Abstract
Anthrax lethal toxin (LeTx) is a virulence factor secreted by Bacillus anthracis and has direct cytotoxic effects on most cells once released into the cytoplasm. The cytoplasmic delivery of the proteolytically active component of LeTx, lethal factor (LF), is carried out by the transporter component, protective antigen, which interacts with either of two known surface receptors known as anthrax toxin receptor (ANTXR) 1 and 2. We found that the cytoplasmic delivery of LF by ANTXR2 was mediated by cathepsin B (CTSB) and required lysosomal fusion with LeTx-containing endosomes. Also, binding of protective antigen to ANXTR1 or -2 triggered autophagy, which facilitated the cytoplasmic delivery of ANTXR2-associated LF. We found that whereas cells treated with the membrane-permeable CTSB inhibitor CA074-Me- or CTSB-deficient cells had no defect in fusion of LC3-containing autophagic vacuoles with lysosomes, autophagic flux was significantly delayed. These results suggested that the ANTXR2-mediated cytoplasmic delivery of LF was enhanced by CTSB-dependent autophagic flux.
Introduction
Anthrax lethal toxin (LeTx)2 and edema toxin are two key virulence factors secreted by Bacillus anthracis, the causative agent of anthrax (1, 2). LeTx and edema toxin are composed of lethal factor (LF) or edema factor (EF), respectively, and protective antigen (PA), which functions as a cytoplasmic transporter of LF or EF. These toxins are main contributors to the clinical manifestations of anthrax and are cytotoxic to host cells once delivered into the cytoplasm. EF is an adenylate cyclase that raises cAMP levels in cells (3). LF is a metalloproteinase that targets the N-terminal end of the mitogen-activated protein kinase kinase 1–7 (MEK1–7) (except MEK5) (4–6) and in certain mouse cells induces pyronecrosis by activating NACHT-leucine-rich repeat and pyrin domain-containing protein 1b (NALP1b) (7).
Incorporation of LF or EF into the cytoplasm is initiated by the binding of PA to the host cell surface through interacting with either of two known receptors: anthrax receptor 1 (ANTXR1, also known as the tumor endothelial marker 8) (8) and ANTXR2 (also known as the capillary morphogenesis gene-2) (9). Both ANTXR1 and -2 are widely distributed in human tissues and share molecular and biochemical similarities in their extracellular PA interacting domains known as von Willebrand factor A or integrin-like inserted (I) domain (10), post-translational modifications such as palmitoylation and ubiquitination of cytoplasmic domains (11), and associations with the co-receptor lipoprotein receptor-related 6 molecule (12, 13). ANTXR1 and -2 are also distinct in that ANTXR1 is highly expressed in tumor endothelial and cancer cells, and ANTXR2 has a higher binding affinity to PA and requires a lower pH to form a transmembrane pore than ANTXR1 (10, 14).
After binding PA to either ANTXR, a furin-like surface protease then cleaves PA at the N-terminal end to release a 20-kDa soluble fragment, yielding membrane-associated 63-kDa PA (PA63) that forms a ring-shape heptameric oligomerization or prepore and binds LF or EF followed by a lipid raft and clathrin-mediated endocytosis (15, 16). The PA63 heptamers then undergo a pH-dependent conformational change and form pores as the carrier endosomes mature into early endosomes, endosomal carrier vesicles (or multivesicular bodies), and late endosomes. The dissociation and release of LF or EF from PA through the PA63 pores was shown to occur in the membrane of intralumenal vesicles rather than in that of early endosomes whereby LF or EF are transferred to the lumen of the carrier vesicle before being delivered into the cytoplasm through a back fusion process (17, 18). The differences in binding affinities and acidity requirements for pore formation between ANTXR1 and -2 suggest that ANXTR1 and -2 utilize separate routes of LF or EF release into the cytoplasm (14). However, details in the delivery of LF or EF by ANTXR1 and -2 and the mechanisms of the intralumenal vesicle back fusion with the limiting membrane are largely unknown.
Cathepsin B (CTSB) is a lysosomal cysteine protease primarily involved in the degradation or processing of lysosomal proteins (19). In certain conditions CTSB has been shown to be involved in additional cellular processes including cell invasion (20), vesicle trafficking (21), inflammasome formation (22–24), and cell death (25). Because CTSB was shown to be involved in NALP3 inflammasome and caspase-1 activation (23, 24, 26), which shares signaling mechanisms with NALP1b pathways, we have examined the role of CTSB in LeTx cytotoxicity. However, unexpectedly, we found an unknown function of CTSB in autophagy vesicle trafficking and the cytoplasmic delivery LeTx. This study suggested that LF associated with ANTXR2, but not ANTXR1, was released into the cytoplasm through an endocytic trafficking route involving lysosomal fusion and CTSB-dependent autophagy flux.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Cathepsin B inhibitor IV (CA074-Me) was purchased from Peptide Institute. MG132, cathepsin K inhibitor, cathepsin G inhibitor, and vinblastine were purchased from Calbiochem. 3-Methyladenine (3-MA) was purchased from Sigma. LF and PA were prepared in the laboratory as previously described (27, 28), and inactive Lethal Factor (mLF) was purchased from List Biological Laboratories. Antibodies for LC3, phospho-ERK, and p38 mitogen-activated protein kinase were obtained from Cell Signaling Technologies (Pickering, ON). Antibody raised against the N terminus of MEK1 was obtained from QED Bioscience Inc. Antibodies toward lethal factor and protective antigen were a gift from Dr. Stephen H. Leppla (National Institutes of Health, Bethesda, MD). Antibodies for Rab-7, EEA1, actin, and SYBL1 were obtained from Abcam (Cambridge, MA). Cathepsin B antibody was purchased from Calbiochem. GFP-LC3 plasmid was provided by Dr. T. Yoshimori (National Institute for Basic Biology, Okazaki, Japan). pCDNA-ANTXR1-SV1-HA and pCDNA-ANTXR2-HA plasmids were provided by Dr. Jeremy Mogridge (University of Toronto).
Cell Culture
RAW264.7 murine macrophages, HEK293 cells, and the lysosome-associated membrane protein 1 (LAMP-1)/LAMP-2 double-deficient or wild type mouse embryonic fibroblasts cell lines were cultured in DMEM medium containing 10% heated-inactivated fetal bovine serum (Sigma), 10 mm MEM nonessential amino acids solution, 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 1 mm sodium pyruvate. DMEM medium containing 8% heat-inactivated fetal bovine serum was used for RAW264.7 murine macrophages. Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. LAMP-1/LAMP-2 double-deficient and wild type murine embryonic fibroblast (MEF) cell lines were obtained from Dr. Paul Saftig (University of Kiel). Mouse bone marrow-derived macrophages from CTSB-deficient (Ctsb−/−) or control strain C57 BL/6 (Ctsb+/+) mice were cultured in RPMI 1640 medium containing macrophage colony-stimulating factor (21). The human monocytic cell line THP-1 was cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (Sigma), 10 mm MEM nonessential amino acids solution, 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 1 mm sodium pyruvate. Cells were grown at 37 °C in a humidified atmosphere containing 5% CO2. CHOR1.1 and CHOR1.1-ANTXR1-SV1 stably transfected cell lines were kind gifts from Dr. Jeremy Mogridge (University of Toronto) and were maintained in DMEM/F-12 (1:1) media. Cells were grown at 37 °C in a humidified atmosphere containing 5% CO2.
Measurement of Cell Viability
RAW264.7 macrophages were cultured for the indicated times in the presence or absence of LeTx in 96-well plates, and MTT was added at a final concentration of 1 mg/ml. After incubating at 37 °C for an additional 2 h, culture media was carefully aspirated, and 100 μl of dimethyl sulfoxide (DMSO) was added to dissolve crystals. Optical densities of the wells were analyzed using an automatic enzyme-linked immunosorbent assay plate reader (Bio-Rad) at a wavelength of 590 nm. The ratio of cell death was determined based on the optical density of the wells compared with those from nontreated cells as no cell death.
Small Interfering RNA
Human vesicle-associated membrane protein 7 (VAMP7; SYBL1) and ANTXR1- and ANTXR2-specific small interference RNAs (siRNAs) were purchased from Thermo Scientific (Dharmacon, for SYBL1; ON-TARGET plus SMART pool L-020864-00-0005, for ANTXR1; ON-TARGET plus SMART pool L-010679-00-0005, for ANTXR2; ON-TARGET plus SMART pool ID# L-015215-01-0005), and the siRNA negative control was purchased from Ambion. Transfections of HEK293 cells or THP-1 cells with siRNAs were performed using Lipofectamine 2000 (Invitrogen). At 42 h post-transfection some of cells were harvested for validation of siRNA using reverse transcription-PCR or Western blots, and others were treated with LeTx for trafficking analysis. Total RNA was isolated using Trizol (Invitrogen), and cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (New England Biolabs) according to the manufacturer's instructions. Oligonucleotide primers for PCR were: for hANTXR1, 5′- ATGCCTTGTGGGTCCTACTG-3′ (forward) and 5′-GAGGTGTGGTAGGCGTTGTT-3′ (reverse); for hANTXR2, 5′-AAGGACGGGAGGATTCTGTT-3′ (forward) and 5′-CAGCCAGCCAGTGACATAAA-3′ (reverse).
Total Cell Lysate Preparation and Immunoblot Analysis
Total cell lysate preparation and immunoblotting procedures were performed as previously described (29). Briefly, cells were lysed in ice-cold lysis buffer (20 mm MOPS, 2 mm EGTA, 5 mm EDTA, 1 mm Na3VO4, 40 mm β-glycerophosphate, 30 mm sodium fluoride, 20 mm sodium pyrophosphate, and 1% Triton X, pH 7.2) containing a protease inhibitor mixture (Roche Applied Science). The cell lysates were incubated on ice for 10 min and centrifuged at 12,500 rpm for 15 min at 4 °C. The supernatants were separated by SDS-polyacrylamide gels followed by transfer onto nitrocellulose membranes (Bio-Rad). The membranes were blocked with 5% (w/v) skim milk for 1 h at room temperature for and then incubated overnight at room temperature with the primary antibody. The membranes were washed, incubated with secondary antibody for 1 h (Pierce), and developed using an enhanced chemiluminescence detection system (ECL) (Pierce).
Subcellular Fractionation
Endosomal fractionations were prepared by sucrose step gradient (30) after preparing subcellular fractions using a lysosome isolation kit (Sigma). Detailed procedures followed the manufacturer's instructions, and all steps were carried out at 4 °C or on ice. Briefly, cells were washed with PBS and resuspended with 1× homogenization buffer containing protease inhibitor mixture and homogenized gently to limit damage to endosomes. The post-nuclear supernatant (PNS) was prepared by centrifugation for 10 min at 1,000 × g, and PNS was centrifuged at 20,000 × g for 20 min. The supernatant was collected as the cytosolic fraction for Western blot, and the pellet was resuspended with 1× homogenization buffer. Optiprep density gradient solutions were prepared according to the manufacturer's instructions for loading on the gradient. The gradient was centrifuged for 10 h at 35,000 rpm in a SW41 swinging bucket rotor (Beckman Instruments), and gradient fractions were collected and analyzed by immunoblots.
Immunofluorescence Staining and Lethal Toxin Trafficking Analysis
HEK293 cells were plated on coverslips and incubated in the presence or absence of CA074-Me for 1 h at 37 °C in 5% CO2. Cells were then treated with the LeTx (250 ng/ml PA and LF) for 1 h and washed twice with normal growth media to remove unbound toxins, and cells were further incubated at 37 °C for 1 h in 5% CO2. Cells were fixed in 4% formaldehyde and blocked with 10% normal goat serum. Endogenous cathepsin B or endocytosed PA or LF were detected by immunofluorescence staining using the Vector Laboratories system and observed through a Bio-Rad Radiance 2000 two-photon fluorescence confocal microscope. For colocalization of GFP-LC3 and PA or LF, RAW264.7 cells were electroporated with the GFP-LC3 plasmid. At 16 h post-transfection, cells were treated with LeTx and immunostained as above. Immunofluorescence images were obtained and analyzed using a Zeiss LSM510 META confocal microscope and ZEN software.
Electron Microscopy
Cells were grown on 100-cm dishes and incubated in the presence or absence of LeTx for 1 h at 37 °C in 5% CO2. Cells were washed with PBS twice and fixed with 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer for 2 h at room temperature. Grids with specimen were prepared by the Transmission Electron Microscope Facility at The University of Western Ontario (Canada), and micrographs were taken with a transmission electron microscope. Briefly, after fixing with 2.5% glutaraldehyde, Cells were washed with 0.1 m cacodylate buffer 3 times, and cells were further fixed with 1% osmium tetroxide in 0.1 m cacodylate buffer for 1 h and then rinsed with 0.1 m cacodylate buffer. Cells were enrobed in 5% Noble Agar and washed with distilled water 5 times, further fixing with 2% uranyl acetate for 2 h, followed by dehydration in 50% (15 min), 70% (16 h), 85% (15 min), 95% (15 min), and 2 changes of 100% ethanol each 15 min. They were then cleared by 2 changes of propylene oxide, each 15 min, and infiltrated with epon resin:propylene oxide (1:1) for 3 h, epon resin:propylene oxide (3:1) for 16 h, and 2 changes with pure epon resin for total 6 h. Thin sections were mounted on grids and examined under the electron microscope (Philips EM410).
Autophagic Flux Analysis
Autophagy flux was analyzed by flow cytometry and confocal microscopy using DQTM Red BSA (self-quenched red BODIPY dye conjugated to BSA; Molecular Probes, Eugene, OR). Red DQ-BSA requires enzymatic cleavage in acidic intracellular lysosomal compartments to generate a highly fluorescent product that can be monitored by confocal microscopy or flow cytometry. GFP-LC3 transiently expressing-RAW264.7 cells were incubated in RPMI media containing DQ-BSA (10 μg/ml) for 30 min and washed twice with PBS. Cells were then treated with LeTx in the presence or absence of CA074-Me for 60 min and fixed with 4% formaldehyde. Colocalization of GFP-LC3 and red fluorescent of DQ-BSA were imaged using a Bio-Rad Radiance 2000 two-photon confocal microscope and LaserSharp 2000 software. For flow cytometry analysis, the human monocytic cell line THP-1 was incubated in RPMI media containing DQ-BSA (10 μg/ml) for 15 min at 37 °C in 5% CO2. Cells were washed twice with PBS and then incubated for 45 min to ensure that DQ-BSA had reached the lysosomal compartment. THP-1 cells were further incubated in the presence or absence of CA074-Me for the indicated times. Cells were harvested, and red-fluorescent of DQ-BSA was analyzed by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences) and CellQuest (BD Biosciences) and FlowJo (Treestar) software. For confocal images analysis, cells were plated on coverslips after treatment as above and fixed with 4% formaldehyde. The fluorescent degradation products of DQ-BSA in lysosomes were imaged using a Bio-Rad Radiance 2000 two-photon confocal microscope and LaserSharp 2000 software.
RESULTS
Cathepsin B Inhibitor CA-074-Me Inhibits LeTx-induced Cell Death of Macrophages
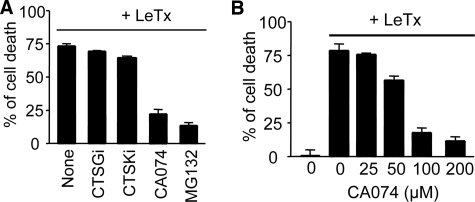
We examined whether CTSB is involved in LeTx-induced cell death using the CTSB-specific chemical inhibitor CA074-Me (CA074). RAW264.7 macrophages, which express NALP1b, were rapidly killed by LeTx, and CA074, but not other cathepsin inhibitors targeting cathepsin G (CTSGi) and cathepsin K (CTSKi), significantly prevented cell death (Fig. 1A). The cell death protective effect of CA074 was dose-dependent (Fig. 1B) and more pronounced in early cell death and at low LeTx doses (supplemental Fig. 1, A and B). As previously shown (31), the proteasome inhibitor MG132 prevented LeTx-induced cytolysis to similar extents of 100 μm CA074 (Fig. 1A).
FIGURE 1.
The cathepsin B inhibitor CA074 prevents cell death induced by anthrax lethal toxin in RAW 264.7 macrophages. A, RAW264.7 murine macrophages were treated with different kinds of cathepsin inhibitors (cathepsin G inhibitor (CTSGi; 100 μm), cathepsin K inhibitor (CTSKi;100 μm), cathepsin B inhibitor (CA074-Me; 100 μm), and proteasome inhibitor (MG132; 10 μm) for 1 h and then treated with LeTx (250 ng/ml LF and 500 ng/ml PA) in the presence of the inhibitors for 5 h. MTT was added 2 h before the end of the experiment for cell death determination. Data are expressed as the mean ± S.D. (n = 3). B, RAW264.7 cells were pretreated with different concentrations of CA074-Me and then treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 5 h. Cell death assays were performed as above in A. Data are expressed as the mean ± S.D. (n = 4).
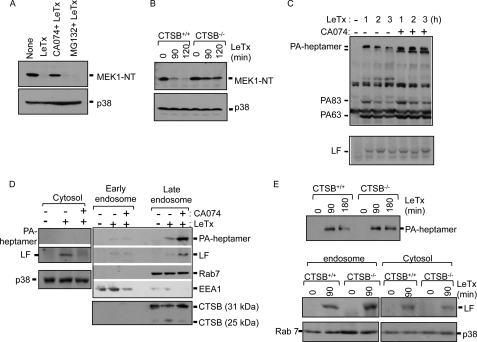
Cathepsin B Inhibitor Inhibits the Delivery of LF into the Cytoplasm from Late Endosomes
We further examined whether CA074 inhibited LeTx-induced cell death before or after the release of LF into the cytoplasm and whether the protective effect was specific to cathepsin B. Based on MEK1 N-terminal cleavage, which indicates the release of LF into the cytoplasm, CA074 appeared to inhibit the cytoplasmic delivery of LF (Fig. 2A). In comparison, the proteasome inhibitor MG132 prevented LeTx-induced cell death to a similar extent as CA074 (Fig. 1A) but had no effects on MEK1 cleavage by LF (Fig. 2A) as previous reported (31). MEK1 cleavage by LF was also greatly suppressed in primary bone marrow-derived macrophages prepared from CTSB-deficient mice (CTSB−/−) (Fig. 2B), indicating that CTSB, but not cathepsin L, which could have been inhibited by CA074 (32), was mainly responsible for the delivery of LF into the cytoplasm.
FIGURE 2.
Inhibition of cathepsin B blocks MEK1 cleavage induced by anthrax lethal toxin through delaying LeTx release into the cytoplasm. A, RAW264.7 cells were pretreated with CA074-Me (100 μm) or MG132 (10 μm) for 1 h and treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 2 h. MEK1 cleavage was analyzed by Western blots. Immunoblotting for p38 was used as a loading control. NT, N terminus. B, bone marrow-derived macrophages from C57BL/6 (Ctsb+/+) or (Ctsb−/−) mice were treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for the indicated times, and MEK1 cleavage was analyzed by Western blots. The immunoblot for p38 was used as a loading control. C, C57 bone marrow-derived immortalized macrophage cells were incubated in the presence or absence of CA074-Me (100 μm) for 1 h and treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 1 h. LeTx-treated cells were washed twice with PBS and incubated for another 1 h or 2 h with fresh media. Cell homogenates were heated at 96 °C for 5 min and separated using SDS-PAGE. PA-heptamer stability was analyzed by immunoblot. D, RAW264.7 macrophages were pretreated with CA074-Me (100 μm) for 1 h and treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 1 h. Macrophages were washed twice with PBS and harvested after another 1 h of incubation at 37°C. Macrophage homogenates were prepared in 1× extraction buffer as described under “Experimental Procedures,” and the gradient was centrifuged for 10 h at 35,000 rpm in the SW41 rotor (Beckman). Gradient fractions were collected and analyzed by Western blots. E, bone marrow-derived macrophages from C57BL/6 (Ctsb+/+) or (Ctsb−/−) mice were treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 90 min, and macrophages were washed twice with PBS. Half of the cells were harvested, and intracellular total endosomal compartments were prepared as under “Experimental Procedures.” The remaining cells were further incubated at 37°C for 90 min, and intracellular total endosomal compartments were prepared as above. PA-heptamer stabilities were analyzed by immunoblots (top panel). Bone marrow-derived macrophages from C57BL/6 (Ctsb+/+) or (Ctsb−/−) mice were treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 90 min. The cytosolic and endosomal compartments were prepared as above, and LF was analyzed by Western blot. Immunoblots for Rab7 and p38 were used as loading controls (bottom panel).
Delivery of LF into the cytoplasm involves the formation of the heptameric PA63 pore, which becomes an SDS-resistant complex in acidified endosomes (28). The SDS-resistant PA63 complex is then transported to lysosomes for rapid degradation (16, 33). Thus, we examined whether CTSB was involved in intracellular uptake of LeTx by analyzing formation of the SDS-resistant PA63 complex. CA074 had no effect on SDS-resistant PA63-heptamer formation but, rather, protected it from lysosomal degradation (Fig. 2C, upper panel). The total amounts of intracellular LF did not change by CA074 (Fig. 2C, lower panel), indicating that CA074 did not interfere intracellular uptake of LeTx. It has also been suggested that internalized LeTx is sequentially transported to early endosomes, intralumenal vesicles (also known as multivesicular bodies), and late endosomes before being released into cytoplasm through back fusion with the limiting membrane (17, 18). We further examined in which endosomal pathways LF delivery was inhibited by CA074. Early and late endosomes were isolated from LeTx-treated RAW264.7 cells in the presence or absence of CA074. The SDS-resis tant PA63 heptamer and LF were highly enriched in late endosomes, which co-fractionated with the late endosomal small GTPase rab7 (34, 35) and CTSB in the presence of CA074 (Fig. 2D, right panel). Unlike PA, overall amounts of LF did not change (Fig. 2C, lower panel), and LF was mostly released into the cytoplasm in the absence of CA074 but retained in late endosomes in the presence of CA074 (Fig. 2D). Similar experiments were repeated in primary bone marrow-derived macrophages prepared from wild type (CTSB+/+) and CTSB−/− mice. Unlike CA074-treated cells, CTSB−/− cells had no significant changes in total amounts of PA63 heptamers (Fig. 2E, upper panel), suggesting that PA was degraded mainly by a CTSB-independent but CA074-sensitive protease such as cathepsin L. However, consistent with data shown in Fig. 2B, more LF was retained in endosomes, and less was released into the cytoplasm of CTSB−/− than CTSB+/+ cells (Fig. 2E, lower panel). These observations suggest that LF is transported to late endosomes and released into the cytoplasm mainly through a CTSB-sensitive pathway.
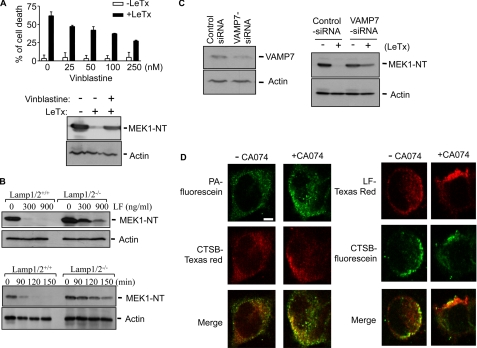
Lysosomal Fusion with Late Endosomes Is Required for the Delivery of LF into the Cytoplasm
Because CTSB is likely compartmentalized in lysosomes, we examined whether lysosomal fusion with LeTx-containing vesicles was required for the cytosolic delivery of LF. Several molecules known to be involved in the lysosomal fusion process were examined for their involvement in the cytosolic delivery of LF. The chemical lysosomal fusion inhibitor vinblastine prevents microtubule-dependent transport of lysosomes to endosomes (36, 37). RAW264.7 macrophages pretreated with vinblastine at various doses were more resistant to LeTx-induced cell death in a dose-dependent manner (Fig. 3A). MEK1 cleavage by LeTx was also inhibited (Fig. 3A, lower panel), suggesting that LF was not released into the cytoplasm. The LAMP1 and -2 play an important role in the fusion of lysosomes with other vacuoles, likely through a dynein-mediated transport of lysosomes and phagolysosome formation, and cells deficient in LAMP1 and LAMP2 showed defects in lysosomal fusion with autophagic vacuoles and phagosomes (38, 39). Within 90 min of LeTx treatment, wild type immortalized MEFs incorporated LF into the cytoplasm, and subsequently, MEK1 was cleaved by LF, whereas MEFs lacking LAMP1 and -2 required more time (>150 min) and higher doses to reach similar extents of MEK1 cleavage (Fig. 3B). The fusion of late endosomes and lysosomes requires the presence of N-ethylmaleimide-sensitive factor (NSF), soluble NSF attachment proteins, and a small GTPase of the Rab family such as Rab7 (40). After formation of tethering between late endosomes and lysosomes, a trans-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex must be formed to fuse the membranes (41). One of the key molecules required for heterotypic late endosome-lysosome fusions is VAMP7, also known as tetanus-neurotoxin-insensitive VAMP or synptobrevin-like-1 (42). We used easily transfected HEK293 cells to specifically knock down VAMP7 mRNA using siRNAs and examined MEK1 cleavage induced by LeTx. About 50% of VAMP7 at mRNA levels were diminished by the siRNAs, based on a real-time reverse transcription-PCR analysis (data not shown), and Western blots also supported the knockdown of VAMP7 by the siRNAs (Fig. 3C, left panel). Indeed, knocking down VAMP7 partially prevented LeTx-induced MEK1 cleavage, and intact MEK1 was readily detectible by Western blots, albeit the level of intact MEK1 was lower than that of nontreated cells (Fig. 3C, right panel). To further address whether CTSB activity is required for the process of, or after the fusion between late endosomes and lysosomes, we examined co-localization of CTSB with PA or LF in the presence or absence of CA074 using confocal microscopy. Immunofluorescent staining in HEK293 cells with CTSB and PA or LF indicated that both PA and LF were co-localized with CTSB-containing vesicles, and the colocalization was more prominent in cells treated with CA074 (Fig. 3D), suggesting that CA074 did not block fusion of LeTx-containing vesicles with CTSB-containing lysosomes.
FIGURE 3.
Late endosome and lysosome fusion is required for release of LeTx into the cytoplasm. A, inhibition of microtubule activity blocked cell death and MEK1 degradation induced by LeTx. RAW264.7 macrophages were incubated in the absence or presence of the microtubule inhibitor vinblastine (0–250 ng/ml) for 1 h and treated with LeTx (125 ng/ml LF and 250 ng/ml PA) for 5 h. Cell death was measured using a MTT assay (top panel). Data are expressed as the mean ± S.D. (n = 3). RAW264.7 macrophages were pretreated with the vinblastine (250 ng/ml) for 1 h and treated with LeTx (125 ng/ml LF and 250 ng/ml PA) for 2 h. MEK1 degradation was analyzed by Western blots (bottom panel). NT, N terminus. B, Lamp1/2 deficiency in MEF cell lines caused delayed MEK1 degradation induced by LeTx. Mouse embryo fibroblast wild type (Lamp1/2+/+) or Lamp1/2 double-deficient (Lamp1/2−/−) cell lines were treated with different doses of LeTx for 2 h, and MEK1 degradation were analyzed by immunoblotting (top panel). Lamp1/2+/+ or Lamp1/2−/− MEF cell lines were treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for the indicated times, and MEK1 degradation was analyzed by Western blot (bottom panel). C, HEK293 cells were transfected with siRNA against VAMP7 or control siRNA. A portion of the transfected cells was harvested at 42 h post-transfection for confirmation of protein knockdown, and the remaining cells were treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 2 h. MEK1 degradation was analyzed by Western blot. D, inhibition of cathepsin B activity caused enhanced colocalization with cathepsin B-containing vesicles and PA or LF. HEK293 cells were pretreated with or without CA074-Me (100 μm) for 1 h and then treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 1 h. Cells were washed twice with media and further incubated at 37°C for 1 h. After blocking with 10% goat serum, cells were immunostained with anti-CTSB, anti-PA or anti-LF antibodies, and visualized using a confocal microscope; scale bar, 5 μm.
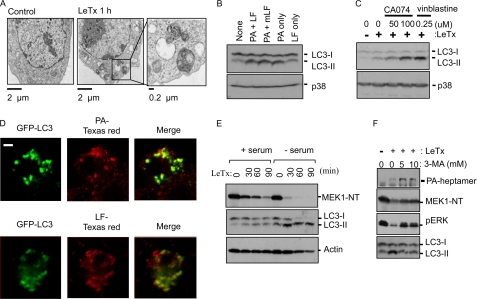
LeTx-induced Autophagy Facilitates the Delivery of LF into the Cytoplasm
Recently, LeTx was shown to induce autophagy, suggested to be a long term (after 24–48 h of LeTx treatment) survival mechanism of LeTx-exposed human promyelocytic leukemia HL-60 cells (43). We also observed the formation of autophagy-related structures, likely sequestered mitochondria or other organelles, in LeTx-treated RAW264.7 cells using electron microscopy (Fig. 4A). During autophagy, a ubiquitin-like microtubule-associated protein 1 light chain 3-I (LC3-I, the mammalian orthologue of yeast autophagy-related gene (Atg8; 18 kDa), converts to a phosphatidylethanolamine-conjugated LC3-II (16 kDa) form, and the amount of LC3-II correlates with the extent of autophagosome formation (44). Thus, we examined the amounts of LC3-II to examine whether autophagy was induced by PA or protease activity of LF. In fact, the amounts of LC3-II formation by PA alone or PA with protease-inactivated mutant LF were similar as those induced by LeTx (Fig. 4B). The amounts of LC3-II were even further enhanced when cells were pretreated with CA074 or vinblastine (Fig. 4C). When we visualized cells using immunofluorescent staining against PA or LF and transiently expressed GFP-LC3 followed by confocal microscopy, both PA (80–90% of PA puncta) and LF (40–50% of LF puncta) were co-localized with LC3-II containing autophagosomes or amphisomes (Fig. 4D), suggesting that the delivery of LF into the cytoplasm was at least in part carried out through autophagy processes.
FIGURE 4.
Induction of autophagy is required for effective trafficking of LeTx. A, LeTx induced the formation of intracellular vacuoles in macrophages. RAW264.7 macrophages were incubated in absence or presence of LeTx (250 ng/ml LF and 500 ng/ml PA) at 37 °C for 1 h and washed twice with ice-cold PBS. Cells were then fixed with 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer, and grids with specimen were prepared as described under “Experimental Procedures.” Micrographs were taken with a transmission electron microscope. Scale bars show 2 μm in length for the lower magnification and 0.2 μm for higher magnification. B, LeTx induced LC3-II formation in a PA-dependent manner. RAW264.7 macrophages were treated with PA and LF (PA + LF, 500 ng/ml PA and 250 ng/ml LF), PA and inactive LF (PA + mLF, 500 ng/ml PA and 250 ng/ml LF), PA only (500 ng/ml PA), or LF only (250 ng/ml PA) for 2 h. Induction of autophagy was analyzed by immunoblotting against LC3-II. An immunoblot for p38 MAPK was used as a loading control. C, RAW264.7 cells were pretreated with CA074-Me (50 or 100 μm) or the microtubule inhibitor vinblastine (0.25 μm) and treated with LeTx (125 ng/ml LF and 250 ng/ml PA) for 2 h. LC3-II accumulation was analyzed by Western blots. D, N-terminal GFP-conjugated LC3 was transiently transfected in RAW264.7 macrophages and treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 1 h at 16 h post-transfection. Cell were washed twice with normal media and further incubated at 37°C for 30 min. Cells were then fixed and immunostained with anti-PA or anti-LF as described under “Experimental Procedures.” GFP-conjugated LC3 and PA (top panel) or LF (bottom panel) were visualized using a Zeiss LSM510 META confocal microscope; scale bar, 5 μm. E, THP-1 cells were incubated with serum or without serum for 4 h and treated with LeTx (125 ng/ml LF and 250 ng/ml PA) for the indicated times. MEK1 degradation or LC3-II formation was analyzed by Western blots. Immunoblot for actin was used as a loading control. NT, N terminus. F, THP-1 cells were treated with different doses of 3-MA for 1 h and exposed by LeTx for 2 h. PA-heptamer and MEK1 degradation, phospho-ERK (pERK), and LC3-II formation were analyzed by Western blots.
To evaluate the role of autophagy in the delivery of LF into the cytoplasm, we examined whether inhibition or enhancement of autophagy processes affects the rate of LF delivery into the cytoplasm. In these experiments we used the human monocytic cell line THP-1 cells, which had slower rates of LeTx uptake and higher sensitivity to the autophagy inhibitor 3-methyladenine (3-MA). During serum starvation conditions, the rates of MEK1 cleavage and LC3-II formation were much faster than those of cells in normal serum conditions (Fig. 4E). In line with these data, 3-MA inhibited LeTx-induced MEK1 cleavage and LC3-II accumulation. Basal levels of phospho-ERK1 and -2, of which phosphorylations are mainly mediated by MEKs, were also diminished by LeTx, and 3-MA was able to restore the basal levels of the phosphorylations (Fig. 4F). The degradation of PA63 heptamer was also suppressed by 3-MA (Fig. 4F, top panel). Altogether, these data support that autophagy induced by PA facilitated the delivery of LF into the cytoplasm.
Cathepsin B Activity Is Required for Autophagic Flux
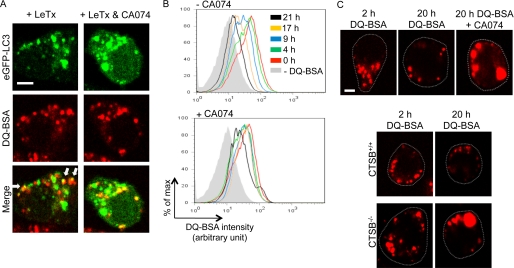
Because previous results suggested that autophagy was required for an efficient delivery of LF into the cytoplasm but CA074 enhanced LC3-II formation, we investigated whether CTSB was involved in the process of autophagy and, if so, in which step of autophagy process using DQTM Red BSA (DQ-BSA). DQ-BSA is a derivative of bovine serum albumin that is labeled with a self-quenched red fluorescent dye and dequenched by lyso somal proteases, releasing red fluorescence. As shown in Fig. 5A, left panels, red fluorescent dequenched DQ-BSA-containing vesicles were partly co-localized with LC3-positive puncta after LeTx treatments. CA074, even at high concentrations (100 μm), had no effect on the co-localization of dequenched DQ-BSA vesicles with LC3-positive puncta (Fig. 5A, right panels), suggesting that CA074 had no effects on the fusion process of endosomes and lysosomes but, rather, enhanced or stabilized the fusion of dequenched DQ-BSA-containing vesicles (endolysosomes) and LC3-containing puncta (autophagosomes). Thus, we examined whether CTSB deficiency or CA074 could prevent autophagy flux. DQ-BSA was pre-loaded and dequenched in THP-1 cells, and the rate of disappearance of dequenched DQ-BSA was monitored. Dequenched DQ-BSA disappeared over the time period of 21 h (Fig. 5B). However, in the presence of CA074 (50 μm), the flux of dequenched DQ-BSA was significantly delayed (Fig. 5B and supplemental Fig. 2). Also, cells treated with CA074 (50 μm) or CTSB-deficient macrophages (CTSB−/−) showed enlarged dequenched DQ-BSA-containing vesicles (Fig. 5C, top and bottom panels). These results consistently suggested that CTSB was required for clearing dequenched DQ-BSA or flux of amphisomes.
FIGURE 5.
Cathepsin B inhibition causes defective autophagy flux. A, RAW264.7 cells were electroporated with GFP-LC3 plasmid and incubated in DMEM containing DQ-BSA (10 μg/ml) for 30 min. Cell were then washed twice with DMEM and treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 60 min in the presence or absence of CA074-Me (100 μm). Cells were fixed, and colocalization of GFP-LC3 and red fluorescent of DQ-BSA was imaged using a Bio-Rad Radiance 2000 two-photon confocal microscope and LaserSharp 2000 software. Arrows show the colocalization of dequenched DQ-BSA vesicles with LC3-positive puncta; scale bar, 5 μm. B, THP-1 cells were incubated in RPMI media containing DQ-BSA (10 μg/ml) for 15 min at 37 °C in 5% CO2. Cells were washed and incubated further in regular growth media for 45 min to ensure that DQ-BSA had reached the lysosomal compartment. Cells were then further incubated in regular growth media in the presence or absence of CA074-Me (50 μm), then harvested at indicated time points. Cells were analyzed using flow cytometry. C, for confocal images analysis, THP-1 cells were plated on coverslips after treatment as above in B, and cells were incubated in RPMI media with or without CA074-Me (50 μm). Cells were then fixed with 4% formaldehyde at the indicated time point. The fluorescent products of DQ-BSA were imaged using the Bio-Rad Radiance 2000 two-photon confocal microscope and LaserSharp 2000 software. Dotted lines indicate the cell margin; scale bar, 5 μm. D, C57 wild type (CTSB+/+) or cathepsin B-deficient (CTSB−/−) cell lines were plated on coverslips and incubated in RPMI media containing DQ-BSA (10 μg/ml) for 15 min at 37 °C in 5% CO2. Cells were washed twice with PBS and incubated in RPMI media for the indicated times. Cells were fixed as above in C, and the fluorescent degradation products of the DQ-BSA in lysosome were imaged using Bio-Rad Radiance 2000 two-photon confocal microscope and LaserSharp 2000 software. Dotted lines indicate the cell margin.
ANTXR2-mediated LeTx Uptake Requires Autophagic Flux for the Cytoplasmic Delivery of LF
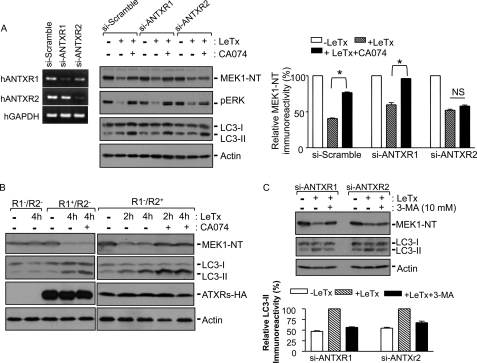
PA binds to one of the two ANTXRs: ANTXR1 and ANTXR2 (45). Because the delivery of LF into the cytoplasm was not completely inhibited by CA074, we investigated whether the involvement of CTSB was receptor-specific. ANTXR1 or -2 was specifically knocked down up to ∼70% at the mRNA level in HEK293 (Fig. 6A, left panel) and THP-1 cells (data not shown) by siRNAs. In ANTXR1-knockdown cells, CA074 almost completely inhibited MEK1 cleavage by LeTx, whereas in ANTXR2-knockdown cells MEK1 cleavage was not significantly inhibited (Fig. 6B). Similarly, basal levels of phosphorylated ERK1 and -2 were correlated with intact MEK1 levels. LC3-II formation by LeTx was induced to similar extents in these cells, and the levels of LC3-II were further enhanced in the presence of CA074 as previously observed in RAW264.7 cells (Fig. 4C). To further corroborate that ANTXR2 utilized a CA074-sensitive or CTSB-mediated process, we investigated the effect of CA074 on LeTx uptake in ANTXR1- and -2-deficient CHO-K1 cells (R1−/R2−) reconstituted with full-length ANTXR1-sv1 (R1+/R2−) or ANTXR2489 (R1−/R2+) (46). Consistent with the data shown in Fig. 6A, CA074 failed to inhibit MEK1 cleavage induced by LeTx in R1+/R2− cells, whereas MEK1 cleavage by LeTx was completely inhibited in R1−/R2+ cells (Fig. 6B). The amounts of LC3-II were also increased in both R1+/R2− and R1−/R2+ cells by LeTx, which was further increased in the presence of CA074. We also examined whether ANTXR2-mediated LF delivery was mediated through autophagy. The autophagy inhibitor, 3-MA, which at least partially prevented LC3-II formation by LeTx in either ANTHR1 or ANTXR2 knockdown cells, was able to prevent MEK1 cleavage in ANTXR1 but not ANTXR2 knockdown cells (Fig. 6C). These results collectively suggest that ANTXR2 but not ANTXR1 delivery of LF into the cytoplasm was mediated through a CTSB- and autophagy-dependent pathway.
FIGURE 6.
Cathepsin B is involved in ANTXR2-dependent LeTx delivery through autophagic flux. A, HEK293 cells were transfected with control siRNA (si-Scramble) or human (h) ANTXR1- or ANTXR2-specific siRNAs (si-ANTXR1 or si-ANTXR2), and at 42 h post-transfection, portions of the cells were harvested for reverse transcription-PCR (left panel). The remaining cells were incubated in the presence or absence of CA074-Me (100 μm), and treated with LeTx (250 ng/ml LF and 500 ng/ml PA) for 2 h. MEK1 degradation, down-regulation of phospho-ERK (pERK), and LC3-II formation were analyzed by Western blots (middle panel) and MEK1-NT immunoreactivities were analyzed using the NIH-Image program. Data are expressed as mean ± S.D. (n = 3; *, significant, p < 0.05; NS, not significant with p > 0.5). NT, N terminus. B, ANTXR1/R2-deficient CHO cells (R1−/R2−), ANTXR1-reconstituted CHO cells (R1+/R2−), or ANTXR2-reconstituted CHO cells (R1−/R2+) were treated with LeTx (500 ng/ml LF and 1000 ng/ml PA) for the indicated times. MEK1 degradation and LC3-II formation were analyzed by Western blot. Immunoblots for HA or actin were used for transfection or loading control. C, human monocytic THP-1 cells were transfected with control siRNA or siANTXRs, and after 42 h cells were treated with LeTx as above. MEK1 degradation and LC3-II formation were analyzed by Western blot (upper panel), and LC3-II immunoreactivities were analyzed using NHI-Image program (lower panel). Data are expressed as the mean ± S.D. (n = 2).
DISCUSSION
This study showed that CTSB is involved in the delivery of ANTXR2-associated LF into the cytoplasm. It appeared that CTSB played an important role in autophagic flux whereby LF was delivered from intralumenal vesicles into the cytoplasm likely through back fusion. The current model of LeTx entry into the cytoplasm involves translocation of LF from the luminal compartment of the multivesicular late endosomes through an undefined back-fusion process with the limiting membrane (17, 47). This study showed that LF accumulated in late endosomes in the presence of CA074 (Fig. 2D) or in CTSB-deficient macrophages (Fig. 2E), and CTSB-containing lysosomes colocalized with LF-containing endosomes (Fig. 3D), suggesting that CTSB activity was required for the delivery of LF from endolysosomes into the cytoplasm. These data are somewhat unexpected, considering that lysosomal proteases can cause early degradation of LF. However, it was postulated that LFs are packaged into endosomal carrier vesicles, and unlike PA, which is exposed to lysosomal proteases and undergoes a rapid degradation, LF is sheltered in the lumen of endosomal carrier vesicles and protected from degradation (48). There are several evidences that late endosomes and lysosomes fuse to form hybrid organelles, which then undergo back fusion (40, 49, 50). Therefore, we speculate that lysosomal fusion is a cue for back fusion of intralumenal vesicles, which can be a safety measure to ensure that endocytosed molecules or phagocytosed particles are degraded by lysosomal proteases before back fusion starts.
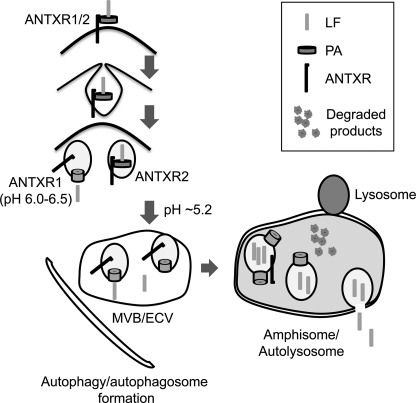
This study also showed that the CTSB-mediated delivery of LF into the cytoplasm was involved only for the ANTXR2 pathway. Although ANTXR1 and -2 share high amino acid sequence homology (∼60% in extracellular domains and ∼68% in the first 145 residues of cytoplasmic domain) and post-translational modifications such as palmitoylation and ubiquitination (10, 11), the pH thresholds for PA63 heptamers to form a transmembrane pore are different in these two receptors (14, 51). It was shown that PA dissociates from ANTXR2 and forms a pore at a lower pH than that required for ANTXR1. The lower pH requirement of ANTXR2 is in line with our observations that the cytoplasmic delivery of LF through ANTXR1 could have occurred in early endosomes and, therefore, was insensitive to CA074 (Fig. 6). However, PA dissociated from ANTXR2 only in mature endosomes with lower pH, resulting in the release of LF into the lumen of intralumenal vesicles followed by CA074-sensitive back fusion with the limiting membrane and release of LF into the cytoplasm (Fig. 7).
FIGURE 7.
A diagram illustrating the proposed pathway involved in the cytoplasmic delivery of LF. Unlike ANTXR1, which can deliver LF into the cytoplasm from early endosomes, ANTXR2-mediated delivery of LF requires autophagy flux, which is triggered by lysosomal fusion.
Autophagy is a dynamic, multistep lysosomal degradation pathway initiated by forming a double-membrane sac called the isolation membrane, which elongates until two leading edges meet to form double membrane-bound structures called the autophagosomes. The outer membrane of autophagosome fuses with endosomes or lysosomes to form an amphisome or autolysosome. It is a cell survival and protective mechanism, particularly during starvation or infections, achieved by procuring energy sources through intracellular organelle turnover (52) or by killing phagocytosed microbes (53, 54). At the same time autophagy can also be utilized by microbes for their survival and evasion of host cell-protective mechanisms (55). This study, consistent with the previous report of Tan et al. (43), showed that PA alone could induce autophagy (Fig. 4, A and B) through either ANTXR1 or -2 (Fig. 6). How ANTXRs triggered autophagy is still to be investigated, but because PA can induce tyrosine phosphorylations of the ANTXR-co-receptor lipoprotein-receptor-related protein 6 (LRP-6) (13), it is possible that the phosphorylation of LRP-6 or possibly other ANTXR-associated proteins triggered autophagy formation. We further showed that ANTXR2-associated LFs took advantage of the host autophagic flux process to reach the cytoplasm (Figs. 4, D–F, and 6B). Both early and late endosomes fuse with autophagic vacuoles (56), which is even necessary for the formation of amphisomes or autolysosomes and subsequent autophagic flux (57). Therefore, the release of LF through autophagic flux is not a surprising proposition.
We also showed that CTSB was required for efficient autophagic flux but not in the fusion processes of autophagosomes and lysosomes, based on the facts that there were no defects in the dequenching process of DQ-BSA in CA074-treated cells (Fig. 5A) and in CTSB-deficient cells (data not shown) and on enhanced colocalization of PA or LF with CTSB (Fig. 3D) as well as dequenched DQ-BSA-containing puncta (Fig. 5A) in the presence of CA074 (Figs. 3D and 5A). However, CA074-treated or CTSB-deficient cells showed more LC3-II accumulation than nontreated (Figs. 4C and 6) or wild type cells (data not shown), respectively, and clearing cleaved DQ-BSA in autolysosomes was delayed by CA074 treatment (Fig. 5, B and C) and in CTSB−/− cells (Fig. 5C, bottom panel). These results suggest that CTSB is involved in the flux of autophagy or amphisomes. Although little is known about the mechanism of autophagosome intralumenal vesicle back fusion, defects in autophagy flux were suggested to be involved in the cytoplasmic delivery of LF. The Niemann Pick Type C disease (NPC) is an autosomal recessive neurodegenerative disease due to mutations in NPC1 or NPC2 genes involved in intracellular cholesterol transport (58). NPC1 cells or cells treated with the drug U18666A, which causes a phenotype identical to NPC1 mutations (59), were shown to have defects in back fusion of intralumenal vesicles and release of LF into the cytoplasm, likely caused by accumulations of cholesterol and, possibly, protein contents in late endosomes (17). Also, Npc1-deficient mice showed significant higher levels of LC3-II and the presence of autophagic vacuole-like structures and multivesicles in brain tissue (60). These studies suggest that alteration of lipid composition and possibly protein contents in late endosomes or autophagic vacuoles could affect intralumenal vesicle back fusion. However, how lysosomal proteases such as CTSB or lipases affect this back fusion still remains to be addressed.
While this manuscript was in preparation, Newman et al. (61) reported that CA074 protected from LeTx-induced cytotoxicity in macrophages. The protection by CA074 was suggested to be mediated by broader inhibitory effects of CA074 and after the release of LF into the cytoplasm. The study also detected a delay in the cleavage of MEKs by CA074; however, the delay was not as prominent as we observed in this study. Because their study used very high doses of LeTx (>1 μg/ml PA and LF) and CA074 (500 μm), most of LF released into the cytoplasm was likely mediated by ANTXR1. At lower doses of CA074 (<100 μm) and LeTx (<500 ng/ml), robust inhibition of LeTx-induced MEK1 cleavage and cytotoxicity were detected in CA074-treated and CTSB−/− cells (Figs. 1 and 2, supplemental Fig. 1).
Physiologically speaking, although ANTXR1 and -2 are ubiquitously expressed in human tissues (9), ANTXR2 has been shown to play a more important role in both anthrax spore-induced macrophage death (62) and LeTx-induced animal lethality (63, 64). ANTXR1, however, is prevalently expressed in cancer cells (65–67), particularly up-regulated in human colorectal cancer endothelium (68), suggesting it as a potential tumor target. Therefore, targeting CTSB or autophagy could be a useful therapeutic strategy for reducing the toxic effects of LeTx without affecting its anti-tumor activity.
Supplementary Material
Acknowledgments
We thank Dr. K. Khazaie for providing CTSB−/− mice and A. Martins for critical reading of the manuscript.
This work was supported by Canadian Institute of Health Research Operating Grant MOP68841 and China-Canada Joint Health Research Initiative Grant CCI82416 (to S. O. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- LeTx
- lethal toxin
- LF
- lethal factor
- EF
- edema factor
- PA
- protective antigen
- MEK
- mitogen-activated protein kinase kinase
- ANTXR
- anthrax receptor 1
- 3-MA
- 3-methyladenine
- MTT
- (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- LAMP
- lysosome-associated membrane protein
- MEF
- murine embryonic fibroblast
- siRNA
- small interference RNA
- LC3-I
- light chain 3-I
- 3-MA
- 3-methyladenine
- NPC
- Niemann Pick Type C disease
- CTSB
- cathepsin B
- ERK
- extracellular signal-regulated kinase
- GFP
- green fluorescent protein
- DMEM
- Dulbecco's modified Eagle's medium
- VAMP
- vesicle-associated membrane protein
- MOPS
- 4-morpholinepropanesulfonic acid
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- CHO
- Chinese hamster ovary.
REFERENCES
- 1.Dixon T. C., Meselson M., Guillemin J., Hanna P. C. (1999) N. Engl. J. Med. 341, 815–826 [DOI] [PubMed] [Google Scholar]
- 2.Mikesell P., Ivins B. E., Ristroph J. D., Dreier T. M. (1983) Infect. Immun. 39, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leppla S. H. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 3162–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopra A. P., Boone S. A., Liang X., Duesbery N. S. (2003) J. Biol. Chem. 278, 9402–9406 [DOI] [PubMed] [Google Scholar]
- 5.Duesbery N. S., Webb C. P., Leppla S. H., Gordon V. M., Klimpel K. R., Copeland T. D., Ahn N. G., Oskarsson M. K., Fukasawa K., Paull K. D., Vande Woude G. F. (1998) Science 280, 734–737 [DOI] [PubMed] [Google Scholar]
- 6.Vitale G., Pellizzari R., Recchi C., Napolitani G., Mock M., Montecucco C. (1998) Biochem. Biophys. Res. Commun. 248, 706–711 [DOI] [PubMed] [Google Scholar]
- 7.Boyden E. D., Dietrich W. F. (2006) Nat. Genet. 38, 240–244 [DOI] [PubMed] [Google Scholar]
- 8.Bradley K. A., Mogridge J., Mourez M., Collier R. J., Young J. A. (2001) Nature 414, 225–229 [DOI] [PubMed] [Google Scholar]
- 9.Scobie H. M., Rainey G. J., Bradley K. A., Young J. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5170–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scobie H. M., Young J. A. (2005) Curr. Opin. Microbiol. 8, 106–112 [DOI] [PubMed] [Google Scholar]
- 11.Abrami L., Leppla S. H., van der Goot F. G. (2006) J. Cell Biol. 172, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W., Lu Q., Chaudry G. J., Leppla S. H., Cohen S. N. (2006) Cell 124, 1141–1154 [DOI] [PubMed] [Google Scholar]
- 13.Abrami L., Kunz B., Deuquet J., Bafico A., Davidson G., van der Goot F. G. (2008) Cell Microbiol 10, 2509–2519 [DOI] [PubMed] [Google Scholar]
- 14.Rainey G. J., Wigelsworth D. J., Ryan P. L., Scobie H. M., Collier R. J., Young J. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13278–13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klimpel K. R., Molloy S. S., Thomas G., Leppla S. H. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10277–10281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abrami L., Liu S., Cosson P., Leppla S. H., van der Goot F. G. (2003) J. Cell Biol. 160, 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobo K., Le Blanc I., Luyet P. P., Fivaz M., Ferguson C., Parton R. G., Gruenberg J., van der Goot F. G. (2007) PLoS One 2, e851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abrami L., Lindsay M., Parton R. G., Leppla S. H., van der Goot F. G. (2004) J. Cell Biol. 166, 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGrath M. E. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 181–204 [DOI] [PubMed] [Google Scholar]
- 20.Yan S., Sloane B. F. (2003) Biol. Chem. 384, 845–854 [DOI] [PubMed] [Google Scholar]
- 21.Ha S. D., Martins A., Khazaie K., Han J., Chan B. M., Kim S. O. (2008) J. Immunol. 181, 690–697 [DOI] [PubMed] [Google Scholar]
- 22.Duncan J. A., Gao X., Huang M. T., O'Connor B. P., Thomas C. E., Willingham S. B., Bergstralh D. T., Jarvis G. A., Sparling P. F., Ting J. P. (2009) J. Immunol. 182, 6460–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halle A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J., Golenbock D. T. (2008) Nat. Immunol. 9, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. (2008) Nat. Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willingham S. B., Bergstralh D. T., O'Connor W., Morrison A. C., Taxman D. J., Duncan J. A., Barnoy S., Venkatesan M. M., Flavell R. A., Deshmukh M., Hoffman H. M., Ting J. P. (2007) Cell Host Microbe 2, 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentze H., Lin X. Y., Choi M. S., Porter A. G. (2003) Cell Death Differ. 10, 956–968 [DOI] [PubMed] [Google Scholar]
- 27.Wesche J., Elliott J. L., Falnes P. O., Olsnes S., Collier R. J. (1998) Biochemistry 37, 15737–15746 [DOI] [PubMed] [Google Scholar]
- 28.Miller C. J., Elliott J. L., Collier R. J. (1999) Biochemistry 38, 10432–10441 [DOI] [PubMed] [Google Scholar]
- 29.Kim S. O., Ono K., Tobias P. S., Han J. (2003) J. Exp. Med. 197, 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aniento F., Emans N., Griffiths G., Gruenberg J. (1993) J. Cell Biol. 123, 1373–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang G., Leppla S. H. (1999) Infect. Immun. 67, 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montaser M., Lalmanach G., Mach L. (2002) Biol. Chem. 383, 1305–1308 [DOI] [PubMed] [Google Scholar]
- 33.Liu S., Leppla S. H. (2003) J. Biol. Chem. 278, 5227–5234 [DOI] [PubMed] [Google Scholar]
- 34.Bottger G., Nagelkerken B., van der Sluijs P. (1996) J. Biol. Chem. 271, 29191–29197 [DOI] [PubMed] [Google Scholar]
- 35.Chavrier P., Parton R. G., Hauri H. P., Simons K., Zerial M. (1990) Cell 62, 317–329 [DOI] [PubMed] [Google Scholar]
- 36.Fengsrud M., Roos N., Berg T., Liou W., Slot J. W., Seglen P. O. (1995) Exp. Cell Res. 221, 504–519 [DOI] [PubMed] [Google Scholar]
- 37.Høyvik H., Gordon P. B., Berg T. O., Strømhaug P. E., Seglen P. O. (1991) J. Cell Biol. 113, 1305–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González-Polo R. A., Boya P., Pauleau A. L., Jalil A., Larochette N., Souquère S., Eskelinen E. L., Pierron G., Saftig P., Kroemer G. (2005) J. Cell Sci. 118, 3091–3102 [DOI] [PubMed] [Google Scholar]
- 39.Huynh K. K., Eskelinen E. L., Scott C. C., Malevanets A., Saftig P., Grinstein S. (2007) EMBO J. 26, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luzio J. P., Pryor P. R., Bright N. A. (2007) Nat. Rev. Mol. Cell Biol. 8, 622–632 [DOI] [PubMed] [Google Scholar]
- 41.Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. (1998) Cell 92, 759–772 [DOI] [PubMed] [Google Scholar]
- 42.Pryor P. R., Mullock B. M., Bright N. A., Lindsay M. R., Gray S. R., Richardson S. C., Stewart A., James D. E., Piper R. C., Luzio J. P. (2004) EMBO Rep. 5, 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan Y. K., Kusuma C. M., St. John L. J., Vu H. A., Alibek K., Wu A. (2009) Biochem. Biophys. Res. Commun. 379, 293–297 [DOI] [PubMed] [Google Scholar]
- 44.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collier R. J., Young J. A. (2003) Annu. Rev. Cell Dev. Biol. 19, 45–70 [DOI] [PubMed] [Google Scholar]
- 46.Go M. Y., Chow E. M., Mogridge J. (2009) Infect. Immun. 77, 52–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abrami L., Reig N., van der Goot F. G. (2005) Trends Microbiol. 13, 72–78 [DOI] [PubMed] [Google Scholar]
- 48.van der Goot F. G., Gruenberg J. (2006) Trends Cell Biol. 16, 514–521 [DOI] [PubMed] [Google Scholar]
- 49.Bright N. A., Reaves B. J., Mullock B. M., Luzio J. P. (1997) J. Cell Sci. 110, 2027–2040 [DOI] [PubMed] [Google Scholar]
- 50.Mullock B. M., Bright N. A., Fearon C. W., Gray S. R., Luzio J. P. (1998) J. Cell Biol. 140, 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfe J. T., Krantz B. A., Rainey G. J., Young J. A., Collier R. J. (2005) J. Biol. Chem. 280, 39417–39422 [DOI] [PubMed] [Google Scholar]
- 52.Klionsky D. J., Emr S. D. (2000) Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 54.Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., Hamada S., Yoshimori T. (2004) Science 306, 1037–1040 [DOI] [PubMed] [Google Scholar]
- 55.Hernandez L. D., Pypaert M., Flavell R. A., Galán J. E. (2003) J. Cell Biol. 163, 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Razi M., Chan E. Y., Tooze S. A. (2009) J. Cell Biol. 185, 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blom T. S., Linder M. D., Snow K., Pihko H., Hess M. W., Jokitalo E., Veckman V., Syvänen A. C., Ikonen E. (2003) Hum. Mol. Genet. 12, 257–272 [DOI] [PubMed] [Google Scholar]
- 59.Koh C. H., Cheung N. S. (2006) Cell. Signal. 18, 1844–1853 [DOI] [PubMed] [Google Scholar]
- 60.Liao G., Yao Y., Liu J., Yu Z., Cheung S., Xie A., Liang X., Bi X. (2007) Am. J. Pathol. 171, 962–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newman Z. L., Leppla S. H., Moayeri M. (2009) Infect. Immun. 77, 4327–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banks D. J., Barnajian M., Maldonado-Arocho F. J., Sanchez A. M., Bradley K. A. (2005) Cell. Microbiol. 7, 1173–1185 [DOI] [PubMed] [Google Scholar]
- 63.Liu S., Crown D., Miller-Randolph S., Moayeri M., Wang H., Hu H., Morley T., Leppla S. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12424–12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scobie H. M., Wigelsworth D. J., Marlett J. M., Thomas D., Rainey G. J., Lacy D. B., Manchester M., Collier R. J., Young J. A. (2006) PLoS Pathog. 2, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonuccelli G., Sotgia F., Frank P. G., Williams T. M., de Almeida C. J., Tanowitz H. B., Scherer P. E., Hotchkiss K. A., Terman B. I., Rollman B., Alileche A., Brojatsch J., Lisanti M. P. (2005) Am. J. Physiol. Cell Physiol. 288, C1402–C1410 [DOI] [PubMed] [Google Scholar]
- 66.Rmali K. A., Al-Rawi M. A., Parr C., Puntis M. C., Jiang W. G. (2004) Int. J. Mol. Med. 14, 75–80 [PubMed] [Google Scholar]
- 67.Carson-Walter E. B., Watkins D. N., Nanda A., Vogelstein B., Kinzler K. W., St. Croix B. (2001) Cancer Res. 61, 6649–6655 [PubMed] [Google Scholar]
- 68.St. Croix B., Rago C., Velculescu V., Traverso G., Romans K. E., Montgomery E., Lal A., Riggins G. J., Lengauer C., Vogelstein B., Kinzler K. W. (2000) Science 289, 1197–1202 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.