Abstract
The vertebrate hypothalamic-pituitary axis (HP) is the main link between the central nervous system and endocrine system. Although several signal pathways and regulatory genes have been implicated in adenohypophysis ontogenesis, little is known about hypothalamic and neurohypophysial development or when the HP matures and becomes functional. To identify markers of the HP, we constructed subtractive cDNA libraries between adult zebrafish hypothalamus and pituitary. We identified previously published genes and ESTs and novel zebrafish genes, some of which were predicted by genomic database analysis. We also analyzed expression patterns of these genes and found that several are expressed in the embryonic and larval hypothalamus, neurohypophysis, and/or adenohypophysis. Expression at these stages makes these genes useful markers to study HP maturation and function.
Keywords: Adenohypophysis, cell differentiation, hormone, hypothalamic-pituitary axis, hypothalamus, neurohypophysis, organ maturation, pituitary, subtractive cDNA library
1. Results and Discussion
The hypothalamic-pituitary axis (HP) is the main link between the central nervous system and endocrine system. The HP regulates hormone secretion during homeostasis, growth, reproduction, and several other physiological functions. Under control of the hypothalamus, the adenohypophysis (anterior pituitary lobe) produces and secretes hormones involved in these functions. This control occurs through the production of hypothalamic hypophysiotropic hormones that stimulate or inhibit synthesis and release of adenohypophysial hormones. The hypothalamus also secretes hormones such as vasotocin and oxytocin (in mammals), and isotocin and vasopressin (in fish) through its neuronal projections into the neurohypophysis (Bentley, 1998; Unger and Glasgow, 2003).
Several genes regulate induction, specification, survival, patterning, and cell differentiation in the adenohypophysis (reviewed in (Dasen and Rosenfeld, 2001; Pogoda and Hammerschmidt, 2007; Zhu et al., 2005). However, little is known about the genetic regulation of later hypothalamic and neurohypophysial development or even when the HP matures and becomes physiologically functional.
To identify developmental markers of the HP, we constructed subtractive cDNA libraries between adult hypothalamus and pituitary and examined expression of these genes in embryos and larvae. We report the construction of these subtractive libraries and the identification of genes represented in them. We also present the expression patterns of some of these genes in the HP during the first 5 days of development. Some of these genes are previously known expressed sequence tags (ESTs), with undocumented or incompletely documented HP expression patterns. Other genes were previously unreported in zebrafish, some of which are novel and some of which are homologous to genes in other species.
1.1. Library construction and clone identification
To identify genes expressed in the HP, we constructed subtractive cDNA libraries between adult zebrafish hypothalamus and pituitary. The hypothalamic cDNA library was obtained by subtracting pituitary cDNAs from hypothalamic cDNAs: 448 clones were isolated (Table 1). Conversely, the hypothalamic cDNAs were subtracted from pituitary cDNAs to construct the pituitary cDNA library: 558 clones were isolated (Table 1). We eliminated clones that contained the most redundant sequences by membrane hybridization. We then characterized the remaining clones by sequencing and comparison with NCBI nucleotide and protein databases and the Sanger Institute Danio rerio genomic database.
Table 1.
Hypothalamus and pituitary library construction.
| hypothalamic library | pituitary library | |||||
|---|---|---|---|---|---|---|
| # total clones (1) | 448 | 558 | ||||
| total unusable clones (2) | 114 (25.4%) | 78 (14.0%) | ||||
| total analyzed clones (3) | 334 (74.6%) | 480 (86.0%) | ||||
| total identified sequences (4) | 348 | 492 | ||||
| # sequences (5) |
# unique genes or genomic sequences (6) |
% of total (7) |
# sequences (5) |
# unique genes or genomic sequences (6) |
% of total (7) |
|
| ➢ mitochondrial sequences | 21 | 5 | 6.0% | 3 | 2 | 0.6% |
| ➢ ribosomal sequences | 4 | 2 | 1.1% | 7 | 6 | 1.4% |
| ➢ known genes | 199 | 22 | 57.2% | 438 | 54 | 89.0% |
| ➢ ESTs and new genes | 83 | 17 | 23.9% | 34 | 23 | 6.9% |
| ➢ genomic DNA sequences | 41 | 12 | 11.8% | 10 | 8 | 2.0% |
(1) Represents the number of clones isolated after subtraction and stored as glycerol stocks. (2) From these clones, some did not survive, did not contain an insert, or were resistant to sequencing because of their base composition. The remaining clones were analyzed by sequencing. Some clones contained more than one insert. Therefore, the total number of identified sequences in row (4) is larger than the number of analyzed clones in row (3). Identified sequences were grouped into categories: mitochondrial sequences, ribosomal sequences, sequences of known genes, published ESTs, predicted genes or genomic DNA sequences. In each category, column (5) lists the number of sequences found, (6) indicates the number of unique genes or unique genomic sequences represented and in (7) the percentage of sequences is listed compared to the total number of identified sequences in column (4).
The subtraction process enriched cDNAs that are differentially expressed in hypothalamus or adenohypophysis. Our initial analysis suggested that the proportion of house keeping genes, such as mitochondrial and ribosomal genes, is low (Table 1). With the exception of the spermatogenesis associated protein 2 gene that we found in both libraries (1 sequence in the pituitary library, 2 similar sequences in the hypothalamic library), no other overlap was observed between the libraries, including mitochondrial and ribosomal sequences (not shown).
The sequences we identified consisted of previously published genes (known genes) and ESTs or novel genes (genes not cloned or identified in zebrafish yet or predicted only by genomic database analysis). Based on studies in other species and analysis of conserved domains, these genes can be classified into functional categories that include structural and physiological functions, cell metabolism, and protein synthesis.
We identified only one transcription factor in the pituitary library: the POU domain class1 transcription factor 1 (pou1f1) that plays a role in somatotrope, lactotrope, and thyrotrope development (Table 2; Gage et al., 1996; Nica et al., 2004). Because transcription factors are expressed at a lower frequency compared to many other mRNAs, the PCR amplification and subtraction method we used might not have been sensitive enough to identify genes expressed at these low levels. Alternatively, it is possible that many transcription factors are expressed in both tissues and were subtracted during the construction of the libraries. Also, some tissue specific transcription factors are probably no longer expressed in differentiated adult tissues.
Table 2.
List of genes expressed in the HP during the first 5d of development.
| Gene | NCBI accession# | expression in HP (1) |
library |
|---|---|---|---|
| known genes | |||
| cellular retinoic acid binding protein 1a | NM 182858 | neuro | pituitary |
| follicle stimulating hormone beta-subunit | NM_205624 | adeno | pituitary |
| glycoprotein hormones, alpha polypeptide | NM_205687 | adeno | pituitary |
| growth hormone | AJ937858 | adeno | pituitary |
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
NM-199809 | adeno | pituitary |
| luteinizing hormone beta-subunit | AY714132 | adeno | pituitary |
| POU domain, class 1, transcription factor 1 | NM 212851 | adeno | pituitary |
| prolactin | NM 181437 | adeno | pituitary |
| proopiomelanocortin | NM 181438 | adeno | pituitary |
| secretogranin III | NM 200757 | adeno | pituitary |
|
solute carrier family 16 (monocarboxylic acid transporters), member 9a |
NM 200410 | adeno | pituitary |
| somatolactin beta | NM_001037674 | adeno | pituitary |
| thyroid stimulating hormone, beta subunit | NM_181494 | adeno | pituitary |
| adenylate cyclase activating polypeptide 1b | NM_214715 | hypo | hypothalamus |
| cytochrome P450, family 19, subfamily A, polypeptide 1b | NM 131642 | hypo | hypothalamus |
| pro-melanin concentrating hormone-like | FJ392645 | hypo | hypothalamus |
| ESTs and new genes | |||
| adrenomedullin 2 | FJ392613 | adeno | pituitary |
| uo:ion002 | FJ392616 | adeno | pituitary |
| uo:ion003 | FJ392618 | adeno+hypo | pituitary |
| calcium/calmodulin-dependent protein kinase 1D | NM 001080658 | hypo | hypothalamus |
| pleckstrin and Sec7 domain containing 3 like | FJ392621 | hypo | hypothalamus |
| potassium channel tetramerization domain containing 4 | FJ392642 | hypo | hypothalamus |
| serpin peptidase inhibitor, clade I (neuroserpin), member 1 | FJ392619 | hypo | pituitary |
| stathmin 1/oncoprotein 18 b | FJ392638 | hypo | hypothalamus |
Column (1) indicates that the gene is expressed in the neurohypophysis (neuro), adenohypophysis (adeno), or hypothalamus (hypotha) during the first 5d of development.
We also identified genomic DNA sequences that do not apparently correspond to genes. Some might represent non-coding RNA. Others that correspond to intronic sequences might have resulted from unspliced mRNA variants, although we did not find corresponding exon sequences of these genes. We can also not entirely exclude the possibility that some of these sequences might derive from genomic DNA contamination. Because our goal was to isolate genes expressed in the HP, we did not analyze these sequences further.
The pituitary library (supplemental Table 1) contained genes encoding most of the adenohypophysial hormones including: follicle stimulating hormone beta-subunit (fshb), glycoprotein hormones alpha polypeptide (cga), growth hormone (gh), luteinizing hormone beta-subunit (lhb), prolactin (prl), proopiomelanocortin (pomc), somatolactin beta (smtlb) and thyroid stimulating hormone beta subunit (tshb). One exception was somatolactin alpha (smtla) that we did not detect. Together, these represent 68.5% of all identified sequences.
Although pomc is expressed in both the pituitary and hypothalamus and should therefore have been subtracted, pomc was represented abundantly (36.4%) in the pituitary library, and was absent from the hypothalamic library. Presumably, this results from very high expression levels of pomc in the pituitary compared to the hypothalamus at the time of tissue dissection. In spite of our precautions to limit stress by MS-222 anesthesia and chilling on ice, pomc encoded ACTH biosynthesis might have been elevated in response to stress (Engelmann et al., 2004).
In the hypothalamic library (supplemental Table 2), ependymin (epd) was the most frequently represented gene (36.7%). This gene encodes a specific CNS glycoprotein, an adhesion molecule involved in memory consolidation (Schmidt, 1995; Schmidt et al., 1995; Shashoua, 1991). We found only one gene that has been reported to regulate pituitary hormone production and/or secretion: adenylate cyclase activating polypeptide 1b (adcyap1b, 1.4%). This gene, also known as pituitary adenylate cyclase activating peptide (pacap), functions in various species as a hormone releasing factor (Gracia-Navarro et al., 2002; Zhou et al., 2005). We also found genes corresponding to neuropeptides, such as proenkephalin-like (1.7%) and prodynorphin (0.3%) that belong to the opioid gene family (Dores et al., 2002), and pro-melanin concentrating hormone-like encoding the hypothalamic hormone, Melanin concentrating hormone (Mch; 0.6%; manuscript in preparation).
1.2. Gene expression patterns during the first 5d of development
The genes represented in these libraries were expressed in a tissue-specific manner in the mature, functional HP of adults. Therefore, the onset of expression during development of the hypothalamus, adenohypophysis, and neurohypophysis would make these genes interesting potential markers for the functional maturation of the HP.
To test whether the genes isolated in the adult pituitary and hypothalamic libraries are expressed in embryos and larvae, we analyzed their expression patterns between Prim-5 (24h) stage and day 5 (5d) of development. We analyzed whole-mount embryos by mRNA in situ hybridization, if the length of the isolated sequence permitted such analysis (45 genes out of 116 total; 39% genes), or consulted the ZFIN gene expression database (http://zfin.org, 57 genes out of 116 total; 49% genes).
Because we subtracted cDNAs exclusively between hypothalamic and pituitary tissues and not from the entire fish, several genes were also expressed in other tissues, for example in the CNS, or throughout the animal (Suppl. Fig. 1–21). In addition, because we constructed the libraries from adult fish, some genes were not expressed between Prim-5 (24h) stage and 5d. Probably, these are expressed later during development and adulthood (see section 1.3).
Our analysis indicates that, among the genes with expression pattern data, 23.5% (24 out of 102 total) are expressed in the HP during the first 5 days of development (Table 2). 63.7% (65 out of 102 total) are expressed outside of the HP and 12.7% (13 out of 102 total) are not expressed during the first 5d of development. Because we analyzed gene expression by whole mount in situ hybridization, it is possible that the sensitivity of this assay was insufficient to detect low expression levels. For instance, some gene expression not detected by in situ hybridization was successfully detected by PCR (see section 1.3; Suppl. Fig. 22).
1.2.1. Genes expressed in the adenohypophysis
All the genes expressed in the adenohypophysis during the first 5d of development were isolated from the pituitary library.
Adenohypophysial hormones and pou1f1 were previously shown to be expressed in the adenohypophysis during the first 5 days of development (Table 2; (Herzog et al., 2003; Lopez et al., 2005; Nica et al., 2004; Pogoda et al., 2006).
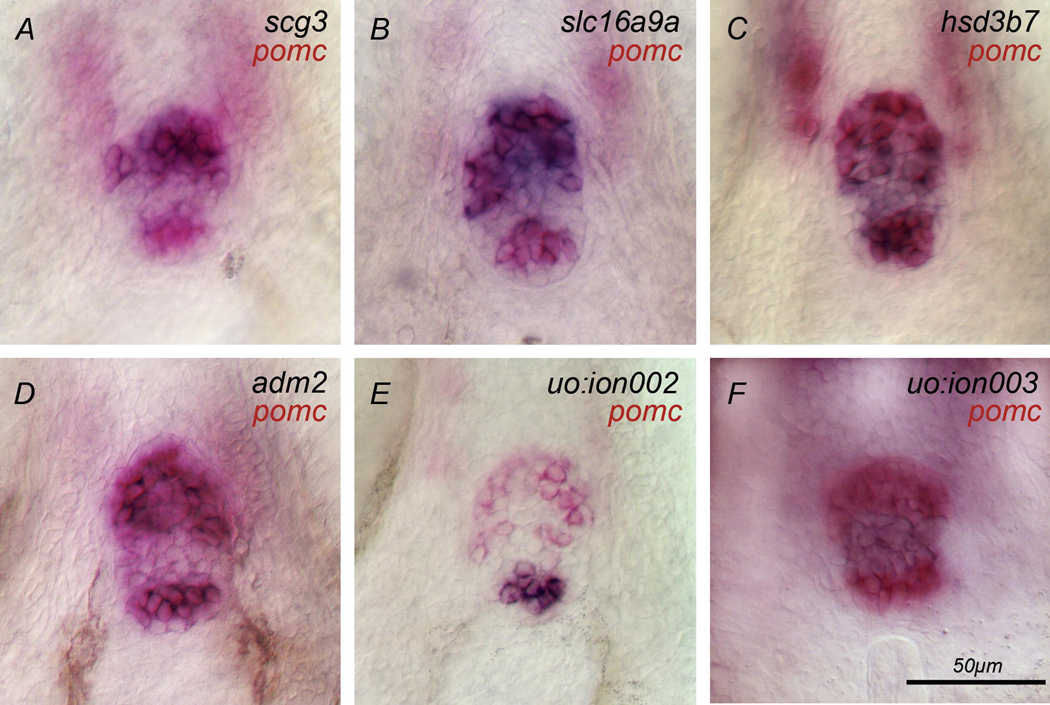
secretogranin III (scg3) belongs to the granin family (chromogranin, secretogranin), a group of acid secretory proteins found in the secretory granules of endocrine cells. Even though their precise role is unclear, proteins of this family are thought to be involved in secretion (Holthuis et al., 1996; Holthuis and Martens, 1996). Consistent with this postulated role, scg3 is found in zebrafish in endocrine organs such as adenohypophysis (Fig. 1A), pancreas, and epiphysis (not shown) at Prim-5 (24h) stage. Additional weak expression is observed in the telencephalon and midbrain. Expression in these domains continues through protruding mouth (72h) stage. scg3 expression also appears in the retina and epiphysis by 5d (ZFIN, http://zfin.org). In the adenohypophysis, cells in the most anterior part of the gland, called the rostral pars distalis (RPD; Fig. 1A), express scg3.
Fig. 1. Genes expressed in the adenohypophysis.
(A) scg3, (B) slc16a9a, (C) hsd3b7, (D) adm2, (E) uo:ion002 and (F) uo:ion003 are expressed (blue) in the adenohypophysis that is delimited by pomc expression (red). scg3 (A) and slc16a9a (B) are expressed in the rostral pars distalis (RPD). (C) hsd3b7 is expressed in the entire gland. (D) The EST adm2 is expressed in the proximal pars distalis (PPD) and pars intermedia (PI). (E) The novel gene uo:ion002 is expressed in the PI. (F) The novel gene uo:ion003 is expressed in the rostral pars distalis (RPD). Ventral views of adenohypophysis at 72h, anterior to the top; scale bar 50 µm.
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase (hsd3b7) plays a role in steroid biosynthesis. It was identified in zebrafish using microarrays to analyze global hematopoietic and vascular endothelial gene expression (Covassin et al., 2006). In addition, hsd3b7 is expressed in the adenohypophysis (Fig. 1C) and in the telencephalon (Suppl. Fig. 3) from Prim-5 (24h) stage to protruding mouth (72h) stage. We find that all adenohypophysial cells express the gene. Additional expression of hsd3b7 outside of the HP is shown in Suppl. Fig. 3.
solute carrier family 16 (monocarboxylic acid transporters) member 9a (slc16a9a) belongs to the monocarboxylic acid transporter family. slc16a9a is expressed from Prim-5 (24h) stage to 5d in the RPD (Fig. 1B), which contains lactotrope, corticotrope, and melanotrope cells. Additional expression outside of the HP is visible from 72h and later (Suppl. Fig. 6).
adrenomedullin 2 (adm2) is a predicted gene that belongs to the calcitonin/calcitonin related gene family. In rat, it is also known as intermedin (imdn) and is expressed in the intermediate lobe of the pituitary and in specific adenocorticotrope cells in the anterior lobe (Lin Chang et al., 2005). Lin Chang et al. showed that PRL release increases in a dose dependent manner in response to Intermedin treatment. In female rats, these results suggested a role of Intermedin in the regulation of PRL release during reproduction.
In zebrafish, adm2 is weakly expressed in the adenohypophysis at 24h. adm2 expression intensifies by long-pec (48h) stage and is reduced at 72h (Fig. 1D). We observed no expression at 4d or 5d. adm2 expressing cells in the adenohypophysis coexpress pomc in the RPD and the posterior part of the adenohypophysis called pars intermedia (PI). Lactotrope, corticotrope, and melanotrope cells are located in these subdomains of the adenohypophysis.
uo:ion002 is a novel gene. A peptide domain of the acyltransferase family was recognized as a putative conserved domain in the protein encoded by this gene, indicating a potential role of this gene in lipid metabolism. Expression of uo:ion002 starts at 48h and is maintained until 5d. It is exclusively expressed in the PI which contains melanotrope cells (Fig. 1E).
1.2.2. Genes expressed in the hypothalamus
Genes expressed in the hypothalamus during the first 5d of development were found in the hypothalamic library except for serpin peptidase inhibitor, clade I (neuroserpin), member 1 which was found in the pituitary library.
cytochrome P450 family 19 subfamily A polypeptide 1b (Lassiter and Linney, 2007) and pro-melanin concentrating hormone like (pmchl) are expressed in the hypothalamus during the first 5 days of development.
adenylate cyclase activating polypeptide 1b (adcyap1b), also known as pituitary adenylate cyclase-activating peptide (pacap) was originally discovered in hypothalamic extracts and was shown to stimulate cAMP accumulation in pituitary cells (Miyata et al., 1989; Miyata et al., 1990). Among several postulated roles, pacap is known as a releasing factor for hormones, including adenohypophysial hormones (Rawlings and Hezareh, 1996). pacap was shown to increase the secretion of Growth Hormone (Gracia-Navarro et al., 2002; Wong et al., 2000; Zhou et al., 2005), Gonadotropin (Petersen et al., 1996; Wong et al., 2000), Thyrotropin (TSH; (Okada et al., 2006), and Prolactin (Rawlings and Hezareh, 1996).
In zebrafish, adcyap1b is expressed in the telencephalon at Prim-5 (24h) stage (Fig. 2A). At 48h (Fig. 2B) and 72h (Fig. 2C), the expression pattern expands throughout the anterior CNS, including the telencephalon, hypothalamus, ventral mesencephalon, and cerebellum. Double in situ hybridization with pomc mRNA probe indicates that the hypothalamic expression is located close to the adenohypophysis (Fig. 2D). This proximity is consistent with a role in pituitary hormone regulation. We did not observe adcyap1b expression in the neurohypophysis.
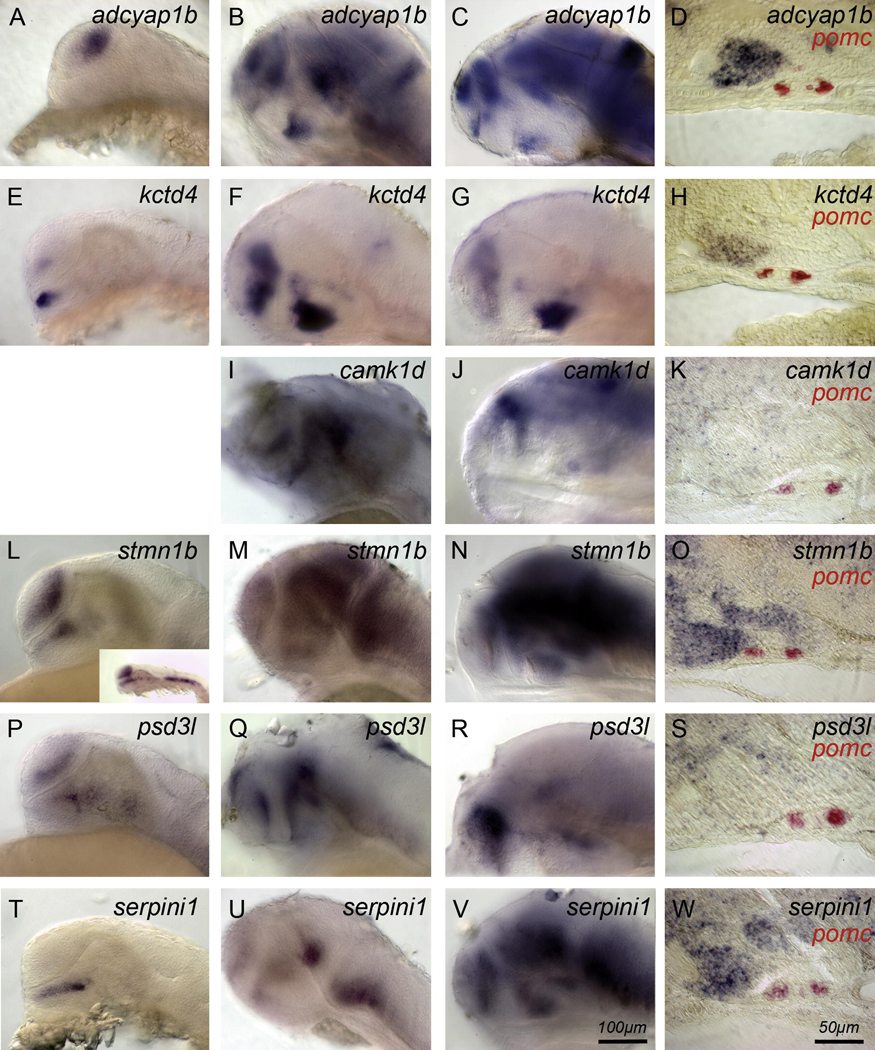
Fig. 2. Genes expressed in the hypothalamus.
(A–D) adcyap1b and kctd4 (E–H) are expressed in the hypothalamus. adcyap1b is expressed in the telencephalon from 24h to 72h (A–C) and in the hypothalamus at 48h and 72h (B–C). (D) adcyap1b and kctd4 (H) are expressed by hypothalamic cells (black) close to the adenohypophysis (demarcated by pomc, red) but not in neurohypophysial cells. (I–K) camk1d is expressed in the dorsal hypothalamus, telencephalon, and midbrain at 48h and 72h. (L–N) stmn1b is expressed in telencephalic, mesencephalic, and hypothalamic cells between 24–72h. (O) The section at 72h suggests that ventral hypothalamic cells and some, but not all neurohypophysial cells express the gene. (P–R) psd3l is also expressed in telencephalon and hypothalamus between 24h–72h. (S) Hypothalamic cells express psd3l. (T–V) serpini1 is expressed in the dorsal hypothalamus at 24h (T), at 48h in telencephalon, diencephalon, and mesencephalon. (W) serpini1 is expressed by hypothalamic cells (blue) close to the adenohypophysis (demarcated by pomc, red) but not in neurohypophysial cells. Side views, anterior to the left. (A,E,L,P,T) 24h; (B,F,I,M,Q,U) 48h; (C,G,J,N,R,V) 72h; Scale bar 100 µm. (D,H,K,O,S,W) 15 µm sagittal sections through the pituitary and adjacent hypothalamus at 72h, scale bar 50 µm.
potassium channel tetramerization domain containing 4 (kctd4) is an EST. kctd4 is expressed in the telencephalon and hypothalamus from Prim-5 (24h) stage through 5d. The hypothalamic expression at 24h is located close to the adenohypophysis (Fig. 2E). Later, this expression domain extends throughout the anterior hypothalamus, close to the adenohypophysis although not in the neurohypophysis (Fig. 2F–H). Weak hindbrain expression starts at 48h (Fig. 2F). kctd4 is an interesting marker because its expression pattern resembles that of the hypophysiotropic gene adcyap1b.
calcium/calmodulin-dependent protein kinase 1D (camk1d) was known as an EST. The calcium/calmodulin-dependent protein kinase best documented in the literature is camkII. It is a multifunctional serine/threonine kinase acting by phosphorylating proteins which functions are Ca2+ dependent. CamkII is involved in a variety of functions, such as cell division, differentiation, synaptic plasticity and transmission, dendritic morphology plasticity, and cardiac contraction (Colbran and Brown, 2004; Griffith, 2004; Hudmon and Schulman, 2002).
In zebrafish, camk1d expression starts at 48h in the hypothalamus, telencephalon and midbrain (Fig. 2I). This expression persists at 72h (Fig. 2J–K). No expression was found in the neurohypophysis (Fig. 2K).
stathmin 1/ oncoprotein 18 b (stmn1b) was known as an EST. It plays a role in microtubule destabilization. In vertebrates, it is expressed in neurons and in cells with a potential to proliferate. The expression is reduced in terminally differentiated cells (Cassimeris, 2002).
In zebrafish, stmn1b is expressed in the hypothalamus, telencephalon and neural crest at 24h (Fig. 2L). stmn1b is expressed in the telencephalon, hypothalamus and mesencephalon at 48h (Fig. 2M) and 72h (Fig. 2 N,O). Expression in the hypothalamus is near the adenohypophysis. Some, but not all neurohypophysial cells express stmn1b (Fig. 2O).
pleckstrin and Sec7 domain containing 3 like (psd3l) is a novel gene. According to conserved domains, it is a guanine nucleotide exchange factor for small GTPases. The Sec7 domain is the central domain.
psd3l is expressed in the hypothalamus and telencephalon at 24h (Fig. 2P). This expression is maintained through 48h (Fig. 2Q) and 72h (Fig. 2R,S). We found no expression in the neurohypophysis (Fig. 2S).
serpin peptidase inhibitor, clade I (neuroserpin), member 1 (serpini1) is a novel gene. It is a member of the serine proteinase inhibitor (serpin) gene family. It was shown in other species that it reacts preferentially with plasminogen activator in the brain. It is released from neurons and is involved in synaptic plasticity (Yepes and Lawrence, 2004).
serpini1 was found in the pituitary library, however we detected no expression in the adenohypophysis during the first 5d of development. Expression in the hypothalamus is visible from 24h (Fig. 2T). At 48h (Fig. 2U) and 72h (Fig. 2V,W), we observed additional expression in telencephalon and midbrain (Fig. 2U–W). Expression in the hypothalamus is close to the adenohypophysis but not in the neurohypophysis (Fig. 2W).
1.2.3. uo:ion003 is expressed in both adenohypophysis and hypothalamus
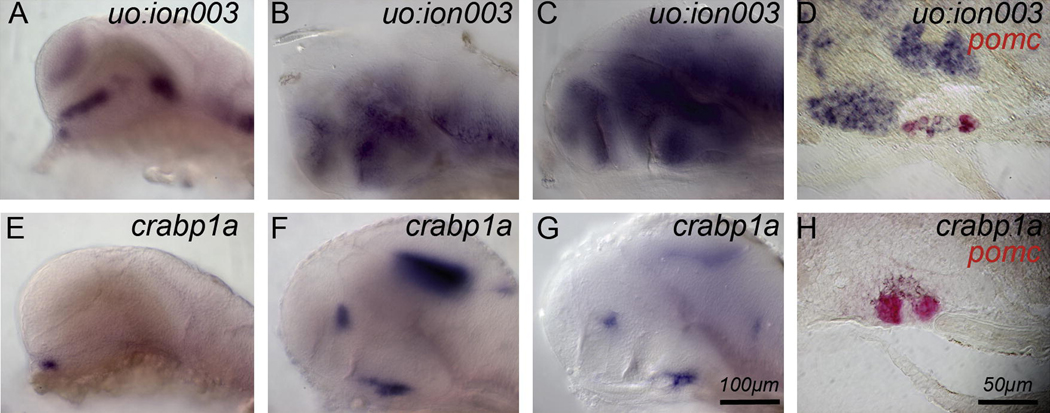
Genes represented in the subtractive libraries were expected to be expressed exclusively in the hypothalamus or in the pituitary. However, one gene, uo:ion003 (from the pituitary library), was expressed both in adenohypophysis and hypothalamus during the first 5d of development and in adult brain (Fig. 3 and 4). uo:ion003 is a novel gene. No putative conserved domains were detected. At 24h, uo:ion003 is expressed in both hypothalamus and adenohypophysis (Fig. 3A). This expression persists through 48h (Fig. 3B) and 72h (Fig. 3C–D and Fig. 1F). In addition, uo:ion003 is expressed in the telencephalon, midbrain and hindbrain at all stages. In the adenohypophysis, uo:ion003 is expressed in the posterior pars distalis (Fig. 1F and Fig. 3D), but not in the area that corresponds to the neurohypophysis (Fig. 3D).
Fig. 3. Genes expressed in hypothalamus and adenohypophysis and in neurohypophysis.
(A–D) uo:ion003 is expressed in telencephalon, diencephalon, and mesencephalon between 24h–72h. (D) The section shows that hypothalamic cells and cells in the posterior pars distalis (APD and PI are demarcated by pomc, red) express uo:ion003. (E–H) crabp1a is expressed in the presumptive neurohypophysis. crabp1a is expressed close to the adenohypophysis from 24h to 72h (E–H) and in the telencephalon and tectum at the later stages. Sagittal sections through the pituitary (pomc, red) show crabp1a expression dorsal to the adenohypophysis in neurohypophysial cells (L). Side views, anterior to the left. (A,E) 24h; (B,F) 48h; (C,G) 72h; scale bar is 100 µm. (D,H) 15 µm sagittal sections, anterior to the left, dorsal up, through the pituitary and adjacent hypothalamus at 72h; scale bar: 50 µm.
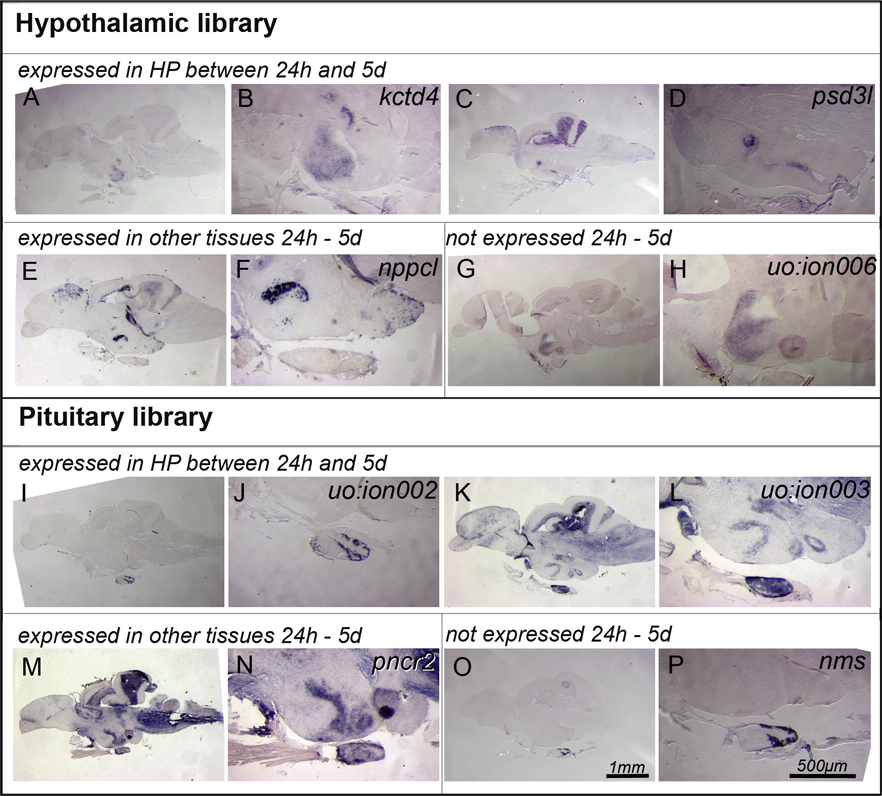
Fig. 4. Genes expressed in adult hypothalamic-pituitary axis.
(A–H) Genes isolated in the Hypothalamic library expressed in the adult. (A–D) kctd4 and psd3l were expressed in adult telencephalon and hypothalamus corresponding to the expression domains we found in 1–5 day old larvae (Fig. 2 E–H, 2 P–S). (E,F) In contrast, nppcl was not expressed in the hypothalamus between 1d and 5d; however, we found expression in a distinct set of cells in the adult hypothalamus. (G,H) The novel gene uo :ion006 was not expressed in embryonic and larval tissues at all, however, we detected hypothalamic expression in the adult. (I–P) We made similar observations for the pituitary library: (I–L) genes such as uo :ion002 and uo :ion003 that were expressed in the embryonic larval HP were also found in the adult pituitary (I,J) or hypothalamus and pituitary (K,L). (M–P) pncr2 that was not detected in embryonic and larval HP, or nms that was not detected in embryos and larvae at all, were both detected in adult HP (M,N) and adult pituitary (O,P). (A–P) 15 µm, sagittal sections of the adult brain, anterior left, dorsal up. (A,C,E,G,I,K,M,O,) scale bar 1 mm; (B,D,F,H,J,L,N,P) scale bar 500 µm.
1.2.4. crabp1a is expressed in the neurohypophysis
In teleosts, neurohypophysis and adenohypophysis do not form morphologically distinct lobes. Hypothalamic neurons project and interdigitate with the adenohypophysis. This interdigitation can be observed morphologically and is defined as the neurohypophysis (Chapman et al., 2005). During dissection, it was not possible to separate the neurohypophysis from the hypothalamus or the adenohypophysis. Consequently, neurohypophysial cDNAs contributed to hypothalamic and pituitary cDNA pools and might have been subtracted. However, depending on the location and expression level of particular mRNAs within the neurons contributing to the neurohypophysis (in the cell body, axon, or dendrites), the corresponding cDNAs might have been abundantly represented in either of the cDNA pools.
We identified cellular retinoic acid binding protein 1a (crabp1a) in the pituitary library. Crabp1a belongs to the intracellular lipid-binding protein family that binds fatty acids, retinoids, and steroids. In mammals, Crabp1 function is not fully understood. Based on its expression in sites of ongoing plasticity that respond to retinoic acid, Crabp1 has been proposed to play a role in retinoic acid targeting, metabolism, and presumably also in neurogenesis (Thompson Haskell et al., 2002; Zetterstrom et al., 1999; Zetterstrom et al., 1994; Zhou and Wei, 2001). Two orthologs of crabp1 exist in the zebrafish genome: crabp1a that we identified in the present study and crabp1b (Liu et al., 2005; Sharma et al., 2003).
crabp1a is expressed at 24h in hypothalamic cells, near the adenohypophysis (Fig. 3E), and in the neurohypophysis at 48h (Fig 3F) and 72h (Fig 3G). Saggital sections of the adenohypophysis confirmed that crabp1a is expressed in the neurohypophysis, dorsal to the adenohypophysis (visualized by pomc expression; Fig. 3H). In addition, crabp1a is also expressed in the telencephalon, in the tegmentum, and in the anterior spinal chord from 48h until 4d (Fig. 3F,G).
1.3. Gene expression in adults
Because we subtracted cDNAs from adult hypothalamus and pituitary tissues, genes we identified that were not expressed in the HP during the first 5d of development might presumably be expressed later during development and adulthood. To analyze expression in the adult, we performed PCR on adult hypothalamus and pituitary cDNAs and in situ hybridized mRNA probes for a subset of genes to adult brain.
We performed PCR on adult hypothalamus and adult pituitary cDNAs using primers for 20 genes represented in each library. Primers were designed to amplify a fragment between 350 and 500 bp (not the full length cDNA). In each library, 80–85% of the genes (17 genes from the pituitary library, 16 genes from the hypothalamic library) were amplified from both adult pituitary and hypothalamic cDNA. 15% of the genes (3 genes) were amplified only from the adult pituitary cDNA (genes from the pituitary library) or from the adult hypothalamic cDNA (genes from the hypothalamic library). Only one gene (from the hypothalamic library) was not amplified from either cDNA set. However, we cannot rule out a failure of the PCR or primers used (Suppl. Fig. 22). Apparently, these results are independent of the number of clones represented in the library for each gene. For example, genes that are represented in a large number of clones in the libraries (for example epd in the hypothalamic library or pomc in the pituitary library) were amplified from both sets of cDNAs, whereas genes represented by 1–3 clones were amplified from only one set of cDNA (Suppl. Fig. 22).
At first sight, these results appear inconsistent with the theoretically predicted outcome of the subtraction method, however, they are probably due to a higher sensitivity to gene expression differences than anticipated.. When most of the genes analyzed were detected by PCR from both hypothalamic and pituitary cDNAs, there was almost no identical sequence represented in both libraries (including mitochondrial and ribosomal). Only spermatogenesis associated protein 2 was represented in both hypothalamus and pituitary library, corresponding to 1 sequence in the pituitary library, 2 identical sequences in the hypothalamic library, out of 840 total sequences.
It is possible that hypothalamus and pituitary express different isoforms of the same gene. If this is true, the sequences represented in each library would be specific for the particular isoform. However, no identical genes were represented in both hypothalamic and pituitary library by different sequences. Interestingly, for some of the genes analyzed, we amplified a fragment of different size from hypothalamic cDNAs and from pituitary cDNAs. In some cases, two fragments of different sizes were amplified from one set of cDNAs, when a single fragment was amplified from the other set of cDNAs (Suppl. Fig. 22).
The fact that most of the analyzed genes were amplified by PCR from both sets of cDNAs can also be explained by a difference in the level of expression in the hypothalamus compared to the pituitary. Accordingly, genes represented in each library are specifically expressed or expressed at higher level in one tissue compared to the other. For example, pomc was represented abundantly only in the pituitary library, even though it is expressed in both pituitary and hypothalamus.
Even though the intensity of the amplified fragment obtained by PCR is obvious in some cases, we based our analyses on the presence or absence of amplification, without taking the intensity of the amplified product into account. Quantitative PCR would be the best way to address a difference in gene expression levels between hypothalamus and pituitary. However, because the goal of this study was to identify useful markers for the HP, we instead analyzed the expression of some genes in adult brain by in situ hybridization.
Consistent with a differential expression level, some cDNAs (6 out of 13) amplified from both tissues were detected by in situ hybridization in only one of the tissues. kctd4, psd3l, natriuretic peptide precursor C - like and uo:ion 006 (from the hypothalamic library) expression was found only in hypothalamus and uo:ion002 and neuromedin s (from the pituitary library) was found only in the adenohypophysis (Fig. 4). Three genes (out of 13) were detected by in situ hybridization in both hypothalamus and adenohypophysis (proline-rich nuclear receptor coactivator 2, uo:ion 001 (not shown) and uo:ion 003, from the pituitary library; Fig. 4). The other 4 genes analyzed (interleukin 1 receptor accessory protein-like 2 from hypothalamic library; adrenomedullin 2, serpini1 and uo:ion005 from the pituitary library) had no expression detectable by in situ hybridization.
Our subtractive library analysis from adult hypothalamus and pituitary identified genes expressed in the embryonic, larval, and adult HP and serves as the basis for a more extended analysis of HP development. Specifically, we will use these genes to analyze development, maturation, and function of the HP.
2. Experimental procedures
2.1. Tissue Dissection and Library Construction
We dissected 50 hypothalami and 179 pituitaries from adult zebrafish (see Suppl. Fig. 23) and isolated total RNA from each tissue. We synthesized and amplified cDNA from 1.5 µg of total RNA using the BD SMART™ PCR cDNA Synthesis Kit (BD Biosciences). The subtraction of pituitary cDNAs from hypothalamic cDNAs, and vice versa, was performed according to the Clontech PCR-Select™ cDNA Subtraction Kit (Clontech) protocol. We cloned the subtracted pituitary and hypothalamic cDNAs into pCRII-TOPO TA vector (Invitrogen) and stored the libraries as glycerol stocks of individual bacterial clones.
2.2. Clone identification and analysis
We screened the libraries by sequencing each clone and analyzing the resulting sequences by comparison against nucleotide and protein databases at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) and against the Sanger Institute zebrafish genome database (Zebrafish Genome Annotation Browser v5 and v6, http://vega.sanger.ac.uk/Danio_rerio/).
After sequence analysis of the first 100 clones, we identified the most redundant sequences: ependymin was the most redundant clone in the hypothalamic library and pomc was the most redundant clone in the pituitary library. To eliminate additional copies of these clones before further sequencing of the libraries, we identified pomc/ependymin positive clones by membrane hybridization.
Glycerol stocks were spotted onto nylon membranes, DNA was UV cross-linked to membranes and hybridized with ependymin and pomc DNA digoxygenin probes (synthesized with PCR dig probe synthesis kit, Roche). Positive clones were detected using Alkaline Phosphatase mediated NBT/BCIP color reaction.
2.3. Expression patterns
The expression patterns of genes identified in the screen were determined between Prim-5 (24h) stage and 5d. We either consulted the ZFIN expression database or analyzed whole-mount embryos or larvae by mRNA in situ hybridization is the size of the fragment allowed (~350bp). We also confirmed whether adrenomedullin 2, kctd4, interleukin 1 receptor accessory protein-like 2, psd3l, nppcl, pncr2, serpini1, nms, uo:ion 001, uo:ion002, uo:ion003, uo:ion005 and uo:ion006 were expressed in adult HP (Fig. 4).
Embryos were fixed and hybridized with one or two mRNA probes according to previously described protocols (Hauptmann and Gerster, 1994; Varga et al., 1999). We used antisense mRNA probes for the gene fragment contained in each library clone and used pomc as a reference for the adenohypophysis. When doubly labeling embryos, we used NBT/BCIP to detect one mRNA probe and Fast Red to detect the other probe. For more detailed morphological analysis, several embryos were cryosectioned at a thickness of 15µm.
Supplementary Material
Acknowledgments
We thank the University of Oregon DNA sequencing facility and the Institute of Neuroscience fish facility for husbandry. We thank Sofie Seibel for technical assistance during construction of the libraries. We thank Christine and Bernard Thisse and the ZFIN database curators for providing gene expression information online. Supported by the Deutsche Forschungsgemeinschaft SFB592 (AG5) and NIH DC04186 and HD22486. S.T. was supported by a fellowship from the “Fonds à la Recherche dans l’Industrie et dans l’Agriculture“ (F.R.I.A.). M.M. is a "Chercheur Qualifié du F.N.R.S".
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bentley PJ. Comparative Vertebrate Endocrinology. Cambridge University Press; 1998. [Google Scholar]
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Sawitzke AL, Campbell DS, Schoenwolf GC. A three-dimensional atlas of pituitary gland development in the zebrafish. J Comp Neurol. 2005;487:428–440. doi: 10.1002/cne.20568. [DOI] [PubMed] [Google Scholar]
- Colbran RJ, Brown AM. Calcium/calmodulin-dependent protein kinase II and synaptic plasticity. Curr Opin Neurobiol. 2004;14:318–327. doi: 10.1016/j.conb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Covassin L, Amigo JD, Suzuki K, Teplyuk V, Straubhaar J, Lawson ND. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol. 2006;299:551–562. doi: 10.1016/j.ydbio.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Rosenfeld MG. Signaling and transcriptional mechanisms in pituitary development. Annu Rev Neurosci. 2001;24:327–355. doi: 10.1146/annurev.neuro.24.1.327. [DOI] [PubMed] [Google Scholar]
- Dores RM, Lecaude S, Bauer D, Danielson PB. Analyzing the evolution of the opioid/orphanin gene family. Mass Spectrom Rev. 2002;21:220–243. doi: 10.1002/mas.10029. [DOI] [PubMed] [Google Scholar]
- Engelmann M, Landgraf R, Wotjak CT. The hypothalamic-neurohypophysial system regulates the hypothalamic-pituitary-adrenal axis under stress: an old concept revisited. Front Neuroendocrinol. 2004;25:132–149. doi: 10.1016/j.yfrne.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Gage PJ, Brinkmeier ML, Scarlett LM, Knapp LT, Camper SA, Mahon KA. The Ames dwarf gene, df, is required early in pituitary ontogeny for the extinction of Rpx transcription and initiation of lineage-specific cell proliferation. Mol Endocrinol. 1996;10:1570–1581. doi: 10.1210/mend.10.12.8961267. [DOI] [PubMed] [Google Scholar]
- Gracia-Navarro F, Castano JP, Malagon MM, Sanchez-Hormigo A, Luque RM, Hickey GJ, Peinado JR, Delgado E, Martinez-Fuentes AJ. Research progress in the stimulatory inputs regulating growth hormone (GH) secretion. Comp Biochem Physiol B Biochem Mol Biol. 2002;132:141–150. doi: 10.1016/s1096-4959(01)00544-9. [DOI] [PubMed] [Google Scholar]
- Griffith LC. Calcium/calmodulin-dependent protein kinase II: an unforgettable kinase. J Neurosci. 2004;24:8391–8393. doi: 10.1523/JNEUROSCI.2888-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Herzog W, Zeng X, Lele Z, Sonntag C, Ting JW, Chang CY, Hammerschmidt M. Adenohypophysis formation in the zebrafish and its dependence on sonic hedgehog. Dev Biol. 2003;254:36–49. doi: 10.1016/s0012-1606(02)00124-0. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Jansen EJ, Martens GJ. Secretogranin III is a sulfated protein undergoing proteolytic processing in the regulated secretory pathway. J Biol Chem. 1996;271:17755–17760. doi: 10.1074/jbc.271.30.17755. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Martens GJ. The neuroendocrine proteins secretogranin II and III are regionally conserved and coordinately expressed with proopiomelanocortin in Xenopus intermediate pituitary. J Neurochem. 1996;66:2248–2256. doi: 10.1046/j.1471-4159.1996.66062248.x. [DOI] [PubMed] [Google Scholar]
- Hudmon A, Schulman H. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu Rev Biochem. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- Lassiter CS, Linney E. Embryonic expression and steroid regulation of brain aromatase cyp19a1b in zebrafish (Danio rerio) Zebrafish. 2007;4:49–57. doi: 10.1089/zeb.2006.9995. [DOI] [PubMed] [Google Scholar]
- Lin Chang C, Roh J, Park JI, Klein C, Cushman N, Haberberger RV, Hsu SY. Intermedin functions as a pituitary paracrine factor regulating prolactin release. Mol Endocrinol. 2005;19:2824–2838. doi: 10.1210/me.2004-0191. [DOI] [PubMed] [Google Scholar]
- Liu RZ, Sharma MK, Sun Q, Thisse C, Thisse B, Denovan-Wright EM, Wright JM. Retention of the duplicated cellular retinoic acid-binding protein 1 genes (crabp1a and crabp1b) in the zebrafish genome by subfunctionalization of tissue-specific expression. Febs J. 2005;272:3561–3571. doi: 10.1111/j.1742-4658.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- Lopez M, Nica G, Motte P, Martial JA, Hammerschmidt M, Muller M. Expression of the somatolactin beta gene during zebrafish embryonic development. Gene Expr Patterns. 2005;6:156–161. doi: 10.1016/j.modgep.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, Jiang L, Culler MD, Coy DH. Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun. 1989;164:567–574. doi: 10.1016/0006-291x(89)91757-9. [DOI] [PubMed] [Google Scholar]
- Miyata A, Jiang L, Dahl RD, Kitada C, Kubo K, Fujino M, Minamino N, Arimura A. Isolation of a neuropeptide corresponding to the N-terminal 27 residues of the pituitary adenylate cyclase activating polypeptide with 38 residues (PACAP38) Biochem Biophys Res Commun. 1990;170:643–648. doi: 10.1016/0006-291x(90)92140-u. [DOI] [PubMed] [Google Scholar]
- Nica G, Herzog W, Sonntag C, Hammerschmidt M. Zebrafish pit1 mutants lack three pituitary cell types and develop severe dwarfism. Mol Endocrinol. 2004;18:1196–1209. doi: 10.1210/me.2003-0377. [DOI] [PubMed] [Google Scholar]
- Okada R, Yamamoto K, Ito Y, Chartrel N, Leprince J, Fournier A, Vaudry H, Kikuyama S. Effects of pituitary adenylate cyclase-activating polypeptide, vasoactive intestinal polypeptide, and somatostatin on the release of thyrotropin from the bullfrog pituitary. Ann N Y Acad Sci. 2006;1070:474–480. doi: 10.1196/annals.1317.064. [DOI] [PubMed] [Google Scholar]
- Petersen B, Buchfelder M, Fahlbusch R, Adams EF. Pituitary adenylate cyclase-activating polypeptide directly stimulates LH and FSH secretion by human pituitary gonadotrophinomas. Exp Clin Endocrinol Diabetes. 1996;104:250–255. doi: 10.1055/s-0029-1211450. [DOI] [PubMed] [Google Scholar]
- Pogoda HM, Hammerschmidt M. Molecular genetics of pituitary development in zebrafish. Semin Cell Dev Biol. 2007;18:543–558. doi: 10.1016/j.semcdb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Pogoda HM, von der Hardt S, Herzog W, Kramer C, Schwarz H, Hammerschmidt M. The proneural gene ascl1a is required for endocrine differentiation and cell survival in the zebrafish adenohypophysis. Development. 2006;133:1079–1089. doi: 10.1242/dev.02296. [DOI] [PubMed] [Google Scholar]
- Rawlings SR, Hezareh M. Pituitary adenylate cyclase-activating polypeptide (PACAP) and PACAP/vasoactive intestinal polypeptide receptors: actions on the anterior pituitary gland. Endocr Rev. 1996;17:4–29. doi: 10.1210/edrv-17-1-4. [DOI] [PubMed] [Google Scholar]
- Schmidt R. Cell-adhesion molecules in memory formation. Behav Brain Res. 1995;66:65–72. doi: 10.1016/0166-4328(94)00126-z. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Brysch W, Rother S, Schlingensiepen KH. Inhibition of memory consolidation after active avoidance conditioning by antisense intervention with ependymin gene expression. J Neurochem. 1995;65:1465–1471. doi: 10.1046/j.1471-4159.1995.65041465.x. [DOI] [PubMed] [Google Scholar]
- Sharma MK, Denovan-Wright EM, Boudreau ME, Wright JM. A cellular retinoic acid-binding protein from zebrafish (Danio rerio): cDNA sequence, phylogenetic analysis, mRNA expression, and gene linkage mapping. Gene. 2003;311:119–128. doi: 10.1016/s0378-1119(03)00580-8. [DOI] [PubMed] [Google Scholar]
- Shashoua VE. Ependymin, a brain extracellular glycoprotein, and CNS plasticity. Ann N Y Acad Sci. 1991;627:94–114. doi: 10.1111/j.1749-6632.1991.tb25916.x. [DOI] [PubMed] [Google Scholar]
- Thompson Haskell G, Maynard TM, Shatzmiller RA, Lamantia AS. Retinoic acid signaling at sites of plasticity in the mature central nervous system. J Comp Neurol. 2002;452:228–241. doi: 10.1002/cne.10369. [DOI] [PubMed] [Google Scholar]
- Unger JL, Glasgow E. Expression of isotocin-neurophysin mRNA in developing zebrafish. Gene Expr Patterns. 2003;3:105–108. doi: 10.1016/s1567-133x(02)00064-9. [DOI] [PubMed] [Google Scholar]
- Varga ZM, Wegner J, Westerfield M. Anterior movement of ventral diencephalic precursors separates the primordial eye field in the neural plate and requires cyclops. Development. 1999;126:5533–5546. doi: 10.1242/dev.126.24.5533. [DOI] [PubMed] [Google Scholar]
- Wong AO, Li WS, Lee EK, Leung MY, Tse LY, Chow BK, Lin HR, Chang JP. Pituitary adenylate cyclase activating polypeptide as a novel hypophysiotropic factor in fish. Biochem Cell Biol. 2000;78:329–343. [PubMed] [Google Scholar]
- Yepes M, Lawrence DA. Neuroserpin: a selective inhibitor of tissue-type plasminogen activator in the central nervous system. Thromb Haemost. 2004;91:457–464. doi: 10.1160/TH03-12-0766. [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Lindqvist E, Mata de Urquiza A, Tomac A, Eriksson U, Perlmann T, Olson L. Role of retinoids in the CNS: differential expression of retinoid binding proteins and receptors and evidence for presence of retinoic acid. Eur J Neurosci. 1999;11:407–416. doi: 10.1046/j.1460-9568.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Simon A, Giacobini MM, Eriksson U, Olson L. Localization of cellular retinoid-binding proteins suggests specific roles for retinoids in the adult central nervous system. Neuroscience. 1994;62:899–918. doi: 10.1016/0306-4522(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Wei LN. Expression of cellular retinoic acid-binding protein I is specific to neurons in adult transgenic mouse brain. Brain Res Gene Expr Patterns. 2001;1:67–72. doi: 10.1016/s1567-133x(01)00010-2. [DOI] [PubMed] [Google Scholar]
- Zhou H, Jiang Y, Ko WK, Li W, Wong AO. Paracrine regulation of growth hormone gene expression by gonadotrophin release in grass carp pituitary cells: functional implications, molecular mechanisms and signal transduction. J Mol Endocrinol. 2005;34:415–432. doi: 10.1677/jme.1.01629. [DOI] [PubMed] [Google Scholar]
- Zhu X, Lin CR, Prefontaine GG, Tollkuhn J, Rosenfeld MG. Genetic control of pituitary development and hypopituitarism. Curr Opin Genet Dev. 2005;15:332–340. doi: 10.1016/j.gde.2005.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.