Abstract
The Escherichia coli chemoreceptors for serine (Tsr) and aspartate (Tar) and several bacterial class III adenylyl cyclases (ACs) share a common molecular architecture; that is, a membrane anchor that is linked via a cytoplasmic HAMP domain to a C-terminal signal output unit. Functionality of both proteins requires homodimerization. The chemotaxis receptors are well characterized, whereas the typical hexahelical membrane anchor (6TM) of class III ACs, suggested to operate as a channel or transporter, has no known function beyond a membrane anchor. We joined the intramolecular networks of Tsr or Tar and two bacterial ACs, Rv3645 from Mycobacterium tuberculosis and CyaG from Arthrospira platensis, across their signal transmission sites, connecting the chemotaxis receptors via different HAMP domains to the catalytic AC domains. AC activity in the chimeras was inhibited by micromolar concentrations of l-serine or l-aspartate in vitro and in vivo. Single point mutations known to abolish ligand binding in Tar (R69E or T154I) or Tsr (R69E or T156K) abrogated AC regulation. Co-expression of mutant pairs, which functionally complement each other, restored regulation in vitro and in vivo. Taken together, these studies demonstrate chemotaxis receptor-mediated regulation of chimeric bacterial ACs and connect chemical sensing and AC regulation.
Keywords: Chemotaxis, Membrane/Enzymes, Signal Transduction/Adenylate Cyclase, Signal Transduction/Cyclic Nucleotides/Cyclic AMP, Adenylate Cyclase (Adenylyl Cyclase), HAMP domain, Tar, Tsr
Introduction
The canonical chemotaxis receptors Tsr for serine and Tar for aspartate of Escherichia coli have been studied extensively as model signaling systems that regulate bacterial swimming behavior (1–5). Ligand binding to the periplasmic domain of the receptor initiates a biochemically defined cascade of events that finally results in reversal of the flagellar beat (4, 6). The chemotaxis receptors are tripartite proteins with a periplasmic receptor anchored in the cytoplasmic membrane by two transmembrane spans followed by a HAMP domain (histidine kinases, adenylyl cyclases, methyl-accepting proteins of chemotaxis, and protein phosphatases (7)) which adjoins the second transmembrane helix and serves as a signal converter followed by an output domain as C terminus (3). Homodimerization of Tsr or Tar chemoreceptors is required for function, and the ligands bind in the periplasmic interface between the subunits (8–11). The ubiquitous HAMP domains are signal converters of 55–60 amino acids with a homodimeric four-helical parallel coiled-coil conformation (12). In the chemotaxis receptors the HAMP domain operates as a C-terminal histidine kinase control unit that has no known intrinsic enzymatic activity. As this molecular architecture is predominant in the 30 known sensor kinases of E. coli, a variety of chimeric sensor proteins has been generated successfully in the past that consist of modules derived from different E. coli sensors (see examples in Refs. 13–15).
A similar tripartite domain architecture as in chemotaxis receptors exists in several class III AC2 isoforms in eubacteria. Class III AC isoforms outnumber all others, are prevalent in bacteria, and include all eukaryotic ACs (16–17). In bacteria, several of these ACs have a hexahelical membrane anchor with a distinctive periplasmic region of unknown function followed by a HAMP domain that abuts the last membrane helix followed by an AC output region. Examples are the four mycobacterial ACs, Rv3645, Rv1318c, Rv1319c, and Rv1320c (18), and an AC from Corynebacterium glutamicum. As is the case with the chemotaxis receptors, homodimerization is required for function because the catalytic center of the AC is formed at the interface of the monomers (19–21). From this it could be deduced that the membrane anchor of these ACs concomitantly serves a receptor function as is the case in Tsr or Tar. This view is bolstered by the fact that HAMP domains usually are inserted between input and output domains of signaling proteins and HAMP domains are considered signal converters (7, 12). It is reasonable to argue that the HAMP domain in bacterial ACs similarly transmits an extracellular signal to the catalytic AC domain. In view of the unidentified nature of such a potential signal, we examined whether the membrane domains of HAMP containing ACs can be functionally replaced by the chemotaxis receptors Tsr and Tar with ensuing regulation of AC activity by the respective amino acids.
In a novel approach to the old, unresolved question of a function for the membrane anchor domains of class III ACs (22), we combined protein modules of chemotaxis receptors and two bacterial ACs that usually are totally unrelated research subjects. We demonstrate that such bi- and tripartite, receptor-coupled ACs are indeed regulated in vitro as well as in vivo by serine and aspartate. Thus, the data uniquely connect chemical sensing and AC regulation.
EXPERIMENTAL PROCEDURES
Materials
An Arthrospira platensis culture was obtained from K. Forchhammer, University of Giessen (Germany). Radiochemicals were from Hartmann Analytik (Braunschweig, Germany) and GE Healthcare. Enzymes were purchased from either Roche Diagnostics or New England Biolabs. Other chemicals were from Merck, Roche Diagnostics, Roth, and Sigma.
Plasmid Construction
Rv3645 (GenBankTM accession number NC_000962) was available in the laboratory (18). Genomic DNA from A. platensis was isolated from a mini-culture by standard procedures and used as a template. The CyaG clone obtained using specific primers had 29 single amino acid exchanges compared with that deposited in GenBankTM accession number D49531.1 (see supplemental Fig. 2). To connect various signaling modules, all sequences were aligned, and borders were determined accordingly (see supplemental Fig. 2 for alignment). The domains were used as follows: Tsr receptor domain, 1–215; Tsr HAMP domain (HAMPTsr), 216–268; Rv3645 HAMP domain (HAMP3645), 278–330; AC Rv3645 catalytic domain, 331–549; Tar receptor domain, 1–213; Tar HAMP domain (HAMPTar), 214–266; AC CyaG HAMP domain (HAMPCyaG), 370–431; AC CyaG catalytic domain, 432–671. The chimeras were cloned using standard molecular biology methods. Silent restriction sites were introduced when necessary. The correctness of all constructs was confirmed by DNA sequencing. All constructs were fitted with a 5′ BamHI site and a 3′ HindIII site and cloned into either pQE30 or pQE80L, thus adding an N-terminal MRGSH6GS tag.
Expression and Purification of Proteins
Expression plasmids were transformed into E. coli BL21(DE3)[pRep4]. Cultures were grown in Lennox L broth at 30 °C containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. Expression was initiated with 100 μm isopropyl thio-β-d-galactoside for 4–5 h at 22 °C or overnight at 18 °C. Bacteria were collected by centrifugation at 5000 × g (10 min), washed once with 50 mm Tris/HCl, 1 mm EDTA, pH 8, and stored at −80 °C. For preparation of membrane fractions, cells were suspended in 25 ml of lysis buffer (50 mm Tris/HCl, 2 mm 3-thioglycerol, 50 mm NaCl, pH 8) and disintegrated using a French press (1000 p.s.i.). Cell debris was removed at 5000 × g for 30 min, and membranes were sedimented at 100,000 × g for 1 h at 4 °C. Membranes were suspended in 40 mm Tris/HCl, pH 8.0, 1.6 mm 3-thioglycerol, 20% glycerol, and assayed for AC activity. Empty vector controls were run routinely. The integrity of expressed recombinant membrane proteins was examined by Western blotting (see insets of Figs. 2–6). Blots were also evaluated by densitometry, and the protein concentration of constructs in the assays was adjusted such that comparable amounts of enzyme were used.
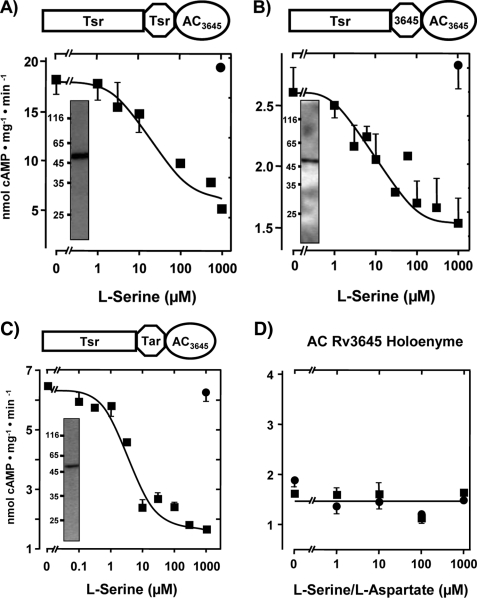
FIGURE 2.
l-Serine inhibits class IIIb AC Rv3645 via the E. coli Tsr chemotaxis receptor. Above each figure a diagram of the chimera is depicted. A, maximal serine inhibition was 71 ± 3%, and the IC50 concentration for serine was 18.5 ± 7 μm (n = 5). Inset, shown is a Western blot with anti-RGSHis6; 4 μg of protein was applied. B, maximal serine inhibition was 45 ± 5%, and IC50 for serine was 15 ± 2 μm (n = 4). Inset, shown is a Western blot with anti-RGSHis6; 3 μg of protein was applied. C, maximal serine inhibition was 73 ± 2%, and IC50 for serine was 4.2 ± 1 μm (n = 6). Inset, shown is a Western blot with anti-RGSHis6; 4 μg of protein was applied. Filled circles are controls with 1 mm l-aspartate. Rectangle, receptor part; octagon, HAMP domain; ellipse, AC catalytic domain; Tsr, serine chemotaxis receptor; Tar, aspartate chemotaxis receptor; AC3645, catalytic AC domain from Rv3645. D, control with expressed AC Rv3645 demonstrates that neither l-serine (filled squares) nor l-aspartate affected activity.
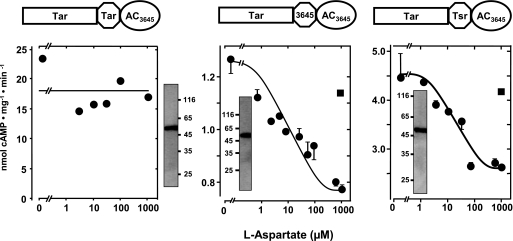
FIGURE 3.
l-Aspartate regulation of AC Rv3645 via the E. coli Tar chemotaxis receptor. Left, aspartate did not regulate Rv3645 in conjunction with the N-terminal Tar receptor including HAMPTar (n = 4). Shown is a Western blot with 4 μg of protein. Center, maximal aspartate inhibition was 42 ± 5%, and the IC50 for aspartate was 10 ± 1 μm (n = 2). Inset, shown is a Western blot with 5 μg of protein. Right, maximal aspartate inhibition was 40 ± 8%, and the IC50 for aspartate was 12 ± 2 μm (n = 2). Inset, shown is a Western blot with 3 μg of protein squares. Filled square, controls with 1 mm serine.
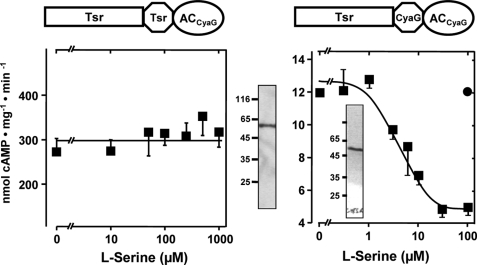
FIGURE 4.
Class IIIa AC CyaG is regulated by l-serine via the E. coli Tsr chemotaxis receptor. Left panel, Tsr linked to CyaG AC from A. platensis via HAMPTsr is unregulated by l-serine. Note the basal activity of 270 ± 76 nmol of cAMP·mg−1·min−1 (n = 8). Shown is a Western blot with 4 μg of protein. Right panel, Tsr linked to CyaG AC via HAMP of CyaG AC is regulated by l-serine. Maximal inhibition was 58 ± 5%, and the IC50 = 6 ± 3 μm (n = 4). Inset, shown is a Western blot with 4 μg of protein. Filled circle, 1 mm serine as a control.
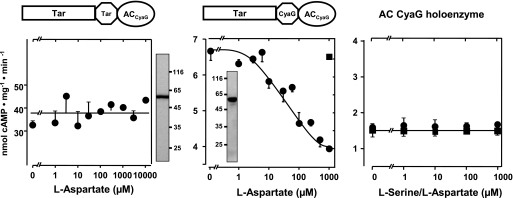
FIGURE 5.
l-Aspartate inhibits class IIIa AC CyaG via the E. coli Tar receptor. Left panel, Tar linked to CyaG AC from A. platensis via HAMPTar is unregulated (basal activity 36 ± 6 nmol of cAMP·mg−1·min−1 (n = 2). Shown is a Western blot with 4 μg of protein. Centre panel, Tar linked to CyaG AC via HAMPCyaG is regulated by aspartate. Maximal inhibition was 45 ± 7%, and the IC50 was 32 ± 8 μm (n = 2). Inset, shown is a Western blot with 5 μg of protein. Filled square, control (1 mm serine). Right panel, the control with recombinant, membrane-bound AC CyaG shows that activity is not affected by l-serine (filled squares) or l-aspartate (filled circles; n = 2)).
FIGURE 6.
Functional complementation of inactivated Tsr and Tar receptor point mutants. A, the rationale for receptor complementation is shown. The numbering refers to Tsr. Neither R69E nor T156K mutant dimers bind ligand. In heterodimers (right) a serine binding site is reconstituted. B, a complementation assay of Tsr AC Rv3645 mutant chimeras is shown. Serine addition to R69E (filled diamond) or T156K (filled triangle) mutant proteins expressed in pQE 30 had no effect on AC activity, i.e. no regulation. Mutant proteins co-expressed from a pETDUET plasmid are regulated by serine (filled squares). AC Inhibition was 35 ± 5%, and IC50 was 15 ± 5 μm (n = 4). At the right, Western blot analysis is shown of assayed proteins with the respective symbols on top (5 μg/lane for individual point mutants and 10 μg lane for jointly expressed proteins were applied). Proteins expressed concomitantly in pETDUET had N-terminal His tags, and the T156K mutant had an additional C-terminal S tag that was used for protein identification and analysis (right). For clarity the respective controls with 1 mm aspartate (in B) and 1 mm serine (in C) were omitted. The controls were negative. C, a complementation assay of Tar CyaG AC mutant chimeras is shown. Aspartate concentration-response curves for the R69E (filled diamond) or T154I (filled triangle) point mutations expressed in pQE 30 had no effect on AC activity, i.e. no regulation. Mutant proteins co-expressed from a pETDUET plasmid are regulated by aspartate (filled circles). AC Inhibition was 20 ± 4%, and IC50 was 20 ± 8 μm (n = 4). At the right, a Western blot analysis of assayed proteins with respective symbols on ths top is shown. Proteins expressed concomitantly in pETDUET had N-terminal His tags, and the T154I mutant had an additional C-terminal S tag that was used for protein identification and analysis (right). Identical amounts of protein applied to gel as in B.
Adenylyl Cyclase Assay
AC activity was determined at 37 °C for 10 min in 100 μl (23). Standard reactions contained 50 mm Tris/HCl, pH 7.5, 22% glycerol, 3 mm MnCl2, 200 μm [α-32P]ATP (Rv3645) or 750 μm [α-32P]ATP (CyaG), and 2 mm [2,8-3H]cAMP. Creatine kinase and creatine phosphate were used as an ATP regenerating system. Data shown are the means ± 1 S.E., with the number of individual experiments indicated in figure legends.
MacConkey Plate Assays
Plates were incubated at 37 °C for 15 h. Constructs were transformed into E. coli cya strains BTH101 or DHM1 depending on the cloning plasmid used (pQE or pET, respectively) (from Euromedex; strain genotypes: BTH101: F−, cya-99, araD139, galE15, galK16, rpsL1 (Strr), hsdR2, mcrA1,mcrB1; DHM1: F−, cya-854, recA1, endA1, gyrA96 (Nalr), thi1, hsdR17, spoT1, rfbD1, glnV44(AS)) and plated on LB plates containing 100 μg/ml ampicillin and incubated at 37 °C for 15 h. Single clones were streaked across agar plates used to detect cAMP-producing strains (40 g/liter MacConkey agar, 28 mm maltose, and 33 μg/ml ampicillin). Strips placed in the center of the plates contained 50 μl of 1 m serine or aspartate. Different E. coli strains were used because strain BTH101 transformed with pQE80Tar-HAMPRv3645-ACRv3645 turned out to be unstable on MacConkey agar plates, whereas transformants in DHM1 cells were stable.
β-Galactosidase Assay
Constructs transformed into cya mutants were grown in LB media (20 g/liter) at 37 °C for 15 h. Cells were collected (15 min, 5000 × g), diluted to an absorbance of 0.2 in M63 medium (24) containing 0.6% glycerol and 30 μg/ml ampicillin, pH 7, and 2.5 ml samples were incubated at 37 °C for 90 min. Induction of β-galactosidase was with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside with/without different concentrations of serine or aspartate. After 90–210 min cells were collected by centrifugation, and pellets were stored at −20 °C. β-Galactosidase activity was determined as described with slight modifications (24). Cell sediments were suspended in 1 ml of assay buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 1 mm MgSO4, and 5 mm dithiothreitol). 10 μl of a 0.1% SDS solution and 20 μl of chloroform were added, and samples were incubated for 5 min at 28 °C. Enzyme reactions were started by the addition of 200 μl of o-nitrophenyl galactose (2.2 mm) and stopped by the addition of 0.5 ml of 1 m Na2CO3. A405 was measured in the supernatant of a 4-min spin (12,000 × g). Data were evaluated in Miller units (24).
RESULTS
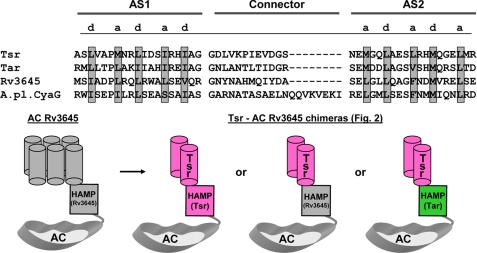
The HAMP domains used in the chimeric constructs originated from respective parent proteins that provided additional subdomains, i.e. the receptor/membrane domains from Tsr or Tar and the AC domains from Rv3645 and CyaG. A HAMP domain alignment demonstrates the characteristically spaced a/d pattern of hydrophobic amino acids embedded within a heptad periodicity, labeled a through g, which is typical for coiled-coils (Fig. 1, top) (25). The general outline of construct design for receptor-linked ACs was similar in all instances (Fig. 1, bottom, schematically shows the construct design for chimeras assayed in Fig. 2). Generally, constructs were expressed in E. coli BL21(DE3)[pRep4] cells, which contributed about 10–40 pmol of cAMP·mg−1·min−1 to total basal AC activity. Throughout our studies this basal activity could be neglected because specific activities of recombinant AC proteins were on the order of nmol of cAMP·mg−1·min−1. All constructs were obtained as intact membrane proteins, as apparent from corresponding Western blots using the N-terminal His tag as an epitope for immunodetection, and degradation products were absent, as shown in Figs. 2–6. Adjustments in the amount of protein were based on densitometric scans of Western blots to ensure that comparable amounts of recombinants protein were used in all experiments.
FIGURE 1.
Alignment of HAMP domains and scheme of chimeras used in this study. top, the heptad periodicity of amino acids a-g is indicated by highlighting residues a and d (gray), which form the hydrophobic core in the coiled-coil of HAMP domains. AC Rv3645, M. tuberculosis gi 81668722; A. platensis CyaG, gi 25535039; Tar, E. coli Tsr, gi 2506837; Tar, gi 89111064. AS1 and AS2 denote borders of the α-helices of the HAMP domains as deduced from the available NMR structure (12). Bottom, general pattern of chimeric ACs used in this study. As examples, the three chimeras examined in Fig. 2 are depicted. In the graph, the membrane anchor of AC Rv3645 with six predicted α-helices (schematically abbreviated as six gray cylinders at the left) was replaced by the E. coli chemotaxis receptor Tsr, which has two predicted membrane-spanning α-helices (depicted as two pink cylinders labeled Tsr at the right). The origin of the HAMP domains is color-coded. The linkers interconnecting the membrane spanning α-helices have been omitted for clarity. Note that the proteins require dimerization for function.
Initially we linked the receptor part of the chemotaxis receptor Tsr to the Rv3645 catalytic domain via the HAMP domains of Tsr, Tar, or AC Rv3645. The chimeras had AC activities that were concentration-dependently inhibited by serine, whereas 1 mm aspartate had no effect (Fig. 2). The construct Tsr/HAMPTsr/Rv3645 AC was also tested at 1 mm with the remaining 18 proteinogenic amino acids for specificity. None inhibited or activated AC activity. Thus, the amino acid specificity is in line with what had been reported in Tsr chemotaxis assays (5, 26). The results established that class III ACs can indeed be directly regulated by the periplasmic Tsr receptor. They further bolstered the idea that HAMP domains as signal converters are at least to some extent functionally interchangeable. Although differences existed in basal AC activities between individual chimeras, the extent and efficacy of serine to inhibit AC Rv3645 were rather similar. With the HAMP domain derived from the Tsr receptor, inhibition was 71 ± 3%, and the IC50 for serine was 18.5 ± 7 μm, whereas these numbers were 73 ± 2% and 4.2 ± 1 μm with the Tar HAMP domain and 45 ± 5% and 15 ± 2 μm for the chimera with the Rv3645 HAMP. In all instances serine inhibition was due to a reduction in Vmax, whereas the Km of ∼400 μm for ATP was unchanged. The Hill coefficients were 1. This indicated that AC inhibition was exclusively due to a defect in generation of the transition state during catalysis. Removal of serine by wash-out showed that AC inhibition was reversible because enzyme activity returned to basal levels and was susceptible to repeated inhibition by serine (not shown). The constructs could be solubilized by 1% CHAPS, and activity and serine inhibition were retained. However, purification efforts failed due to loss of activity (not shown). As a control we ensured that the mycobacterial AC Rv3645 holoenzyme expressed in E. coli was not affected by up to 1 mm l-serine or l-aspartate (Fig. 2D). Furthermore, d-aspartate or d-serine had no effect (see supplemental Fig. 1).
Next we used the Tar receptor as an input domain, the AC Rv3645 as the output domain, and as signal converters, the HAMP domains from Tar, Tsr, and Rv3645, respectively (Fig. 3). The boundaries between individual building blocks were fashioned according to the Tsr constructs outlined above. The results with the Tar receptor differed from those with Tsr. The bipartite chimera with Tar and HAMPTar in front of Rv3645 AC had an activity of 18 ± 3 nmol of cAMP·mg−1·min−1, i.e. it dimerized properly and formed a productive catalytic cleft, yet it was unaffected by aspartate (Fig. 3). In contrast, constructs that consisted of the Tar receptor in conjunction with HAMP domains from either the chemotaxis receptor Tsr or the Rv3645 AC were efficiently and specifically regulated by l-aspartate (d-aspartate was inactive; see supplemental Fig. 1), albeit maximal inhibition of 42 ± 5 and 40 ± 8 fell short of the respective constructs using Tsr as a receptor (compare Figs. 2 and 3). The IC50 concentration for aspartate was 10 ± 1 μm, i.e. within a physiologically meaningful range. The data indicate that as yet poorly understood structural features of different HAMP domains affect their suitability as molecular converters in differing functional contexts.
Consequently, we examined whether we could use another AC in such constructs. The HAMP-containing mycobacterial ACs Rv1318c, Rv1319c, Rv1320c, and Rv3645 are subclassified as class IIIb AC isoforms that normally use a lysine/threonine couple for substrate definition, whereas class IIIa ACs, which include all mammalian membrane-bound ACs, use lysine/aspartate residues for substrate definition (17). The cyanobacterium A. platensis contains a class IIIa AC, CyaG, that has a HAMP domain at the exit of a predicted transmembrane domain. Thus, we explored whether a class IIIa AC can be regulated via the Tsr and Tar receptor domains. We used CyaG and the HAMP domains from either the respective receptor, Tsr or Tar, or from CyaG. The chimeras were expressed as undegraded membrane-bound proteins (Fig. 4). Surprisingly, a chimera consisting of Tsr + HAMPTsr in front of CyaG was not affected by serine (Fig. 4). This failure of signaling could not be ascribed to protein misfolding because the dimerized chimera had high AC activity (270 ± 76 nmol of cAMP·mg−1·min−1 (n = 8); Fig. 4). Because the amount of recombinant protein used in the assay as evaluated by densitometry was comparable with other AC assays, it appeared that the AC was unhooked from the regulatory scaffold and freely folded into a very active dimer. This was reminiscent of the Tar-HAMPTar-Rv3645 construct described above. When the HAMP domain in the unregulated chimera was replaced by that from CyaG, serine inhibition was prominent, specific, and potent (IC50 = 6 ± 3 μm, 58 ± 5% inhibition (n = 4); Fig. 4, right). Next we generated corresponding Tar receptor chimeras, i.e. using the Tar chemotaxis receptor, AC CyaG, and the HAMP domain from either Tar or CyaG. Both constructs were well expressed as determined by a Western blot and had AC activity (Fig. 5).
When the HAMP domain was derived from Tar receptor, AC activity (38 ± 7 nmol of cAMP·mg−1·min−1 (n = 2)) was unresponsive to aspartate. In contrast, with the HAMP from CyaG, AC activity was inhibited by aspartate with an IC50 of 32 ± 8 μm, and maximal inhibition was 45 ± 7% (Fig. 5, right). Taken together, this emphasized the importance of the origin of the HAMP domains in signal conversion between input and output domains.
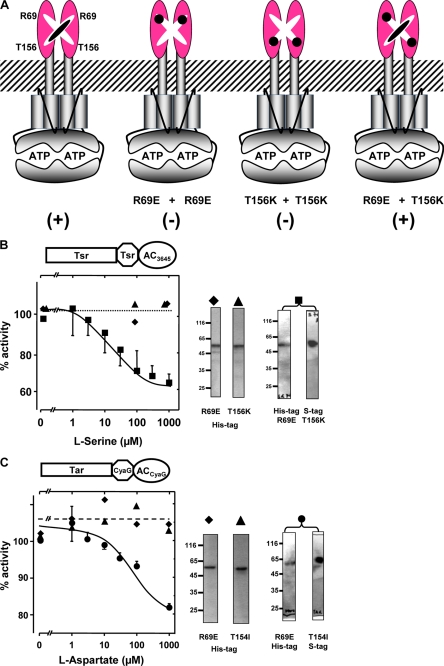
To exclude the possibility that signaling may not originate from Tsr or Tar receptor activation but may be due to unrecognized, unspecific interactions of serine and aspartate with the chimeric proteins, we made use of the fact that the chemotaxis receptors Tsr and Tar can be inactivated by single point mutations. It has been demonstrated that such mutants are unable to signal as homodimers, yet when two of such point mutants heterodimerize, they complement each other, and chemotaxis is restored (27–30) (see the Fig. 6A scheme). We used such an approach to unequivocally establish that regulation of AC activity is receptor-mediated. Using the Tsr-HampTsr-Rv3645 AC construct, we generated two individual mutants, R69E and T156K. When expressed separately, the respective chimeric proteins showed AC activity, yet they were not regulated by serine (Fig. 6B). When both mutants were expressed concomitantly from a single plasmid, heterodimerization resulted in restoration of signaling, i.e. AC regulation by serine (Fig. 6B).
Expectedly, the extent of serine-mediated inhibition was reduced compared with the unmutated constructs (80 versus 40%) because dimerization of individual point mutants should result in unregulated homodimers as well as regulated heterodimers (Fig. 6A). A similar pair of complementary mutants was generated using Tar as the receptor unit and the HAMP domain and cyclase catalytic domain from CyaG (Fig. 6C), i.e. we generated individual point mutants R69E and T154I of Tar. Such point mutants were not regulated by aspartate as homodimers, whereas concomitant, equimolar expression of both mutant proteins from a single plasmid resulted in restoration of aspartate regulation of cyclase activity at the expected reduced level (40 versus 27%, Fig. 6C). Serine and aspartate regulation was mediated exclusively by the l-amino acids; the d-amino acids had no effect. AC activity of the isolated catalytic domains of Rv3645 or CyaG was not affected by serine or aspartate. We generated chimeras consisting of Tsr (1–215) and AC Rv3645 (331–549) devoid of a HAMP domain. The recombinant proteins had AC activity but were not regulated by serine. This demonstrated the requirement for the HAMP domains as signal converters. So far we examined AC regulation via HAMP domains in vitro. Next we asked whether these signaling ensembles actually operate in vivo.
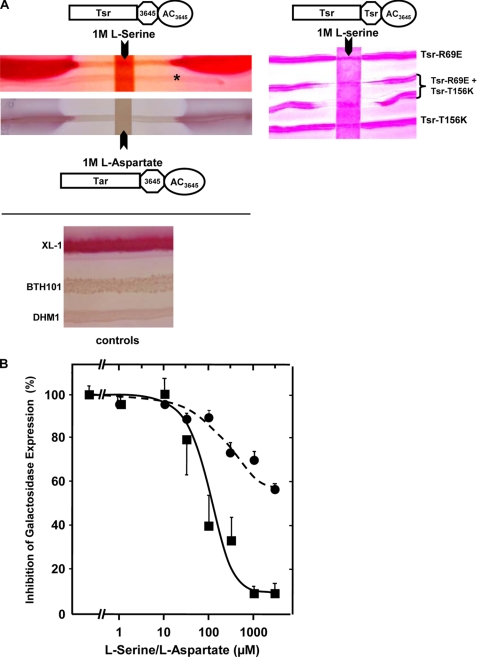
The cAMP receptor protein (CRP), also known as catabolite gene activator protein (31), controls the expression of more than 150 genes in E. coli including genes responsible for the catabolism of alternative carbon sources like lactose and maltose. Efficient use of lactose by E. coli requires activation of the lactose operon via the cAMP/CRP signaling system and dissociation of the Lac repressor protein from the operator upon binding of an inducer molecule. Fermentation of lactose results in production of acids such as lactate, acetate, succinate, and formate, which are secreted and lower the pH of the medium. This is visualized on MacConkey agar plates with phenol red as a pH indicator by the appearance of red colonies. In wild-type E. coli this system operates via its intrinsic class I AC, Cya, which is unrelated in sequence to class III ACs (16, 32). We used AC-deficient E. coli strains (cya-99 or cya-854), which could not use lactose, maltose, and other carbohydrates as the carbon source due to the lack of cAMP production. Consequently, when grown on MacConkey agar, colonies appear whitish. We transformed the Tsr-HAMP3645-Rv3645 AC construct into E. coli BTH101 (cya-99) and plated the cells on MacConkey maltose agar plates. The AC chimera produced cAMP, induced the mal regulon, and restored the capability for maltose fermentation, i.e. it rescued the AC-deficient strain (Fig. 7).
FIGURE 7.
In vivo inhibition of cAMP formation in E. coli by serine and aspartate via mutated and reconstituted Tsr and Tar receptor chimeras. A, MacConkey maltose agar plates with E. coli streaked across and a filter strip soaked with 50 μl of either 1 m serine (above) or 1 m aspartate (below) placed on top. Tsr-HAMP3645-Rv3645 AC and Tar-HAMP3645-Rv3645 AC were transformed in BTH101 and DHM1 cells, respectively (see “Experimental Procedures”). The asterisk indicates the faint streak of an E. coli cya strain transformed with a control plasmid (left). At the right, single point mutants expressed individually or concomitantly are probed on a MacConkey agar plate as indicated using DHM-1 cells. The paper strip contained 50 μl of 1 m l-serine. Bottom, control cultures of E. coli wild type and E. coli cya− strains BTH101- and DHM1 are shown. B, β-galactosidase expression is inhibited in cultures by the concomitant presence of either serine (squares) or aspartate (circles) in cells transformed with Tsr-HAMP3645-Rv3645 AC (squares) or Tar-HAMP3645-Rv3645 AC (circles), respectively. Activity of 100% corresponds to β-galactosidase activities without amino acid addition to the culture. Expression was for 90 min. Data were normalized to permit presentation of different β-galactosidase activities in a single graph (n = 3).
Placing a filter strip soaked with 50 μl of 1 m serine across the MacConkey plate inhibited maltose fermentation within a large serine diffusion zone (Fig. 7A). This demonstrated that serine inhibited cAMP formation and lowered intracellular cAMP concentrations to a level insufficient to turn on the mal genes. Although the data are strictly qualitative, the extent of the inhibition zone from the 1 m serine strip indicated that serine concentrations required for AC inhibition may be rather low. This was addressed in the next in vivo experiment. E. coli cya cultures transformed with Tsr-HAMP3645-Rv3645 AC were grown for 90 min in the presence of various serine concentrations. After cell lysis β-galactosidase was determined spectrophotometrically. Serine concentration-dependently (IC50 of 100 μm) inhibited expression of β-galactosidase. When cultures were run for up to 210 min, in all instances serine inhibition of cAMP was observed; however, the IC50 concentrations drifted toward somewhat higher amino acid concentrations (not shown). This probably was due to the observation that cAMP is constantly exported by E. coli into the medium, which has complicated exact determinations of intracellular cAMP for decades (33–35). Therefore, we have refrained from attempts to measure cAMP levels in E. coli under our experimental conditions because even without such measurements the data unequivocally demonstrated that the Tsr-HAMP3645-Rv3645 protein was properly inserted into the E. coli cell membrane and that cAMP biosynthesis was inhibited by serine in vivo. cAMP level estimations would not have added additional knowledge. Similar experiments were carried out with a corresponding Tar receptor construct in E. coli DHM1 (cya-854) on MacConkey maltose agar plates and in planktonic cultures with various aspartate concentrations. Comparable results were obtained (Fig. 7, A and B). The IC50 concentration for aspartate was 110 μm. It is noteworthy that the diminished extent of aspartate inhibition in vitro was also evident in a reduced inhibition of β-galactosidase expression. Finally, we transformed cells with the single mutants Tsr-R69E and Tsr-T156K and plated the cells on MacConkey agar plates (Fig. 7A). These cells produced sufficient cAMP for mal regulon induction and did not respond to serine (Fig. 7A). However, transformation with both mutant proteins restored serine sensitivity as expected, i.e. the inhibition zones caused by the 1 m serine strip were pronounced. Data were similar with the corresponding Tar single mutants R69E and T154I (data not shown).
DISCUSSION
ACs generate the universal second messenger cAMP and, thus, affect metabolic pathways in all cells. Accordingly, AC regulation is of paramount importance in all organisms. Many bacterial class III ACs, like their mammalian congeners, lack recognizable regulatory domains and are membrane-anchored by mostly six predicted membrane-spanning α-helices (17, 22, 36). The function of the oversized membrane anchors remains enigmatic and a matter of speculation since the first mammalian AC was cloned in 1989 (22). Several bacterial ACs have a 6TM anchor that is connected to the catalytic domain via a HAMP domain. HAMP domains are considered universal signal converters (>12,000 HAMP domains are in the EMBL SMART data base) that are located between transmembrane and cytoplasmic signaling regions of known or putative membrane receptors (4). Therefore, the molecular architecture of AC Rv3645 or CyaG may indicate that at least in these instances the membrane anchor may actually be a sensory receptor. We started to probe into this possibility and used a well established homodimeric signal transduction system with an identical domain composition, the bacterial chemotaxis receptors, for generation of signaling chimeras. Thus, we experimentally brought together two totally unrelated cellular signaling systems, which appear mechanistically connected by a HAMP signal converter in the midst of a periplasmic sensing and a cytoplasmic output domain.
We joined the intramolecular networks of these two proteins across their signal transmission sites such that the sensor part of Tsr or Tar is connected via a HAMP domain to a catalytic domain of either Rv3645, a class IIIb AC isoform, or CyaG, a class IIIa AC isoform. Surprisingly, replacement of the AC membrane anchors by Tsr or Tar resulted in serine- and aspartate-regulated ACs Rv3645 and CyaG, i.e. the chemotaxis receptors coupled ligand recognition and binding to AC control in vitro. This was proof that the uniform construct design of the chimeras preserved register and packing of the signaling ensembles. The dose-response curves for serine or aspartate yielded IC50 concentrations in the lower micromolar range that were in good agreement with those derived from chemotaxis experiments (26, 37).
Expression of the recombinant proteins for in vitro experiments was in E. coli BL21 cells, which contributed about 10–40 pmol of cAMP·mg−1·min−1 to total basal AC activity. This negligible background activity was not figured into calculations. Further control experiments included the use of d-amino acids, which were inactive, tests of amino acid specificity, which were high, and a lack of serine or aspartate to affect AC activity of isolated catalytic domains. The receptor-mediated signaling was proven unequivocally by complementation of the heterodimeric Tsr and Tar receptors, which were inactive as R69E or T156K homodimers but active as heterodimers (numbers correspond to Tsr). The point mutants were designed according to those successfully employed in chemotaxis studies (27, 30). In addition, Tsr or Tar-mediated AC regulation was fully operating in vivo using E. coli cya cells and MacConkey maltose agar plates or β-galactosidase determinations with IC50 concentrations comparable with those established in physiological assays. This was not necessarily to be expected because AC inhibition by serine or aspartate in vitro was limited to about 75 and 50%. Either the extent of AC inhibition was sufficient to lower intracellular cAMP concentrations below the threshold needed for mal-regulon or lac-operon induction or the chimeras were inhibited to a larger extent in vivo. Expectedly, the R69E and T156K (Tsr receptor) in vitro mutant data were reproducible in respective in vivo experiments (Fig. 7B and data not shown).
Undoubtedly then, regulation of AC activity was initiated when the extracellular ligands bound to the periplasmic chemotaxis receptor, and intracellular signal conversion probably proceeded in a mechanistically highly similar manner as in bacterial chemotaxis. The structural changes that will occur in different output domains may, however, differ. In chemotaxis, serine and aspartate are attractants, and an increase in concentration shifts receptors to the “histidine-kinase-off” state by stabilizing an expanded arrangement of the four-helix HAMP signal converter, whereas a drop in attractant concentration shifts the receptors to the “histidine-kinase-on” position or compact arrangement (38). Our experiments show a similar situation; that is, an increase in serine or aspartate shifts the AC into the off-state and vice versa. The data do not address the contested question of whether the mechanical output of HAMP domain-mediated signaling is rotation, a piston movement, or a combination of both. Yet our chimeras provide a novel system to examine HAMP-mediated signaling quantitatively in vitro compared with scoring swimming behavior in vivo.
The components of the nine chimeras that we investigated originated from rather distant bacteria. Tsr and Tar were from Gram-negative E. coli, class IIIb AC Rv3645 was from the Gram-positive Mycobacterium tuberculosis, and class IIIa AC CyaG from the cyanobacterium A. platensis. The receptor modules Tar and Tsr are similar in sequence yet specifically bind either serine or aspartate. In the four HAMP domains used in this study, the α-helices show the typically spaced a/d pattern of hydrophobic amino acids characteristic for coiled-coils (25) and are by necessity of identical length. However, the non-helical connector of the CyaG HAMP domain (20 amino acids) was 8 amino acids longer than in Tar or Tsr. This difference probably did not affect signal conversion because in separate experiments length variations introduced into the connector did not affect signaling, whereas targeted proteolytic cleavage at predestined sites of the connector abolished regulation (data not shown).
The catalytic domains of class IIIa and IIIb ACs differ in several structural features, e.g. a substrate-defining aspartate in class IIIa is replaced by a threonine in class IIIb isoforms. This turned out to be irrelevant for the successful construction of regulated chimeras. A potentially more important difference is a 25-amino acid-length variation between the C terminus of the HAMP domain and the first catalytic amino acid, a metal binding aspartate that is part of a conserved FXD motif in class III ACs. In Rv3645 the distance is 38 amino acids, and in CyaG it is 63. In both isoforms this stretch is predicted to be largely α-helical. It is likely that this linker between HAMP and AC participates in signal transmission because chimeras using the CyaG AC from A. platensis were only regulated by Tsr or Tar when the corresponding HAMP domain and linker regions were from CyaG as well, whereas regulation was not accomplished when the respective HAMP domains originated from the receptor module (Figs. 4 and 5). Although we can conclude that amino acid sensing and signal conversion by the HAMP domains resulted in the designated conformational motions, these could not be translated properly into the required dimerization steps in the unregulated constructs. Perhaps these linker helices in conjunction with the adjoining HAMP domains serve as a spreader that holds the AC monomers fixed in a less productive state when the receptor is occupied by a ligand. Removal of ligand would result in a conformational change of the HAMP domain, and the α-helical linker between the HAMP and AC catalytic domain may melt into a flexible structure and allow the catalytic to align in a productive conformation. Such a scenario was visualized for the mycobacterial AC Rv1264, in which the active and inactive states were interconvertible via the melting of a long α-helical switch helix resulting in assembly/disassembly of the catalytic domains (39).
The surprising results highlight an unexpected and remarkable example of the modularity of signaling systems connecting chemical sensing and AC regulation. The HAMP-AC ensembles perfectly served as signal converter-signal output couples for the chemotaxis receptors Tsr and Tar. This may be taken as highly circumstantial evidence that the corresponding AC membrane anchors may serve additional functions on top of being anchoring devices as has already been speculated earlier (22). In fact, in 1960 Sutherland and Rall (40) hypothesized that in mammals a single protein would both recognize a hormone molecule and synthesize cAMP. This prediction, which at the time was made without a shred of structural knowledge, has not been fulfilled. Although we have no evidence yet of a ligand for any membrane anchor of bacterial or mammalian ACs, the data presented here may support the idea to look for such ligands and possibly add a novel facet to the firmly established sequels of hormone-regulated class III ACs.
Supplementary Material
Acknowledgment
We thank U. Kurz for expert technical support.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 766-B8).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- AC
- adenylyl cyclase
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Yoshida T., Phadtare S., Inouye M. (2007) Methods Enzymol. 423, 184–202 [DOI] [PubMed] [Google Scholar]
- 2.Ames P., Parkinson J. S. (1994) J. Bacteriol. 176, 6340–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd A., Kendall K., Simon M. I. (1983) Nature 301, 623–626 [DOI] [PubMed] [Google Scholar]
- 4.Hazelbauer G. L., Falke J. J., Parkinson J. S. (2008) Trends Biochem. Sci. 33, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adler J. (1965) Cold Spring Harbor Symp. Quant. Biol. 30, 289–292 [DOI] [PubMed] [Google Scholar]
- 6.Falke J. J., Hazelbauer G. L. (2001) Trends Biochem. Sci. 26, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravind L., Ponting C. P. (1999) FEMS Microbiol. Lett. 176, 111–116 [DOI] [PubMed] [Google Scholar]
- 8.Milligan D. L., Koshland D. E., Jr. (1988) J. Biol. Chem. 263, 6268–6275 [PubMed] [Google Scholar]
- 9.Kim K. K., Yokota H., Kim S. H. (1999) Nature 400, 787–792 [DOI] [PubMed] [Google Scholar]
- 10.Kim S. H., Wang W., Kim K. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11611–11615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr., Kim S. H. (1991) Science 254, 1342–1347 [DOI] [PubMed] [Google Scholar]
- 12.Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M. (2006) Cell 126, 929–940 [DOI] [PubMed] [Google Scholar]
- 13.Jin T., Inouye M. (1994) J. Mol. Biol. 244, 477–481 [DOI] [PubMed] [Google Scholar]
- 14.Appleman J. A., Chen L. L., Stewart V. (2003) J. Bacteriol. 185, 4872–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appleman J. A., Stewart V. (2003) J. Bacteriol. 185, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bârzu O., Danchin A. (1994) Prog. Nucleic Acid Res. Mol. Biol. 49, 241–283 [DOI] [PubMed] [Google Scholar]
- 17.Linder J. U., Schultz J. E. (2003) Cell. Signal. 15, 1081–1089 [DOI] [PubMed] [Google Scholar]
- 18.Linder J. U., Hammer A., Schultz J. E. (2004) Eur. J. Biochem. 271, 2446–2451 [DOI] [PubMed] [Google Scholar]
- 19.Tesmer J. J., Sunahara R. K., Gilman A. G., Sprang S. R. (1997) Science 278, 1907–1916 [DOI] [PubMed] [Google Scholar]
- 20.Zhang G., Liu Y., Ruoho A. E., Hurley J. H. (1997) Nature 386, 247–253 [DOI] [PubMed] [Google Scholar]
- 21.Linder J. U. (2006) Cell. Mol. Life Sci. 63, 1736–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krupinski J., Coussen F., Bakalyar H. A., Tang W. J., Feinstein P. G., Orth K., Slaughter C., Reed R. R., Gilman A. G. (1989) Science 244, 1558–1564 [DOI] [PubMed] [Google Scholar]
- 23.Salomon Y., Londos C., Rodbell M. (1974) Anal. Biochem. 58, 541–548 [DOI] [PubMed] [Google Scholar]
- 24.Miller J. H. (1972) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25.Mason J. M., Arndt K. M. (2004) Chembiochem 5, 170–176 [DOI] [PubMed] [Google Scholar]
- 26.Mesibov R., Adler J. (1972) J. Bacteriol. 112, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y., Park H., Inouye M. (1993) J. Mol. Biol. 232, 493–498 [DOI] [PubMed] [Google Scholar]
- 28.Ames P., Zhou Q., Parkinson J. S. (2008) J. Bacteriol. 190, 6676–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mowbray S. L., Koshland D. E., Jr. (1990) J. Biol. Chem. 265, 15638–15643 [PubMed] [Google Scholar]
- 30.Gardina P. J., Manson M. D. (1996) Science 274, 425–426 [DOI] [PubMed] [Google Scholar]
- 31.Shenoy A. R., Capuder M., Draskovic P., Lamba D., Visweswariah S. S., Podobnik M. (2007) J. Mol. Biol. 365, 211–225 [DOI] [PubMed] [Google Scholar]
- 32.Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bettenbrock K., Sauter T., Jahreis K., Kremling A., Lengeler J. W., Gilles E. D. (2007) J. Bacteriol. 189, 6891–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matin A., Matin M. K. (1982) J. Bacteriol. 149, 801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterkofsky A., Gazdar C. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 2794–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shenroy A. R., Visweswariah S. S. (2004) FEBS Lett. 561, 11–21 [DOI] [PubMed] [Google Scholar]
- 37.Mesibov R., Ordal G. W., Adler J. (1973) J. Gen. Physiol. 62, 203–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khursigara C. M., Wu X., Zhang P., Lefman J., Subramaniam S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16555–16560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tews I., Findeisen F., Sinning I., Schultz A., Schultz J. E., Linder J. U. (2005) Science 308, 1020–1023 [DOI] [PubMed] [Google Scholar]
- 40.Sutherland E. W., Rall T. W. (1960) Acta Endocrinol. Suppl. (Copenh) 34, 171–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.