Abstract
We studied the effect of N-cadherin, and its free or membrane-anchored cytoplasmic domain, on the level and localization of β-catenin and on its ability to induce lymphocyte enhancer-binding factor 1 (LEF-1)-responsive transactivation. These cadherin derivatives formed complexes with β-catenin and protected it from degradation. N-cadherin directed β-catenin into adherens junctions, and the chimeric protein induced diffuse distribution of β-catenin along the membrane whereas the cytoplasmic domain of N-cadherin colocalized with β-catenin in the nucleus. Cotransfection of β-catenin and LEF-1 into Chinese hamster ovary cells induced transactivation of a LEF-1 reporter, which was blocked by the N-cadherin-derived molecules. Expression of N-cadherin and an interleukin 2 receptor/cadherin chimera in SW480 cells relocated β-catenin from the nucleus to the plasma membrane and reduced transactivation. The cytoplasmic tails of N- or E-cadherin colocalized with β-catenin in the nucleus, and suppressed the constitutive LEF-1-mediated transactivation, by blocking β-catenin–LEF-1 interaction. Moreover, the 72 C-terminal amino acids of N-cadherin stabilized β-catenin and reduced its transactivation potential. These results indicate that β-catenin binding to the cadherin cytoplasmic tail either in the membrane, or in the nucleus, can inhibit β-catenin degradation and efficiently block its transactivation capacity.
β-catenin is an adherens junction and signaling molecule with diverse roles in the regulation of cell structure and fate. In adherens-type junctions, β-catenin interacts with the cytoplasmic domains of cadherin molecules and links them, through α-catenin, to the actin cytoskeleton (1, 2). In addition, β-catenin can translocate into the nucleus (3–7), where it is involved, together with transcription factors of the lymphocyte enhancer-binding factor 1 (LEF)/T cell factor (TCF) family, in the expression of specific genes (8, 9). By playing this dual role—a structural one in the junction and a regulatory outside the junction—changes in cell adhesion and junction formation can affect transmembrane signaling and gene expression. This intriguing hypothesis gained strong support from recent studies on wg/wnt signaling in a variety of organisms. It was shown that the Drosophila segment polarity gene product armadillo, a homologue of vertebrate plakoglobin and β-catenin, is part of a signaling pathway driven by the secreted molecule wingless (wg) (10). Wnt, the vertebrate homologue of wg, regulates morphogenetic events in Xenopus (11, 12) and adhesion-related responses in mammalian cells (13, 14). In both Drosophila and Xenopus, it was demonstrated that the signaling activity of armadillo/β-catenin is independent of cadherin-based adhesion (15, 16). On the other hand, this signaling is strongly affected by the levels of cadherin because overexpression of cadherin mimics the wg phenotype in Drosophila (16) and blocks β-catenin signaling in Xenopus (15), suggesting that cadherin may be a negative regulator of armadillo/β-catenin signaling.
β-catenin-mediated signaling also can be affected by the association of β-catenin with the tumor suppressor molecule adenomatous polyposis coli (APC) (17, 18) and their further interaction with axin (19, 20) and glycogen synthase kinase 3β (21). Phosphorylation of β-catenin by the APC-axin–glycogen synthase kinase 3β complex (22, 23) leads to its degradation by the ubiquitin–proteasome system (6, 24). The failure of this degradation in cells expressing mutant APC or β-catenin leads to the accumulation of β-catenin and is common in human colon cancer and melanoma (25–27). Of interest, apart from the increase in β-catenin levels in certain tumors, a reduction in E-cadherin levels also was found in many carcinomas (28–30), and the invasiveness of these tumor cells could be suppressed by overexpression of E-cadherin (31, 32).
In this study, we investigated the ability of the cytoplasmic domains of N- and E-cadherin to modulate β-catenin localization, stability, and transactivation potential. We show that expression of the cytoplasmic tail of cadherin, either membrane bound or soluble, protects endogenous β-catenin from degradation and blocks its transactivation capability. In colon cancer cells containing mutant APC (and hence high levels of β-catenin), expression of cadherin derivatives, especially its soluble cytoplasmic tail, strongly suppressed β-catenin-mediated transactivation.
MATERIALS AND METHODS
Plasmid Constructions.
Full length chicken N-cadherin (33) was cloned into the pECE expression vector as described (34). The N-cadherin cytoplasmic tail was fused to the interleukin 2 receptor (IL-2R) α extracellular and transmembrane domains in the pCDNA3 expression vector to form the IL-2R/N-cadherin chimera (35). The N-cadherin tail cDNA was isolated from the IL-2R/N-cadherin chimera by using HindIII and was recloned into Bluescript (Stratagene). The insert was isolated by EcoRI and XhoI digestion and was ligated in frame to a flag epitope in the pECE plasmid. Truncations in the N- and E-cadherin (36) cDNAs tails were generated by PCR by using oligonucleotide primers. The 5′ primers contained EcoRI and the 3′ primers XbaI sites. The PCR products were fused to the C terminus of the green fluorescent protein (GFP) in the pEGFP C1 plasmid (CLONTECH). A stable mouse β-catenin (ΔN57) (6) also was fused to the C terminus of GFP. The fidelity of the constructs was verified by DNA sequencing. HA-LEF-1, mouse β-catenin, and TOPFLASH/FOPFLASH vectors were provided by R. Kemler (Max-Planck Institute of Immunobiology, Freiburg, Germany) and M. van de Wetering and H. Clevers (University Hospital, Utrecht, Netherlands), respectively.
Cell Culture and Transfections.
Chinese hamster ovary (CHO), 293, and SW480 colon carcinoma cells were cultured in DMEM containing 10% fetal calf serum, with 8% CO2, at 37°C. Stably transfected CHO clones were generated by transfection, using lipofectamine (GIBCO/BRL), of the cadherin derivatives and the puromycin resistance gene. Two days after transfection, the cells were replated in the presence of 10 μg/ml puromycin (Sigma, Holon, Israel). Clones were isolated after 2 weeks and were tested for transgene expression by Western blotting.
Immunoblotting and Immunoprecipitation (IP).
Polyclonal pan-cadherin, monoclonal anti-cadherin (CH-19), and polyclonal anti-β-catenin antibodies were from Sigma. Monoclonal anti-β-catenin, (5H10) was from M. Wheelock (University of Toledo, Toledo, Ohio), anti-IL-2Rα was from Upstate Biotechnology, anti-flag (M2) was from Kodak, anti-HA (clone 12CA5) was from Boehringer Mannheim, anti-GFP (polyclonal) from CLONTECH, and anti-LEF-1 was from R. Grossched (University of California, San Francisco). Immunoreactive bands on blots were visualized by the enhanced chemiluminescence method (ECL, Amersham). Cells were harvested in Laemmli’s sample buffer, and equal amounts of protein were separated by SDS/PAGE, were electrotransferred to nitrocellulose, and were incubated with different antibodies. For IP, cells were harvested in a buffer containing 20 mM Tris (pH 8.0), 1% Triton X-100, 140 mM NaCl, 10% glycerol, 1 mM EGTA, 1.5 mM MgCl2, 1 mM DTT, 1 mM sodium vanadate, 50 μg/ml PMSF, and equal amounts of protein incubated with 1 μl of antibody for 2 h at 4°C followed by incubation with 20 μl of protein A+G/agarose beads (Santa Cruz Biotechnology) for 2 h at 4°C. The beads were washed with 20 mM Tris (pH 8.0), 150 mM NaCl, and 0.5% Nonidet P-40, and the immune complexes were recovered by boiling in Laemmli’s sample buffer and were resolved by SDS/PAGE.
Triton X-100 Fractionation.
Cells cultured on 35-mm plates were extracted at 25°C with 200 μl of 0.5% Triton X-100, 2.5 mM EGTA, 5 mM MgCl2, and 50 mM MES (pH 6.0) for 2 min. The Triton-soluble fraction was collected, and the plates were washed twice with the same buffer. The insoluble fraction was scraped into 200 μl of the same buffer. Equal volumes of these fractions were analyzed by SDS/PAGE and immunoblotting by using anti-cadherin and β-catenin antibodies.
Immunofluorescence Microscopy.
Cells cultured on glass coverslips were fixed with 3% paraformaldehyde in PBS, were permeabilized with 0.5% Triton X-100, and were labeled immunofluorescently as described (34). The fluorescence was examined by using a Zeiss Axiophot microscope. For triple fluorescence, GFP-tagged β-catenin and Cy3 and Cy5 secondary antibodies (Jackson ImmunoResearch) were used.
Transactivation Assays.
Cells were transfected with TOPFLASH or FOPFLASH plasmids (9) and pCDNA3 coding for β-galactosidase to evaluate transfection efficiency. In addition, either β-catenin, LEF-1, or cadherin derivatives in different combinations were cotransfected. After 48 h, luciferase and β-galactosidase activities were determined after washing the cells with PBS resuspension in 33 mM NaH2PO4, 66 mM Na2HPO4, 0.1 mM MnCl2, 2 mM MgSO4, 40 mM β-mercaptoethanol, and lysis. Equal amounts of protein were incubated with O-nitrophenyl-β-d-galactopyranoside at 37°C until a yellow color appeared. The reactions were stopped with 1 M Na2CO3, and absorbance at 420 nm was determined. Luciferase activity was measured in equal aliquots by using luciferine buffer [100 mM Tris-O-acetate, pH 7.8/10 mM Mg-O-acetate/1 mM EDTA/74 μM luciferine (Boehringer Mannheim)/2.22 mM ATP, pH 7.0] and was determined with a TD-20e luminometer (Turner, Palo Alto, CA). Values were normalized to β-galactosidase activity.
RESULTS
The Effects of Cadherin Derivatives on the Stability of β-Catenin in CHO Cells.
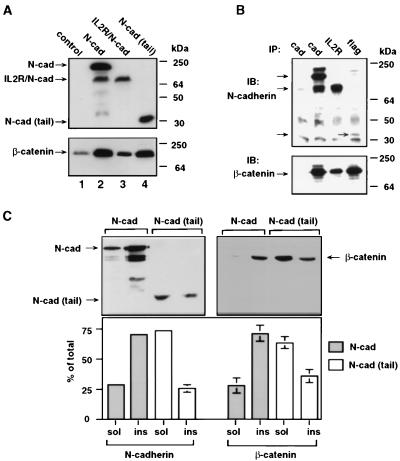
To study the effect of cadherin and cadherin derivatives on β-catenin organization and signaling, cDNA constructs encoding N-cadherin, its cytoplasmic domain fused to the transmembrane and extracellular parts of IL-2R, and the soluble cytoplasmic domains of N- and E-cadherin (or parts of them) were prepared (Fig. 1). Each of the cadherin derivatives contained an antigenic tag (IL-2R or flag) or the autofluorescent GFP that enabled visualization of the transfected protein. CHO cells that contain very low levels of N-cadherin (35) were stably transfected with either full length N-cadherin, the IL-2R/N-cadherin chimera, or the cytoplasmic tail of N-cadherin. As shown in Fig. 2A, the expression of each of the three cadherin derivatives induced an increase in β-catenin levels, probably by complexing with and protecting β-catenin from degradation. This was supported by showing a direct interaction of these cadherin derivatives with β-catenin by co-IP by using antibodies against cadherin, IL-2R, and flag followed by immunoblotting with anti-β-catenin antibody (Fig. 2B).
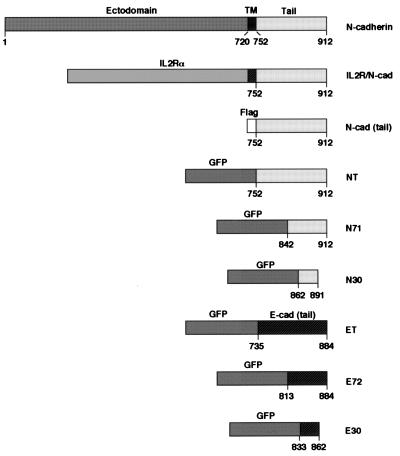
Figure 1.
Schematic representation of cadherin constructs used in this study. Full length N-cadherin and a chimera consisting of the extracellular and transmembrane domains of IL-2Rα and the intracellular domain of N-cadherin (amino acids 752–912) (ILR/N-cad) are shown. The cytoplasmic domain of N-cadherin was tagged with flag [N-cad (tail)] or was fused to the C terminus of GFP (NT). Fragments of the cytoplasmic N-cadherin tail (N71, amino acids 842–912; N30, amino acids 862–891), the E-cadherin tail (ET, amino acids 735–844), and two fragments from this domain (E72, amino acids 813–844; E30, amino acids 833–862) were fused to the C terminus of GFP.
Figure 2.
Stabilization of β-catenin by N-cadherin derivatives. (A) Western blot of proteins from CHO cells (control), and cells stably expressing N-cadherin (N-cad), the IL-2R/N-cadherin chimera (IL-2R/N-cad), or the N-cadherin cytoplasmic tail [N-cad (tail)] with anti-N-cadherin (lanes 1 and 2), the IL-2R (lane 3), and Flag (lane 4). An identical blot was probed with anti-β-catenin antibody. (B) Co-IP with anti-cadherin (cad) from CHO cell extracts (lane 1) and from cells transfected with N-cadherin (lane 2), with anti-IL-2R from cells transfected with IL-2R/N-cadherin (IL-2R, lane 3), or anti-flag from cells transfected with the N-cadherin tail (flag, lane 4). Shown is an immunoblot (IB) of the IPs with anti-cadherin or anti-β-catenin antibodies. The bands around 25 kDa and 50 kDa in all lanes are light and heavy Ig chains from the IP. (C) Triton X-100 solubility of proteins from CHO cells expressing N-cadherin and the N-cadherin cytoplasmic tail. Triton X-100-soluble (sol) and -insoluble (ins) fractions of cell extracts were immunoblotted with anti-cadherin or anti-flag antibodies.
The association of each of the cadherin derivatives with the cytoskeleton was determined by detergent extraction followed by immunoblot analysis. Triton X-100 fractionation indicated that 71% of the full length N-cadherin was associated with the Triton X-100-insoluble fraction (Fig. 2C). In contrast, 74% of the cadherin tail was Triton-soluble in CHO cells stably expressing these molecules (Fig. 2C). Similarly, 72% of β-catenin was detergent-insoluble in CHO cells transfected with N-cadherin whereas 64% of β-catenin was Triton-soluble in the N-cadherin tail expressing CHO cells (Fig. 2C). The detergent solubility of these molecules in the IL-2R/N-cadherin chimera-expressing cells was similar to that of cells expressing full length cadherin (results not shown).
Inhibition of β-Catenin-Driven Transactivation by Cadherin Derivatives.
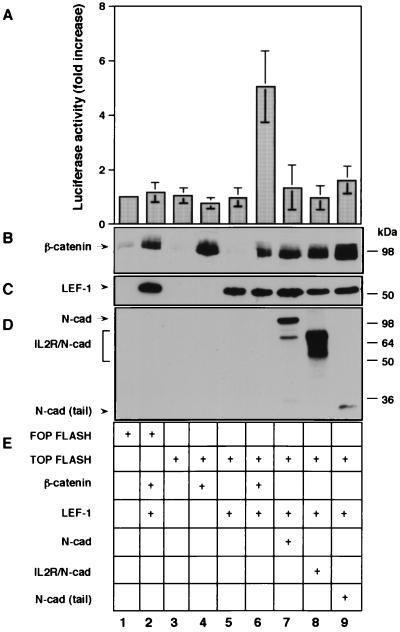
β-Catenin was shown to associate with transcription factors of the LEF/TCF family, forming a bipartite complex that can transactivate genes containing a LEF/TCF binding sequence near their promoter. To study the effect of the different cadherin derivatives on β-catenin-mediated transactivation, a construct containing a multimeric synthetic LEF-1 binding site and a control, mutated LEF-1 binding site upstream of a luciferase reporter gene (9), were transfected into CHO cells, together with either LEF-1 and β-catenin or with LEF-1 and each of the cadherin constructs. The results presented in Fig. 3A show a 5-fold increase in luciferase activity when β-catenin was cotransfected with LEF-1 compared with transfection without β-catenin (Fig. 3A, compare lane 6 to lane 5). In contrast, transfection of LEF-1 with each of the three cadherin-derived molecules (Fig. 3D, lanes 7–9) was inefficient in elevating luciferase activity (Fig. 3A, lanes 7–9), despite the high levels of β-catenin in these cells after transfection (Fig. 3B, lanes 7–9).
Figure 3.
The effect of cadherin derivatives on β-catenin-mediated transactivation in CHO cells transfected with various plasmid combinations. Transactivation was examined (A) as the level of luciferase activity driven by TOPFLASH. The levels of β-catenin (B), LEF-1 (C), and cadherin derivatives (D) were determined by Western blotting.
Localization of N-Cadherin Derivatives and Their Effect on β-Catenin Transactivation in SW480 Cells.
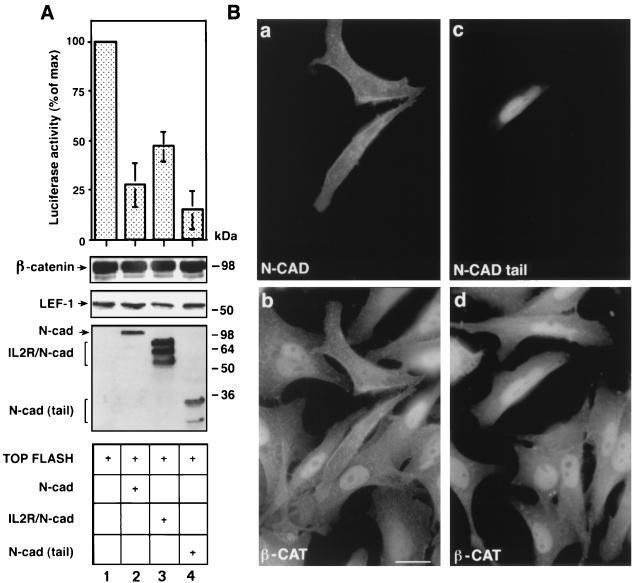
SW480 colon carcinoma cells express mutant APC, low levels of E-cadherin, and relatively high levels of free β-catenin (5, 7, 37). These cells also display constitutively significant β-catenin-mediated transcription after transfection with a LEF-1-responsive reporter (Fig. 4A, lane 1). To determine the effect of cadherin on this β-catenin-driven transcription, each of the three N-cadherin derivatives was cotransfected into SW480 cells with the LEF-1 responsive reporter, and luciferase activity determined. The results shown in Fig. 4A (lanes 2–4) demonstrate that the different cadherin constructs reduced LEF-1-responsive transcription. The most efficient inhibition of this transactivation was obtained with the cadherin tail (Fig. 4A, lane 4), which inhibited the expression of the reporter gene by ≈85%. Full length N-cadherin and the IL-2R/N-cadherin chimera decreased the reporter activation by 70 and 50%, respectively. Immunofluorescence staining of SW480 cells transfected with N-cadherin (Fig. 4Ba) showed that N-cadherin transfection resulted in binding of the endogenous β-catenin to the plasma membrane (Fig. 4Bb) whereas the transfected cytoplasmic tail of N-cadherin (Fig. 4Bc) colocalized with β-catenin in the nucleus (Fig. 4Bd).
Figure 4.
Inhibition of the constitutive LEF-1 responsive transactivation in SW480 cells by cadherin derivatives. (A) SW480 cells were transfected with TOPFLASH and the different cadherin constructs. The levels of β-catenin, LEF-1, and cadherins were determined by Western blotting. (B) Subcellular localization of N-cadherin (N-CAD) (a), the N-CAD tail (c), and β-catenin (b and d) in SW480 cells transfected with N-cadherin or the cadherin tail. (Bar = 10 μm.)
A Region in the C-Terminal Tail of Cadherin Stabilizes β-Catenin and Inhibits Transactivation.
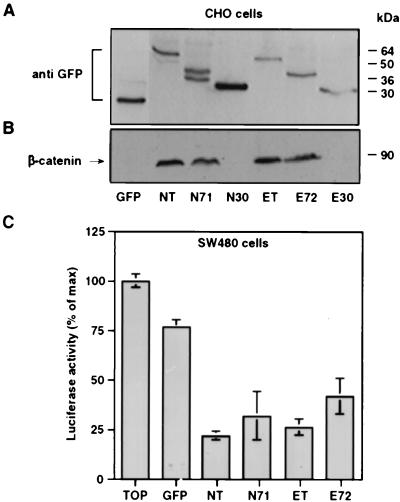
To identify the region in the cadherin cytoplasmic tail that is involved in the suppression of LEF-1-directed transcription, the ability to inhibit transactivation was examined for GFP fusions (Fig. 1) of the cytoplasmic tails of E- and N-cadherin and two deletion tail mutants. The constructs were transfected into CHO cells to determine their effect on the level of β-catenin and into SW480 cells to examine the inhibition of transactivation (Fig. 5). The expressed proteins were analyzed by SDS/PAGE, and the expected molecular weights were obtained for these constructs (Fig. 5A). Immunoblotting of the same extracts with β-catenin antibodies indicated that the N- (NT) and E-cadherin (ET) tails were both efficient in stabilizing the endogenous β-catenin against degradation (Fig. 5B). Furthermore, the C-terminal 71 amino acids of N-cadherin (N71, amino acids 842–912, Fig. 1) and E-cadherin (E72, amino acids 813–884, Fig. 1) both stabilized β-catenin in CHO cells (Fig. 5B). In contrast, shorter fragments of the cadherin cytoplasmic tails consisting of ≈30 amino acids of the N- (N30, amino acids 860–891, Fig. 1) and E-cadherin tails (E30, amino acids 833–862, Fig. 1), corresponding to the middle part of N71 and E72 (see Fig. 1), were ineffective in stabilizing β-catenin against degradation (Fig. 5B).
Figure 5.
The effect of cadherin tail constructs on β-catenin level and transactivation. (A) CHO cells transiently transfected with GFP-cadherin constructs (Fig. 1) were analyzed by Western blotting by using anti-GFP antibody. (B) The blot was reprobed with anti-β-catenin antibody. (C) In SW480 cells, N71 and E72 inhibited transactivation when compared with control (TOP), albeit less efficiently than the full length cytoplasmic tails of N- and E-cadherin.
Analysis of transactivation in SW480 cells (Fig. 5C) showed that the GFP-constructs containing the N- and E-cadherin tails inhibited luciferase activity by 80 and 75%, respectively, whereas GFP only slightly reduced this activity. The shorter GFP-N71 and GFP-E72 constructs were less efficient and reduced transactivation by 70 and 60%, respectively (Fig. 5C). The 30-aa E- and N-cadherin tail fragments did not affect β-catenin-mediated transactivation in SW480 cells (data not shown).
The N-Cadherin Tail Inhibits LEF-1/β-Catenin Complex Formation.
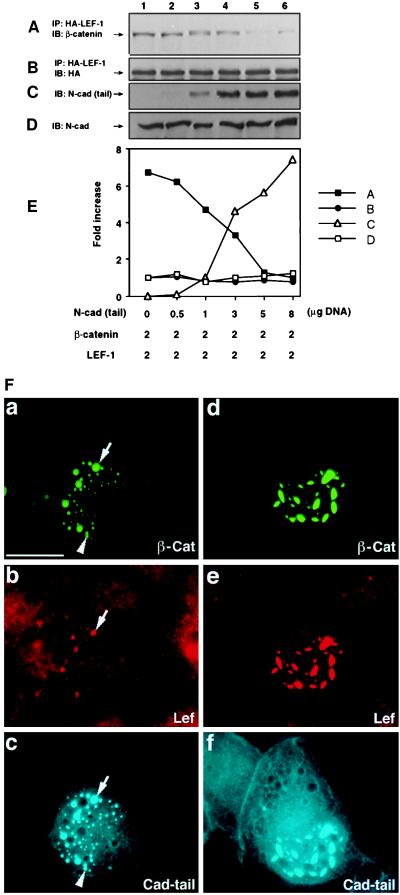
The inhibition of β-catenin-driven transactivation by N-cadherin could result from either displacement of LEF-1 binding to β-catenin by N-cadherin or the formation of a ternary complex (cadherin-LEF-1-β-catenin) that has no transactivation potential. To distinguish between these possibilities, 293 cells were transfected with constant amounts of HA-tagged LEF-1 and β-catenin and increasing amounts of the N-cadherin tail (Fig. 6C, lanes 2–6). After transfection, LEF-1 was immunoprecipitated with anti-HA antibody, and the associated β-catenin was detected by Western blot analysis (Fig. 6A). The results in Fig. 6 A and E show that the N-cadherin tail inhibited β-catenin binding to LEF-1 in a dose-dependent manner whereas the level of the endogenous N-cadherin remained unchanged (Fig. 6D).
Figure 6.
Competition between the N-cadherin tail and LEF-1 for β-catenin binding. 293 cells were transfected at 1:1 ratio with β-catenin and HA-tagged LEF-1 cDNA together with increasing amounts of the N-cadherin tail. LEF-1 was immunoprecipitated by using anti-HA antibody, and β-catenin (A) and LEF-1 (B) levels determined by blotting with anti-β-catenin and -HA antibodies. N-cadherin tail (C) and N-cadherin (D) levels were determined by immunoblotting of cell extracts with anti-cadherin. (E) Quantitative determination of changes in the levels of the proteins shown in A–D. (F) Triple fluorescence analysis of cells transfected with GFP-tagged β-catenin and N-cadherin tail (at 0.5 and 5 μg, respectively) (a–c), or 5 μg and 5 μg (d–f). Endogenous LEF-1 was visualized by Cy3-labeled secondary antibodies (b and e), and the cadherin tail was visualized with Cy5-labeled secondary antibodies (c and f). The arrow marks aggregates containing β-catenin, LEF-1, and cadherin tail whereas the arrowhead marks an aggregate deficient in LEF-1.
In a recent study (7), it was demonstrated that transient overexpression of β-catenin results in its accumulation in the nucleus in either small (Fig. 6Fa) or large aggregates (Fig. 6Fd). These aggregates also contained the endogenous LEF-1 (7). When the cadherin tail was cotransfected with GFP-β-catenin, it colocalized with the large speckles (Fig. 6Fc and Ff) that also contained LEF-1 (Fig. 6Fe). When higher levels of the cadherin tail were expressed, LEF-1 colocalized with the cotransfected β-catenin only partially, in the large speckles, but not in the small ones (Fig. 6Fb). This implies that the N-cadherin tail is effective in competing with LEF-1 for complexing with β-catenin in the nucleus.
DISCUSSION
β-Catenin interacts with three major subcellular systems that affect its activities and fate: adherens-type junctions, where β-catenin forms a complex with the cytoplasmic domain of different cadherins and links them to the actin cytoskeleton (1, 2); a unique degradation pathway that regulates the level of β-catenin and includes binding to APC, axin, phosphorylation by glycogen synthase kinase 3β, and degradation by the ubiquitin-proteasome system (24); and the transcriptional machinery, where β-catenin interacts with LEF/TCF transcription factors and activates the expression of specific target genes (3–5, 7–9).
In this study we investigated the cross-talk between these systems and characterized the effect of cadherin and cadherin derivatives on β-catenin stability and signaling. Elevated expression of cadherin could protect β-catenin from degradation, increasing its level in the cytoplasm, and subsequently was expected to stimulate LEF-1-responsive transcription. However, this “protective cadherin” could block β-catenin-mediated transactivation either by sequestering β-catenin to the plasma membrane (away from the nucleus) and/or by competing with transcription factors of the LEF/TCF family that interact with β-catenin. Indeed, we showed here that intact cadherin can translocate β-catenin to junctional sites and stabilize it against degradation in cells in which a “normal” (rapid) turnover of β-catenin takes place. This recruitment of β-catenin to junctions was accompanied by a strong inhibition of LEF-1-directed transcription. The presence of organized junctions, however, was not essential for membrane translocation and stabilization of β-catenin nor for inhibition of β-catenin-mediated transactivation. A chimeric receptor consisting of the cadherin cytoplasmic domain and an inert transmembrane anchor (IL-2R) was fully effective, despite being unable to participate in junction formation and despite its capacity to inhibit junction assembly in cadherin-containing cells (35). Of interest, the free cytoplasmic tails of both E- and N-cadherin were most effective in protecting β-catenin from degradation and inhibiting its transactivating potential without affecting its subcellular distribution. Thus, in cells expressing high levels of both β-catenin and the cadherin tail, the two proteins colocalized in the nucleus, and β-catenin-driven transcription was suppressed strongly. The most likely explanation for this effect is that binding of the cadherin tail (free or membrane-bound, junctional or extrajunctional) to β-catenin inhibited the binding of β-catenin to LEF-1 and blocked its transactivation capacity. This notion is supported by the co-IP and triple fluorescence labeling experiments, demonstrating that increased levels of the cadherin tail suppressed β-catenin-LEF-1 complex formation in the nucleus. The mechanism whereby N-cadherin or its tail confer β-catenin stabilization also could result from an effective competition with the binding of β-catenin to APC (38, 39) or other components of the APC-axin-glycogen synthase kinase 3β-ubiquitin-proteasome systems.
The binding sites on β-catenin for cadherin, APC, and LEF-1 involve multiple overlapping armadillo repeats: Repeats 4–13 are important for E-cadherin binding (39, 40), repeats 1–10 are important for APC binding (39), and repeats 3–8 are important for interaction with dTCF (9) whereas repeats 1–14 are involved in β-catenin-LEF-1 interaction (3). Taken together with the present study, the association of β-catenin with each of these components appears to be mutually exclusive.
In this study, we also demonstrated that sequestration to the membrane or the cytoplasm is not necessary for protecting β-catenin from degradation because the soluble N-cadherin tail did not affect the nuclear localization of β-catenin while being efficient in stabilizing β-catenin in CHO cells (similar to full length cadherin). As we found that the soluble cadherin tail was capable of efficiently competing with LEF-1 for binding to β-catenin in the nucleus, β-catenin translocation into the nucleus may not require LEF-1, in agreement with a recent report showing a role for the importin β/karyophilin β system in this process (41). Because shorter fragments of the cadherin tail could inhibit the constitutive transcriptional activity of β-catenin in SW480 cells, the antagonistic effect of cadherin on β-catenin signaling in these human cancer cells is most probably independent of adherens junction formation, similarly to results obtained for β-catenin signaling in Drosophila and Xenopus (15, 16).
The results presented here are also relevant to novel approaches aiming to suppress β-catenin-driven oncogenesis. It was demonstrated that mutations in colon carcinoma that render APC unable to participate in β-catenin degradation result in high levels of β-catenin and in the activation of genes (the nature of which is unknown) that are probably involved in the transformation of these cells (25, 26). Thus, the cytoplasmic cadherin tail and fragments derived from it (that contain the β-catenin-binding site) may prove to be useful in blocking the expression of such target genes and therefore in suppressing tumorigenicity. Our study indicates that the C-terminal region of E- and N-cadherin corresponding to ≈70 amino acids retains the transactivation suppressive capacity. Additional attempts are currently underway to define the minimal effective region on the cadherin tail that can inhibit β-catenin-driven transcriptional activity and thus suppress β-catenin-mediated tumorigenicity.
Acknowledgments
We are grateful to R. Kemler, R. Grosschedl, M. Wheelock, H. Clevers, and M. van de Wetering for sending reagents. These studies were supported by grants from the Israel Science Foundation, the U.S.–Israel Binational Foundation, the German–Israeli Foundation for Scientific Research and Development, the Forchheimer Center for Molecular Genetics, and the Cooperation Program in Cancer Research between the German Cancer Research Center and Israel Ministry of Science. A.B.-Z. holds the Lunenfeld-Kunin Chair in Cell Biology and Genetics, and B.G. holds the E. Netter Chair in Cell and Tumor Biology.
ABBREVIATIONS
- APC
adenomatous polyposis coli
- IL-2R
interleukin 2 receptor
- GFP
green fluorescent protein
- CHO
Chinese hamster ovary
- IP
immunoprecipitation
- LEF-1
lymphocyte enhancer-binding factor 1
- TCF
T cell factor
References
- 1.Geiger B, Yehuda-Levenberg S, Bershadsky A D. Acta Anat. 1995;154:46–62. doi: 10.1159/000147751. [DOI] [PubMed] [Google Scholar]
- 2.Kemler R. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 3.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 4.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 5.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Mech Dev. 1996;59:3–11. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 6.Salomon D, Sacco P A, Guha Roy S, Simcha I, Johnson K R, Wheelock M J, Ben-Ze’ev A. J Cell Biol. 1997;139:1325–1335. doi: 10.1083/jcb.139.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, Ben-Ze’ev A. J Cell Biol. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 9.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 10.Peifer M. Trends Cell Biol. 1995;5:224–229. doi: 10.1016/s0962-8924(00)89015-7. [DOI] [PubMed] [Google Scholar]
- 11.Gumbiner B M. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 12.Miller J R, Moon R T. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 13.Bradely R S, Cowin P, Brown A M. J Cell Biol. 1993;123:1857–1865. doi: 10.1083/jcb.123.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinck L, Nelson W J, Papkoff J. J Cell Biol. 1994;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagotto F, Funayama N, Glück U, Gumbiner B M. J Cell Biol. 1996;132:1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanson B, White P, Vincent J P. Nature (London) 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- 17.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 18.Rubinfeld B, Souza B, Albert I, Müller O, Chamberlain S F, Masiarz R, Munemitsu S, Polakis P. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 19.Hart M, de los Santos R, Albert I, Rubinfeld B, Polakis P. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 20.Behrens J, Jerchow B A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 21.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 22.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1372–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 26.Korinek V, Backer N P, Morin J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 27.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 28.Birchmeier W, Behrens J. Biochim Biophys Acta. 1994;1198:11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Ze’ev A. Curr Opin Cell Biol. 1997;9:99–108. doi: 10.1016/s0955-0674(97)80158-5. [DOI] [PubMed] [Google Scholar]
- 30.Perl A-K, Wilgenbus P, Dahl U, Semb H, Christofori G. Nature (London) 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 31.Vleminckx K, Vakaet L J, Mareel M, Fiers W, Van Roy F. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 32.Frixen U H, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Löchner D, Birchmeier W. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salomon D, Ayalon O, Patel-King R, Hynes R O, Geiger B. J Cell Sci. 1992;102:7–17. doi: 10.1242/jcs.102.1.7. [DOI] [PubMed] [Google Scholar]
- 34.Levenberg S, Katz B-Z, Yamada K M, Geiger B. J Cell Sci. 1998;111:347–357. doi: 10.1242/jcs.111.3.347. [DOI] [PubMed] [Google Scholar]
- 35.Katz B-Z, Levenberg S, Yamada K M, Geiger B. Exp Cell Res. 1998;243:415–424. doi: 10.1006/excr.1998.4194. [DOI] [PubMed] [Google Scholar]
- 36.Butz S, Stappert J, Weissing H, Kemler R. Science. 1992;257:1142–1144. doi: 10.1126/science.257.5073.1142-a. [DOI] [PubMed] [Google Scholar]
- 37.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinfeld B, Souza B, Albert I, Munemitsu S, Polakis P. J Biol Chem. 1995;270:5549–5555. doi: 10.1074/jbc.270.10.5549. [DOI] [PubMed] [Google Scholar]
- 39.Hülsken J, Birchmeier W, Behrens J. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orsulic S, Peifer M. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagotto F, Glück U, Gumbiner B M. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]