Abstract
Dysregulation of β-catenin levels and localization and constitutive activation of β-catenin/TCF (T cell factor)-regulated gene expression occur in many cancers, including the majority of colorectal carcinomas and a subset of ovarian endometrioid adenocarcinomas. Based on the results of microarray-based gene expression profiling we found the insulin receptor substrate 1 (IRS1) gene as one of the most highly up-regulated genes upon ectopic expression of a mutant, constitutively active form of β-catenin in the rat kidney epithelial cell line RK3E. We demonstrate expression of IRS1 can be directly activated by β-catenin, likely in part via β-catenin/TCF binding to TCF consensus binding elements located in the first intron and downstream of the IRS1 transcriptional start site. Consistent with the proposal that β-catenin is an important regulator of IRS1 expression in vivo, we observed that IRS1 is highly expressed in many cancers with constitutive stabilization of β-catenin, such as colorectal carcinomas and ovarian endometrioid adenocarcinomas. Using a short hairpin RNA approach to abrogate IRS1 expression and function, we found that IRS1 function is required for efficient de novo neoplastic transformation by β-catenin in RK3E cells. Our findings add to the growing body of data implicating IRS1 as a critical signaling component in cancer development and progression.
Introduction
Much progress has been made in identifying conserved signaling pathways that are recurrently deranged in cancer. The canonical Wnt signaling pathway or β-catenin-dependent Wnt pathway is one of the pathways most commonly altered in human cancer (1, 2). In the absence of activating Wnt signals, glycogen synthase kinase 3β (GSK3β)3 functions in concert with the AXIN and APC (adenomatous polyposis coli) tumor suppressor proteins and other factors to phosphorylate β-catenin at defined serine and threonine residues in its amino (N)-terminal domain. The phosphorylated β-catenin is recognized and ubiquitinated by a complex containing β-transducin repeat-containing protein (TrCP), and β-catenin is then degraded by the proteasome. Wnt ligand binding to the Frizzled-low density lipoprotein-related protein-5/6 co-receptor complex on the cell surface leads to inhibition of the AXIN·GSK3β complex and stabilization of the “free” cytosolic and nuclear pools of β-catenin. In the nucleus, β-catenin can bind to members of the T cell factor (TCF) transcription factor family, and β-catenin·TCF complexes modulate transcription of an array of genes, several of which play roles in effecting changes in cell fate, proliferation, and other processes (1).
Mutational mechanisms with major contributing roles in constitutively stabilizing β-catenin in human cancer include inactivation of the APC or AXIN1 tumor suppressor proteins or activating (oncogenic) mutations in the N-terminal domain of β-catenin (3). These oncogenic mutations lead to reduced phosphorylation and ubiquitination of β-catenin. In cancer cells, the net consequence of these mutations in APC, AXIN1, or β-catenin is that the β-catenin protein is constitutively stabilized in the absence of Wnt signals, with resultant altered transcription of downstream β-catenin/TCF-regulated target genes (4).
Many candidate β-catenin/TCF-regulated target genes have been proposed. However, the functional significance in cancer development of most candidate targets remains uncertain. We performed microarray analyses comparing the expression of genes in epithelial cell lines engineered to express an oncogenic stabilized form of β-catenin. Among the genes expressed at higher levels in cell lines with activated β-catenin, we further investigated the gene for insulin receptor substrate 1 (IRS1).
Insulin receptor substrates are adaptor molecules that serve to couple receptor tyrosine kinase activation to downstream effector cascades. There are three human IRS genes (IRS1, IRS2 and IRS4). IRS1 and IRS2 seem to mediate the major metabolic, proliferative, and antiapoptotic functions of the insulin receptor and the insulin-like growth factor receptor by relaying signals from the activated receptors to downstream effector cascades, such as the phosphatidylinositol 3-kinase and Ras-Raf pathways (5–8). Of interest, insulin-like growth factor signaling appears to play a prominent role in colorectal carcinogenesis. For instance, it was noted that loss of imprinting of the IGF2 gene is significantly associated with an increased colorectal carcinoma risk (6). Mice heterozygous for an inactivating mutation in the APC gene develop fewer polyps in an IGF2-deficient background and more polyps in a background of increased IGF2 expression (9). To what extent this effect is actually mediated by IRS1 is unclear. However, several lines of evidence implicate IRS1 in the development of cancer and cellular transformation. Epidemiological studies have shown that a polymorphism in the coding region of IRS1 is associated with an increased incidence of colorectal cancers (10). In several cell lines, ectopic expression of IRS1 can promote cellular transformation (7, 11–16). It has also been noted that IRS1 is highly expressed in a variety of cancers and that overexpression of a dominant-negative IRS1 mutant inhibits neoplastic characteristics of several human cancer cell lines (17, 18). In addition to its adaptor function in the cytosol and membrane compartment, a function for nuclear IRS1 has recently been described. Transduction of murine fibroblast lines with different oncogenes such as large T and src, or treatment of cells with insulin-like growth factor leads to a nuclear translocation of IRS1 (19–23). Once in the nucleus, IRS1 seems to be able to augment transcription of ribosomal RNA, a process that seems to involve activation of a nuclear form of phosphatidylinositol 3-kinase and AUF (20). Interestingly, there is also evidence that IRS1 can facilitate the nuclear translocation of β-catenin and might even be required in this process (24, 25). Last, but not least, in a recent study by Ramocki and colleagues (26) it was noted that mice with a constitutional inactivating mutation in the murine Apc gene (ApcMin) develop significantly fewer intestinal adenomatous polyps when the Irs1 gene is inactivated (Irs1−/− mice). We present data to suggest that IRS1 might in fact be an important downstream target gene of the Wnt/β-catenin pathway that plays a role in the initiation of neoplastic transformation by β-catenin.
EXPERIMENTAL PROCEDURES
Cell Lines and Plasmids
Unless mentioned otherwise, all cell lines were obtained from American Type Culture Collection (Rockville, MD). The amphotropic Phoenix packaging cell line was obtained from G. Nolan (Stanford University School of Medicine); Gli-transformed RK3E cells were obtained from J. M. Ruppert (University of Alabama, Birmingham, AL) (27). All cells were grown in 5% CO2 with medium containing 10% fetal bovine serum and penicillin/streptomycin. LS174T cells were grown in α minimum Eagle's medium (Invitrogen); RKO, HT29, cells were cultured in McCoy medium (Invitrogen). All other cell lines were grown in Dulbecco's modified Eagle's medium. A polyclonal RK3E cell line expressing the β-catenin S33Y-ER fusion protein was obtained after retroviral transduction of RK3E cells with supernatants from amphotrophic Phoenix cells transfected with pBabe-S33Yβ-ER-puro. Drug selection on the pBabe-S33Yβ-ER-puro-transduced RK3E cells was carried out in puromycin (Sigma) at a concentration of 2.0 μg/ml. To activate the S33Yβ-ER fusion protein, the RK3E/S33Yβ-ER cells were treated with 0.5 μm 4-hydroxytamoxifen (4-OHT) (Sigma), made from a stock concentration of 500 μm 4-OHT in 100% ethanol. To inhibit new protein synthesis in RK3E/S33Y-ER cells, the medium was supplemented with cycloheximide (Sigma) at a concentration of 1.5 μg/ml. To assess the effects of dominant-negative TCF-4 on IRS1 gene expression, a retroviral TCF-4ΔN31 expression construct was used to transduce the RK3E/S33Y-ER cell line (28). Empty vector (pPGS-Neo) control transductions were carried out in parallel. The TCF4ΔN31- and empty vector-transduced cells were subsequently selected for 7–10 days in 1.5 mg/ml of G418 (Sigma).
Activity of GSK3β in the ovarian cancer cell line MDAH-2774 was inhibited by treating 50% confluent cells with a final concentration of 10 nm SB216763 (Sigma). RNA was collected at the indicated time points after initiation of treatment. Rat intestinal epithelial cell line IEC6 and immortalized ovarian surface epithelial cells were treated with 50 ng/ml of mouse Wnt3a (R&D Systems, Minneapolis, MN) for 8 h.
Stable RNA interference of IRS1 transcripts was performed by retrovirally transducing polymerase III expression cassettes driving expression of short hairpins specifically targeting IRS1 transcripts, or scrambled control hairpins. The following nucleotides were used for ligation into pSUPERIOR-RETRO-PURO (Oligoengine, Seattle, WI): human IRS1 A4 (sense 5′-GAT CCC GAG CAT GTA CAA ATG CTT CTC TTC CTG TCA AGA AGC GTT TGT GCA TGC TCT TTT TA-3′, antisense 5′-AGC TTA AAA AGA GCA TGC ACA AAC GCT TCT TGA CAG GAA GAG AAG CAT TTG TAC ATG CTC GG), human IRS1 B1 (sense 5′-GAT CCC GCT ATG CTG ACA TGT GAA TAC TTC CTG TCA TGT TCG CAT GTC AGC ATA GCT TTT TA-3′), rat Irs1 shRNA1 (sense 5′-GAT CCC GCC TGG AGT ATT GTG AGA ATG TGT GCT GTC CGT TCT CAT AAT ACT CCA GGC TTT TTA-3′, reverse 5′-AGC TTA AAA AGC CTG GAG TAT TAT GAG AAC GGA CAG CAC ACA TTC TCA CAA TAC TCC AGG CGG-3′), rat Irs1 shRNA2 (sense 5′-GAT CCC GAT TGT TGA GAT GGT GCC TGC TGT GCT GTC GCA GGT ATC ATC TTA ATA GTC TTT TTA-3′, reverse 5′-AGC TTA AAA AGA CTA TTA AGA TGA TAC CTG CGA CAG CAC AGC AGG CAC CAT CTC AAC AAT CGG-3′), and scrambled (sense 5′-GAT CCC TTC TCC GAA CGT GTC ACG TTT CAA GAG AAC GTG ACA CGT TCG GAG AAT TTT TA-3′, reverse 5′-AGC TTA AAA ATT CTC CGA ACG TGT CAC GTT CTC TTG AAA CGT GAC ACG TTC GGA GAA GG-3′).
Focus formation and soft agar growth assays were performed as described previously with the only modification that cells were plated in 12-well plates (28). Mouse xenograft tumor growth was assayed twice weekly after subcutaneous injection of 5 × 106 cells in the flanks of Nu/Nu mice (number 088, Charles River, Wilmington, MA). Volume was estimated using the formula: (length × height × width)/2.
Microarray Analysis
Microarray analysis of derivatives of RK3E samples was performed. Specifically, parental RK3E cells, S33Y-β-catenin-transformed RK3E cells, as well as S33Y-β-catenin-transformed RK3E cells expressing a dominant-negative TCF4 construct (pPGS dnTCF4) or the control vector (pPGS-NEO) were used. To assess for acute inducibility of genes by β-catenin, samples from RK3E/S33Yβ-ER cells treated for 24 h in the presence or absence of 4-OHT were examined. Samples were analyzed on rat RAE_230A and RAE_230B oligonucleotide microarrays containing 31256 probe sets (Affymetrix, Santa Clara, CA). cRNA preparation, hybridization, scanning, and image analysis were performed according to the manufacturer's protocols. Probe-set intensities were obtained and normalized as previously described (29, 30).
Western Blot Analysis
Whole cell lysates were prepared in radioimmunoprecipitation assay buffer (Tris-buffered saline, pH 7.4, 0.5% deoxycholic acid, 0.1% SDS, and 1% Nonidet P-40 with Complete® protease inhibitors (Roche Molecular Biochemicals), 2 mm Na3VO4). Protein concentration was determined by the bicinchoninic acid assay (Pierce Biochemicals), and 30–50 μg of total protein from each sample was separated on SDS-polyacrylamide gels. Proteins were transferred to Immobilon P membranes (Millipore, Bedford, MA) by electroblotting. Immunoblot analyses were carried out with the affinity purified polyclonal rabbit anti-IRS1 antibody (Cell Signaling Technology, Beverly, MA), at a 1:2000 dilution in 1× Tris-buffered saline with 3% bovine serum albumin and 0.5% Tween or a mouse monoclonal antibody against the FLAG epitope (M2, Sigma). To verify equal loading of the samples, membranes were incubated with a mouse monoclonal antibody against β-actin (Sigma). Secondary antibody incubations were performed with horseradish peroxidase-conjugated donkey anti-rabbit IgG or goat anti-mouse IgG antibody (Pierce Biochemicals). Antibody complexes were detected with the ECL Western blot kit (Amersham Biosciences) and exposure to Blue Basic Autorad film (ISC Bioexpress, Kaysville, UT).
Northern Blot Analysis
Total cellular RNA was extracted with TRIzol reagent (Invitrogen). 10 μg of total RNA was separated on a 1.2% formaldehyde-agarose gel and transferred to Zeta-Probe GT membranes (Bio-Rad) by capillary action. cDNA probes to detect IRS1 and glyceraldehyde-3-phosphate dehydrogenase expression in rat and human were generated by reverse transcriptase-PCR, using primers derived from sequences in GenBank™. The sequences of all PCR product probes were confirmed by automated sequencing. All probes were random labeled with [α-32P]dCTP using Redi-prime (Amersham Biosciences) and hybridized to the membrane with RapidHyb buffer (Amersham Biosciences) according to the manufacturer's protocol. All Northern blots were stripped and hybridized to a rat glyceraldehyde-3-phosphate dehydrogenase cDNA probe to control for RNA loading and transfer efficiency.
Immunohistochemistry
Immunohistochemical analysis of β-catenin and IRS1 expression in human OEAs and CRCs and mouse adenomas was performed essentially as described previously (31). In brief, 5-μm sections of formalin-fixed, paraffin-embedded tissues were mounted on Probe-On slides (Fisher Scientific, Hanover Park, IL), deparaffinized in xylene, and then rehydrated into distilled water through graded alcohols. Antigen retrieval was enhanced by microwaving the slides in citrate buffer (pH 6.0, Biogenex, San Ramon, CA) for 15 min. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide in phosphate-buffered saline. Blocking and antibody incubations were performed using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA) according to the manual. The concentration used was 1:50 for the affinity purified rabbit antiserum, from Cell Signaling, and incubating overnight at 4 °C. Immunostained sections were dehydrated, counterstained with hematoxylin, and then examined by light microscopy.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed using SimpleChIP™ Enzymatic Chromatin IP kit with magnetic beads according to the manufacturer (Cell Signaling). Antibodies used were rabbit monoclonal TCF4 and rabbit polyclonal β-catenin antibody (Cell signaling). Primers for the quantitative PCR are available upon request.
Reporter Gene Assays
TOPFLASH and FOPFLASH luciferase constructs contain three consensus β-catenin binding sites or three mutant β-catenin binding sites, respectively, before a minimal promoter driving a firefly luciferase cassette (Upstate). Three separate genomic fragments encompassing three chromatin immunoprecipitation sites were cloned into the NheI site of the negative control FOPFLASH plasmid. Mutations were introduced using the Stratagene QuikChange kit (Stratagene). Transfections were performed with 293T cells in 12-well plates using FuGENE and 0.4 μg of pCDNA3 or pCDNA3 S33Y-β-catenin plasmid, 0.4 μg of the indicated reporter constructs, and 0.1 μg of SV40 Renilla luciferase construct (Promega, Madison, WI). Cells were harvested 48 later. Luciferase activities were measured using a Dual-luciferase kit (Promega).
RESULTS
Regulation of IRS1 by Wnt/β-Catenin Signaling
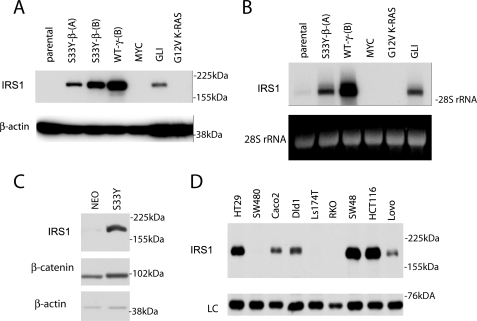
In an effort to identify novel β-catenin target genes we performed microarray analyses of epithelial cells expressing constitutive and conditional oncogenic β-catenin alleles. Previous studies from our laboratory have shown that the rat RK3E epithelial cell line can be neoplastically transformed by ectopic expression of cancer-derived mutant β-catenin alleles (e.g. a codon 33 substitution of tyrosine for serine; S33Y-β-catenin) (28). As such, the RK3E cell line represents a useful experimental system for the identification of genes involved in neoplastic transformation of cells by β-catenin. Among the probe sets with the highest increase in signal intensity in RK3E cells stably expressing the S33Y-β-catenin protein (versus vector control cells) was a probe set predicted to measure the transcript abundance of the Irs1 gene (1369771_at, supplemental Table S1). Upon further inspection, we noted two more probe sets (1374060_at and 1390429_at) targeting a putative extended 3′-untranslated region of Irs1 similarly regulated. To pursue further the role of the Wnt/β-catenin pathway in activating Irs1 expression, we analyzed the expression of IRS1 in derivatives of the RK3E cell line that were neoplastically transformed by selected genes, including S33Y-β-catenin, wild-type γ-catenin, GLI1, MYC, and an oncogenic KRAS cDNA (G12V-K-Ras). Significant increases in the Irs1 gene and protein expression were observed in S33Y-β-catenin and WT-γ-catenin transformed RK3E lines as well as RK3E cells transformed by GLI (Fig. 1, A and B) (27). Notably, MYC and G12V KRAS-transformed RK3E lines did not show increased Irs1 levels, suggesting that Irs1 up-regulation was not merely a consequence of neoplastic transformation per se. Prior work from our laboratory demonstrated that γ-catenin activates a subset of β-catenin target genes and could potentially mediate part of the transcriptional program activated upon inactivation of the APC tumor suppressor (32). Work from Ruppert and colleagues has suggested that GLI1 mediates its oncogenic effects in RK3E cells via β-catenin/TCF and the Ruppert laboratory (33, 34) also reported that Irs1 is up-regulated by GLI1.
FIGURE 1.
IRS1 expression levels are increased by β-catenin signaling in epithelial cells and IRS1 is expressed in the majority of CRC cell lines. A, Western blot and B, Northern blot analyses of parental rat kidney epithelial cell line RK3E and transformed cell lines derived from RK3E cells by transduction of a stabilized mutant of β-catenin (S33Y), wild-type γ-catenin (WTγ), c-MYC, GLI, and G12V mutant KRAS. C, Western blot analysis of IRS1 expression in rat intestinal epithelial cell line IEC18 after transduction of activated β-catenin (S33Y) or empty vector (NEO). D, Western blot analysis of IRS1 expression in selected colorectal carcinoma cell lines. In all panels, equal loading is demonstrated by immunoblotting for β-actin or ethidium bromide staining of 28 S ribosomal RNA, respectively.
To assess the ability of β-catenin/TCF to activate Irs1 gene expression in other settings, we ectopically expressed a stabilized mutant of β-catenin (S33Y-β-catenin) by retroviral transduction in rat intestinal epithelial cell line IEC18 and selected a polyclonal cell population of transduced cells. As shown by Western blot analysis, we observed a significant increase in Irs1 expression levels in the β-catenin-transduced IEC18 cells (Fig. 1C). We then analyzed whether IRS1 is expressed in human colorectal cancer cell lines. By Western blot analysis, IRS1 protein expression was clearly detectable in six of nine colorectal cancer cell lines (Fig. 1D). Notably, in RKO cells, the only one of the nine colorectal cancer cell lines studied that lacks activation of the APC/Wnt/β-catenin signaling pathway, IRS1 levels were not elevated. The basis for why IRS1 was not highly expressed in the SW480 and Ls174T cell lines is not known. It is, however, not uncommon for β-catenin target genes to be variably expressed in colorectal cancers with β-catenin dysregulation (35, 36).
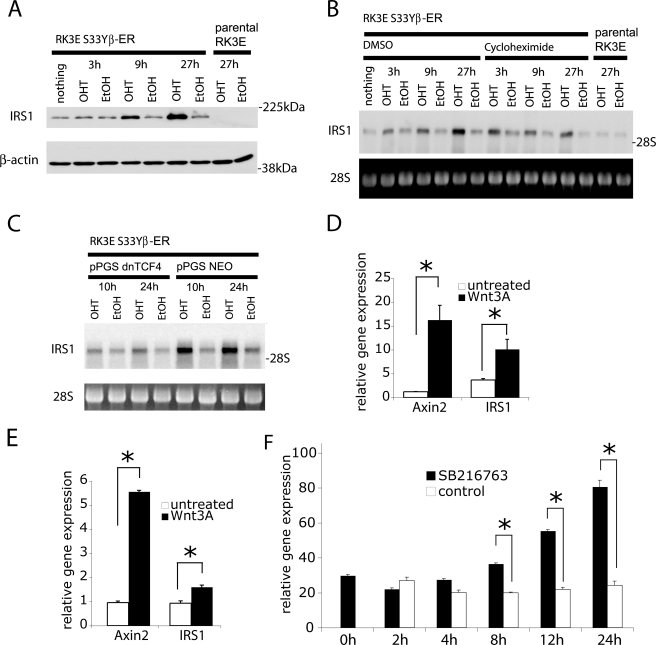
To further characterize the role of β-catenin in activation of Irs1 gene expression, we used a polyclonal cell line stably expressing a chimeric β-catenin protein in which estrogen receptor hormone binding domain sequences were fused in-frame to the full-length sequences for stabilized mutant S33Y-β-catenin (RK3E/S33Y-ER) (37). In the absence of the ligand 4-OHT, this fusion protein resides mostly in an inactive state as a result of its binding to heat shock family proteins. Upon addition of the ligand 4-OHT, the fusion protein is released from this sequestration and can activate downstream target genes of β-catenin. Because activation of the fusion protein does not require de novo protein synthesis, the ER fusion protein system offers a powerful method to determine whether activation of downstream transcriptional targets is independent of protein synthesis and therefore likely to be mediated directly by a transcription factor or co-factor of interest.
In a first step, we pursued Western blot assays to assess effects on Irs1 expression levels in these cell lines upon addition of 4-OHT. As shown in Fig. 2A, addition of 4-OHT led to a robust increase in Irs1 protein levels within 9 h. Prior studies have shown that β-catenin/TCF transcription is not activated immediately upon 4-OHT addition, but that several hours are needed for substantial accumulation of the β-catenin-ER fusion protein in the nucleus following 4-OHT addition (37). We observed further increases in Irs1 levels at 27 h after 4-OHT addition. We then analyzed the transcript levels of Irs1 in cell lines that had been pretreated with the inhibitor of protein synthesis cycloheximide or DMSO as solvent control 15 min before treatment with 4-OHT. We observed that Irs1 transcript levels began to be increased as early as 3 h after 4-OHT (Fig. 2B). Inhibition of protein synthesis by cycloheximide treatment did not prevent the induction of IRS1 transcripts, indicating that regulation of IRS1 gene expression by β-catenin does not require new protein synthesis. Hence, our findings imply that IRS1 is a target gene directly regulated by β-catenin action.
FIGURE 2.
The IRS1 gene is a direct target of β-catenin signaling and IRS1 transcript induction by β-catenin does not require protein synthesis. A, Western blot analysis of a polyclonal RK3E cell line expressing a fusion protein of the hormone binding domain of the murine estrogen receptor (RK3E-S33Y-ER) and a stabilized mutant of β-catenin. IRS1 protein levels were determined at the indicated time points after addition of 4-hydroxytamoxifen (4OHT, 500 nm) or a solvent control (Ethanol). B, Northern blot analysis of samples from parental RK3E cells and the RK3E-S33Y-ER cell line stimulated with 4-OHT for the indicated times. Cycloheximide (1.5 μg/ml) or DMSO (i.e. the solvent for cycloheximide) were added 15 min. before treatment with 4-OHT. C, Northern blot analysis of samples from the RK3E-S33Y-ER cell line transduced with a retrovirus to express a dominant-negative mutant of TCF4 (dnTCF4) or control vector. Irs1 mRNA were determined after treatment with 4-OHT or ethanol for 10 and 24 h. In all panels, equal loading is demonstrated by immunoblotting for β-actin or ethidium bromide staining of 28 S ribosomal RNA, respectively. D, IEC6 rat intestinal epithelial cell and E, immortalized ovarian surface epithelium cell lines were treated for 8 h with 50 ng/ml of Wnt3a. Expression of AXIN2 and IRS1 was measured by quantitative PCR and normalized to the expression levels of U6 (D) or HPRT (E), respectively. F, MDAH-2774 cells were treated for the indicated times with the GSK3 inhibitor SB216763. Expression of IRS1 was measured by quantitative PCR and normalized to the expression level of HPRT. Asterisks denote p < 0.05 in Student's t test, and error bars denote S.D.
Although expression of mutant β-catenin alleles clearly up-regulates IRS1 expression, we were also interested to see whether treatment of cells with Wnt ligands could have a similar effect. To this end, we treated an immortalized rat intestinal epithelial cell line (IEC6) and immortalized ovarian surface epithelial cells with 50 ng/ml of recombinant mouse Wnt3a. In both cases, we observed a significant up-regulation of IRS1 mRNA levels after 8 h of exposure to Wnt ligand (Fig. 2, D and E). Given the central role of GSK3β in the destruction machinery of β-catenin, we also inhibited GSK3β activity using the specific inhibitor SB216763 (38). Again we observed a time-dependent and significant increase of IRS1 mRNA levels. Overall, the data suggest that IRS1 is indeed a direct target of the Wnt/β-catenin signaling pathway.
We therefore set out to further study the mechanism of this regulation. Because β-catenin is known to mediate effects on target genes in part via its interaction with TCF/LEF transcription factors, we analyzed whether ectopic expression of a dominant-negative form of TCF4 would antagonize β-catenin-mediated induction of IRS1 expression. However, we found no significant inhibition of IRS1 expression following ectopic expression of dominant-negative TCF4 in IEC18 or RK3E cell lines that were already stably transformed by β-catenin or in colorectal cancer cells with mutational stabilization of β-catenin (data not shown). These results are consistent with the observation from Clevers and co-workers (39) microarray analyses of colorectal carcinoma cell lines DLD1 and LS174T carrying an inducible dominant-negative TCF4 mutant, where no inhibition of IRS1 gene expression was observed following expression of the dominant-negative TCF4 protein. At first sight, this result might suggest that IRS1 is independent of TCF/LEF factors. However, it leaves the possibility that activation of IRS1 transcription by β-catenin and TCF/LEFs could occur at early stages of the transformation process, but β-catenin and TCF/LEFs might not be required following the initial transcriptional activation, perhaps because epigenetic changes (e.g. chromatin modification and/or remodeling) lead to stable patterns of IRS1 expression. We therefore expressed a dominant-negative form of TCF4 in the RK3E-S33Y-ER cell line. Upon addition of 4-OHT we observed a significantly lowered, albeit not totally inhibited induction of IRS1, consistent with the notion that TCF/LEF factors are in fact contributing to the initial induction of IRS1 in non-transformed cell lines (Fig. 2C).
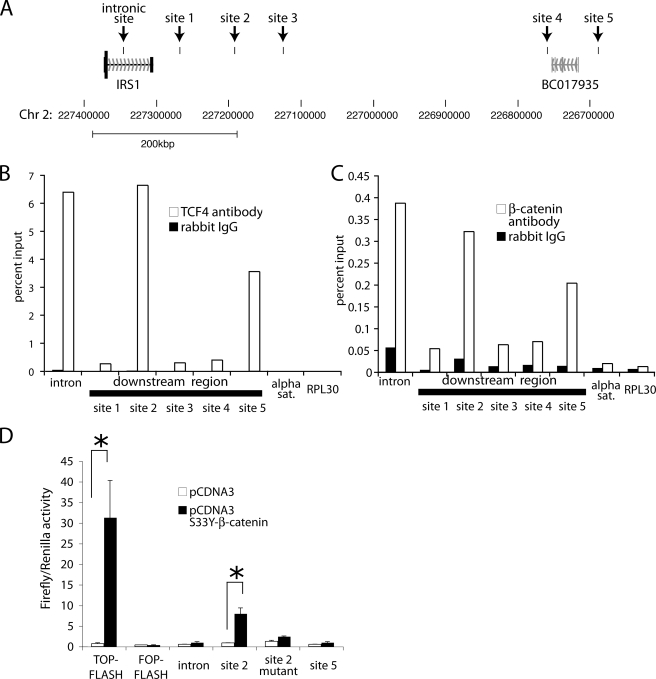
In an effort to delineate the location of the TCF/LEF responsive elements at the IRS1 locus, a 14-kb promoter fragment was cloned upstream of a luciferase expression cassette. This construct showed considerable activation of the luciferase gene transcription at baseline. However, upon ectopic expression of S33Y β-catenin, no further increase in reporter gene activity was observed (data not shown). The unprocessed IRS1 transcript comprises two exons separated by a large intron of 58.7 kbp (Fig. 3A). The entire coding sequence is contained in the first exon. However, the second exon is nevertheless highly conserved in various species. In a recent genome-wide chromatin immunoprecipitation study using antibodies for TCF4, several genomic binding sites for TCF4 were observed in a 500-kb region downstream of the IRS1 gene as well in the first intron of IRS1 (Fig. 3A) (40). Given the fact that no known genes are transcribed in this genomic region downstream of IRS1, we hypothesized that IRS1 expression might be regulated by the combined action of enhancers localized in its first intron and downstream of its transcriptional unit. To investigate this possibility we first validated that TCF4 in fact binds to the sites that were identified in the genome wide study of Hatzis et al. (40). As shown in Fig. 3B, TCF4 showed strong binding and allowed robust detection by PCR after chromatin immunoprecipitation for three of the six regions tested. Notably, these three regions also show significant binding to β-catenin (Fig. 3C). Predicted TCF/LEF binding sites were present in these genomic fragments. We therefore cloned the genomic regions encompassing the strongest TCF/LEF/β-catenin binding sites (i.e. the intronic site, site 2, and site 5) in a luciferase reporter construct containing a minimal promoter, but no functional known or predicted TCF/LEF binding sites. Cotransfection of one of these reporter constructs (downstream site 2) with a stabilized mutant of β-catenin led to a strong transcriptional activation (×9) (Fig. 3D). Site-directed mutagenesis of the putative TCF binding site in the constructs allowed us to demonstrate that this activation is largely dependent on one TCF/LEF binding site (i.e. site 2 mutant). It is unclear why the remaining two reporter constructs only showed marginal activation upon cotransfection with β-catenin. It is, however, conceivable that the reporter constructs might not adequately represent the endogenous IRS1 locus, where genomic DNA is packaged into chromatin and bound by numerous other transcription factors. Taken together, our data suggest that acute activation of β-catenin rapidly activates IRS1 expression without the requirement of protein synthesis in a process involving TCF/LEF binding to a downstream enhancer (with possible contribution of intronic and further downstream sites).
FIGURE 3.
IRS1 directly binds to downstream enhancers of IRS1. A, schematic representation of the IRS1 locus. Black boxes mark the tested IRS1 binding sites in the intron and downstream of the IRS1 transcript. B, chromatin immunoprecipitation of DLD1 cells using an antibody against TCF4 shows binding to intronic and regions downstream of the IRS1 gene. Relative recovery of the immunoprecipitation was measured by quantitative PCR and is normalized to the amount in the chromatin immunoprecipitation input. Chromatin immunoprecipitation using IgG and amplification of irrelevant multicopy (α-sat) or single copy (RPL30) locus were used demonstrate specificity. Recovery of RPL30 and α-satellite with TCF4 antibody was 0.048 and 0.040%, respectively, which is not visible in this scale. C, chromatin immunoprecipitation using an antibody against β-catenin was performed as described in B. D, 293T cells were transfected with the indicated reporter plasmids containing a minimal promoter and the genomic region encompassing the intronic as well as the strongest binding sites of β-catenin downstream of the IRS1 gene. Firefly luciferase activity in the absence or presence of S33Y β-catenin plasmid were normalized to the activity of a cotransfected constitutively active Renilla luciferase construct. Asterisks denote significance with p < 0.05 in Student's t test, and error bars denote S.D.
IRS1 Is Overexpressed in Tumors with Wnt/β-Catenin Pathway Deregulation
To determine the expression level of IRS1 in primary CRCs, we performed an immunohistochemical analysis using a tissue microarray consisting of 44 colorectal carcinoma specimens and three normal mucosa samples. The normal colorectal mucosa tissues showed low to moderate expression levels of IRS1 (supplemental Fig. S1). Of the 44 colorectal carcinoma samples studied, seven showed high levels of IRS1 expression, 21 showed moderate levels, and 16 showed low levels. Interestingly, four of the seven tumors with high IRS1 expression were mucinous tumors (57%), whereas only four mucinous tumors were represented in the remaining moderately and weakly staining tumors (10.8%).
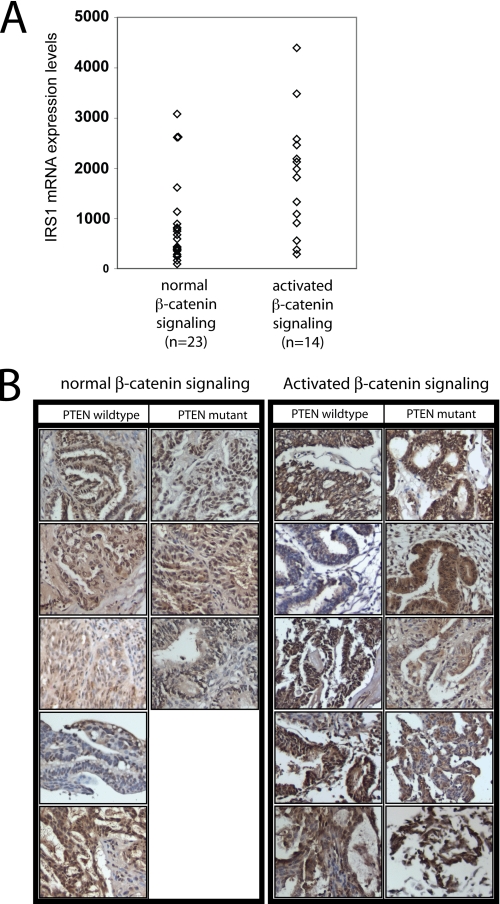
Because most CRCs show dysregulation of β-catenin, it is difficult to judge whether increased IRS1 expression is a consequence of β-catenin signaling or whether it might be due to alterations in unrelated pathways in CRC. The situation is a bit different in ovarian endometrioid adenocarcinomas (OEAs), where 30% show β-catenin dysregulation due to mutations in β-catenin, APC, or AXIN1 and 70% have no apparent dysregulation of β-catenin or mutations in key Wnt/β-catenin components (31). We analyzed IRS1 expression initially using a previously published highly annotated Affymetrix gene expression profiling dataset of OEAs (Gene Expression Omnibus GSE 6008) (31). As shown in Fig. 4A, average IRS1 gene expression levels are 2.07-fold higher in OEAs with β-catenin dysregulation than in other tumors, suggesting that β-catenin signaling indeed plays a role for the endogenous expression levels of IRS1 (p = 0.003, t test on log-transformed values).
FIGURE 4.
IRS1 expression correlates with β-catenin activation in ovarian endometrioid adenocarcinomas. A, expression level of IRS1 as assessed by Affymetrix GeneChip hybridization of ovarian endometroid adenocarcinomas (GSE6008) with or without nuclear β-catenin staining. Significance was assessed using the two-sample t test of log-transformed values. B, immunohistochemical staining for IRS1 expression in ovarian endometrioid adenocarcinomas with or without the activated β-catenin signaling pathway. Tumor samples were subdivided into samples showing inactivation of PTEN or not.
IRS1 protein stability and translation is highly regulated (41–44). It is therefore not immediately obvious that a difference of IRS1 mRNA levels between these tumor types would be paralleled by an increase in IRS1 protein levels. To address this, we performed IRS1 IHC studies in a collection of OEAs where the β-catenin mutational status was previously defined. Strong staining for IRS1 was observed in 6 of 10 carcinomas with β-catenin mutational dysregulation. Only 2 of 8 OEAs lacking β-catenin dysregulation showed similar IRS1 staining results (Fig. 4B). Due to the small numbers of available OEAs, the comparison was not statistically significant, although the trend is consistent with the notion that IRS1 protein and RNA levels are dysregulated in human OEAs with β-catenin dysregulation.
IRS1 Is Required for Transformation of RK3E Cells by Oncogenic Variants of β-Catenin
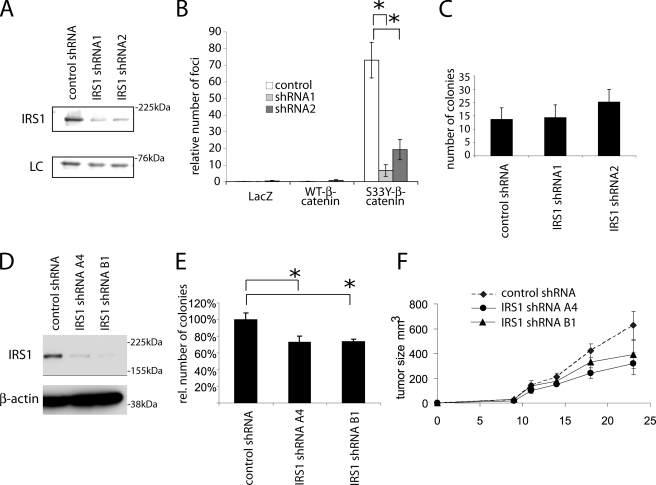
To characterize the role of IRS1 in neoplastic transformation of epithelial cells, we created short hairpin RNA expression cassettes that specifically target the rat Irs1 transcript. Two of these allowed efficient knockdown of Irs1 expression in RK3E cells transformed by S33Y-β-catenin (Fig. 5A). We subsequently transduced parental RK3E cells with these expression cassettes. After 7 days of selection, this polyclonal population of cells was super-infected with retroviruses allowing the expression of WT-β-catenin, S33Y-β-catenin, and LacZ. As previously described, upon transduction with S33Y-β-catenin, RK3E cells formed numerous transformed foci (Fig. 5B and supplemental Fig. S2). The number of these foci was reduced by 70–90% in cell lines expressing short hairpin RNAs against Irs1, implicating IRS1 expression and function as a critical factor in transformation of RK3E cells by β-catenin (Fig. 5B). Of course, this raises the question whether it is really the β-catenin-dependent increase of Irs1 that is required for the neoplastic transformation process or whether just some baseline Irs1 might be required for the transformation process. To exclude that Irs1 knockdown generally inhibits neoplastic transformation of RK3E cell lines, in parallel we infected the Irs1 knockdown cell lines with retroviruses encoding the mutant K-Ras G12V protein. Focus formation by the mutant K-Ras in a monolayer of RK3E cells generates very large and more poorly demarcated foci that are difficult to assess by quantitative methods. However, introduction of mutant KRAS into RK3E cells leads to robust colony formation in soft agar (28, 45, 46). We therefore reasoned that if fully intact basal levels of Irs1 are required for initiation of RK3E transformation, the possible effects of Irs1 inhibition on KRAS transformation might possibly be even more pronounced in soft agar growth assays, because additional neoplastic capacities are required for cells to grow in soft agar. When analyzing neoplastic transformation efficiency by KRAS, we did not observed similar inhibition suggesting that knockdown of Irs1 does not generally block neoplastic transformation (Fig. 5C).
FIGURE 5.
Down-regulation of IRS1 expression by RNA interference reduces β-catenin-dependent neoplastic transformation. A, Western blot analysis of IRS1 protein levels in the S33Y-β-catenin-transformed RK3E cell lines, upon retroviral transduction with constructs driving expression of two different shRNAs targeting Irs1 (shRNA1 and shRNA2) and a nonsilencing control shRNA. To demonstrate equal loading a nonspecific band at 66 kDa is shown. B, transformation of RK3E cells expressing different Irs1 shRNAs after transduction with retroviruses driving expression of a stabilized mutant of β-catenin (S33Y), wild-type β-catenin (WT-β), or β-galactosidase (LacZ). Cells were transduced with the corresponding retroviruses and focus formation was observed for 3 weeks. Bars denote the number of foci and standard deviation from three independent experiments. Representative methylene blue-stained plates are shown in supplemental Fig. S2. C, soft agar colony formation of RK3E cells expressing two different IRS1 shRNAs or a negative control shRNA after transduction with G12V mutant K-Ras. Cells were plated in 0.3% soft agar 2 days after transduction (without selection) and cultured for 2 weeks. Values represent the colony numbers of triplicates, mean ± S.D. Asterisks denote p < 0.05 in Student's t test. D, Western blot analysis of IRS1 protein levels in HT29 colorectal cancer cells upon transduction with retroviruses driving expression of two different shRNAs targeting IRS1 (shRNA-A4 and shRNA-B1) or a nonsilencing control shRNA. To demonstrate equal loading β-actin levels are shown. E, soft agar colony formation of the cells from D. Colony numbers are represented relative to the number of colonies in the cell line expressing a nonsilencing shRNA. Values are mean and S.D. of three experiments performed in triplicates. F, xenograft tumor growth after subcutaneous injection of the cells from C in immunocompromised nude mice. Five mice for each group were injected on both flanks (i.e. 10 tumors per group). Asterisks denote significance with p < 0.05 in Student's t test, and error bars represent mean ± S.E.
In light of the high levels of IRS1 expression in numerous CRC cell lines, we also studied the role of IRS1 in the maintenance of the neoplastic phenotype. We focused on cell line HT29, which expresses high levels of IRS1 and has previously been shown to express high levels of insulin-like growth factor receptor on the cell surface. Expression of two independent shRNAs strongly down-regulated IRS1 levels (Fig. 5D). When grown in soft agar, we observed a small but reproducible reduction in colony formation when IRS1 expression was inhibited (Fig. 5E). To verify that IRS1 inhibition affected HT29 tumorigenicity in vivo, we injected the HT29 polyclonal shRNA cells lines subcutaneously into nude mice. Over the next 4 weeks we assessed tumor size every 4 days. Tumor cell lines expressing short hairpin RNAs against IRS1 showed slower growth in nude mice, albeit not to statistical significance (Fig. 4F). In contrast to the striking effect of IRS1 shRNA expression on the initiation of the neoplastic transformation process, high levels of IRS1 expression do not seem to be essential for the maintenance of the neoplastic phenotype in CRC cells.
DISCUSSION
Context-dependent Direct Regulation of IRS1 by β-Catenin
We found ectopic expression of a stabilized oncogenic form of β-catenin or acute activation of a hormone-regulated mutant β-catenin protein leads to induction of IRS1 expression. Inhibition of protein synthesis by cycloheximide does not prevent induction of IRS1 transcripts, suggesting that IRS1 transcription is likely to be directly regulated by β-catenin and not by further downstream β-catenin-regulated transcription factors. β-Catenin is believed to exert its transcriptional effects mainly by binding to members of the TCF/LEF family (e.g. TCF4/TCF7L2). Indeed, ectopic expression of a dominant-negative form of TCF4/TCF7L2 (dnTCF) inhibited the induction of IRS1 by S33Y-ER-β-catenin in RK3E cells, suggesting β-catenin activates IRS1 expression at least in part via TCF/LEF transcription factors.
Our investigation of sequence elements in the IRS1 transcriptional unit that might be responsive to β-catenin suggests that regulation of IRS1 by β-catenin/TCF may differ to some degree from previously characterized β-catenin/TCF target genes. A 14-kb region upstream of the transcriptional start site did not confer β-catenin responsiveness in reporter assays. However, given the rather unusual genomic organization of the IRS1 locus, with the complete open reading frame in the first exon and a conserved noncoding second exon downstream of a large intron of >50 kb, we wondered whether the IRS1 gene might be regulated by downstream or intronic enhancers. We demonstrated that three of six of the regions identified as potential genomic TCF4 binding sites in a recent genome-wide approach, in fact bind both TCF4 and β-catenin (40). Interestingly, recent work by Yochum et al. (47) suggests that a major context-dependent β-catenin-responsive enhancer in MYC is also localized downstream of its transcriptional unit. Several well characterized β-catenin target genes (e.g. AXIN2, ENC1, BMP4, and c-MYC), show numerous TCF binding sites distributed over large genomic regions and not simply a localization of key TCF elements to the putative transcriptional start site region (40, 47).
The existence of a genomic insulator sequence between the IRS1 transcriptional start site and the TCF/β-catenin binding sites in the gene might account for the failure of IRS1 expression to be repressed upon overexpression of the dominant-negative mutant of TCF in cancer cell lines with constitutively active β-catenin (48). For instance, β-catenin might play a key role in mediating the initial activation of the IRS1 promoter and remodeling of chromatin for further access by other transcription factors (49). After this initial activation and remodeling, perhaps β-catenin is not continuously required for maintenance of IRS1 expression. However, other explanations are also possible. Several TCF/LEF factors are transcribed from alternative transcriptional start sites and show numerous splice variants (50–52). There is strong evidence that TCF4 and TCF1 are essential binding partners for β-catenin during intestinal development (53), although the roles of TCF4 and TCF1 in mediating the effects of β-catenin in the nucleus in colorectal cancer are potentially more ambiguous. In fact, recent data suggest that in colorectal cancer cells TCF4 has an inhibitory effect on β-catenin-dependent transcription (54). As such, in the context of the regulation of IRS1 by β-catenin and TCF/LEF proteins, it is conceivable that TCF/LEF factors other than TCF4 might be responsible for regulating IRS1 gene expression. Overexpression of a dominant-negative TCF4 mutant protein in colorectal cancer cells (i.e. once these sites are occupied by higher affinity alternative TCF/LEF factors) might not be sufficient to antagonize IRS1 expression. Further studies are needed to elucidate the differential expression repertoire of TCF/LEF factor splice variants in colorectal carcinoma and normal tissue, as well as their functional contribution to the activation of particular β-catenin target genes, such as IRS1.
IRS1 and the Cooperation of Wnt/β-Catenin and IGF in Neoplastic Transformation
The Wnt/β-catenin pathway is activated in the majority of colorectal carcinomas (CRC). Several lines of evidence demonstrate that a cooperation of Wnt/β-catenin and IGF signaling exists, and might contribute to colorectal carcinogenesis. As described in the Introduction, loss of imprinting of the IGF2 gene and the ensuing bi-allelic expression appears to be a significant risk factor for colorectal cancer (6), and mice carrying one truncating allele of the APC tumor suppressor gene (ApcMin) develop fewer polyps in an IGF2+/− background and develop more polyps when IGF2 is transgenically overexpressed (9). The extent of the involvement of IRS1 in these effects is not clear. However, several observations implicate IRS1 in the development of cancer and cellular transformation induced by the Wnt/β-catenin pathway. Epidemiological studies have shown that a polymorphism in the coding region of IRS1 is associated with an increased incidence of colorectal cancers (10). Although its function as a cytosolic adaptor protein is best characterized, several articles (19–25) have contributed to our understanding that IRS1 might also act in the nucleus, where it increases transcription of ribosomal RNA and might interact with β-catenin. Finally, in a recent study by Ramocki et al. (26) it was noted that mice carrying one inactivating mutation in the murine Apc gene (ApcMin) develop significantly fewer intestinal adenomatous polyps when the Irs1 gene is mutated (Irs1−/− mice), suggesting that IRS1 is required for efficient intestinal tumorigenesis in vivo. Irs1−/− mice are often runted (55). Hence, general growth defects and secondary effects in addition to the observed increased level of apoptosis might have contributed to the observed phenotype.
In the current study we investigated the effect of shRNA-mediated down-regulation of IRS1 on the neoplastic transformation of the epithelial cell line RK3E by S33Y-β-catenin and K-Ras G12V. We observed that upon down-regulation of IRS1, transformation by S33Y-β-catenin was strongly inhibited, whereas this was not the case for the oncogenic mutant K-Ras G12V (Fig. 4). A general growth defect therefore does not seem to be responsible for the reduced transformation potential of β-catenin when IRS1 levels are reduced. Notably, this effect of IRS1 knockdown on de novo transformation was much stronger than the phenotypic effect of down-regulation of IRS1 in cells that have already been transformed. Our data therefore support the notion that changing IRS1 levels can modulate the initiation of the epithelial neoplastic transformation process by S33Y-β-catenin. This is also in line with the observation of Ramocki et al. (26) that the number of APCMIN adenomas in Irs1−/− mice is more strongly reduced than their size.
These observations may be somewhat reminiscent of the roles for IRS1 in mammary tumorigenesis. Transgenic overexpression of IRS1 is capable of inducing tumors, however, persistent IRS1 expression in some settings might inhibit further tumor progression (25, 56).
IRS1 Is Expressed in Cancers That Show β-Catenin Activation
We observed a significantly higher expression of IRS1 mRNA in ovarian endometrioid adenocarcinomas with β-catenin dysregulation versus cases without these changes (fold-change 2.07, p = 0.003). When analyzing the IRS1 protein levels by immunohistochemistry, the effect were less clearcut. Although the percentage of cases with strong staining for IRS1 was higher in cases with β-catenin activation (60%) versus the remaining cases (25%), this did not reach statistical significance. A likely explanation for this divergence lies in the existence of significant feedback mechanisms. IRS1 protein levels are strongly regulated post-transcriptionally (42, 43). Most notably, activation of mTOR signaling downstream of IRS1, phosphatidylinositol 3-kinase, and protein kinase B leads to phosphorylation and subsequent destabilization of IRS1 (42). In the case of OEAs, mutational activation of β-catenin and phosphatase and tenson homolog (PTEN) inactivation are significantly correlated (31). Because PTEN inactivation, in turn, activates mTOR activity, the post-translational reduction of IRS1 protein levels due to PTEN inactivation might dampen the increase of IRS1 induced by transcriptional activation through β-catenin. In fact, it is conceivable that selective pressure to inactivate both PTEN and β-catenin together might in part reflect the requirement of β-catenin to increase IRS1 mRNA expression in a setting where the IRS1 protein is destabilized (e.g. due to PTEN inactivation). Due to the limited availability of OEA tissues with these distinct genetic alterations, we were not able to further investigate this hypothesis.
Taken together, we have provided evidence that IRS1 is a direct β-catenin target gene whose expression can be regulated by a distal enhancer. It is strongly expressed in several cancers that carry constitutive β-catenin signaling. Our data in the RK3E epithelial cell line and the recent report by Ramocki et al. (26) suggest that IRS1 might be an important regulator of the initiation of the neoplastic transformation of the epithelial cell by β-catenin.
Supplementary Material
Acknowledgment
We thank Dr. Yali Zhai for help with immunohistochemistry.
This work was supported, in whole or in part, by National Institutes of Health Grants CA085463 (to E. R. F.), CA094172 and RO1CA94172 (to K. R. C.), and CA046592 (to the University of Michigan Comprehensive Cancer Center core facility support), and the Region Bruxelles-Capitale Grant IRSIB BB2B 2008-1-01 (to G. T. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- GSK3
- glycogen synthase kinase 3β
- IEC
- immortalized rat intestinal epithelial cell
- OEA
- ovarian endometrioid adenocarcinoma
- CRC
- colorectal carcinoma
- APC
- adenomatous polyposis coli
- TCF
- T cell factor
- IRS1
- insulin receptor substrate 1
- ER
- endoplasmic reticulum
- 4-OHT
- 4-hydroxytamoxifen
- WT
- wild-type
- IEC
- intestinal epithelial cell
- shRNA
- short hairpin RNA
- IGF
- insulin-like growth factor
- PTEN
- phosphatase and tensin homolog.
REFERENCES
- 1.Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 3.Polakis P. (2007) Curr. Opin. Genet. Dev. 17, 45–51 [DOI] [PubMed] [Google Scholar]
- 4.Städeli R., Hoffmans R., Basler K. (2006) Curr. Biol. 16, R378–R385 [DOI] [PubMed] [Google Scholar]
- 5.Sakatani T., Kaneda A., Iacobuzio-Donahue C. A., Carter M. G., de Boom Witzel S., Okano H., Ko M. S., Ohlsson R., Longo D. L., Feinberg A. P. (2005) Science 307, 1976–1978 [DOI] [PubMed] [Google Scholar]
- 6.Cui H., Cruz-Correa M., Giardiello F. M., Hutcheon D. F., Kafonek D. R., Brandenburg S., Wu Y., He X., Powe N. R., Feinberg A. P. (2003) Science 299, 1753–1755 [DOI] [PubMed] [Google Scholar]
- 7.Dearth R. K., Cui X., Kim H. J., Hadsell D. L., Lee A. V. (2007) Cell Cycle 6, 705–713 [DOI] [PubMed] [Google Scholar]
- 8.Tseng Y. H., Ueki K., Kriauciunas K. M., Kahn C. R. (2002) J. Biol. Chem. 277, 31601–31611 [DOI] [PubMed] [Google Scholar]
- 9.Hassan A. B., Howell J. A. (2000) Cancer Res. 60, 1070–1076 [PubMed] [Google Scholar]
- 10.Slattery M. L., Samowitz W., Curtin K., Ma K. N., Hoffman M., Caan B., Neuhausen S. (2004) Cancer Epidemiol. Biomark. Prev. 13, 1206–1214 [PubMed] [Google Scholar]
- 11.Cristofanelli B., Valentinis B., Soddu S., Rizzo M. G., Marchetti A., Bossi G., Morena A. R., Dews M., Baserga R., Sacchi A. (2000) Oncogene 19, 3245–3255 [DOI] [PubMed] [Google Scholar]
- 12.Sun H., Baserga R. (2008) J. Cell. Physiol. 215, 725–732 [DOI] [PubMed] [Google Scholar]
- 13.Valentinis B., Baserga R. (2001) Mol. Pathol. 54, 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka S., Wands J. R. (1996) Cancer Res. 56, 3391–3394 [PubMed] [Google Scholar]
- 15.Surmacz E., Burgaud J. L. (1995) Clin. Cancer Res. 1, 1429–1436 [PubMed] [Google Scholar]
- 16.D'Ambrosio C., Keller S. R., Morrione A., Lienhard G. E., Baserga R., Surmacz E. (1995) Cell Growth & Differ. 6, 557–562 [PubMed] [Google Scholar]
- 17.Tanaka S., Wands J. R. (1996) J. Clin. Invest. 98, 2100–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang Q., Li Y., White M. F., Fletcher J. A., Xiao S. (2002) Cancer Res. 62, 6035–6038 [PubMed] [Google Scholar]
- 19.Wu A., Chen J., Baserga R. (2008) Oncogene 27, 397–403 [DOI] [PubMed] [Google Scholar]
- 20.Drakas R., Tu X., Baserga R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9272–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu A., Sciacca L., Baserga R. (2003) J. Cell. Physiol. 195, 453–460 [DOI] [PubMed] [Google Scholar]
- 22.Tu X., Wu A., Maiorana A., Baserga R. (2003) Horm. Metab. Res. 35, 734–739 [DOI] [PubMed] [Google Scholar]
- 23.Prisco M., Santini F., Baffa R., Liu M., Drakas R., Wu A., Baserga R. (2002) J. Biol. Chem. 277, 32078–32085 [DOI] [PubMed] [Google Scholar]
- 24.Chen J., Wu A., Sun H., Drakas R., Garofalo C., Cascio S., Surmacz E., Baserga R. (2005) J. Biol. Chem. 280, 29912–29920 [DOI] [PubMed] [Google Scholar]
- 25.Dearth R. K., Cui X., Kim H. J., Kuiatse I., Lawrence N. A., Zhang X., Divisova J., Britton O. L., Mohsin S., Allred D. C., Hadsell D. L., Lee A. V. (2006) Mol. Cell. Biol. 26, 9302–9314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramocki N. M., Wilkins H. R., Magness S. T., Simmons J. G., Scull B. P., Lee G. H., McNaughton K. K., Lund P. K. (2008) Endocrinology 149, 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster K. W., Ren S., Louro I. D., Lobo-Ruppert S. M., McKie-Bell P., Grizzle W., Hayes M. R., Broker T. R., Chow L. T., Ruppert J. M. (1999) Cell Growth & Differ. 10, 423–434 [PubMed] [Google Scholar]
- 28.Kolligs F. T., Hu G., Dang C. V., Fearon E. R. (1999) Mol. Cell. Biol. 19, 5696–5706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giordano T. J., Kuick R., Thomas D. G., Misek D. E., Vinco M., Sanders D., Zhu Z., Ciampi R., Roh M., Shedden K., Gauger P., Doherty G., Thompson N. W., Hanash S., Koenig R. J., Nikiforov Y. E. (2005) Oncogene 24, 6646–6656 [DOI] [PubMed] [Google Scholar]
- 30.Shedden K., Chen W., Kuick R., Ghosh D., Macdonald J., Cho K. R., Giordano T. J., Gruber S. B., Fearon E. R., Taylor J. M., Hanash S. (2005) BMC Bioinformatics 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu R., Hendrix-Lucas N., Kuick R., Zhai Y., Schwartz D. R., Akyol A., Hanash S., Misek D. E., Katabuchi H., Williams B. O., Fearon E. R., Cho K. R. (2007) Cancer Cell 11, 321–333 [DOI] [PubMed] [Google Scholar]
- 32.Kolligs F. T., Kolligs B., Hajra K. M., Hu G., Tani M., Cho K. R., Fearon E. R. (2000) Genes Dev. 14, 1319–1331 [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Deng W., Lobo-Ruppert S. M., Ruppert J. M. (2007) Oncogene 26, 4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louro I. D., Bailey E. C., Li X., South L. S., McKie-Bell P. R., Yoder B. K., Huang C. C., Johnson M. R., Hill A. E., Johnson R. L., Ruppert J. M. (2002) Cancer Res. 62, 5867–5873 [PubMed] [Google Scholar]
- 35.Herbst A., Bommer G. T., Kriegl L., Jung A., Behrens A., Csanadi E., Gerhard M., Bolz C., Riesenberg R., Zimmermann W., Dietmaier W., Wolf I., Brabletz T., Göke B., Kolligs F. T. (2009) Gastroenterology 137, 639–648, 648.e1–9 [DOI] [PubMed] [Google Scholar]
- 36.Batlle E., Bacani J., Begthel H., Jonkheer S., Gregorieff A., van de Born M., Malats N., Sancho E., Boon E., Pawson T., Gallinger S., Pals S., Clevers H. (2005) Nature 435, 1126–1130 [DOI] [PubMed] [Google Scholar]
- 37.Kolligs F. T., Nieman M. T., Winer I., Hu G., Van Mater D., Feng Y., Smith I. M., Wu R., Zhai Y., Cho K. R., Fearon E. R. (2002) Cancer Cell 1, 145–155 [DOI] [PubMed] [Google Scholar]
- 38.Coghlan M. P., Culbert A. A., Cross D. A., Corcoran S. L., Yates J. W., Pearce N. J., Rausch O. L., Murphy G. J., Carter P. S., Roxbee Cox L., Mills D., Brown M. J., Haigh D., Ward R. W., Smith D. G., Murray K. J., Reith A. D., Holder J. C. (2000) Chem. Biol. 7, 793–803 [DOI] [PubMed] [Google Scholar]
- 39.van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A. P., Tjon-Pon-Fong M., Moerer P., van den Born M., Soete G., Pals S., Eilers M., Medema R., Clevers H. (2002) Cell 111, 241–250 [DOI] [PubMed] [Google Scholar]
- 40.Hatzis P., van der Flier L. G., van Driel M. A., Guryev V., Nielsen F., Denissov S., Nijman I. J., Koster J., Santo E. E., Welboren W., Versteeg R., Cuppen E., van de Wetering M., Clevers H., Stunnenberg H. G. (2008) Mol. Cell. Biol. 28, 2732–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Easton J. B., Kurmasheva R. T., Houghton P. J. (2006) Cancer Cell 9, 153–155 [DOI] [PubMed] [Google Scholar]
- 42.Shah O. J., Wang Z., Hunter T. (2004) Curr. Biol. 14, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 43.Shi B., Sepp-Lorenzino L., Prisco M., Linsley P., deAngelis T., Baserga R. (2007) J. Biol. Chem. 282, 32582–32590 [DOI] [PubMed] [Google Scholar]
- 44.Shi B., Prisco M., Calin G., Liu C. G., Russo G., Giordano A., Baserga R. (2006) J. Cell. Physiol. 207, 706–710 [DOI] [PubMed] [Google Scholar]
- 45.Feng Y., Bommer G. T., Zhai Y., Akyol A., Hinoi T., Winer I., Lin H. V., Cadigan K. M., Cho K. R., Fearon E. R. (2007) Cancer Res. 67, 482–491 [DOI] [PubMed] [Google Scholar]
- 46.Feng Y., Lee N., Fearon E. R. (2003) Cancer Res. 63, 8726–8734 [PubMed] [Google Scholar]
- 47.Yochum G. S., Cleland R., Goodman R. H. (2008) Mol. Cell. Biol. 28, 7368–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V., Ren B. (2007) Cell 128, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H. (2001) EMBO J. 20, 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prokunina-Olsson L., Welch C., Hansson O., Adhikari N., Scott L., Usher N., Tong M., Sprau A., Swift A., Bonnycastle L., Erdos M., He Z., Saxena R., Harmon B., Kotova O., Hoffman E., Altshuler D., Groop L., Boehnke M., Collins F., Hall J. L. (2009) Hum. Mol. Genet. 20, 3795–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hovanes K., Li T. W., Munguia J. E., Truong T., Milovanovic T., Lawrence Marsh J., Holcombe R. F., Waterman M. L. (2001) Nat. Genet. 28, 53–57 [DOI] [PubMed] [Google Scholar]
- 52.Roose J., Huls G., van Beest M., Moerer P., van der Horn K., Goldschmeding R., Logtenberg T., Clevers H. (1999) Science 285, 1923–1926 [DOI] [PubMed] [Google Scholar]
- 53.Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P. J., Clevers H. (1998) Nat. Genet. 19, 379–383 [DOI] [PubMed] [Google Scholar]
- 54.Tang W., Dodge M., Gundapaneni D., Michnoff C., Roth M., Lum L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Araki E., Lipes M. A., Patti M. E., Brüning J. C., Haag B., 3rd, Johnson R. S., Kahn C. R. (1994) Nature 372, 186–190 [DOI] [PubMed] [Google Scholar]
- 56.Ma Z., Gibson S. L., Byrne M. A., Zhang J., White M. F., Shaw L. M. (2006) Mol. Cell. Biol. 26, 9338–9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.