Abstract
The E2A-HLF fusion transcription factor generated by t(17;19)(q22;p13) translocation is found in a small subset of pro-B cell acute lymphoblastic leukemias (ALLs) and promotes leukemogenesis by substituting for the antiapoptotic function of cytokines. Here we show that t(17;19)+ ALL cells express Survivin at high levels and that a dominant negative mutant of E2A-HLF suppresses Survivin expression. Forced expression of E2A-HLF in t(17;19)− leukemia cells up-regulated Survivin expression, suggesting that Survivin is a downstream target of E2A-HLF. Analysis using a counterflow centrifugal elutriator revealed that t(17;19)+ ALL cells express Survivin throughout the cell cycle. Reporter assays revealed that E2A-HLF induces survivin expression at the transcriptional level likely through indirect down-regulation of a cell cycle-dependent cis element in the promoter region. Down-regulation of Survivin function by a dominant negative mutant of Survivin or reduction of Survivin expression induced massive apoptosis throughout the cell cycle in t(17;19)+ cells mainly through caspase-independent pathways involving translocation of apoptosis-inducing factor (AIF) from mitochondria to the nucleus. AIF knockdown conferred resistance to apoptosis caused by down-regulation of Survivin function. These data indicated that reversal of AIF translocation by Survivin, which is induced by E2A-HLF throughout the cell cycle, is one of the key mechanisms in the protection of t(17;19)+ leukemia cells from apoptosis.
Introduction
The E2A-HLF fusion transcription factor, which is generated by the t(17;19)(q22;p13) translocation, is found in a small subset of pro-B cell acute lymphoblastic leukemias (ALLs)2 that occurs in older children and adolescents (1, 2). In this chimeric molecule, the trans-activation domain of E2A is fused to the basic region and leucine zipper domain of HLF, which mediates DNA binding and dimerization. Patients with this chimera share distinct clinical features such as hypercalcemia and coagulopathy and very poor prognosis because of resistance to intensive chemotherapy, including aggressive conditioning for bone marrow transplantation (3–5), all of which are unusual for pro-B cell ALLs. Thus, these features may be a direct consequence of aberrant gene expression induced by E2A-HLF fusion transcription factor, rather than a consequence of the nature of B cell progenitors.
We previously demonstrated that inhibition of the trans-activation potential of the E2A-HLF chimera by a dominant negative mutant results in apoptosis in t(17;19)+ ALL cells but does not affect the cell cycle (6). Moreover, E2A-HLF blocks apoptosis normally induced by cytokine deprivation in murine interleukin (IL)-3-dependent B precursor lines such as Baf-3 or FL5.12 cells, suggesting that this fusion protein contributes to leukemogenesis through modification of apoptosis regulatory pathways normally controlled by cytokines (6, 7). We speculated that the target genes of E2A-HLF involved in the inhibition of apoptosis are those regulated via Ras pathways in IL-3-dependent cells, because activation of Ras pathways is indispensable for long term survival of Baf-3 cells in cytokine-free medium (8, 9). Moreover, we previously identified E4BP4/NFIL3, a related basic region and leucine zipper factor with antiapoptotic function, as a possible physiological counterpart of E2A-HLF (10), and we found that E4BP4 expression is induced by IL-3 through Ras-phosphatidylinositol 3-kinase and Ras-Raf-MAPK pathways in IL-3-dependent cells (9).
The survivin gene may be a good candidate for a target gene of E2A-HLF involved in the inhibition of apoptosis in t(17;19)+ ALL cells. Survivin, at 142 amino acids, is the smallest member of the inhibitor of apoptosis protein family and significantly prolongs the viability of cytokine-deprived IL-3-dependent cells (11). The expression of Survivin is controlled by oncogenic c-H-ras, and up-regulation of Survivin depends on functional Ras/phosphatidylinositol 3-kinase and Ras-Raf-MAPK signaling pathways (12). Overexpression of Survivin can protect cells from both extrinsically and intrinsically induced apoptosis (13, 14), whereas inhibition of Survivin expression by antisense ribozyme or RNA interference leads to increased spontaneous apoptosis (15, 16).
A unique feature of Survivin as an apoptosis regulator is its involvement in cell cycle progression (17). survivin expression is transcriptionally induced in the G2/M phase through cell cycle- dependent cis elements located near the transcription initiation site (16). These elements, including the cell cycle-dependent element (GGCGG) and the cell cycle homology region (CHR; ATTTGAA), are implicated in G1 transcriptional repression in S/G2-regulated genes, such as cyclin A, cdc25C, and cdc2 (18). In addition, Survivin is activated through phosphorylation of Thr-34 by mitotic kinase CDC2-cyclin-B1 (14). Enforced expression of a phosphorylation-defective Survivin T34A mutant (Survivin-T34A) initiates mitochondrial dependent apoptosis in a variety of tumor cell lines (14, 16).
Here, we show that Survivin expression is induced by the E2A-HLF chimera, and down-regulation of Survivin induces caspase-independent massive apoptosis in t(17;19)+ ALL cell lines. These findings indicate that Survivin contributes to leukemogenesis by subverting genetic pathways responsible for the apoptosis of B cell progenitors.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Human ALL cell lines that express E2A-HLF (UOC-B1, HAL-O1, YCUB-2, and Endo-kun) and other leukemia cell lines (Nalm-6, RS4;11, REH, 697, 920, HL-60, NB-4, and Jurkat) were cultured in RPMI 1640 medium containing 10% fetal bovine serum. Establishment of Nalm-6 human pro-B cell leukemia cells that express zinc-inducible E2A-HLF (Nalm-6/E2A-HLF) using the pMT-CB6+ eukaryotic expression vector (a gift from Dr. F. Rauscher III, Wistar Institute, Philadelphia) has been described previously (19). UOC-B1/E2A-HLF(dn) cells transfected with a dominant negative mutant of E2A-HLF, which lacks the AD1 transactivation domain of E2A and contains a mutated HLF DNA-binding domain with an intact leucine-zipper domain, were prepared as described previously (6). UOC-B1, Endo-kun, REH, and Jurkat cells that were transfected with either the pMT/Survivin-T34A vector or the empty pMT-CB6+ vector were designated as UOC-B1/Survivin(dn), UOC-B1/pMT, Endo-kun/Survivin(dn), Endo-kun/pMT, REH/Survivin(dn), REH/pMT, Jurkat/Survivin(dn), and Jurkat/pMT, respectively.
Counterflow Centrifugal Elutriations
Counterflow centrifugal elutriations were performed using the SRR6Y elutriation system and rotor equipped with a 4.5-ml chamber (Hitachi Koki Co., Ltd., Tokyo, Japan) (20). Target cells were resuspended at 1–2 × 108 cells in 50 ml of PBS containing 1% fetal bovine serum and injected into the elutriation system at 4 °C using an initial flow rate of 16 ml/min and rotor speed of 2,000 rpm. The flow rate was incrementally increased, and cell fractions were collected serially as follows: fraction 1, 200 ml at 16 ml/min; fraction 2, 200 ml at 18 ml/min; fraction 3, 200 ml at 20 ml/min; fraction 4, 200 ml at 22 ml/min; fraction 5, 200 ml at 24 ml/min; fraction 6, 200 ml at 26 ml/min; and fraction 7, 200 ml at 28 ml/min. Cell cycle analysis was performed on each fraction by staining DNA with propidium iodide (PI) in preparation for flow cytometry with the FACScan/CellFIT system (BD Biosciences).
Gene Silencing by RNA Interference
Short hairpin/short interfering RNA (shRNA/siRNA) was introduced into UOC-B1 or UOC-B1/Survivin(dn) cells to down-regulate the expression of Survivin or apoptosis-inducing factor (AIF) by the shRNA lentivirus system (21, 22). Oligonucleotides were chemically synthesized, annealed, terminally phosphorylated, and inserted into the vector pLL3.7 (Addgene, Cambridge, MA). Oligonucleotides containing siRNA target for survivin sequences (23) were as follows: 5′-TGAAGCGTCTGGCAGATACTTTCAAGAGAAGTATCTGCCAGACGCTTCTTTTTTC-3′ (forward 1) and 5′-TCGAGAAAAAAGAAGCGTCTGGCAGATACTTCTCTTGAAAGTATCTGCCAGACGTTCA-3′ (reverse 1); 5′-TGTGGATGAGGAGACAGAATTTCAAGAGAATTCTGTCTCCTCATCCACTTTTTTC-3′ (forward 3) and 5′-TCGAGAAAAAAGTGGATGAGGAGACAGAATTCTCTTGAAATTCTGTCTCCTCATCCACA-3′ (reverse 3); 5′-TGGATACTTCACTTTAATAATTCAAGAGATTATTAAAGTGAAGTATCCTTTTTTC-3′ (forward 4) and 5′- TCGAGAAAAAAGGATACTTCACTTTAATAATCTCTTGAATTATTAAAGTGAAGTATCCA-3′ (reverse 4); 5′-TGCTTCCTCGACATCTGTTATTCAAGAGATAACAGATGTCGAGGAAGCTTTTTTC-3′ (forward 5) and 5′-TCGAGAAAAAAGCTTCCTCGACATCTGTTATCTCTTGAATAACAGATGTCGAGGAAGCA-3′ (reverse 5). Oligonucleotides containing siRNA target for AIF sequences were as follows: 5′-TGGAGGAGTCTGCGTAATGTTTCAAGAGAACATTACGCAGACTCCTCCTTTTTTC-3′ (forward 1) and 5′-TCGAGAAAAAAGGAGGAGTCTGCGTAATGTTCTCTTGAAACATTACGCAGACTCCTCCT-3′ (reverse 1); 5′- TGCAGGAAGGTAGAAACTGATTCAAGAGATCAGTTTCTACCTTCCTGCTTTTTTC-3′ (forward 2) and 5′-TCGAGAAAAAAGCAGGAAGGTAGAAACTGATCTCTTGAATCAGTTTCTACCTTCCTGCT-3′ (reverse 2); 5′-TGCATGCTTCTACGATATAATTCAAGAGATTATATCGTAGAAGCATGCTTTTTTC-3′ (forward 3) and 5′-TCGAGAAAAAAGCATGCTTCTACGATATAATCTCTTGAATTATATCGTAGAAGCATGCT-3′ (reverse 3); the nucleotide sequences corresponding to the siRNA are underlined. The resulting plasmids or the parental pLL3.7, along with lentiviral packaging mix (ViraPower, Invitrogen), was transfected into 293FT cells (Invitrogen) to produce recombinant lentivirus, and the UOC-B1 or UOC-B1/Survivin(dn) cells were infected with the virus. Enhanced green fluorescent protein (GFP)-positive cells were purified by FACSAria (BD Biosciences) as shRNA-transfected cell populations.
Reporter Assay
Fragments of the 5′-flanking region of the human survivin gene spanning 147, 213, 288, 503, or 698 bp were generated by PCR using Pfu polymerase from genomic DNA of human placenta. The positions of the forward (5′) primers with respect to the translational initiation codon (according to NCBI GenBankTM sequence U75285) are −124 (−124 forward primer, 5′-ACTCCCAGAAGGCCGCGGGGGGTG-3′), −190 (5′-ACCACGGGCAGAGCCACGCGGCGGG-3′), −265 (5′-GTTCTTTGAAAGCAGTCGAGGGGGC-3′), −480 (5′-CGGGTTGAAGCGATTCTCCTGCCT-3′), and −675 (5′-CGATGTCTGCACTCCATCCCTC-3′). The reverse (3′) primer used for these amplifications was at position 23 (+23-reverse primer, 5′-GGGGGCAACGTCGGGGCAagCtTGC-3′) and was constructed based on the genomic sequence with a modification (lowercase) to create a HindIII site. The PCR products were cloned into a pGL3-basic vector (Promega, Madison, WI). The resulting reporter plasmids were designated as pGL3-124, pGL3-190, pGL3-265, pGL3-480, and pGL3-675, respectively. The pGL3-124mut1 vector containing two mutated cell cycle-dependent elements (−6 and −12) was generated by PCR using the −124 forward primer and a reverse primer (5′-GCAAGCTTGtcactGtcactACCTCTG-3′); pGL3-124mut2 vector containing mutated CHR (−42) in addition to two mutated cell cycle-dependent elements (−6 and −12) was generated by the −124 forward primer and a reverse primer (5′-GCAAGCTTGtcactGtcactACCTCTGCCAACGGGTCCCGCGATTCgggTCTGG-3′); and pGL3-124mut3 vector containing a mutated CHR (−42) was generated by the −124 forward primer and a reverse primer (5′-GCAAGCTTGCCGCCGCCGCCACCTCTGCCAACGGGTCCCGCGATTCgggTCTGG-3′) (lowercase indicates mutations).
For transfection with a pMT-CB6+/E2A-HLF construct, Nalm-6 cells (6 × 104) were seeded into 24-well plates, cotransfected with pGL3-survivin promoter construct plus pRL-TK vector, which contains the Renilla luciferase gene, by Lipofectamine 2000 (Invitrogen), and harvested 24 h later. E2A-HLF expression was induced in Nalm-6 cells by the addition of 100 μm ZnCl2 24 h after transfection. Firefly luciferase and Renilla luciferase as a transfection efficiency control were detected with Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions and measured in a Veritas Microplate Luminometer (Promega).
Electrophoretic Mobility Shift Assays (EMSA)
EMSA were performed by incubating 12 μg of nuclear protein lysate at 30 °C for 15 min with a 32P-end-labeled DNA oligonucleotide probe (2 × 104 cpm) containing the CHR-binding site sequence in the survivin promoter (5′-CATTAACCGCCAGATTTGAATCGCGG-3′) in a solution of 12% glycerol, 12 mm HEPES (pH 7.9), 4 mm Tris (pH 7.9), 133 mm KCl, 1.5 μg of sheared calf thymus DNA, and 300 μg of bovine serum albumin per ml as described previously (24). In the competitive inhibition experiments, excess of the unlabeled CHR-consensus sequence probe, i.e. oligonucleotide containing the candidate-binding sites of CHR in the survivin gene promoter or its 3-bp mismatched oligonucleotide (5′-CATTAACCGCCAGAcccGAATCGCGG-3′) was added to the reaction mixture. The entire mixture was incubated at 30 °C for 15 min. Nondenaturing polyacrylamide gels containing 4% acrylamide and 2.5% glycerol were prerun at 4 °C in a high ionic strength Tris-glycine buffer for 30 min and run at 50 mA for ∼45 min. The gel was then dried under vacuum and analyzed by autoradiography.
Other Experimental Procedures
For visualization of intracellular AIF, cytospinned cells were fixed with 1% paraformaldehyde in PBS for 10 min, and permeabilized with 0.5% Triton X-100 in PBS for 5 min. Cells were rinsed twice with PBS (5 min for each rinse), blocked with 5% goat serum in PBS for 30 min, and incubated with anti-AIF antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4 °C in a humidified chamber. Cells were incubated with a secondary antibody, fluorescein isothiocyanate (FITC)-labeled anti-goat IgG (1:500; Santa Cruz Biotechnology), at 37 °C for 30 min.
For Northern blot analysis, 1 μg of poly(A)-selected RNA was separated by electrophoresis in 1% agarose gels containing 2.2 m formaldehyde, transferred to nylon membranes, and hybridized with the appropriate probe according to standard procedures as described previously (5). For immunoblot analysis, the primary antibodies used were anti-Survivin polyclonal (R & D Systems, Minneapolis, MN), anti-α-tubulin monoclonal (Sigma), anti-caspase 3 polyclonal (Cell Signaling Technology, Beverly, MA), anti-caspase 9 polyclonal (BD Biosciences), anti-PARP monoclonal (BD Biosciences), and anti-AIF polyclonal antibodies (Santa Cruz Biotechnology). Anti-HLF(C) antibody for detection of the E2A-HLF chimeric protein was described previously (24).
Cell viability was determined by trypan blue dye exclusion. Early apoptotic events were detected by flow cytometric measurement of externalized phosphatidylserine with the annexin-V-FITC apoptosis detection kit I (BD Biosciences) in preparation for flow cytometry with the FACScan/CellFIT system (BD Biosciences). For caspase inhibition, 20 μm benzyloxycarbonyl-VAD-fluoromethyl ketone (BD Biosciences) was added to the cells 1 h before the addition of zinc. Terminal deoxynucleotidyltransferase-mediated dUTP nick-end-labeling (TUNEL) was performed using the apo-BrdUrd TUNEL assay kit (Molecular Probes, Eugene, OR). Briefly, cells fixed with paraformaldehyde and ethanol were incubated with BrdUrd and TdT for 1 h at 37 °C. BrdUrd uptakes were detected by Alexa dye-leveled anti-BrdUrd antibodies. Cells were stained by PI just before analysis using FACScan/CellFIT system.
RESULTS
E2A-HLF Regulates Survivin Expression
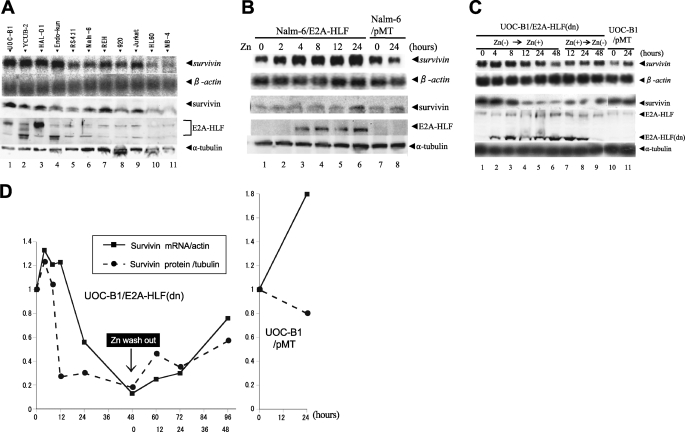
Cell lines were used in this study instead of primary patient samples, because t(17;19)+ ALLs constitute only ∼1% of childhood B-precursor ALLs (1–3). Four t(17;19)+ ALL cell lines (UOC-B1, YCUB-2, HAL-O1, and Endo-kun) expressed the E2A-HLF chimeric protein on immunoblot analysis (Fig. 1A, 4th panel, lanes 1–4) either as a slower (lanes 1 and 3) or a faster migration band (lanes 2 and 4) corresponding to difference in the fusion junction, as described previously (3). Of the seven t(17;19)− leukemia cell lines tested (RS4;11, Nalm-6, REH, 920, Jurkat, HL-60 and NB-4), none expressed the E2A-HLF chimera (Fig. 1A, lanes 5–11). We performed Northern blot and immunoblot analyses to test human leukemia cell lines for the expression of survivin. Survivin mRNA and protein were expressed at uniformly high levels in the four t(17;19)+ ALL cell lines (Fig. 1A, top and 3rd panels). By contrast, survivin mRNA levels varied among the t(17;19)− leukemia cell lines and appeared to determine Survivin protein expression levels in each line (Fig. 1A, lanes 5–11).
FIGURE 1.
Expression of Survivin in human leukemia cell lines and induction of Survivin by E2A-HLF in human ALL cells. A, top 2 panels, Northern blot analysis of poly(A)+ RNA isolated from human leukemia cell lines. The blot was hybridized with a survivin cDNA probe and then rehybridized with a β-actin probe. Lower three panels, immunoblot analysis using whole-cell lysates. Survivin, E2A-HLF, and α-tubulin proteins were detected with specific antibodies. Lanes 1–4, the UOC-B1, YCUB-2, HAL-O1, and Endo-kun t(17;19)-positive pro-B ALL cell lines; lanes 5–8, the RS4;11, Nalm-6, REH, and 920 pro-B ALL cell lines without t(17;19); lane 9, the Jurkat T-ALL cell line; lane 10, the HL-60 AML cell line; and lane 11, the NB-4 APL cell line. B, Nalm-6 cells with zinc-inducible expression of E2A-HLF (Nalm-6/E2A-HLF) and control Nalm-6/pMT cells were cultured in medium containing 100 μm zinc for the indicated length of time. C and D, UOC-B1 cells with zinc-inducible expression of E2A-HLF(dn) (UOC-B1/E2A-HLF(dn)) and control UOC-B1/pMT cells were cultured in medium containing 100 μm zinc for the indicated length of time (Zn(−) → Zn(+)) and removal of zinc from the growth medium (Zn(+) → Zn(−)). C, upper two panels, Northern blot analysis of poly(A)+ RNA. The blot was hybridized with a survivin cDNA probe and then rehybridized with a β-actin probe. Lower three panels, immunoblot analysis for Survivin, E2A-HLF, or α-tubulin proteins. D, quantification of intensity of each band.
Next, we tested whether E2A-HLF induces the expression of Survivin. For these experiments, Nalm-6 cells were transfected with a pMT-CB6+/E2A-HLF construct to generate clones (Nalm-6/E2A-HLF) with zinc-inducible expression of E2A-HLF (Fig. 1B, 4th panel). Ectopic expression of E2A-HLF in Nalm-6 cells induced survivin mRNA by 5-fold within 24 h after the addition of zinc (Fig. 1B, top panel). Accordingly, Survivin protein expression increased within 24 h after induction of E2A-HLF (Fig. 1B, 3rd panel). In control Nalm-6/pMT cells, which contained the empty vector, Survivin expression was unaffected by zinc (Fig. 1B, lanes 7 and 8), confirming that the observed changes in Survivin expression were induced by E2A-HLF and not by zinc.
Induction of Survivin by E2A-HLF was further confirmed using UOC-B1/E2A-HLF(dn) cells, which express zinc-inducible E2A-HLF(dn), a dominant negative mutant of E2A-HLF (see under “Experimental Procedures”) (6, 19). Survivin mRNA and protein expression in UOC-B1/E2A-HLF(dn) cells were high in the absence of zinc (Fig. 1C, top and 3rd panels, lane 1; see also Fig. 1D) but decreased within 24 h after the addition of zinc (Fig. 1C, lane 5), coincident with expression of E2A-HLF(dn) protein (4th panel). Removal of zinc from the growth medium restored Survivin expression within 48 h, again coincident with a decline in the E2A-HLF(dn) protein level (Fig. 1C, lane 9). These data suggested that E2A-HLF induces Survivin mRNA expression. Down-regulation of Survivin protein preceded the reduction of Survivin mRNA (Fig. 1D), suggesting the involvement of post-transcriptional mechanism(s).
Cell Cycle-independent Induction of Survivin by E2A-HLF
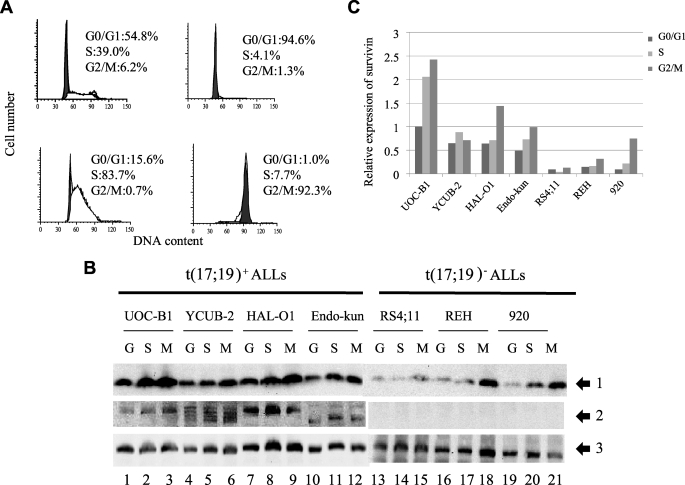
The Survivin mRNA and protein levels at the G2/M phase of the cell cycle are more than 10-fold higher than those at the G1 phase in NIH3T3 murine fibroblasts synchronized by serum starvation and in drug-synchronized HeLa cells (17, 25). Because it is difficult to synchronize leukemia cells by serum starvation or by reagents inhibiting cell cycle progression at a specific phase, we performed counterflow centrifugal elutriation to enrich cells at each phase of the cell cycle. The purity of the preparations was typically more than 90% for G0/G1-phase cells, more than 80% for S-phase cells, and ∼90% for G2/M-phase cells (Fig. 2A). We performed immunoblot analysis to measure Survivin expression in the enriched fractions. In t(17;19)− ALL cell lines (RS4;11, REH, and 920), Survivin expression was most evident at the G2/M-phase (Fig. 2, B, lanes 13–21, and C). In particular, 920 cells at the G2/M phase showed ∼11- and 4-fold higher expression than those at the G1 and S phase, respectively. By contrast, the four cell lines harboring the E2A-HLF chimeric protein expressed Survivin at high levels throughout the cell cycle (Fig. 2, B, lanes 1–12, and C).
FIGURE 2.
Cell cycle-dependent and -independent expression of Survivin in human leukemia cells. Fractions enriched with cells at each phase of the cell cycle were separated by counterflow centrifugal elutriation. A, representative DNA histogram of each fraction subjected to flow cytometry after staining DNA with PI. Upper left, no fractionation; upper right, G0/G1 phase-enriched fraction; lower left, S-phase-enriched fraction; lower right, G2/M-phase-enriched fraction. B, immunoblot analysis of fractions of t(17;19)+ ALL cells or t(17;19)− ALL cells enriched with cells in the G0/G1- (G), S- (S), or G2/M (M)-phase. Survivin (arrow 1), E2A-HLF (arrow 2), and α-tubulin (arrow 3) proteins were detected with specific antibodies. C, levels of Survivin and α-tubulin proteins were determined by the band intensity on autoradiograms from B. Levels of Survivin were normalized to levels of α-tubulin, and amounts shown are relative to amounts in UOC-B1 cells in the G0/G1-phase.
E2A-HLF Enhances the Promoter Activity of the Survivin Gene
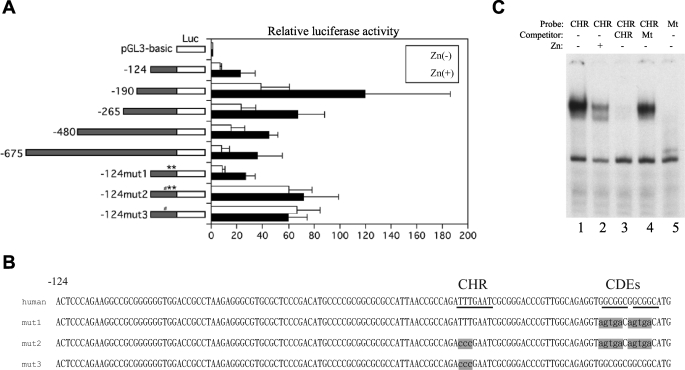
To elucidate how E2A-HLF induces expression of the survivin gene, we analyzed the effects of E2A-HLF on the function of the survivin promoter. We initially generated reporter plasmid vectors (pGL3-124, -190, -265, -480, and -675), each of which contained a different length of human survivin promoter. These vectors were analyzed for luciferase activity in transiently transfected Nalm-6/E2A-HLF cells. When cells were cultured without zinc, luciferase activity was low in cells transfected with pGL3-124 (Fig. 3A). Transfection of pGL3-190 resulted in the highest luciferase activity; it was nearly 6-fold higher than that which resulted from transfection of pGL3-124. However, transfection of survivin constructs longer than pGL3-265 resulted in significantly less activity compared with that of pGL3-190, suggesting the presence of enhancer elements in the region from nt −124 to −190 and repressor elements in the region upstream of nt −190. When cells were cultured with zinc for 24 h, the luciferase activity of each reporter construct, including the shortest pGL3-124, increased by ∼3-fold compared with the respective cells cultured without zinc, suggesting that E2A-HLF induces survivin transcription through cis elements in the region from nt 0 to −124.
FIGURE 3.
Effect of E2A-HLF on survivin promoter activity in transiently transfected t(17;19)− ALL cells. A, Nalm-6/E2A-HLF cells cotransfected with pRL-TK vector and the pGL3-survivin promoter constructs indicated at the left were cultured in the absence (open bars) or presence (black bars) of zinc for 24 h. Firefly luciferase (Luc) activity was normalized to Renilla luciferase as a transfection efficiency control. The level of activity of the promoterless Renilla plasmid luciferase was defined as 1. The results depicted are the averages of three independent experiments; error bars indicate S.D. # indicates mutation of CHR, and ** indicates mutation of CDE. B, nucleotide sequences of the human survivin promoter and three mutants. Underlines indicate CHR or CDE region. Shaded characters indicate mutation (mut). C, EMSA. Nuclear lysates extracted from Nalm-6/E2A-HLF cells cultured without (lanes 1 and 3–5) or with zinc (lane 2) were incubated with a 32P-end-labeled oligonucleotide probe containing the CHR sequence (lanes 1–4) or mutated CHR sequence (lane 5). An excess of unlabeled CHR sequence competitor (lane 3) or mutant competitor (lane 4) was added to the reaction mixture. Mt, mutant.
To further investigate the mechanism through which E2A-HLF induces transcription of the survivin gene, we used luciferase reporter constructs with mutated cell cycle-dependent cis elements. These elements, including the cell cycle-dependent element (CDE; GGCGG) and the cell cycle homology region (CHR; ATTTGAA), are implicated in G1 transcriptional repression in S/G2-regulated genes, such as cyclin A, cdc25C, and cdc2 (18). A previously published report demonstrated two CDEs (−6 and −12) and one CHR (−42) in the human survivin promoter between nt 0 and −124 (Fig. 3B) (18). When pGL3-124mut1, which contained mutated CDE-6 and CDE-12 but had intact CHR-42, was transfected in Nalm-6/E2A-HLF cells, the level of luciferase activity was virtually the same as that of pGL3-124 regardless of the presence of zinc, suggesting that CDE-6 and CDE-12 do not contribute to regulation of survivin transcription in Nalm-6 cells (Fig. 3A). By contrast, transfection of pGL3-124mut2, which contained mutated CHR-42 in addition to mutated CDE-6 and CDE-12, resulted in 10-fold higher luciferase activity in the absence of zinc and 3-fold higher luciferase activity in the presence of zinc compared with transfection of pGL3-124. As a result, there was virtually no difference in the level of luciferase activity between the presence or absence of zinc in cells transfected with pGL3-124mut2. Transfection of pGL3-124mut3, in which only CHR-42 was mutated, show similar results as transfection of pGL3-124mut2. These results suggested that E2A-HLF directly or indirectly up-regulates transcription of survivin through a CHR-42 silencer.
To elucidate transcription factors that bind to CHR-42, we performed EMSA. Smear-looking CHR probe-protein complexes were readily detected (Fig. 3C, lane 1) and were ablated by the addition of an excess amount of cold competitor (lane 3) but not by mutated CHR competitor (lane 4). These complexes were not detected when using mutated CHR as a probe (Fig. 3C, lane 5), suggesting that this complex represents specific binding between transcription factor(s) and the CHR sequence. When E2A-HLF was induced by the addition of zinc, the intensity of the smear decreased (Fig. 3C, lane 2), further supporting that E2A-HLF up-regulates expression of survivin via a CHR-42 silencer.
Specific Inhibition of Survivin-induced Apoptosis in t(17;19)+ ALL Cell Lines
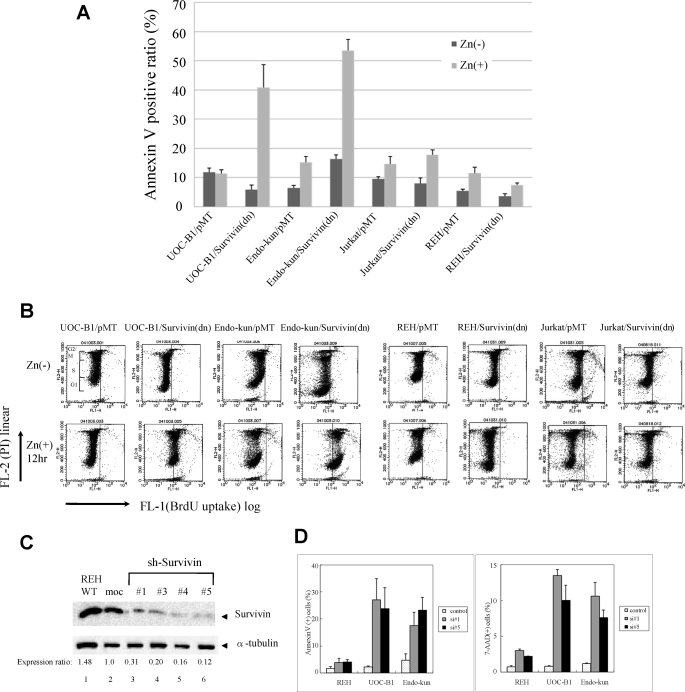
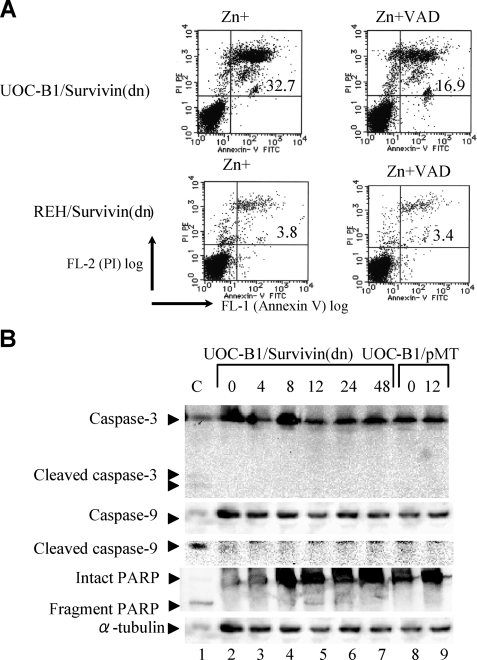
To test whether induction of Survivin by E2A-HLF is essential for the survival of t(17;19)+ leukemia cells, we initially used zinc-inducible expression of a phosphorylation-defective Survivin mutant (Survivin-T34A) that functions as a dominant negative inhibitor. An annexin-V binding assay was used to measure externalization of phosphatidylserine, an indicator of cell death. Ectopic expression of Survivin-T34A in two t(17;19)+ ALL cell lines (UOC-B1 and Endo-kun) caused a rapid increase in the fraction of annexin-V-positive cells within 24 h after the addition of zinc (Fig. 4A). In control UOC-B1/pMT and Endo-kun/pMT cells, which contained the empty vector, less than 20% of cells were positive for annexin-V regardless of the presence of zinc. By contrast, Survivin-T34A did not induce massive cell death in two t(17;19)− leukemia cell lines (REH and Jurkat), which express relatively high levels of Survivin (Fig. 1A). The basis for the altered survival of UOC-B1 and Endo-kun cells expressing Survivin-T34A was investigated by TUNEL analysis using flow cytometry. BrdUrd uptake (Fig. 4B, x axis) by TdT that reflects a number of DNA ends in each cell was markedly increased in UOC-B1 and Endo-kun cells expressing Survivin-T34A. Interestingly, intensities of BrdUrd signals increased equally in cells at each cell cycle phase (y axis), suggesting that down-regulation of Survivin function induces apoptosis in a cell cycle-independent manner. By contrast, expression of Survivin-T34A did not induce apoptosis in REH cells and induced apoptosis in Jurkat cells only at the G2/M phase (Fig. 4B).
FIGURE 4.
Effect of enforced overexpression of Survivin-T34A and introduction of Survivin-shRNA in ALL cells. UOC-B1, Endo-kun, Jurkat, and REH cells inducibly expressing Survivin-T34A (UOC-B1/Survivin(dn), Endo-kun/Survivin(dn), Jurkat/Survivin(dn) and REH/Survivin(dn) cells, respectively) were compared with control UOC-B1/pMT, Endo-kun/pMT, Jurkat/pMT, and REH/pMT cells, respectively. A, externalization of phosphatidylserine as determined by annexin-V binding. Cells cultured in medium with or without 100 μm zinc for 24 h were simultaneously stained with FITC-annexin-V and PI. The FITC-annexin-V-positive ratios were determined by representative flow cytometric plots. B, cells cultured in medium with or without 100 μm zinc for 12 h were simultaneously stained with PI and BrdUTP in a TdT-catalyzed reaction and then subjected to flow cytometric analysis. DNA ends labeled with BrdUTP (abscissa) are shown as a function of cellular DNA content of PI-stained nuclei (ordinate). Cells to the right of the vertical line had free DNA ends labeled with TdT, indicating apoptosis. Range of each cell cycle was shown in the panel of UOC-B1/pMT, Zn(−). C, immunoblot analyses using Survivin (upper panel) and α-tubulin (lower panel) antibodies. REH cells without treatment (lane 1) or infected with lentivirus (lanes 2–6) were sorted by GFP expression. moc indicates control sh-RNA. Ratios of intensity are shown below. WT, wild type. D, ratios of annexin-V-phycoerythrin (PE) (left) or 7-AAD (right)-positive cells in the GFP-positive fraction of REH, UOC-B1, or Endo-kun cells infected with lentivirus expressing GFP alone (control) or GFP and Survivin shRNA1 or -5 (si#1 or si#5, respectively). Mean values from three independent experiments are shown with standard error.
We next down-regulated Survivin by lentivirally introduced short hairpin (sh) RNA. The Survivin protein expression level in cells sorted by expression of GFP (as an indicator of infection) was significantly reduced by Survivin-shRNA1 and -3–5 compared with that in cells infected with control-shRNA (Fig. 4C). We introduced shRNA1 or -5 into REH, UOC-B1, and Endo-kun cells. Twenty four hours later, when about 10% of the cells were GFP-positive, dead cells were determined by annexin-V and 7-AAD staining. Marked increases in annexin-V- and 7-AAD-positive cells were detected in the GFP-positive population of UOC-B1 or Endo-kun cells compared with those in GFP-positive REH cells (Fig. 4D).
Caspase-dependent and -independent Cell Death Are Induced by Survivin-T34A in t(17;19)+ Cells
To elucidate the molecular mechanisms through which Survivin protects t(17;19)+ ALL cells from apoptosis, we initially examined caspase- dependent pathways. A pan-caspase inhibitor, benzyloxycarbonyl-VAD-fluoromethyl ketone, partially blocked cell death induced by Survivin-T34A (Fig. 5A). Immunoblot analysis revealed fragmentation of PARP within 8 h after induction of Survivin-T34A, although cleavage of caspase-3 and -9 was barely detectable up through 48 h (Fig. 5B). These results suggested that caspase-independent pathways contribute to cell death induced by Survivin-T34A in t(17;19)+ ALL cells.
FIGURE 5.
PARP activation in Survivin(dn)-expressing cells and effect of caspase inhibitor. A, flow cytometric analysis stained with annexin-V (abscissa) and PI (ordinate). UOC-B1/Survivin(dn) and REH/Survivin(dn) cells were cultured in medium containing 100 μm zinc 1 h after treatment with or without 20 μm benzyloxycarbonyl-VAD-fluoromethyl ketone (VAD), a pan-caspase inhibitor. B, UOC-B1/Survivin(dn) or UOC-B1/pMT cells were cultured in medium containing 100 μm zinc for the indicated times. Immunoblot analysis of UOC-B1/Survivin(dn) cells was performed to detect caspase-3, cleaved caspase-3, caspase-9, cleaved caspase-9, intact PARP, fragmented PARP, and α-tubulin proteins. As a positive control (C), Jurkat cells were treated with etoposide.
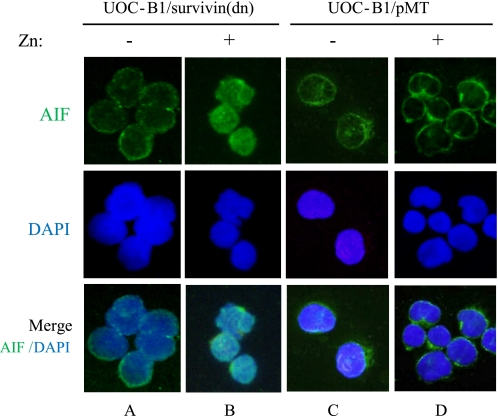
The association of Survivin targeting both preceding and independent of caspase activation suggested to us a potential role for AIF, given its capacity to mediate DNA fragmentation and cytochrome c release in a caspase-independent fashion (28, 29). We analyzed the nuclear translocation of AIF after induction of Survivin-T34A in t(17;19)+ ALL cells. In the UOC-B1/Survivin(dn) cells without induction of Survivin-T34A, AIF signals were found in the cytoplasm in ∼75% of the total cell population (Fig. 6A), consistent with a previous report showing the presence of AIF in mitochondria (27). By contrast, expression of Survivin-T34A for 12 h induced nuclear translocation of AIF signals in more than 90% of cells (Fig. 6B). Nuclear translocation of AIF was induced in only a small percentage (∼ 4%) of the control UOC-B1/pMT cells treated with zinc (Fig. 6D).
FIGURE 6.
Effect of Survivin(dn) on nuclear translocation of AIF. UOC-B1/Survivin(dn) cells (A and B) or UOC-B1/pMT cells (C and D) were cultured for 12 h in the absence of zinc (A and C) or in the presence of 100 μm zinc (B and D). Cells were immunostained with an anti-AIF polyclonal antibody (upper panels). Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize the nuclei (middle and lower panels).
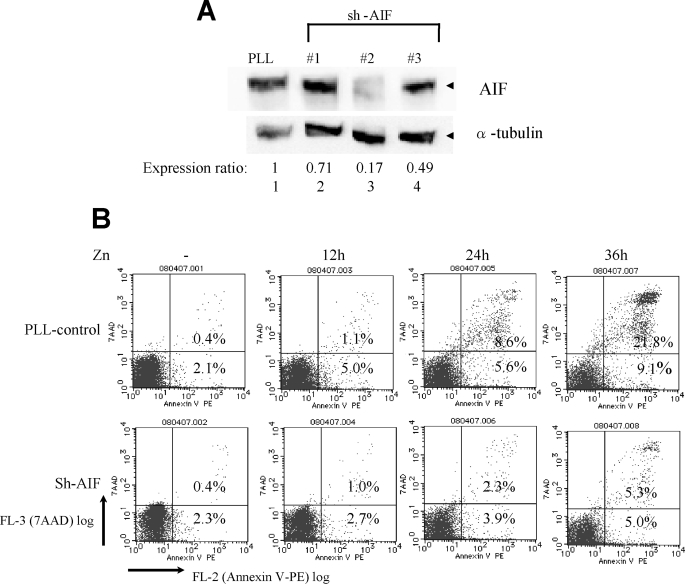
To test the role of AIF in cell death induced by Survivin-T34A in t(17;19)+ ALL cells, we down-regulated AIF expression by lentivirally expressed AIF-shRNA. The AIF protein expression level in UOC-B1/Survivin(dn) cells was significantly reduced by AIF-shRNA2 compared with that in cells infected with control PLL-shRNA sorted by expression of GFP (Fig. 7A). The number of cells undergoing cell death by induction of Survivin-T34A was monitored by annexin-V and 7-AAD staining in GFP-positive cells. Cells treated with AIF-shRNA2 were significantly resistant to cell death compared with those treated with control PLL-shRNA (Fig. 7B), suggesting that AIF plays critical roles in Survivin-mediated cell death of t(17;19)+ ALL cells.
FIGURE 7.
Inhibition of Survivin-T34A-induced apoptosis by knockdown of AIF. A, immunoblot analysis using AIF (upper panel) or α-tubulin (lower panel) antibodies. UOC-B1/Survivin(dn) cells were infected with lentivirus expressing the shRNA indicated above each panel, and GFP-positive cells were sorted. Ratios of intensity are shown below. B, UOC-B1/Survivin(dn) cells were infected with lentivirus expressing control-shRNA (PLL, upper) or AIF-shRNA2 (lower), cultured with 100 μm zinc for the indicated length of time, and stained with annexin-V-phycoerythrin (PE) (abscissa) and 7-AAD (ordinate). The data show the ratio of annexin-V-phycoerythrin- and 7-AAD-positive cells in the GFP-positive fraction as determined by flow cytometric analysis. Numbers indicate the percentage of apoptotic cells.
DISCUSSION
We previously demonstrated that E2A-HLF contributes to leukemogenesis of t(17;19)-positive ALL through inhibition of apoptosis (6). Here, we demonstrate that E2A-HLF induces Survivin expression through transcriptional regulation. Down-regulation of Survivin function by a dominant negative mutant of Survivin (Survivin-T34A) or reduction of Survivin expression by shRNA induced massive apoptosis in t(17;19)+ leukemia cells throughout the cell cycle. Down-regulation of Survivin induced apoptosis via both caspase-dependent and -independent pathways, and AIF was involved in the latter pathways. These findings indicate that Survivin plays critical roles in E2A-HLF-mediated leukemogenesis.
E2A-HLF, known as a trans-activator (24), could either directly or indirectly enhance survivin transcription. However, there is no potential binding site of E2A-HLF (GTTACGTAAT) in the promoter region of survivin, and indeed, no binding activity of E2A-HLF was detected by EMSA in the immediate upstream region (124 bp) of the initial ATG, including a region that contains CHR-42 sequence (ATTTGAA) (negative data not shown). Thus, E2A-HLF most likely inhibits the silencer activity of CHR-42 (Fig. 3A) by down-regulating a certain amount of hypothetical trans-repressor X that binds to CHR-42 (Fig. 3C, lane 2). Theoretically, E2A-HLF may induce another trans-repressor that down-regulates the expression of trans-repressor X. Alternatively, a downstream target factor of E2A-HLF may reduce the DNA binding potential of trans-repressor X. It is of interest to note that whether or not the mechanism through which E2A-HLF induces survivin transcription is common to that, Ras pathways regulate Survivin expression. As we reported previously (30), because downstream targets of Ras enhance Survivin expression through enhancer(s) between −124 to −190, E2A-HLF likely induces Survivin through distinctive pathways.
Previous reports indicated that Survivin inhibits apoptosis through both caspase-dependent and caspase-independent pathways, although detailed mechanisms are not yet understood (31–34). In t(17;19)+ ALL cells undergoing apoptosis by Survivin-T34A, activation of the caspase cascade is likely a secondary event, because activated caspase-3 and -9 were not detectable up through 48 h after induction of Survivin-T34A (Fig. 5B), even though the cells were positive on annexin-V staining and TUNEL analysis within 12 h (Fig. 4, A and B). We observed rapid PARP activation within 8 h that is required for translocation of AIF to the nucleus from mitochondria, followed by morphological changes such as cell shrinkage and chromatin condensation (27, 35). Moreover, knockdown of AIF in UOC-B1/Survivin(dn) cells protected cells from apoptosis induced by Survivin-T34A (Fig. 7B). Therefore, reversal of AIF translocation by Survivin, which is induced by E2A-HLF throughout the cell cycle, appears to be the key mechanism in the protection of t(17;19)+ leukemia cells from apoptosis.
In earlier studies, we identified SLUG as a target gene of E2A-HLF (36). SLUG is a transcription factor closely related to Ces-1, a cell death regulator in Caenorhabditis elegans (36, 37). Importantly, ces-1 is a downstream target gene of ces-2, which is closely related to E2A-HLF (6, 38). The apparent convergence of cell death pathways, including CES-2/CES-1 in the worm and E2A-HLF/SLUG in human pro-B leukemia (6, 36), suggests that SLUG may have an important regulatory role in the survival of lymphoid cells. However, the lack of expression of Slug by normal pro-B cells suggests that E2A-HLF acts not by invoking a normal survival pathway in B lymphocytes but rather by aberrantly activating a Slug-mediated survival pathway normally used by more primitive hematopoietic cell progenitors (39). Therefore, it is still uncertain whether only the E2A-HLF/SLUG pathway inhibits apoptosis in leukemia pro-B cell progenitors (36). Perhaps E2A-HLF has multiple apoptosis-inhibiting pathways to coordinate leukemogenesis.
t(17;19)+ ALL almost always proves refractory to intensive chemotherapy, even to the aggressive conditioning for bone marrow transplantation (3–5). Survivin is an attractive therapeutic target in t(17;19)+ ALLs because of its differential expression in tumors versus normal tissues and because it may be required for maintaining cell viability in this leukemia (14, 16). The efficacy of Survivin antisense oligonucleotides has been demonstrated in vivo (40, 41), and clinical grade antisense Survivin oligonucleotides are currently under development (42, 43). Although Survivin is not a cancer-specific molecule in regulating normal cell function particularly in the hematopoietic stem cell and immune systems, anti-Survivin therapies developed to date have not revealed major systemic toxicities in animal models and are encouraging (44). Our results provide further evidence that Survivin inhibitors may be an effective therapeutic strategy for this refractory ALL.
Acknowledgments
We thank M. Eguchi for helpful discussions, support, and encouragement throughout this study and Y. Sato for support and encouragement. We thank F. J. Rauscher III for providing the pMT-CB6+ expression vector; K. Harada, H. Aoyama, and M. Ishiguchi for excellent technical assistance; K. Ohyashiki and K. Toyama for HAL-O1 cell lines; and M. Endo for Endo-kun cell lines.
This work was supported by Grant-in-aid for Scientific Research (C) 18591201 from the Japan Society for Promotion of Science (to H. K.) and a young investigator award from Dokkyo Medical University (to M. O.).
- ALL
- acute lymphoblastic leukemia
- AIF
- apoptosis-inducing factor
- IL
- interleukin
- MAPK
- mitogen-activated protein kinase
- CHR
- cell cycle homology region
- PBS
- phosphate-buffered saline
- FITC
- fluorescein isothiocyanate
- shRNA
- short hairpin RNA
- siRNA
- short interfering RNA
- GFP
- green fluorescent protein
- EMSA
- electrophoretic mobility shift assay
- BrdUrd
- bromodeoxyuridine
- TdT
- terminal deoxynucleotidyltransferase
- PARP
- poly(ADP-ribose) polymerase
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick-end-labeling
- PI
- propidium iodide
- dn
- dominant negative
- nt
- nucleotide
- 7-AAD
- 7-amino-actinomycin D
- PLL
- plenti-Lox3.7.
REFERENCES
- 1.Inaba T., Roberts W. M., Shapiro L. H., Jolly K. W., Raimondi S. C., Smith S. D., Look A. T. (1992) Science 257, 531–534 [DOI] [PubMed] [Google Scholar]
- 2.Hunger S. P., Ohyashiki K., Toyama K., Cleary M. L. (1992) Genes Dev. 6, 1608–1620 [DOI] [PubMed] [Google Scholar]
- 3.Hunger S. P. (1996) Blood 87, 1211–1224 [PubMed] [Google Scholar]
- 4.Inukai T., Hirose K., Inaba T., Kurosawa H., Hama A., Inada H., Chin M., Nagatoshi Y., Ohtsuka Y., Oda M., Goto H., Endo M., Morimoto A., Imaizumi M., Kawamura N., Miyajima Y., Ohtake M., Miyaji R., Saito M., Tawas A., Yanai F., Goi K., Nakazawa S., Sugita K. (2007) Leukemia 21, 288–296 [DOI] [PubMed] [Google Scholar]
- 5.Matsunaga T., Inaba T., Matsui H., Okuya M., Miyajima A., Inukai T., Funabiki T., Endo M., Look A. T., Kurosawa H. (2004) Blood 103, 3185–3191 [DOI] [PubMed] [Google Scholar]
- 6.Inaba T., Inukai T., Yoshihara T., Seyshab H., Ashmun R. A., Canman C. E., Laken S. J., Kastan M. B., Look A. T. (1996) Nature 382, 541–544 [DOI] [PubMed] [Google Scholar]
- 7.Inukai T., Inaba T., Okushima S., Look A. T. (1998) Mol. Cell. Biol. 18, 6035–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinoshita T., Yokota T., Arai K., Miyajima A. (1995) EMBO J. 14, 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuribara R., Kinoshita T., Miyajima A., Shinjyo T., Yoshihara T., Inukai T., Ozawa K., Look A. T., Inaba T. (1999) Mol. Cell. Biol. 19, 2754–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikushima S., Inukai T., Inaba T., Nimer S. D., Cleveland J. L., Look A. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2609–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambrosini G., Adida C., Altieri D. C. (1997) Nat. Med. 3, 917–921 [DOI] [PubMed] [Google Scholar]
- 12.Sommer K. W., Stumberger C. J., Schmidt G. E., Sasgary S., Cerni C. (2003) Oncogene 22, 4266–4280 [DOI] [PubMed] [Google Scholar]
- 13.Tamm I., Wang Y., Sausville E., Scudiero D. A., Vigne N., Oltersdorf T., Reed J. C. (1998) Cancer Res. 58, 5315–5320 [PubMed] [Google Scholar]
- 14.Li F. (2003) J. Cell. Physiol. 197, 8–29 [DOI] [PubMed] [Google Scholar]
- 15.Li F., Ling X. (2006) J. Cell. Physiol. 208, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altieri D. C. (2003) Nat. Rev. Cancer 3, 46–54 [DOI] [PubMed] [Google Scholar]
- 17.Li F., Ambrosini G., Chu E. Y., Plescia J., Tognin S., Marchisio P. C., Altieri D. C. (1998) Nature 396, 580–584 [DOI] [PubMed] [Google Scholar]
- 18.Li F., Altieri D. C. (1999) Biochem. J. 344, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurosawa H., Goi K., Inukai T., Inaba T., Chang K. S., Shinjyo T., Rake straw K. M., Naeve C. W., Look A. T. (1999) Blood 93, 321–332 [PubMed] [Google Scholar]
- 20.Kikuchi J., Furukawa Y., Iwase S., Terui Y., Nakamura M., Kitagawa S., Kitagawa M., Komatsu N., Miura Y. (1997) Blood 89, 3980–3990 [PubMed] [Google Scholar]
- 21.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. (2003) Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi J., Shimizu R., Wada T., Ando H., Nakamura M., Ozawa K., Furukawa Y. (2007) Stem Cells 25, 2439–2447 [DOI] [PubMed] [Google Scholar]
- 23.Gu C. M., Zhu Y. K., Ma Y. H., Zhang M., Liao B., Wu H. Y., Lin H. L. (2006) Neoplasm 53, 206–212 [PubMed] [Google Scholar]
- 24.Inaba T., Shapiro L. H., Funabiki T., Sinclair A. E., Jones B. G., Ashmun R. A., Look A. T. (1994) Mol. Cell. Biol. 14, 3403–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi K., Hatano M., Otaki M., Ogasawara T., Tokuhisa T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1457–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deleted in proof
- 27.Moubarak R. S., Yuste V. J., Artus C., Bouharrour A., Greer P. A., Menissier-de Murcia J., Susin S. A. (2007) Mol. Cell. Biol. 27, 4844–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Susin S. A., Lorenzo H. K., Zamzami N., Marzo I., Snow B. E., Brothers G. M., Mangion J., Jacotot E., Costantini P., Loeffler M., Larochette N., Goodlett D. R., Aebersold R., Siderovski D. P., Penninger J. M., Kroemer G. (1999) Nature 397, 441–446 [DOI] [PubMed] [Google Scholar]
- 29.Modjtahedi N., Giordanetto F., Madeo F., Kroemer G. (2006) Trends Cell Biol. 16, 264–272 [DOI] [PubMed] [Google Scholar]
- 30.Shinjyo T., Kurosawa H., Miyagi J., Ohama K., Masuda M., Nagasaki A., Matsui H., Inaba T., Furukawa Y., Takasu N. (2008) Tohoku J. Exp. Med. 216, 25–34 [DOI] [PubMed] [Google Scholar]
- 31.Carter B. Z., Kornblau S. M., Tsao T., Wang R. Y., Schober W. D., Milella M., Sung H. G., Reed J. C., Andreeff M. (2003) Blood 102, 4179–4186 [DOI] [PubMed] [Google Scholar]
- 32.Liu T., Brouha B., Grossman D. (2004) Oncogene 23, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T., Biddle D., Hanks A. N., Brouha B., Yan H., Lee R. M., Leachman S. A., Grossman D. (2006) J. Invest. Dermatol. 126, 2247–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croci D. O., Cogno I. S., Vittar N. B., Salvatierra E., Trajtenberg F., Podhajcer O. L., Osinaga E., Rabinovich G. A., Rivarola V. A. (2008) J. Cell. Biochem. 105, 381–390 [DOI] [PubMed] [Google Scholar]
- 35.Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. (2002) Science 297, 259–263 [DOI] [PubMed] [Google Scholar]
- 36.Inukai T., Inoue A., Kurosawa H., Goi K., Shinjyo T., Ozawa K., Mao M., Inaba T., Look A. T. (1999) Mol. Cell 4, 343–352 [DOI] [PubMed] [Google Scholar]
- 37.Metzstein M. M., Horvitz H. R. (1999) Mol. Cell 4, 309–319 [DOI] [PubMed] [Google Scholar]
- 38.Metzstein M. M., Hengartner M. O., Tsung N., Ellis R. E., Horvitz H. R. (1996) Nature 382, 545–547 [DOI] [PubMed] [Google Scholar]
- 39.Inoue A., Seidel M. G., Wu W., Kamizono S., Ferrando A. A., Bronson R. T., Iwasaki H., Akashi K., Morimoto A., Hitzler J. K., Pestina T. I., Jackson C. W., Tanaka R., Chong M. J., McKinnon P. J., Inukai T., Grosveld G. C., Look A. T. (2002) Cancer Cell 2, 279–288 [DOI] [PubMed] [Google Scholar]
- 40.Tu S. P., Jiang X. H., Lin M. C., Cui J. T., Yang Y., Lum C. T., Zou B., Zhu Y. B., Jiang S. H., Wong W. M., Chan A. O., Yuen M. F., Lam S. K., Kung H. F., Wong B. C. (2003) Cancer Res. 63, 7724–7732 [PubMed] [Google Scholar]
- 41.Kanwar J. R., Shen W. P., Kanwar R. K., Berg R. W., Krissansen G. W. (2001) J. Natl. Cancer Inst. 93, 1541–1552 [DOI] [PubMed] [Google Scholar]
- 42.Schimmer A. D. (2004) Cancer Res. 64, 7183–7190 [DOI] [PubMed] [Google Scholar]
- 43.Altieri D. C. (2008) Nat. Rev. Cancer 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 44.Fukuda S., Pelus L. M. (2006) Mol. Cancer Ther. 5, 1087–1098 [DOI] [PubMed] [Google Scholar]