA
- bovine serum albumin
- MFI
- mean fluorescence intensity
- GFP
- green fluorescent protein.
REFERENCES
- 1.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Ginsberg M. H., Partridge A., Shattil S. J. (2005) Curr. Opin. Cell Biol. 17, 509–516 [DOI] [PubMed] [Google Scholar]
- 3.Calderwood D. A. (2004) J. Cell Sci. 117, 657–666 [DOI] [PubMed] [Google Scholar]
- 4.Luo B. H., Carman C. V., Springer T. A. (2007) Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderwood D. A., Zent R., Grant R., Rees D. J., Hynes R. O., Ginsberg M. H. (1999) J. Biol. Chem. 274, 28071–28074 [DOI] [PubMed] [Google Scholar]
- 6.Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., Calderwood D. A. (2003) Science 302, 103–106 [DOI] [PubMed] [Google Scholar]
- 7.Goksoy E., Ma Y. Q., Wang X., Kong X., Perera D., Plow E. F., Qin J. (2008) Mol. Cell 31, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Critchley D. R. (2005) Biochem. Soc. Trans. 33, 1308–1312 [DOI] [PubMed] [Google Scholar]
- 9.Banno A., Ginsberg M. H. (2008) Biochem. Soc. Trans. 36, 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouaouina M., Lad Y., Calderwood D. A. (2008) J. Biol. Chem. 283, 6118–6125 [DOI] [PubMed] [Google Scholar]
- 11.Helsten T. L., Bunch T. A., Kato H., Yamanouchi J., Choi S. H., Jannuzi A. L., Féral C. C., Ginsberg M. H., Brower D. L., Shattil S. J. (2008) Mol. Biol. Cell 19, 3589–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrams C., Deng Y. J., Steiner B., O'Toole T., Shattil S. J. (1994) J. Biol. Chem. 269, 18781–18788 [PubMed] [Google Scholar]
- 13.Buensuceso C., de Virgilio M., Shattil S. J. (2003) J. Biol. Chem. 278, 15217–15224 [DOI] [PubMed] [Google Scholar]
- 14.Cluzel C., Saltel F., Lussi J., Paulhe F., Imhof B. A., Wehrle-Haller B. (2005) J. Cell Biol. 171, 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J., Lim C. J., Watanabe N., Soriani A., Ratnikov B., Calderwood D. A., Puzon-McLaughlin W., Lafuente E. M., Boussiotis V. A., Shattil S. J., Ginsberg M. H. (2006) Curr. Biol. 16, 1796–1806 [DOI] [PubMed] [Google Scholar]
- 16.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. (2007) Molecular Biology of the Cell, 5th Ed., p. 1172, Garland Science, New York, NY [Google Scholar]
- 17.Shattil S. J., Hoxie J. A., Cunningham M., Brass L. F. (1985) J. Biol. Chem. 260, 11107–11114 [PubMed] [Google Scholar]
- 18.Hato T., Pampori N., Shattil S. J. (1998) J. Cell Biol. 141, 1685–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunch T. A., Helsten T. L., Kendall T. L., Shirahatti N., Mahadevan D., Shattil S. J., Brower D. L. (2006) J. Biol. Chem. 281, 5050–5057 [DOI] [PubMed] [Google Scholar]
- 20.Frojmovic M. M., O'Toole T. E., Plow E. F., Loftus J. C., Ginsberg M. H. (1991) Blood 78, 369–376 [PubMed] [Google Scholar]
- 21.Hughes P. E., Diaz-Gonzalez F., Leong L., Wu C., McDonald J. A., Shattil S. J., Ginsberg M. H. (1996) J. Biol. Chem. 271, 6571–6574 [DOI] [PubMed] [Google Scholar]
- 22.Calderwood D. A., Yan B., de Pereda J. M., Alvarez B. G., Fujioka Y., Liddington R. C., Ginsberg M. H. (2002) J. Biol. Chem. 277, 21749–21758 [DOI] [PubMed] [Google Scholar]
- 23.O'Toole T. E., Katagiri Y., Faull R. J., Peter K., Tamura R., Quaranta V., Loftus J. C., Shattil S. J., Ginsberg M. H. (1994) J. Cell Biol. 124, 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konstandin M. H., Sester U., Klemke M., Weschenfelder T., Wabnitz G. H., Samstag Y. (2006) J. Immunol. Methods 310, 67–77 [DOI] [PubMed] [Google Scholar]
- 25.Taub R., Gould R. J., Garsky V. M., Ciccarone T. M., Hoxie J., Friedman P. A., Shattil S. J. (1989) J. Biol. Chem. 264, 259–265 [PubMed] [Google Scholar]
- 26.Mould A. P., Askari J. A., Barton S., Kline A. D., McEwan P. A., Craig S. E., Humphries M. J. (2002) J. Biol. Chem. 277, 19800–19805 [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Salas A., Springer T. A. (2003) Nat. Struct. Biol. 10, 995–1001 [DOI] [PubMed] [Google Scholar]
- 28.Litvinov R. I., Nagaswami C., Vilaire G., Shuman H., Bennett J. S., Weisel J. W. (2004) Blood 104, 3979–3985 [DOI] [PubMed] [Google Scholar]
- 29.Du X. P., Plow E. F., Frelinger A. L., 3rd, O'Toole T. E., Loftus J. C., Ginsberg M. H. (1991) Cell 65, 409–416 [DOI] [PubMed] [Google Scholar]
- 30.Hantgan R. R., Paumi C., Rocco M., Weisel J. W. (1999) Biochemistry 38, 14461–14474 [DOI] [PubMed] [Google Scholar]
- 31.Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 32.Brown N. H., Gregory S. L., Rickoll W. L., Fessler L. I., Prout M., White R. A., Fristrom J. W. (2002) Dev. Cell 3, 569–579 [DOI] [PubMed] [Google Scholar]
- 33.Petrich B. G., Marchese P., Ruggeri Z. M., Spiess S., Weichert R. A., Ye F., Tiedt R., Skoda R. C., Monkley S. J., Critchley D. R., Ginsberg M. H. (2007) J. Exp. Med. 204, 3103–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieswandt B., Moser M., Pleines I., Varga-Szabo D., Monkley S., Critchley D., Fässler R. (2007) J. Exp. Med. 204, 3113–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe N., Bodin L., Pandey M., Krause M., Coughlin S., Boussiotis V. A., Ginsberg M. H., Shattil S. J. (2008) J. Cell Biol. 181, 1211–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marguerie G. A., Edgington T. S., Plow E. F. (1980) J. Biol. Chem. 255, 154–161 [PubMed] [Google Scholar]
- 37.Fox J. E., Shattil S. J., Kinlough-Rathbone R. L., Richardson M., Packham M. A., Sanan D. A. (1996) J. Biol. Chem. 271, 7004–7011 [DOI] [PubMed] [Google Scholar]
- 38.Peerschke E. I. (1999) J. Lab. Clin. Med. 134, 398–404 [DOI] [PubMed] [Google Scholar]
- 39.Luo B. H., Carman C. V., Takagi J., Springer T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isenberg W. M., McEver R. P., Phillips D. R., Shuman M. A., Bainton D. F. (1987) J. Cell Biol. 104, 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons S. R., Sims P. A., Albrecht R. M. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 3311–3320 [DOI] [PubMed] [Google Scholar]
- 42.Xiong J. P., Mahalingham B., Alonso J. L., Borrelli L. A., Rui X., Anand S., Hyman B. T., Rysiok T., Müller-Pompalla D., Goodman S. L., Arnaout M. A. (2009) J. Cell Biol. 186, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]