Abstract
MDM2 and MDMX function as key regulators of p53 by binding to its N terminus, inhibiting its transcriptional activity, and promoting degradation. MDM2 and MDMX overexpression or hyperactivation directly contributes to the loss of p53 function during the development of nearly 50% of human cancers. Recent studies showed that disrupting p53-MDM2 and p53-MDMX interactions can lead to robust activation of p53 but also revealed a need to develop novel dual specific or MDMX-specific inhibitors. Using phage display we identified a 12-residue peptide (pDI) with inhibitory activity against MDM2 and MDMX. The co-crystal structures of the pDI and a single mutant derivative (pDI6W) liganded with the N-terminal domains of human MDMX and MDM2 served as the basis for the design of 11 distinct pDI-derivative peptides that were tested for inhibitory potential. The best derivative (termed pDIQ) contained four amino acid substitutions and exhibited a 5-fold increase in potency over the parent peptide against both MDM2 (IC50 = 8 nm) and MDMX (IC50 = 110 nm). Further structural studies revealed key molecular features enabling the high affinity binding of the pDIQ to these proteins. These include large conformational changes of the pDIQ to reach into a hydrophobic site unique to MDMX. The findings suggest new strategies toward the rational design of small molecule inhibitors efficiently targeting MDMX.
Keywords: Diseases/Cancer, Diseases/Cancer/Therapy, Peptides/Conformation, Protein/Conformation, Protein/Post-translational Modification, Protein/Protein-Protein Interactions, Protein/Structure, Protein/Targeting
Introduction
The p53 tumor suppressor is a potent inducer of cell cycle arrest, apoptosis, cellular senescence, and innate immunity. It is activated in response to oncogenic transformation, extrinsic stress, and viral infection to protect higher organisms from cancer (1–3). p53 also facilitates maternal reproduction through induction of the growth factor leukemia inhibitory factor (LIF) that promotes embryo implantation (4). p53 activity is kept at minimal levels in unstressed cells by interactions with MDM2 and MDMX. MDM2 is an ubiquitin E3 ligase for p53 and an important regulator of p53 stability by forming a negative feedback loop (5, 6). The MDM2 homolog MDMX also binds to p53 and inhibits p53-dependent transcription (7). Loss of MDM2 or MDMX leads to embryonic lethality (8–10). Therefore, the expression of MDM2 and the expression of MDMX are both necessary for regulation of p53 during normal development.
Genetic or functional inactivation of p53 is an obligatory step during cancer development. In human tumors that retain wild type p53, amplification of MDM2 or MDMX serves as an alternative mechanism of p53 inactivation in a subset of tumors (11, 12). Furthermore, MDM2 activity is controlled by the tumor suppressor ARF (alternative reading frame) encoded by the INK4a locus (2). Deletion/epigenetic silencing of ARF occur in most tumors expressing wild type p53, resulting in hyperactive MDM2 and lack of p53 response to oncogenic stress in malignant tumors (13, 14). ARF has also been shown to promote MDMX degradation by MDM2 (15). Loss of ARF expression may result in MDMX stabilization that further inactivates p53. Therefore, MDM2 and MDMX are directly involved in p53 functional inactivation in ∼50% of tumors, making them attractive drug targets.
Both MDM2 and MDMX regulate p53 by binding to a short amphipathic α-helix in its N-terminal transactivation domain. Earlier studies of MDM2-p53 binding and determination of MDM2-p53 crystal structure formed the foundation for recent development of small molecule disruptors of MDM2-p53 binding (16–20). These compounds, such as Nutlin 3a and MI-219, provided proof-of-concept for the anti-tumor potential of MDM2 inhibitors (21, 22). Importantly, Nutlin 3a and MI-219 do not inhibit MDMX, and the efficacy of Nutlin 3a is compromised in cells overexpressing MDMX (23–25). Furthermore, even in tumor cells without MDMX amplification, knockdown of MDMX by small interfering RNA still showed anti-tumor potential and cooperative effects with Nutlin in activating p53 (23, 24, 26). Therefore, MDMX expression contributes to p53 inactivation, suggesting that targeting both MDM2 and MDMX is needed to achieve optimal activation of p53.
The small molecules developed against MDM2 are generally inactive for MDMX (22, 27). Recent structural studies using humanized zebra fish MDMX and human MDMX in complex with the p53 N-terminal peptide revealed extensive similarity between the p53-binding domains of MDM2 and MDMX in overall folding and the shapes of their p53-binding pockets. However, a few sequence differences result in a smaller hydrophobic cleft in MDMX that prevents efficient binding by Nutlin (28, 29). Future development of inhibitors against MDMX can be facilitated by identification of a high affinity artificial ligand that target MDMX and/or MDM2. We recently identified a peptide (pDI) using phage display that selects for sequence with maximal binding to MDM2 and MDMX (30). pDI is 300-fold more potent than p53 peptide in disrupting MDM2-p53 and MDMX-p53 binding. Using a similar phage display strategy, Pazgier et al. (31) recently also identified a different, more potent peptide inhibitor (pMI) in their screen. The co-crystal structures of pMI and pDI in complex with the MDM2 and MDMX N-terminal domains have recently been reported (31, 32), revealing the structural basis for the inhibitory action of these two different peptides.
Here, we determined the crystal structures of MDM2 and MDMX in complex with the pDI and derivatives thereof. Based on the structural information obtained from the pDI and a single mutant peptide (pDI6W), we designed a quadruple mutant peptide (pDIQ) that displays high affinity for MDMX and is the most potent inhibitor against MDM2 reported to date. The findings provide important clues about the molecular basis for the potency and selectivity of MDM2 and MDMX inhibitors and should inspire new strategies toward the design of drug-like small molecule inhibitors specifically targeting MDMX.
EXPERIMENTAL PROCEDURES
Materials
The chemicals and reagents were purchased from Sigma unless otherwise noted. The peptides (>95% purity) were supplied by Genscript (Piscataway, NJ).
Expression and Purification of Human MDM2 and MDMX
The MDM2 N-terminal domain spanning residues 17–125 was subcloned into pGEX2T; a shorter construct (residues 24–109) was subcloned into a pDEST-His-MBP vector provided by Dr. David S. Waugh (33). The pDEST-His-MBP vector was also used to express the N-terminal domain of human MDMX (residues 23–111). The MDM2 and MDMX N-terminal constructs were expressed in Escherichia coli BL21-Gold (DE3). The proteins were purified by affinity chromatography using glutathione-Sepharose (GE Healthcare) for MDM2 (17–125) or nickel-nitrilotriacetic acid Superflow (Qiagen) for MDM2 (24–109) and MDMX (23–111). Eluted proteins were cleaved with thrombin or tobacco etch virus protease (33) and then mixed with the pDI, pDI6W, or pDIQ peptide. The respective MDM2/X-peptide complexes were further purified using SP Sepharose (GE Healthcare; elution buffer: 50 mm HEPES, 0.015–0.5 m NaCl, 5% glycerol, 5 mm dithiothreitol, pH 6.8) followed by size exclusion chromatography on Superdex 75 (GE Healthcare; elution buffer: 50 mm Tris, 150 mm NaCl, 5 mm dithiothreitol, 1 mm EDTA, pH 7.5).
Crystallization
The MDM2-X peptide complexes were crystallized at 19 °C by the hanging drop vapor diffusion method at a protein concentration of ∼10 mg/ml, supplemented with an additional half molar equivalent of the respective peptide. The MDMX (23–111)-pDI and pDI6W complexes were crystallized from 1.4 m sodium/potassium phosphate, pH 8.2; 15% ethylene glycol was included for cryo-protection. The MDMX (23–111)-pDIQ complex was crystallized from 2.1 m ammonium sulfate, 10 mm Tris-HCl, pH 7.4, 5% propanol; 25% glycerol was included for cryo-protection. The MDM2 (17–125)-pDI6W complex crystallized from 2.2 m dl-Malic acid, 100 mm Tris-HCl, pH 8.0; no cryo-protectant was added. The MDM2 (24–109)-pDIQ crystallized from 30% PEGmme2000, 100 mm Tris-HCl, pH 8.5, 200 mm MgCl2; 10% ethylene glycol was included for cryo-protection.
Data Collection, Structure Solution, and Refinement
X-ray diffraction data were recorded at −180 °C using the oscillation method on single flash-frozen crystals (detector, Rigaku HTC image plate; x-rays, CuKα, focused by mirror optics; generator, Rigaku Micro-Max 007-HF). The data were processed with HKL2000 (HKL Research, Inc., Charlottesville, VA). The structures were determined by molecular replacement using the coordinates of human MDM2-p53 (Protein Data Bank entry 1T4F) and human MDMX-p53 (Protein Data Bank entry 3DAB) as search models. The program package CNS (34) was employed for phasing and refinement; model building was performed with O (35). Refinement cycles were performed using data to the highest resolution with no sigma cut-off applied. Several rounds of minimization, simulated annealing (starting temperature, 2500 K), and restrained individual B-factor refinement were carried out. The data collection and refinement statistics are summarized in Table 2. Figs. 1–5 and 7 were drawn with Pymol (DeLano Scientific, Palo Alto, CA).
TABLE 2.
Summary of data collection and structure refinement
The values in parentheses refer to the highest resolution shell.
| MDMX·pDI | MDMX·pDI6W | MDMX·pDIQ | MDM2·pDI6W | MDM2·pDIQ | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C2 | C2 | P212121 | I222 | P21212 |
| Unit cell dimensions (Å) | a = 70.2 b = 27.2 c = 51.8 | a = 70.4 b = 27.6 c = 52.1 | a = 44.9 b = 53.7 c = 87.3 | a = 37.2 b = 68.8 c = 92.4 | a = 43.8 b = 50.6 c = 39.2 |
| α = γ = 90° β = 124.4° | α = γ = 90° β = 124.6° | α = β = γ = 90° | α = β = γ = 90° | α = β = γ = 90° | |
| Resolution range | 25–1.8 (1.86–1.8) | 30–1.73 (1.79–1.73) | 35–1.8 (1.86–1.8) | 46–2.1 (2.18–2.1) | 29–1.78 (1.84–1.78) |
| Unique reflections | 7677 (748) | 8289 (664) | 20193 (1953) | 7261 (714) | 8640 (833) |
| Completeness (%) | 97.2 (94.70) | 93.9 (77.00) | 99.7 (99.00) | 99.8 (100.00) | 98.1 (99.30) |
| I/σI | 27.4 (15.10) | 21.2 (13.20) | 27.5 (3.70) | 30.1 (8.40) | 25.9 (9.50) |
| Rmerge (%)a | 4.4 (8.90) | 4.9 (9.10) | 6.2 (47.60) | 11.9 (31.80) | 3.7 (11.60) |
| Structure refinement | |||||
| Protein atoms | 699 | 699 | 2 × 699 | 793 | 690 |
| Average B-factor (Å2) | 15.0 | 15.7 | 27.3 | 25.0 | 32.2 |
| Ligand atoms | 107 | 109 | 2 × 112 | 109 | 112 |
| Average B-factor (Å2) | 14.1 | 15.5 | 29.2 | 23.6 | 31.5 |
| Solvent molecules | 116 | 123 | 139 | 116 | 55 |
| Average B-factor (Å2) | 27.9 | 29.7 | 37.4 | 34.3 | 42.8 |
| RMSD bonds (Å)b | 0.011 | 0.01 | 0.016 | 0.008 | 0.015 |
| RMSD angles (°) | 1.4 | 1.5 | 1.5 | 1.1 | 1.6 |
| Rcryst (%)c | 16.2 | 16.0 | 20.7 | 19.6 | 21.1 |
| Rfree (%)d | 20.7 | 21.7 | 22.5 | 22.9 | 24.9 |
| Cross-validated estimated | |||||
| Coordinate error | |||||
| From Luzzati plot (Å) | 0.21 | 0.21 | 0.24 | 0.28 | 0.28 |
| From SigmaA (Å) | 0.14 | 0.13 | 0.22 | 0.23 | 0.19 |
a Rmerge = 100 × ΣhΣi |Ihi − Ih|/ΣhiIhi, where Ihi is the observed intensity and Ih is the average intensity of multiple (i) observations of symmetry-related reflections.
b RMSD, root mean square deviation from ideal values.
c Rcryst = 100 × Σ|Fobs − Fmodel|/ΣFobs, where Fobs and Fmodel are the observed and calculated structure factor amplitudes, respectively.
d Rfree is Rcryst calculated for randomly chosen unique reflections, which were excluded from the refinement (818 for MDMX·pDI, 526 for MDMX·pDI6W, 965 for MDMX·pDIQ, 528 for MDM2·pDI6W, and 829 for MDM2·pDIQ).
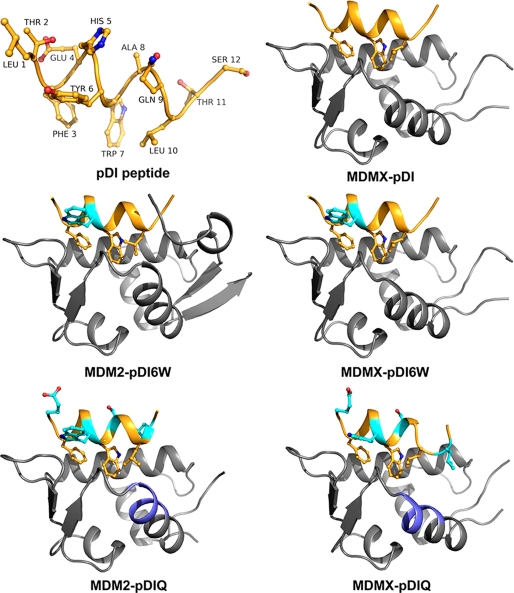
FIGURE 1.
Crystal structures of MDMX and MDM2 liganded with the pDI peptide and derivatives thereof. The structures of the pDI, pDI6W, and pDIQ peptides (shown in orange) were determined in complex with MDMX and MDM2 (shown in gray). Indicated in cyan are the amino acid substitution sites. The major structural difference between MDMX and MDM2 is the α2′-helix, part of which constitutes the peptide-binding site (highlighted in blue in the pDIQ liganded structures). The MDMX-pDIQ complex crystallized with two monomers in the asymmetric unit, and only chain A is displayed.
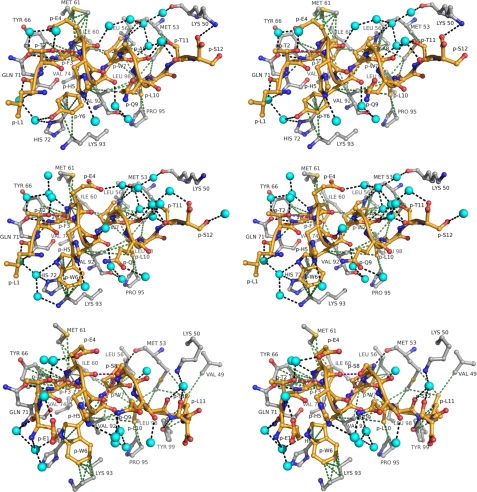
FIGURE 2.
Interaction of pDI peptides with MDMX in atomic detail (stereo views). The peptide-binding site of the respective MDMX peptide complexes is displayed in a 4.2-Å radius around the pDI (top panel), pDI6W (middle panel), and pDIQ (chain A, bottom panel). Protein residues are shown in gray, the peptide is shown in yellow, and water molecules are presented as cyan spheres. Intermolecular hydrogen bonding (d ≤ 3.3 Å) and van der Waal's (d ≤ 4.2 Å) interactions are represented as black and green dotted lines, respectively. The purple dotted lines indicate intrapeptidic side chain/main chain hydrogen bonds.
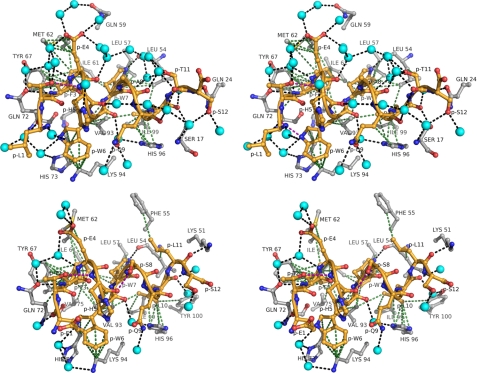
FIGURE 3.
Interaction of pDI peptides with MDM2 in atomic detail (stereo views). The peptide-binding site of the respective MDM2 peptide complexes is displayed in a 4.2-Å radius around the pDI6W (top panel) and pDIQ (bottom panel). The color scheme is the same as in Fig. 2.
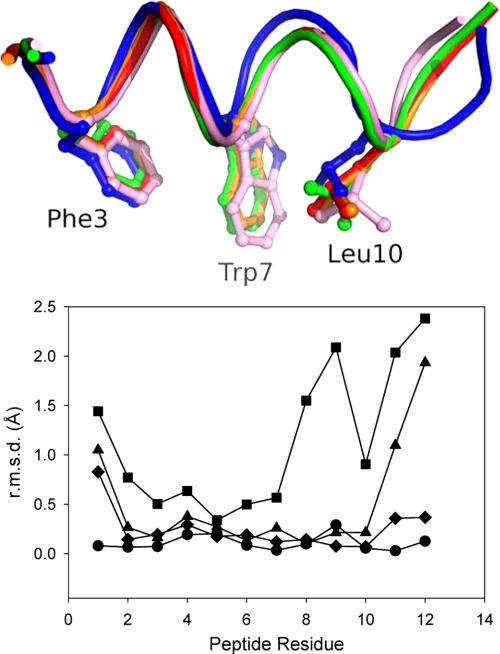
FIGURE 4.
Structural changes in the pDI peptides. Top panel, superposition of the backbone structure of the peptides pDI MDMX (orange), pDI6WMDMX (red), pDIQMDMX (blue), pDI6WMDM2 (green), and pDIQMDM2 (pink). Bottom panel, root mean square deviation (r.m.s.d.) of the C-α atoms of the liganded peptides pDIQMDMX (■), pDIQMDM2 (▴), pDI6WMDM2 (♦), and pDI6WMDMX (●). The C-α atoms of the respective coordinate sets were aligned to the pDIMDMX structure using LSQkab (41, 42).
FIGURE 5.
Molecular basis for the high affinity binding of the pDIQ peptide (stereo views). Top panel, in its complex with MDM2, the C-terminal part of the pDIQ peptide (orange) is helical, and Leu(P)11 points away from the α2′-helix (blue), interacting exclusively with residues of the α2-helix. Bottom panel, upon binding to MDMX the C-terminal helical turn of the pDIQ peptide unwinds positioning Leu(P)11 between the α2′- and α2-helices. Hydrogen bonding interactions (d ≤ 3.3 Å) are indicated by black dotted lines, and van der Waal's interactions (d ≤ 4.2 Å) are indicated by green dotted lines.
FIGURE 7.
Strategies toward the rational design of MDM2 and MDMX inhibitors. Top panel, in complex with MDM2, Nutlin (cyan) aligns well with the side chains of the FYL-triplet of the pDIQ peptide (orange). The Nutlin molecule is roughly centered on the Trp(P)7 site and extends to the sites occupied by Phe(P)3 and Leu(P)10. Bottom panel, in MDMX, the binding site of Leu(P)10 is altered, and the Tyr99 side chain prevents the efficient binding of Nutlin because of potential steric clashes. Consequently, small molecule inhibitors of MDMX may achieve maximum binding potential by centering on the Leu(P)10 site and extending to the sites occupied by Trp(P)7 and Leu(P)11, ignoring the Phe(P)3 site to keep the molecule size small. The Nutlin-MDM2 complex (Protein Data Bank entry 1RV1) was aligned with the complexes of pDIQ-MDM2 and pDIQ-MDMX using LSQkab.
Enzyme-linked Immunosorbent Assay and Fluorescence Polarization Assays
GST-MDM2–1-1504 and GST-MDMX-1–200, and full-length His6-p53 were expressed in E. coli and affinity-purified under nondenaturing conditions. ELISA plates were incubated with 2.5 μg/ml His6-p53 in phosphate-buffered saline (PBS) for 16 h. After washing with PBS + 0.1% Tween 20 (PBST), the plates were blocked with PBS + 5% nonfat dry milk + 0.1% Tween 20 (PBSMT) for 0.5 h. GST-HDM2 and MDMX (5 μg/ml) were mixed with peptides in PBSMT+10% glycerol + 10 mm dithiothreitol and added to the wells. The plates were washed with PBST after incubating for 1 h at room temperature and incubated with MDM2 antibody 4B2 and MDMX antibody 8C6 in PBSMT for 1 h, followed by washing and incubation with horseradish peroxidase rabbit anti-mouse Ig antibody for 1 h. The plates were developed by incubation with TMB peroxidase substrate (KPL) and measured by absorbance at 450 nm. Fluorescence polarization assay was performed using N-terminally conjugated fluorescein isothiocyanate-p53 (LSQETFSDLWKLLPEN) and fluorescein isothiocyanate-pDI peptides. Proteins (2–4000 nm) and peptides (2 nm) were mixed in fluorescence polarization buffer (25 mm HEPES, pH 7.4, 0.1% Tween 20, 150 mm NaCl, 10 mm dithiothreitol). The mixtures were incubated at 25 °C for 1 h and analyzed for fluorescence polarization. The Kd values were determined from fluorescence polarization data using the method described by Zhang et al. (36).
CD Analysis
Circular dichroism measurements were made using an Aviv Model 215 spectrometer. The peptides were prepared at a concentration of 0.1 mm in 10 mm Tris-HCl, pH 7.5, 5% methanol, and 10% trifluoroethanol. The measurements were performed at 23 °C. Blank scan (buffer) was subtracted from the spectra, and values of ellipticity were expressed in units of deg·cm2·mol−1. The amphipathic α-helical peptide melittin was used as a standard.
RESULTS AND DISCUSSION
Characterization of a High Affinity MDM2- and MDMX-binding Peptide
We have recently identified a 12-residue peptide inhibitor of MDM2 and MDMX by screening a phage display library for peptides that bind to GST-MDM2–1-150 and GST-MDMX-1–200 (30). Both screens selected the same peptide sequence (1LTFEHYWAQLTS12) as the highest affinity ligand for MDM2 and MDMX. This peptide was named pDI for peptide dual inhibitor. The pDI is distinct from the equivalent p53 sequence (p53p, 17ETFSDLWKLLPE28) but retains the key p53 residues Phe19, Trp23, and Leu26, which bind to three distinct hydrophobic pockets of MDM2 (16). The binding affinity of pDI to MDM2 and MDMX was analyzed using fluorescence polarization yielding dissociation constants of 1 nm for the MDM2-pDI and 3 nm for the MDMX-pDI interaction. By comparison, the Kd values for p53p binding to MDM2 and MDMX were 160 and 260 nm, respectively, which is in good agreement with the previously determined values obtained by isothermal titration calorimetry (Kd = 130–340 nm) (37). Inhibition of full-length p53 binding to MDM2 or MDMX by the pDI (and all other peptides studied here) was assessed by ELISA, yielding IC50 values of 44 and 550 nm, respectively (Table 1). The only other high affinity peptide also previously identified by phage display is the pMI peptide (31), which under our assay conditions displayed IC50 values of 20 and 40 nm against MDM2 and MDMX, respectively (Table 1).
TABLE 1.
Structure-activity relationship of pDI derivative peptides
The IC50 values were determined by ELISA. The p53 and pMI peptides served as controls.
| Name | Sequence | IC50 |
|
|---|---|---|---|
| MDM2 | MDMX | ||
| nm | nm | ||
| p53 | ETFSDLWKLLPE | 2000 | 6000 |
| pMI | TSFAEYWNLLSP | 20 | 40 |
| pDI | LTFEHYWAQLTS | 44 | 550 |
| 6W | LTFEHWWAQLTS | 36 | 250 |
| 6S | LTFEHSWAQLTS | Inactive | Inactive |
| 1E6N | ETFEHNWAQLTS | Inactive | Inactive |
| 6N | LTFEHNWAQLTS | 400 | 4000 |
| 6W9S | LTFEHWWASLTS | 125 | 500 |
| 6W8S9S | LTFEHWWSSLTS | 130 | 800 |
| 4T6W | LTFTHWWAQLTS | Inactive | Inactive |
| 1E6W | ETFEHWWAQLTS | 20 | 200 |
| 6W8S | LTFEHWWSQLTS | 24 | 180 |
| 6W11L | LTFEHWWAQLLS | 20 | 140 |
| pDIQ | ETFEHWWSQLLS | 8 | 110 |
Structural Characterization of the MDM2 and MDMX pDI Peptide Complexes
In an attempt to understand the differences in binding affinities for MDM2 and MDMX, we first determined the crystal structure of MDMX liganded with the pDI peptide (Fig. 1 and Table 2). Subsequently, both MDM2 and MDMX were co-crystallized with a derivative peptide, pDI6W, which contained a single Y6W mutation and displayed increased inhibitory properties over the parent peptide (Table 1). The structures of the pDI6W peptide served as a template for the design of the pDIQ peptide. The overall structures of the N-terminal domains of MDM2 and MDMX liganded with the pDI and derivative peptides are very similar, as expected from the previously determined MDM2/X structures (16, 38). The major structural difference between MDM2 and MDMX is the orientation of the C-terminal helix (α2′, residues 96–106 for MDM2 and 95–105 for MDMX) relative to the binding site of p53 and the pDI peptide (Fig. 1; see also Fig. 5). The pDI peptides all bind to MDM2 and MDMX in a helical conformation via residues Phe3-Trp7-Leu10 to the same groove that harbors the Phe19-Trp23-Leu26 triplet of the p53 helix (16). The interaction pattern between the triplet residues and their binding sites in MDM2/X is dominated by hydrophobic interactions. In addition, the indole nitrogen of Trp(P)7 establishes a hydrogen bond with the carbonyl oxygen of Leu54MDM2 or Met53MDMX (Figs. 2 and 3).
Structure-Activity Relationship Studies and Design of the pDIQ Peptide
The co-crystal structures of MDM2 and MDMX liganded with the pDI6W peptide provided the basis for our structure-activity relationship studies. Our aim was to design peptides with improved inhibitory potential, in particular against MDMX, by substituting residues other than the conserved Phe3-Trp7-Leu10 triplet. The conformation and binding pattern of the pDI and the pDI6W peptides are very similar (Figs. 2 and 3). Residues Leu1, Thr2, Glu4, His5, Ala8, and Gln9 are completely solvent-exposed; Trp6 and Thr11 are partly solvent-exposed but also interact with protein residues through hydrophobic forces. Consequently, in the respective crystal structures a large portion of the liganded peptide is bound to well defined water molecules, some of which are conserved between the respective complexes of MDM2 and MDMX. The side chains of both Tyr(P)6 and Trp(P)6 are sandwiched between MDMX residues His72 and Lys93 (MDM2 residues His73 and Lys94) and the peptide residue His(P)5. The hydroxyl group of Tyr(P)6 and the indole nitrogen of Trp(P)6 are hydrogen-bonded with water molecules. Both the Tyr(P)6 and Trp(P)6 ring systems interact with the aliphatic portion of Lys93 and the C-β atom of His72 through hydrophobic forces. However, the indole ring of Trp(P)6 establishes more van der Waal's interactions (d < 4 Å) than Tyr(P)6, which possibly contributes to the improved binding potential of the pDI6W over the pDI parent peptide.
Amino acid substitutions were introduced into the pDI6W peptide and analyzed by ELISA to determine their potency in disrupting MDM2-p53 and MDMX-p53 interactions. Replacing Trp(P)6 with Ser or Asn (6S, 6N, and 1E6N of Table 1) reduced or even abolished inhibitory potency, corroborating the above mentioned notion that ring systems in this position confer stability through an increased number of van der Waal's interactions with the protein. Leu(P)1 was replaced by Glu (1E6W of Table 1) to increase solubility of the peptide in aqueous solutions; this substitution also improved by 2-fold the inhibitory potency on both MDM2 and MDMX (compare pDI6W to pDIE6W). Gln(P)9 was replaced with Ser to promote a hydrogen bond between the serine hydroxyl group and the carbonyl oxygen of Trp(P)6. Unexpectedly, the resulting peptides (6W9S and 6W8S9S of Table 1) were less active than the pDI6W. Because Gln(P)9 is too far away from His(P)5 to establish a strong hydrogen bonding interaction (the average distance between the amide group and the imidazole is 3.5 Å), its side chain probably acts as a spacer to keep His(P)5 in place, thereby contributing to the overall stability of the α-helix. Glu(P)4 was replaced with Thr to test the importance of conformational flexibility in this region of the peptide. In the five co-crystal structures determined here, Glu(P)4 adopts distinctly different conformations because of the close proximity of Met61MDMX/Met62MDM2. The resulting peptide (4T6W) was inactive presumably because of steric hindrance between its hydroxyl group and the Met61 side chain and/or between its methyl group and the side chain of Thr(P)2. Notably, the p53 peptide has a Ser, and the recently determined pMI peptide has an Ala in this position, both of which would not clash with Met61 or Thr(P)2. Ala(P)8 was replaced with Ser to establish an intrapeptide hydrogen bond between the Ser hydroxyl group and the carbonyl oxygen of His(P)5. The resulting peptide (6W8S) displayed significantly improved inhibitory activity. Thr(P)11 was replaced with Leu to increase hydrophobic interactions with residues Met53MDMX and Leu54MDM2; this peptide (6W11L) was also considerably more active than the parent peptide.
Based on these results, we designed a peptide with four residues of the original pDI peptide substituted, i.e. L1E, Y6W, A8S, and T11L. This peptide (termed pDIQ for quadruple mutant) displayed 5-fold increased inhibitory potency over the pDI parent peptide against both MDM2 (IC50 = 8 nm) and MDMX (IC50 = 110 nm). In a side-by-side comparison, the pDIQ peptide was 2-fold more potent against MDM2 than the recently reported pMI peptide (31). Thus, the combination of phage display selection and rational design resulted in the most potent peptide inhibitor of MDM2 reported to date.
Molecular Basis for the High Affinity of the pDIQ Peptide
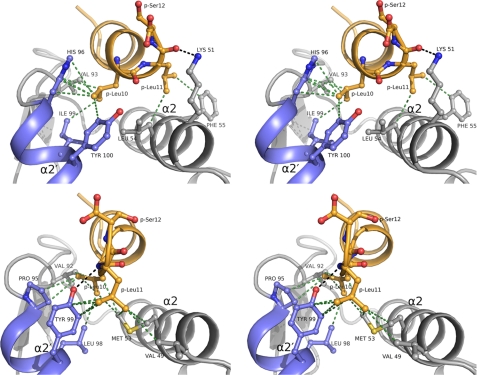
The co-crystal structures of MDM2 and MDMX liganded with pDIQ revealed substantial structural changes of this peptide upon binding to MDMX, with up to 2 Å displacement shifts of the C terminus from residues 8–12 (Fig. 4). This is accompanied by a loss of helical structure of residues 10 and 11 (Figs. 1 and 4), which in turn allows Leu(P)11 to interact with Tyr99 of the MDMX α2′-helix and the side chains of Met53 and Val49 of the α2-helix (Fig. 5). It appears that the increase in hydrophobicity of the Leu11 side chain in the pDIQ versus the Thr11 side chain in pDI6W causes this structural change upon binding to MDMX. By contrast, the pDIQ in complex with MDM2 retains complete helical conformation, directing Leu(P)11 away from the α2′-helix and toward the α2-helix. Here, Leu(P)11 interacts with the side chains of Leu54 and Phe55 through van der Waal's forces, whereas the ϵ-amino group of Lys51 is hydrogen-bonded to its carbonyl oxygen (Fig. 5).
The conformational changes in the pDIQ peptide appear to be induced mainly by residue Tyr99MDMX/Tyr100MDM2 of the α2′-helix (Fig. 5; see also Fig. 7). The orientation of this helix with respect to the peptide-binding site differs considerably between MDMX and MDM2. As a result, this tyrosine residue appears to exert a major role in the binding of the pDIQ to MDM2 and MDMX. In MDM2, Tyr100 prevents Leu(P)11 from reaching into the hydrophobic groove formed between the α2′-helix and the α2-helix. As a consequence, the C terminus of the pDIQ peptide retains helical conformation, positioning Leu(P)11 over the α2-helix. By contrast, Tyr99 of MDMX accommodates the binding of Leu(P)11 to the hydrophobic groove. It appears that the conformational flexibility in the C-terminal region of the pDIQ peptide ensures the high affinity binding to both MDM2 and MDMX. Remarkably, the pDIQ is 5-fold more active against MDMX than the parent pDI peptide, despite the partial loss of the energetically favored helical structure. This indicates the high binding potential of the groove between the α2′ and α2 helices of MDMX attracting the Leu(P)11 side chain. The binding energy gained from this interaction appears to compensate for the energy spent to disrupt the helical turn.
A similar observation was made recently for the interaction of the pMI peptide with MDMX, in which the peptide C-terminal Pro12 residue was found to occupy the same site as Leu(P)11 of the pDIQ peptide (31). The pMI and pDIQ peptides share little sequence homology, and only the FWL triplet residues are strictly conserved (Table 1). The pMI peptide is two or three times more potent against MDMX than pDIQ but two times less active against MDM2. The differential inhibition is reflected in the distinct interaction patterns of the peptide C-terminal residues with the α2′/α2-helices of both proteins (Fig. 5 and supplemental Fig. S3). For the MDM2-pMI interaction, neither Ser(P)11 nor Pro(P)12 is involved in hydrogen bonding or hydrophobic interactions with the target protein, whereas Leu(P)11 of pDIQ establishes multiple noncovalent interactions with residues of the α2-helix. This suggests a significant difference in binding potential for both peptides in MDM2. However, the slightly different overall conformation of the pMI peptide allows the formation of a hydrogen bond between the carbonyl oxygen of Leu(P)10 and the hydroxyl group of Tyr100, which is not seen with the pDIQ peptide. Although the lack of binding potential of Ser(P)11 and Pro(P)12 of the pMI is compensated by the formation of this hydrogen bond with Tyr100, it seems that the distinct interaction pattern of Leu(P)11 renders the pDIQ peptide more effective against MDM2. By contrast, in the MDMX-pMI complex, the Pro(P)12 residue reaches deep into the interface between the α2′/α2 helices, establishing more van der Waal's interactions with MDMX residues than Leu(P)11 of the pDIQ peptide. This presumably accounts for the higher affinity of the pMI peptide toward MDMX.
Optimized Peptides Adopt α-Helical Conformation in Solution
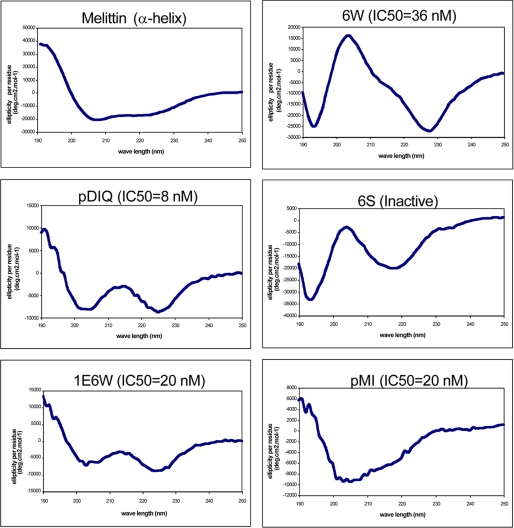
For the pDIQ peptide, the Tyr(P)6 to Trp and the Thr(P)11 to Leu substitutions increased the number of intermolecular noncovalent interactions between the peptide and MDM2/X. The other amino acid substitutions were intended to increase the solubility of the peptide, at the same time strengthening the helical conformation through intrapeptide hydrogen bonds or through bridging water molecules. It has been suggested that the p53 N-terminal region is unstructured in solution but forms an α-helix upon binding to the hydrophobic pockets of MDM2 or MDMX (39). To test whether the inhibitory potency of our derivative peptides is reflected by a higher degree of helical conformation in solution, circular dichroism studies were performed (Fig. 6). The most potent peptides, pDIQ and 1E6W (Table 1), exhibited the strong double-well characteristics expected for α-helices, with pDIQ being most pronounced. The α-helical feature was less prominent in the 6W peptide and absent in the inactive 6S peptide. These results indicate that the improved inhibitory action of peptides is not only due to an increase in noncovalent interactions between the peptide and the protein (as seen for the Y6W and T11L substitutions) but also due to the enhanced stability of the α-helical conformation in solution.
FIGURE 6.
High affinity pDI peptides adopt helical conformation in solution. The peptides were analyzed by circular dichroism spectroscopy at 0.1 mm concentration at 23 °C. Melittin is a representative α-helical peptide from bee venom that served as a positive control. The IC50 values for each peptide against MDM2 are indicated.
Implications for the Design of Small Molecule Inhibitors against MDMX
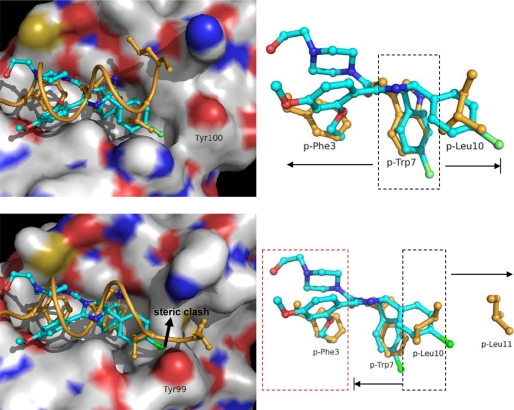
Currently, small molecule inhibitors of MDM2 with anti-tumor activity such as Nutlin and MI- 219 are generally ineffective against MDMX (24, 25, 30, 40). Superposition of the complexes of MDM2-pDIQ and MDM2-Nutlin2 reveals the almost perfect fit of the three hydrophobic residues of Nutlin with the side chains of the peptidic FWL triplet (Fig. 7). However, the structural differences of the α2′ helices of both proteins directly impact the binding site of Leu(P)10. For MDM2, Tyr100 accommodates the binding of Nutlin, whereas Tyr99 in MDMX causes a potential steric clash with the Nutlin molecule. Although the efficient binding of the pDIQ peptide to MDMX is made possible through structural flexibility of the C-terminal region of the pDIQ peptide, small molecules such as Nutlin cannot undergo these conformational changes. Therefore, the design of p53 antagonists that selectively target MDMX over MDM2 must take into account these distinct differences of the peptide-binding site. One strategy could be the design of small molecules centering on the Leu(P)10 site and extending to the Trp(P)7 and Leu(P)11 sites, rather than centering on the Trp(P)7 site and extending to the Phe(P)3 and Leu(P)10 sites as in the case for Nutlin. To keep their size small, scaffolds designed in this manner will have to lack the functionality to bind to the Phe(P)3 site (Phe(P)3 is ∼10 Å away from Leu(P)10). This will likely result in a loss of binding affinity for MDM2 but is expected to gain affinity toward MDMX by exploiting new binding partners in the Leu(P)11 site.
In conclusion, the results from this work demonstrate that peptides identified by phage display as MDM2 antagonists do not necessarily exhibit maximum binding affinity. Rather, such peptides may serve as ideal starting points for the structure-based design of inhibitors with substantially improved potency. In this study, the structure-activity analysis of a limited number of amino acid substitutions resulted not only in a 5-fold increase in inhibitory activity over the parent peptide but also revealed large conformational changes in the peptide required to bind efficiently to MDMX. These structural changes are induced by a hydrophobic site in MDMX, which probably has not yet received due attention in the design of small molecule inhibitors specifically targeting MDMX. The findings reported here should facilitate the rational design of novel MDMX-specific or MDM2/MDMX dual specific inhibitors as potential cancer therapies.
Supplementary Material
Acknowledgments
We thank the Moffitt Molecular Biology Core for DNA sequence analyses, the Moffitt Structural Biology Core for X-ray crystallographic experiments, and Dr. Ted Gauthier (University of South Florida, Tampa, FL) for help with the circular dichroism analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA109636 (to J. C.) and CA118210 (to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- GST
- glutathione S-transferase
- ELISA
- enzyme-linked immunosorbent assay
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Takaoka A., Hayakawa S., Yanai H., Stoiber D., Negishi H., Kikuchi H., Sasaki S., Imai K., Shibue T., Honda K., Taniguchi T. (2003) Nature 424, 516–523 [DOI] [PubMed] [Google Scholar]
- 2.Sherr C. J. (2006) Nat. Rev. Cancer 6, 663–673 [DOI] [PubMed] [Google Scholar]
- 3.Prives C., Hall P. A. (1999) J. Pathol. 187, 112–126 [DOI] [PubMed] [Google Scholar]
- 4.Hu W., Feng Z., Teresky A. K., Levine A. J. (2007) Nature 450, 721–724 [DOI] [PubMed] [Google Scholar]
- 5.Wu X., Bayle J. H., Olson D., Levine A. J. (1993) Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 6.Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 7.Shvarts A., Steegenga W. T., Riteco N., van Laar T., Dekker P., Bazuine M., van Ham R. C., van der Houven van Oordt W., Hateboer G., van der Eb A. J., Jochemsen A. G. (1996) EMBO J. 15, 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- 8.Jones S. N., Roe A. E., Donehower L. A., Bradley A. (1995) Nature 378, 206–208 [DOI] [PubMed] [Google Scholar]
- 9.Montes de Oca Luna R., Wagner D. S., Lozano G. (1995) Nature 378, 203–206 [DOI] [PubMed] [Google Scholar]
- 10.Parant J., Chavez-Reyes A., Little N. A., Yan W., Reinke V., Jochemsen A. G., Lozano G. (2001) Nat. Genet. 29, 92–95 [DOI] [PubMed] [Google Scholar]
- 11.Oliner J. D., Kinzler K. W., Meltzer P. S., George D. L., Vogelstein B. (1992) Nature 358, 80–83 [DOI] [PubMed] [Google Scholar]
- 12.Laurie N. A., Donovan S. L., Shih C. S., Zhang J., Mills N., Fuller C., Teunisse A., Lam S., Ramos Y., Mohan A., Johnson D., Wilson M., Rodriguez-Galindo C., Quarto M., Francoz S., Mendrysa S. M., Guy R. K., Marine J. C., Jochemsen A. G., Dyer M. A. (2006) Nature 444, 61–66 [DOI] [PubMed] [Google Scholar]
- 13.Zindy F., Eischen C. M., Randle D. H., Kamijo T., Cleveland J. L., Sherr C. J., Roussel M. F. (1998) Genes Dev. 12, 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stott F. J., Bates S., James M. C., McConnell B. B., Starborg M., Brookes S., Palmero I., Ryan K., Hara E., Vousden K. H., Peters G. (1998) EMBO J. 17, 5001–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Y., Chen J. (2003) Mol. Cell. Biol. 23, 5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 17.Lin J., Chen J., Elenbaas B., Levine A. J. (1994) Genes Dev. 8, 1235–1246 [DOI] [PubMed] [Google Scholar]
- 18.Chen J., Marechal V., Levine A. J. (1993) Mol. Cell. Biol. 13, 4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliner J. D., Pietenpol J. A., Thiagalingam S., Gyuris J., Kinzler K. W., Vogelstein B. (1993) Nature 362, 857–860 [DOI] [PubMed] [Google Scholar]
- 20.Lawrence H. R., Li Z., Yip M. L., Sung S. S., Lawrence N. J., McLaughlin M. L., McManus G. J., Zaworotko M. J., Sebti S. M., Chen J., Guida W. C. (2009) Bioorg. Med. Chem. Lett. 19, 3756–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 22.Shangary S., Qin D., McEachern D., Liu M., Miller R. S., Qiu S., Nikolovska-Coleska Z., Ding K., Wang G., Chen J., Bernard D., Zhang J., Lu Y., Gu Q., Shah R. B., Pienta K. J., Ling X., Kang S., Guo M., Sun Y., Yang D., Wang S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3933–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B., Gilkes D. M., Farooqi B., Sebti S. M., Chen J. (2006) J. Biol. Chem. 281, 33030–33035 [DOI] [PubMed] [Google Scholar]
- 24.Wade M., Wong E. T., Tang M., Stommel J. M., Wahl G. M. (2006) J. Biol. Chem. 281, 33036–33044 [DOI] [PubMed] [Google Scholar]
- 25.Patton J. T., Mayo L. D., Singhi A. D., Gudkov A. V., Stark G. R., Jackson M. W. (2006) Cancer Res. 66, 3169–3176 [DOI] [PubMed] [Google Scholar]
- 26.Gilkes D. M., Chen L., Chen J. (2006) EMBO J. 25, 5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia M., Knezevic D., Tovar C., Huang B., Heimbrook D. C., Vassilev L. T. (2008) Cell Cycle 7, 1604–1612 [DOI] [PubMed] [Google Scholar]
- 28.Popowicz G. M., Czarna A., Holak T. A. (2008) Cell Cycle 7, 2441–2443 [DOI] [PubMed] [Google Scholar]
- 29.Popowicz G. M., Czarna A., Rothweiler U., Szwagierczak A., Krajewski M., Weber L., Holak T. A. (2007) Cell Cycle 6, 2386–2392 [DOI] [PubMed] [Google Scholar]
- 30.Hu B., Gilkes D. M., Chen J. (2007) Cancer Res. 67, 8810–8817 [DOI] [PubMed] [Google Scholar]
- 31.Pazgier M., Liu M., Zou G., Yuan W., Li C., Li C., Li J., Monbo J., Zella D., Tarasov S. G., Lu W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4665–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czarna A., Popowicz G. M., Pecak A., Wolf S., Dubin G., Holak T. A. (2009) Cell Cycle 8, 1176–1184 [DOI] [PubMed] [Google Scholar]
- 33.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 34.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 35.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 36.Zhang R., Mayhood T., Lipari P., Wang Y., Durkin J., Syto R., Gesell J., McNemar C., Windsor W. (2004) Anal. Biochem. 331, 138–146 [DOI] [PubMed] [Google Scholar]
- 37.Yu G. W., Rudiger S., Veprintsev D., Freund S., Fernandez-Fernandez M. R., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fasan R., Dias R. L., Moehle K., Zerbe O., Obrecht D., Mittl P. R., Grutter M. G., Robinson J. A. (2006) Chembiochem 7, 515–526 [DOI] [PubMed] [Google Scholar]
- 39.Bernal F., Tyler A. F., Korsmeyer S. J., Walensky L. D., Verdine G. L. (2007) J. Am. Chem. Soc. 129, 2456–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wade M., Rodewald L. W., Espinosa J. M., Wahl G. M. (2008) Cell Cycle 7, 1973–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W. (1976) Acta Crystallogr. Sect. A 32, 922–923 [Google Scholar]
- 42.Collaborative Computational Project N. (1994) Acta Crystallogr. Sect. D 50, 760–76315299374 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.