Abstract
Exponentially growing biological and bioinformatics data sets present a challenge and an opportunity for researchers to contribute to the understanding of the genetic basis of phenotypes. Due to breakthroughs in microarray technology, it is possible to simultaneously monitor the expressions of thousands of genes, and it is imperative that researchers have access to the clinical data to understand the genetics and proteomics of the diseased tissue. This technology could be a landmark in personalized medicine, which will provide storage for clinical and genetic data in electronic health records (EHRs). In this paper, we explore the computational and ethical challenges that emanate from the intersection of bioinformatics and healthcare informatics research. We describe the current situation of the EHR and its capabilities to store clinical and genetic data and then discuss the Genetic Information Nondiscrimination Act. Finally, we posit that the synergy obtained from the collaborative efforts between the genomics, clinical, and healthcare disciplines has potential to enhance and promote faster and more advanced breakthroughs in healthcare.
Key words: bioinformatics, healthcare informatics, data mining, electronic health records, Genetic Information Nondiscrimination Act, privacy
Introduction
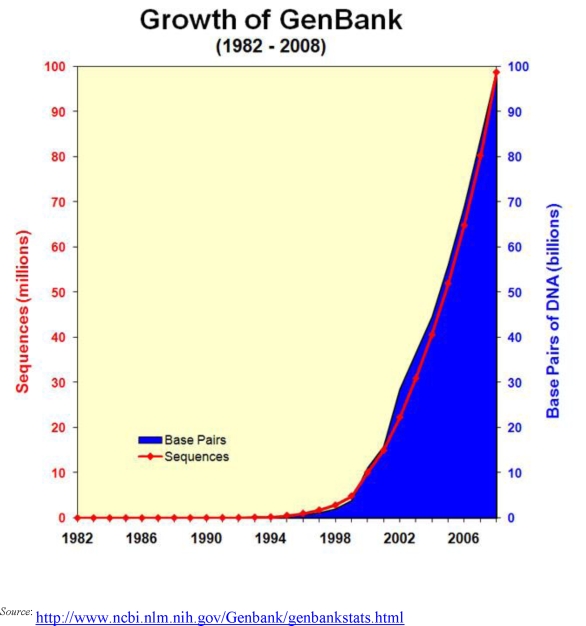
The completion of Human Genome Project in 2003 brought together biologists, statisticians, and computer scientists, collectively known as bioinformaticians, to analyze the genomic data and to further pharmacogenetic research. Because of this collaboration, researchers began to better understand the genes associated with certain diseases and discover their complex biological processes.1 The interdisciplinary area of bioinformatics, which involves managing, analyzing, and interpreting information from biological sequences and structures, has opened doors for sophisticated technology that continues to support the automation and miniaturization of modern instruments that bear large-scale biomedical data. The advancement in technology has eventually led to exponential growth in the DNA and protein sequence databases. The three major publicly available databases that serve as central repositories for DNA and protein data are GenBank, maintained by the National Center for Biotechnology Information (NCBI), DNA Databank of Japan, and European Molecular Biology Laboratory (EMBL), maintained by European Bioinformatics Institute. Each year, the number of sequences submitted to these databases grows. Figure 1 shows the exponential growth of the GenBank database. The GenBank release notes for release 162.0 state that between 1982 and 2007 the size of GenBank doubled roughly every 18 months.2 A similar phenomenon was observed with the growth in the number of transistors in personal computer chips in 1965 (Moore's Law).3
Figure 1.
Growth of GenBank
As biology and medicine move from bench-based to computer-based science, models replace some experiments and complement others. Advancing biomedical research requires an overlap of genomic and clinical research. The assimilation of information at the molecular, cellular, tissue, organ, and personal level will not only enhance the collaborative research agenda but also lead to the development of innovative tools and technologies for enhanced patient care. The aim of translational bioinformatics is to utilize computational tools and techniques for the analysis of large biological databases and to fully comprehend disease mechanisms by not only understanding the genetics and the proteomics but also by associating them with the clinical data. Translational bioinformatics is defined by American Medical Informatics Association (AMIA) as “the development of storage, analytic, and interpretive methods to optimize the transformation of increasingly voluminous biomedical data into proactive, predictive, preventative, and participatory health.”4 As computational capabilities and resources continue to develop, the use of “informatics” by the biomedical community is increasing. One landmark in this area is the development of an electronic health record (EHR) that provides a data-rich repository and the potential to discover genotype-phenotype associations, thereby assisting clinicians in the use of genomic results for better patient care. However, to realize this aim there are hurdles consisting of computational demands associated with analyzing the data and societal and ethical issues that cause public outcry about genetic discrimination. The revolution in bioinformatics not only brought challenges for statisticians, computer scientists, biologists, and allied health informatics professionals but also has impacted the field of health information management (HIM), in which practitioners have developed ethical and policy-based solutions for the protection of genetic data in EHRs. The Genetic Information Nondiscrimination Act (GINA) became effective in May 2009 and governs the collection, use, and disclosure of genetic information by health plans and insurers. A triad model proposed by Kesh and Raghupathi outlines the roles and responsibilities of computer scientists, molecular biologists, and the general public that arise due to the growth in the field of bioinformatics.5 In this article, our purpose is to explore the current trends and challenges faced by computer scientists and HIM professionals who analyze and manage genetic data. We describe the pros and cons of GINA. We also describe the current situation of the EHR and its capabilities to store clinical and genetic data. We conclude that the synergy obtained from collaborative efforts between the genomics and the clinical and healthcare disciplines will eventually enhance the healthcare agenda of the nation and the personal well-being of its citizens.
Computational Challenges Associated with Genetic Data
The main objective of bioinformatics is to increase the understanding of complex biological processes. Statistical and data mining and machine-learning approaches apply computationally intensive approaches for the comprehension of intricate biological details. These approaches are important to bioinformatics, which is data rich but lacks molecular-level direction in the characterization of diseases.6 Since data mining approaches help to develop precise, accurate, and functionally robust analyses of gene expression data, they have a possible impact on drug design and other emerging problems in “omics” technology. This particular area of biomedical data and knowledge will be explored in more depth than the other areas given the emphasis of this paper. The fundamental challenge for genomics is to determine how gene variations are linked to a certain disease and, on a broader perspective, to determine how the interactions of genes vary with environment and lifestyle.
The breakthrough use of microarray technology has made it possible to monitor the expressions of thousands of genes at the same time. By studying such a large number of genes at once, it is possible to study the effect of certain diseases, treatments, or changes in the expression profile over a period of time. Consequently, this technology has allowed researchers to obtain a “global” view of the cell for the first time.7 Although researchers can use microarray technology to potentially study the behavior of thousands of genes simultaneously, microarray technology has led to an exponential growth in the amount of data generated. The computational complexity of analyzing microarray data is further enhanced because a large number of genes can correspond to different time sequences or tissue types, having dimensionality that is several orders of magnitude more than the evaluated samples. Due to the enormous amount of data produced, the extraction of biologically significant knowledge from the genes and their protein counterparts poses a growing computational challenge. Hence, specialized dimensionality reduction methods are needed to reduce the number of attributes (genes) and, consequently, to avoid the infamous “curse of dimensionality.”8 The implications of such research in translational medicine efforts is significant, and the specific computational challenges are mining low abundance proteins as they are most likely to be the biologically relevant markers of a diseased state and integrating proteomics with genomic data to give researchers a better picture of the cellular state.
The major computational techniques required for the analysis of biomedical data include but are not limited to the following:
- Feature selection or dimensionality reduction: These techniques select a set of relevant genes (which behave as discriminatory features) that are most strongly related to a particular class of sample.9–11
- Supervised classification: Challenges in microarray mining include predicting the class label of the unknown sample on the basis of its gene expression profile.12–14 An example in proteomics is the functional classification of proteins for the prediction of cellular function.15
- Unsupervised clustering: This technique reveals groups of genes or conditions based on gene expression patterns or rapid structural/topological clustering of proteins.16–19
- Prediction analysis: This technique predicts the three-dimensional structure of a protein from its amino acid sequence.20
- Associative pattern mining: This refers to finding associations between genes that are similar in behavior, such as genes that are up-regulated or down-regulated throughout the identified conditions.21–23
- Text mining and natural language processing (NLP): These approaches extract important biological relationships in retrieving associated documents. These techniques provide insight into the disease being studied and further enhance the research.24
The object of bioinformatics is to design and deliver novel methodologies and tools that track and predict the future course of a disease based on its genetic mutations and protein interactions. While progress has been achieved in the genomic-level data mining methods, their penetration in healthcare records and clinical benefits has hardly been realized. A comprehensive reform agenda is needed to make that intersection possible. For data mining to reach its potential impact in healthcare and clinical records, stakeholders must agree on standards and approaches for the utilization of these data.
The Current Scenario of Genetic Data and EHRs
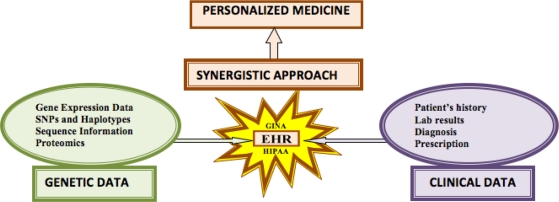
Over the last few decades, life-threatening diseases, such as cancer, and the escalating cost of drug development have combined to increase patient suffering. The growing healthcare burden can be significantly reduced by the design and development of novel methods in translational bioinformatics and allied health information-driven sciences. The advances in genomics and the growing impact of genetics in healthcare are already impacting the medical world. The inclusion of genetic information in EHRs has the potential to provide patients with valuable risk assessment based on their family history and genetic profile, and to carve a niche for personalized medicine. Figure 2 shows a framework representing a synergistic approach toward personalized medicine. The unique combination and availability of genotype-phenotype data has provided opportunities for medical informatics researchers to explore the feasibility of using such data for genetic research.25 For example, the Electronic Medical Records and Genomics (eMERGE) project funded by the National Institutes of Health (NIH) is a five-institution network in the United States that develops a consortium of biorepositories and links them to EHRs for high-throughput genome-wide association analysis.26 Another project launched by the Department of Veterans Affairs links patients' genetic information with the medical history stored in their EHRs.27 The collaboration between Mayo Clinic and IBM is yet another step forward to harness the potential of linking the genetic and clinical data.28
Figure 2.
Framework for a Synergistic Approach toward Personalized Medicine
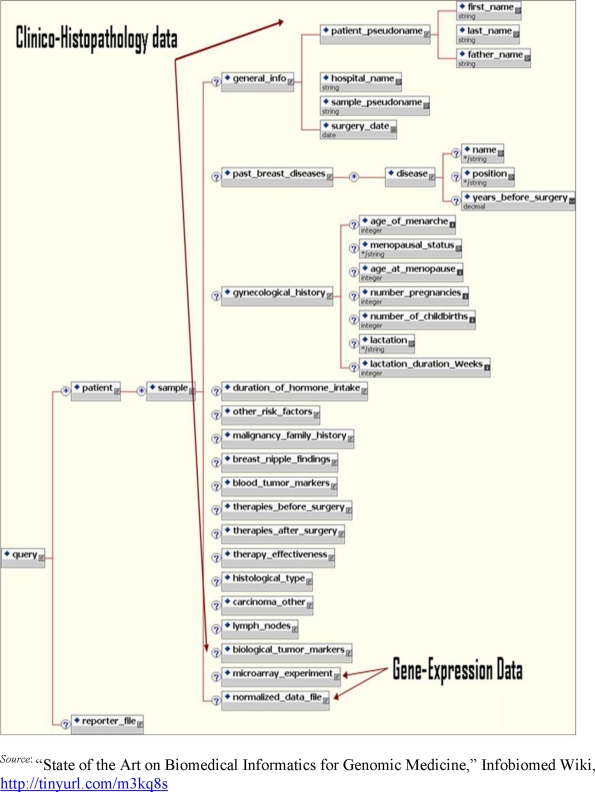
New data models and structures that can integrate genomic data in EHRs as well as interface mechanisms that can link genotype-phenotype data are needed to realize the benefits of new genome-based technologies for the benefits of patients. Biomedical ontologies such as SNOMED (Systematized Nomenclature of Medicine) for clinical information and GO (Gene Ontologies) for genomics are available. With the advancement in efforts for the integration of genomic and clinical data, it is imperative to have semantic interoperability for mapping between the two ontologies. Further, the Clinical Genomics special interest group at Health Level Seven proposed specifications for linking the individual's genomic data with the individual's clinical data.29 However reasonable it may seem to include genetic data in the EHR, our current health information systems are not yet ready to handle genomic data. Figure 3 shows a schematic representation of the clinical and genetic data that can be stored in an EHR.30 The questions of how to retain the genetic and clinical data together with proper standards for interoperability have not yet been answered. In the white paper that emanated from the conference “Synergy between Research in Medical Informatics, Bioinformatics and Neuroinformatics” organized by the Belgian presidency of the European Union, the authors outline four key areas where the challenges for the interoperability standards for genetic and clinical data need to be addressed:31
- Data and knowledge (structure, representation, terminology, etc.)
- Technique (architecture, hardware, topology)
- Presentation of data and knowledge
- Security for systems, health care professionals, and patients
Figure 3.
Schematic Representation of Clinical and Genetic Data Stored in an EHR
“Best practices” can be achieved when all four of the above standards are objectively met in a consistent and timely manner.
Ethical Challenges Associated with Genetic Data
In recent years, discrimination based on genetic information has become a major concern for patients and physicians. The privacy of genetic data is more sensitive than the privacy of clinical data, as it involves discrimination not only to the individual but also to relatives who have not been genetically tested. Further, predictions about disease conditions or medical risks, including the risk of genetic discrimination, can be made for an individual who has been genetically tested. A National Library of Medicine handbook states that genetic testing can cost from $100 to $2,000 or more depending upon the nature and scope of the test.32 Given the likelihood that genetic tests will become more affordable in the future, there is an increased chance that more people will obtain their genetic profile, and the issues related to the privacy of genetic data cannot be ignored.
Recent court cases have emerged as evidence of the challenges and risks associated with the collection and use of genetic information. Modern courts have recognized the sensitive nature of genetic information, and their recent decisions reflect a perceived need for additional protection of this type of information. Unauthorized access to medical records has been classified as unlawful invasion of privacy. Additionally, in 2000, state legislatures introduced more than 100 bills related to use and misuse of genetic information. Various workgroups, committees, and institutes have studied the prevalence of discrimination and have made recommendations, which culminated with the congressional passage of GINA.
Genetic Information Nondiscrimination Act
When GINA was signed into law on May 21, 2008, it became the legal standard for the collection, use, and disclosure of genetic information. Considering new developments in the availability of genetic information and the use of treatment, healthcare professionals will now be faced with managing this data according to applicable laws. All new health plans taking effect on or after May 21, 2009, must comply with the guidelines set forth in GINA. Although GINA specifically addresses genetic information, it is vital for HIM professionals to be aware of how this legislation amends current laws, including HIPAA.33
In order to prohibit discrimination based on genetic information, Title I of GINA sets forth strict requirements regarding the use of such information in making decisions about health insurance coverage. Specifically, health plans and group health insurers are prohibited from using genetic information to adjust premium amounts for a group covered under a plan, from requiring or requesting that a family member or individual take a genetic test, and from requiring, requesting, or purchasing genetic information for underwriting purposes or with respect to any individual before the individual enrolls in the plan.34 In addition, GINA changes the term “Protected Health Information” (PHI) coined by HIPAA to now include genetic information. Consequently, genetic information now becomes PHI and must be protected accordingly.
In order to determine what qualifies as genetic information according to GINA, it is necessary to analyze the term. GINA defines genetic information as “information about (i) such individual's genetic tests, (ii) the genetic tests of family members of such individual, and (iii) the manifestation of a disease or disorder in family members of such individual.” Also included is “any request for, or receipt of, genetic services, or participation in clinical research which includes genetic services, by such individual or any family member of such individual.” Furthermore, any genetic information relative to a fetus carried by a pregnant woman or to an embryo legally held by an individual or family member using assisted reproductive technology is also considered protected. “Genetic tests” include an analysis of human DNA, RNA, chromosomes, proteins, or metabolites that detects genotypes, mutations, or chromosomal changes. “Genetic services” includes genetic counseling or genetic education.35
Health plan providers and insurers will continue to have a legitimate need for the minimum information necessary for payment of health care claims, including claims for genetic tests. The documents required will vary depending on the type of claim; however, it can be assumed that genetic information may be included for payment of claims related to testing, treatment, or prophylactic procedures. In these instances, the provided information will be protected as PHI and subject to the same regulations. Patients' family medical histories are traditionally collected during annual wellness exams as well as during enrollment prescreening physicals. Considering the broad definition of “genetic information,” collecting family history may present a risk to health insurers. Family history information obtained regarding a patient currently enrolled in a wellness program is also specifically protected and may not be used for underwriting purposes.36
Title II of GINA applies to all employers, employment agencies, labor organizations, and training programs. Healthcare professionals collecting employment information should be mindful of the potential impacts on their business practices. GINA specifically prohibits discriminating against employees based on genetic information in hiring, termination, referral, or other decisions regarding compensation, terms, conditions, or privileges of employment. If an employer possesses any genetic information in an employee file, that information must be kept separate and treated as a confidential medical record. Title II of GINA becomes effective November 21,2009.37
Since GINA has amended HIPAA to include genetic information, the penalties for noncompliance with HIPAA regulations now include misuse of genetic information. Further complicating the law structure, the American Recovery and Reinvestment Act (ARRA) of 2009 has significantly increased the penalties assessed for a violation of HIPAA regulations. Those changes will also be applied to GINA violations, creating a significant risk for healthcare entities who fail to change policies and procedures accordingly. Currently, civil monetary penalties for HIPAA violations equal $100 per violation, up to a maximum amount of $25,000 for multiple violations in the same year. After ARRA regulations are finalized, the penalties will increase to a range of $1,000 to $50,000 per violation, with a cap of $25,000 to $1.5 million per year for violations of an identical requirement during the same calendar year.38 Finally, ARRA grants a portion of the penalty assessed to the individuals harmed by the violation. In order to stay current with changes to HIPAA, laws such as GINA and ARRA must be examined and applied.39
Health plans are currently required to revise their policies and procedures as well as their Notice of Privacy Practices documents to reflect these changes. Business associate agreements may also need to be reviewed in order to indicate compliance with the guidelines and limitations established by GINA. Managing genetic information will be a priority for HIM professionals as well. Health information protection in the EHR environment will become more challenging as genetic information becomes more readily available and further protected. First, HIM professionals must identify sources of genetic information in the patient record. These sources could range from documentation provided in the family history portion of the history and physical to laboratory results and others. All sources must then be closely monitored throughout the release of information to ensure proper protection. Perhaps steps similar to those currently taken to protect HIV/AIDS status in records could be employed to ensure appropriate safeguard of genetic status or history.
Legislatures and researchers have voiced criticism of GINA due to its strict limitations on genetic information. With these limitations, many wonder if enough information will be available to continue progressive research and make further advancements in this field. Still fresh in the minds of researchers is the nightmare effects HIPAA had on research and the costs associated with compliance. When HIPAA went into effect, researchers were faced with strict authorization requirements and incurred costs associated with obtaining consent for many research subjects.40 Concerns linger regarding whether GINA will have the same effect on genetic research. The executive vice president for governmental affairs of the U.S. Chamber of Commerce officially opposed GINA, citing fears of frivolous litigation and the inconsistent nature of the current law infrastructure. He further expressed concerns regarding the confusing patchwork of state and federal laws and the burdensome overhaul of medical privacy laws before GINA was signed into law.41
Although fairly comprehensive, loopholes exist in the enforcement and application of GINA regulations. Some have suggested that amending HIPAA to include genetic information as PHI does little to ensure the privacy of that information, given the relative few instances of actual penalties handed out for noncompliance with HIPAA. To date, the number of HIPAA civil penalties pale in comparison to the number of claims, making enforcement seem lax. In addition, life, disability, and long-term care insurances are excluded from GINA, therefore making it acceptable to deny any or all of those types of insurance policies to individuals with preexisting genetic conditions or family histories.42 With those specific exceptions, patients may remain hesitant to obtain genetic testing due to fear of discrimination in those areas.
Although privacy and confidentiality are the major concerns associated with GINA, healthcare professionals should also consider other ethical dilemmas that may arise with respect to disclosing genetic information. Availability of genetic information may present physicians with an opportunity as well as a conundrum. Patients' genetic disposition may reveal a great deal about not only about their physical illnesses, characteristics, and risks but also those of their relatives and potential offspring. If a dangerous, life-threatening disease is diagnosed as a result of genetic testing, should the physician be expected to withhold that information from others who may be affected (such as the patient's immediate family or children) simply to maintain privacy? If a genetic disorder is found, will the physician be held liable for not warning of the chance of passing that disorder on to children? Does a duty exist between the physician and the patient's family? These questions may require solutions in the form of court decisions, thus leaving interpretation to our judicial system. Furthermore, physicians will be confronted with the issue of withholding genetic information from a patient who may be mentally unstable if that information could pose a risk to the patient. While justified normally, this type of withholding could affect not only the patient but third parties as well. Although some of these ethical challenges have been present as long as genetic information has been available, now those challenges must be balanced with the strict privacy laws handed down.
Conclusions
In this article, we have attempted to highlight the successes, challenges, and limitations related to the current and future status of the integration of genetic and clinical data.
We summarize our key conclusions as follows:
- Significant strides have already been achieved in genomic- and proteomic-level data mining methods. These accomplishments have promoted the understanding of the genes associated with certain diseases and the identification of new target drugs.
- A number of international and national workgroups and committees are promoting the inclusion of genetic information in EHRs. These efforts will have a broad impact on personalized medicine.
- Congress has passed GINA to protect citizens against discrimination based on their genetic information.
- The adoption of sound privacy and security policies is imperative to protect the personalized nature of genomic data. The current incarnation of GINA has loopholes that may bar citizens from obtaining their genetic tests and consequently affect biomedical research.
- Standards that enable interoperability standards among different formats of health records for genetic and clinical data are needed to realize the benefits of new genome-based technologies.
- A collaborative research agenda for translational bioinformatics and healthcare informatics should be prioritized. Such a synergistic approach will promote faster and more advanced breakthroughs in medicine and healthcare.
The growth in genetic research has carved the way for the futuristic vision of personalized medicine, which uses a patient's genetic makeup to help to determine the right medicine for the patient. Approximately 10 percent of pharmacy labels already contain information on how the drug will respond to an individual's genetic variation, and this situation has represented a large portion of the discussion about personalized medicine.43 As the field of systems biology moves forward with personalized medicine, researchers will be able to apply tools and technology to understand the disease mechanism that progresses from the molecular, cellular, tissue, and organ levels to the personal and, finally, the population levels. President Obama recently introduced the first comprehensive legislation designed to advance progress toward personalized medicine.44 The American Health Information Community (AHIC) recommends that the federal government include genetic information in EHRs and research databases.45 However, the challenge on hand is to maintain the privacy and security of patient genetic data and to simultaneously promote research for enhanced patient care. Special emphasis needs to be given to the protection of genetic data, which can lead to potential discrimination against not only an individual but his or her family as well.
Advancing the field of translational bioinformatics will require collaboration between the genomics, clinical, and healthcare disciplines. The past director of the NIH initiated the NIH Roadmap, which outlines the directions for the future of biomedical research and calls for a synergistic approach between NIH research domains that cross boundaries.46 Involving genetic data in addition to clinical data for medical decision making will lead to a paradigm shift toward evidence-based research—a realization for which scientists are hoping. The scope of EHRs should be extended from storing clinical data to storing genetic data. Further, the architecture should be robust enough to extract information from patient-centric to population-centric facilities, including knowledge discovery processes and the potential for evidence-based research for advanced epidemiological research.47 This synergistic approach will promote faster and more advanced breakthroughs in medicine and healthcare, making the United States the healthiest nation in the world.
Acknowledgments
This research was made possible by National Institutes of Health Grant Number P20 RR16456 from the INBRE Program of the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Contributor Information
Prerna Sethi, Department of Health Information Management and an adjunct assistant professor of research in Biological Sciences at Louisiana Tech University in Ruston, LA..
Kimberly Theodos, Department of Health Information Management at Louisiana Tech University in Ruston, LA..
Notes
- 1.Muller U.R, Nicolau D.V, editors. Microarray Technology and Its Applications. Berlin: Springer; 2004. pp. 361–374. [Google Scholar]
- 2.GenBank release notes. Available at http://www.ncbi.nlm.nih.gov/Genbank/
- 3.Moore Gordon E. “Cramming More Components onto Integrated Circuits.”. Electronics Magazine. 1965;38(no. 8):114–117. [Google Scholar]
- 4.American Medical Informatics Association. AMIA Strategic Plan 2006 Available at http://www.amia.org/inside/stratplan
- 5.Kesh S, Raghupathi W. “Critical Issues in Bioinformatics and Computing.”. Perspectives in Health Information Management. 2004;1(no. 9) [PMC free article] [PubMed] [Google Scholar]
- 6.Kuonen D. “Challenges in Bioinformatics for Statistical Data Miners.”. Bulletin of the Swiss Statistical Society. 2003;46:10–17. [Google Scholar]
- 7.Piatetsky-Shapiro G, Tamayo P. “Microarray Data Mining: Facing the Challenges.”. AC M SIGKDD Explorations Newsletter. 2003;5(no. 2) [Google Scholar]
- 8.Sethi P. “Gene Selection through Association Rule Filtering for Supervised Classification.”. Pr oceedings of the Biotechnology and Bioinformatics Symposium. 2008;5:65–72. [Google Scholar]
- 9.Sethi P. “Gene Selection through Association Rule Filtering for Supervised Classification.”. Pr oceedings of the Biotechnology and Bioinformatics Symposium. 2008;5:65–72. [Google Scholar]
- 10.Voulgaris Z, Magoulas G.D. “Dimensionality Reduction for Feature and Pattern Selection in Classification Problems.”. ICCGI Proceedings of the Third International Multi-Conference on Computing in the Global Information Technology. 2008:160–65. [Google Scholar]
- 11.Ding C, Peng H. “Minimum Redundancy Feature Selection from Microarray Gene Expression Data.”. Journal of Bioinformatics and Computational Biology. 2005;3(2):185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
- 12.Golub T.R, et al. “Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring.”. Science. 1999;286(5439):531–37. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Dor A, et al. “Tissue Classification with Gene Expression Profiles.”. Journal of Computation Biology. 2000;7:559–83. doi: 10.1089/106652700750050943. [DOI] [PubMed] [Google Scholar]
- 14.Califano A, Stolovitzky Gustavo, Tu Yuhai. “Analysis of Gene Expression Microarrays for Phenotype Classification.”. Proceedings of the International Conference on Intelligent Systems for Molecular Biology. 2000;8:75–85. [PubMed] [Google Scholar]
- 15.Brun C, et al. “Functional Classification of Proteins for the Prediction of Cellular Function from a Protein-Protein Interaction Network.”. Genome Biology. 2003;5(no. 1):R6. doi: 10.1186/gb-2003-5-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen M.B, Spellman Paul T, Brown Patrick O, Botstein David. “Cluster Analysis and Display of Genome-Wide Expression Patterns.”. Proceedings of the National Academy of Sciences. 1998;95:14863–68. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeung K.Y, Ruzzo W.L. “Principal Component Analysis for Clustering Gene Expression Data.”. Bioinformatics. 2001;17(9):763–74. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Dor A, Shamir Ron, Yakhini Zohar. “Clustering Gene Expression Patterns.”. Comp utational Biology. 1999;6(nos. 3–4):281–97. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- 19.Mulheran P.A. “Mechanisms and Dynamics of Protein Clustering on a Solid Surface.”. Phys ical Review Letters. 2002;100(no. 6) doi: 10.1103/PhysRevLett.100.068102. [DOI] [PubMed] [Google Scholar]
- 20.Moult J. “Predicting Protein Three-Dimensional Structure.”. Current Opinion in Biotechnology. 1999;10(no. 6) doi: 10.1016/s0958-1669(99)00037-3. [DOI] [PubMed] [Google Scholar]
- 21.Creighton C, Hanash S. “Mining Gene Expression Databases for Association Rules.”. Bioinformatics. 2003;19(1):79–86. doi: 10.1093/bioinformatics/19.1.79. [DOI] [PubMed] [Google Scholar]
- 22.Becquet C, Blachon Sylvian, Jeudy Baptiste, Boulicaut Jean-Francois, Gandrillon Olivier. “Strong-Association-Rule Mining for Large-Scale Gene-Expression Data Analysis: A Case Study on Human SAGE Data.”. Genome Biology. 2002;3(no. 12) doi: 10.1186/gb-2002-3-12-research0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmona-Saez P, Chagoyen Monica, Rodriguez Andres, Trelles Oswaldo, Carazo Jose M, Pascual-Montano Alberto. “Integrated Analysis of Gene Expression by Association Rules Discovery.”. BMC Bioinformatics. 2006;7(no. 54) doi: 10.1186/1471-2105-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen Aaron M, William R. Hersh. “A Survey of Current Work in Biomedical Text Mining.”. Briefings in Bioinformatics. 2005;6(1):57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 25.Council for Responsible Genetics. Genetic Testing, Discrimination & Privacy Available at http://www.genewatch.org/
- 26.The eMERGE Network. Available at https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/in dex.php/Main_Page
- 27.U.S. Department of Veterans Affairs. “Studies Under Way in VA to Advance Genomic Medicine.” April 20, 2008. Available at http://www.research.va.gov/news/research_highlights/genomics-042008.cfm
- 28.Mayo Clinic. “Mayo Clinic, IBM Aim to Drive Medical Breakthroughs.” Available at http://www.mayoclinic.org/feature-articles/mayoibmcollaboration.html
- 29.Health Level Seven HL7 Clinical Genomics Special Interest Group. 2005. Available at http://www.hl7.org/Special/committees/clingenomics
- 30.Infobiomed Wiki. “State of the Art on Biomedical Informatics for Genomic Medicine.” Available at http://tinyurl.com/m3kq8s
- 31.Martin-Sanchez F, et al. “Synergy between Medical Informatics and Bioinformatics: Facilitating Genomic Medicine for Future Healthcare.”. Journal of Biomedical Informatics. 2004;37(1):30–42. doi: 10.1016/j.jbi.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 32.U.S. National Library of Medicine. Genetics Home Reference. Available at http://ghr.nlm.nih.gov/handbook/testing/costresults
- 33.U.S. Congress. The Genetic Information Nondiscrimination Act of 2008. H.R.493.ENR. Available at http://www.govtrack.us/congress/bill.xpd?bill=h110-493
- 34.U.S. Congress. The Genetic Information Nondiscrimination Act of 2008. H.R.493.ENR. Available at http://www.govtrack.us/congress/bill.xpd?bill=h110-493
- 35.U.S. Congress. The Genetic Information Nondiscrimination Act of 2008. H.R.493.ENR. Available at http://www.govtrack.us/congress/bill.xpd?bill=h110-493
- 36.Baruch S., J. Gruber, and K. Pollitz. GINA Q&A Transcript, Genetics & Public Policy Center Transcript of Webinar hosted by Genetic Alliance. Available at http://www.dnapolicy.org/resources/MoreQandAforGINAwebsite01.09.pdf
- 37.U.S. Congress. The Genetic Information Nondiscrimination Act of 2008.
- 38.Stamer C.M, et al. “Genetic Information Nondiscrimination Act of 2008.”. ABA HEALTH eSOURCE. 2008;4(no. 10) [Google Scholar]
- 39.Boardman Law Firm Reading Room. “FYI: HIPAA Privacy—GINA and Stimulus Law Changes.” Available at http://www.boardmanlawfirm.com/fyi/02_27_09.php
- 40.Norrgard K. “Protecting Your Genetic Identity: GINA and HIPAA.”. Nature Education. 2008;1(no. 1) [Google Scholar]
- 41.U.S. Chamber of Commerce. “Letter Opposing H.R. 493, the ‘Genetic Information Nondiscrimination Act of 2007.’” March 27, 2007. Available at http://www.uschamber.com/issues/letters/2007/070327_genetic_info.htm
- 42.McClain, B. “Genetic Information Nondiscrimination Act: Will It Protect You?” May 22, 2008. Available at http://cpab.info/Documents/Gina_bam_5-22-08.htm
- 43.Frueh F.W, et al. “Pharmacogenomic Biomarker Information in Drug Labels Approved by the United States Food and Drug Administration: Prevalence of Related Drug Use.”. Pharmacoth erapy. 2008;28(8):992–98. doi: 10.1592/phco.28.8.992. [DOI] [PubMed] [Google Scholar]
- 44.National e-health Collaborative. Laying the Foundation for a Secure, Interoperable, Nationwide Health Information Network Executive Summary. Available at http://www.nationalehealth.org/WorkArea/DownloadAsset.aspx?id=226 [DOI] [PubMed]
- 45.Glaser J, Henley D.E, Downing G, Brinner K.M. “Advancing Personalized Health Care through Health Information Technology: An Update from the American Health Information Community's Personalized Health Care Workgroup.”. Journal of the American Medical Informatics Association. 2008;15(4):391–96. doi: 10.1197/jamia.M2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.National Institutes of Health. NIH Roadmap for Medical Research Available at http://nihroa dmap.nih.gov/
- 47.Potamias George. “Mining Distributed and Heterogeneous Clinical Data Sources.”. ERCIM News. 2005;60:33–35. [Google Scholar]