SUMMARY
Inhibition of BCR-ABL by imatinib induces durable responses in many patients with chronic myeloid leukemia (CML), but resistance attributable to kinase domain mutations can lead to relapse and a switch to second-line therapy with nilotinib or dasatinib. Despite three approved therapeutic options, the cross-resistant BCR-ABLT315I mutation and compound mutants selected on sequential inhibitor therapy remain major clinical challenges. We report design and pre-clinical evaluation of AP24534, a potent, orally available multi-targeted kinase inhibitor active against T315I and other BCR-ABL mutants. AP24534 inhibited all tested BCR-ABL mutants in cellular and biochemical assays, suppressed BCR-ABLT315I-driven tumor growth in mice, and completely abrogated resistance in cell-based mutagenesis screens. Our work supports clinical evaluation of AP24534 as a pan-BCR-ABL inhibitor for treatment of CML.
Keywords: dasatinib, nilotinib, imatinib resistance, compound mutation
INTRODUCTION
The judicious use of tyrosine kinase inhibitors that target BCR-ABL constitutes an effective strategy for sustained disease control in chronic myeloid leukemia (CML). The exemplar of targeted therapy in CML is the BCR-ABL inhibitor imatinib (Gleevec; STI571), a safe and effective first-line therapy for most patients diagnosed with chronic phase disease (Druker et al., 2006). Although most patients attain a durable complete cytogenetic response, minimal residual disease persists in nearly all patients, and active disease recurs if treatment is discontinued. More importantly, discontinuation of imatinib due to intolerance or resistance is necessary in up to 30% of patients within the first five years on therapy. Also, durable responses are uncommon in patients with advanced CML or Philadelphia chromosome (Ph)-positive acute lymphoblastic leukemia (ALL).
Resistance to imatinib usually involves point mutations in the kinase domain of BCR-ABL that impair inhibitor binding. A broad spectrum of kinase domain mutations that confer resistance to the drug have been reported (Hughes et al., 2006). Clinically, identification of a BCR-ABL kinase domain mutation provides a potential explanation for imatinib resistance and suggests a clear treatment strategy: second-line therapy with an ABL kinase inhibitor active against the particular BCR-ABL mutant present in the patient (Jabbour et al., 2009). To date, two ABL kinase inhibitors have achieved regulatory approval for second-line use: the imatinib family member nilotinib (Tasigna; AMN107) and the multi-targeted kinase inhibitor dasatinib (Sprycel; BMS-354825) (Shah et al., 2004; Weisberg et al., 2005). With the availability of these three oral BCR-ABL inhibitors, most patients are successfully matched to an appropriate and effective drug, leading to retained or recaptured response. However, several kinase domain mutations confer high-level resistance to one or more of these therapies, in particular the BCR-ABLT315I mutation, which confers resistance to all three (reviewed by (O’Hare et al., 2007)).
Given the location of the T315 residue in the gatekeeper region of the ATP-binding site, the T315I mutant has proven difficult to inhibit with ATP mimetics. Modeling analysis indicates that the mutation eliminates a critical hydrogen bonding interaction required for high-affinity binding of imatinib, nilotinib, and dasatinib and alters the topology of the ATP-binding pocket (Tokarski et al., 2006; Weisberg et al., 2005). Although several reports have described approaches to overcome this, compound-to-clinic progress has been slow (reviewed by (Quintas-Cardama and Cortes, 2008)). Several ATP competitive inhibitors originally designed to target the Aurora kinase family have been found to be active against ABLT315I, including MK-0457, PHA-739358, AT9283, and XL-228 (Quintas-Cardama and Cortes, 2008). These molecules have been formulated for intravenous administration in the clinic, and MK-0457 has shown some activity as salvage therapy for advanced phase CML patients harboring the T315I mutation (Giles et al., 2007), but clinical development has been halted due to toxicity concerns.
The importance of controlling mutation-mediated resistance is underscored by recent reports on the potential for sequential ABL kinase inhibitor therapy to select for compound (multiple mutations in same BCR-ABL allele) mutants resistant to all current ABL inhibitors, including some that do not involve T315I (Shah et al., 2007). Therefore, an optimal next-generation ABL inhibitor capable of exerting a high level of disease control in CML would incorporate potent activity against BCR-ABLT315I and the full range of BCR-ABL kinase domain mutations as well as the native (unmutated) enzyme, while matching the pharmacologic advantages of the currently approved therapies. Here we report on the design and pre-clinical testing of AP24534, an orally active pan-inhibitor of BCR-ABL, including BCR-ABLT315I.
RESULTS
Design of AP24534 and Crystallographic Analysis of AP24534:ABLT315I
Recent X-ray crystallographic studies on the ABL kinase domain reveal that the threonine to isoleucine gatekeeper mutation, T315I, acts as a simple point mutant without significant perturbation of the overall protein structure (Zhou et al., 2007). Thus, as imatinib, nilotinib, and dasatinib each form a hydrogen bond with the side chain of T315 in native ABL, we designed ligands devoid of this interaction by introducing vinyl and ethyl linkages into a purine-based inhibitor scaffold (Wang et al., 2008) targeting both DFG-in (active) and DFG-out (inactive) binding modes. One DFG-out targeted compound also inhibited ABLT315I in biochemical and cellular assays (M. Azam, personal communication; (Huang et al., 2009)). Subsequent structure-guided design experiments led to AP24534 (Figure 1A), which accommodates the T315I side chain by virtue of a carbon-carbon triple bond (ethynyl) linkage.
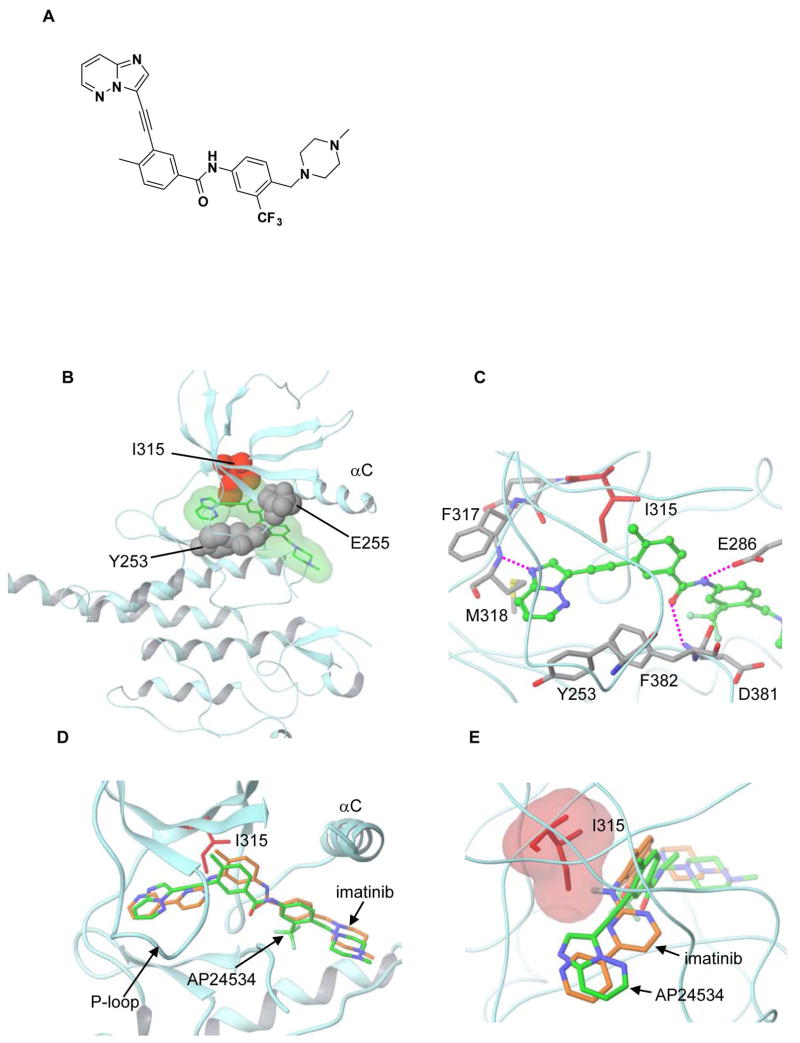
Figure 1. Chemical Structure of AP24534 and Co-Crystal Structure with ABLT315I.
(A) Chemical structure of AP24534.
(B) Crystal structure of AP24534 in complex with the ABLT315I mutant kinase. AP24534 is shown in green with translucent molecular surface. The side chain of the mutated gatekeeper residue Ile315 is shown in red. The side chains of Y253 and E255, locations of point mutations appearing in the resistant outgrowth screen of AP24534 are shown in grey. The C-helix is labeled (αC).
(C) Key interactions of AP24534 with ABLT315I at the ATP binding site. Hydrogen bonds are highlighted with pink dashed lines. Residues making critical contact with the imidazo[1,2b]pyridazine core and the ethynyl linker group of AP24534 are also labeled.
(D) and (E) Superposition of imatinib and AP24534 highlighting the effect of the Thr to Ile mutation. The superposition was based on the C positions of ABL residues 312–321 in the T315I mutant and in native ABL kinase complexed with imatinib (shown in brown; PDB code 1IEP).
X-ray crystallographic analysis of AP24534 in complex with the murine ABLT315I kinase domain confirmed that AP24534 binds in the DFG-out mode (Figure 1B and 1C) and maintains a network of protein contacts similar to imatinib (Figure 1D and 1E). Specifically, the imidazo[1,2b]pyridazine core of AP24534 occupies the adenine pocket of the enzyme, the methylphenyl group occupies the hydrophobic pocket behind the gatekeeper residue, the trifluoromethylphenyl group binds tightly to the pocket induced by the DFG-out conformation of the protein, and the ethynyl linkage of AP24534 makes favorable van der Waals interactions with the I315 mutated residue. A total of five hydrogen bonds are made between the inhibitor and the protein, including: with the backbone of M318 in the hinge region, with the backbone of D381, and with the side chain of E286 (Figure 1C), as well as two hydrogen bonds from the methylpiperazine group (not shown). The P-loop of the kinase is collapsed in this conformation, bringing Y253 into van der Waals contact with AP24534. Additional favorable contacts are made between the inhibitor and F382 of the DFG motif, displaced outwards into the ligand-binding site in the DFG-out mode. Although the methylphenyl groups occupying the hydrophobic pocket and hinge hydrogen bonding moieties of AP24534 and imatinib are placed similarly (Figure 1D), superposition of the two inhibitors shows AP24534 engaging in productive van der Waals interactions with I315, while steric clash between imatinib and the I315 side chain is evident (Figure 1E).
AP24534 Inhibits the Catalytic Activity of ABLT315I
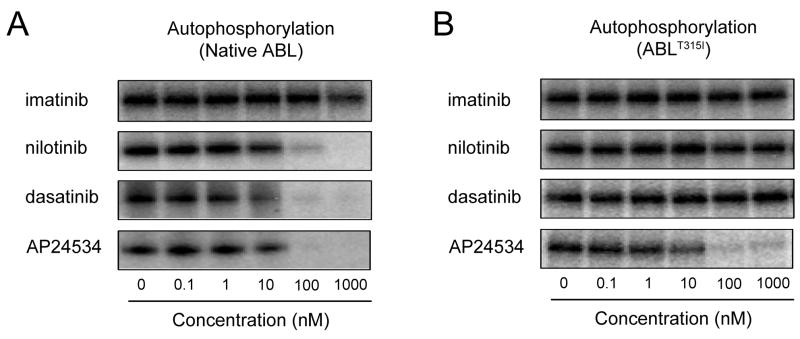
We tested the activity of AP24534, imatinib, nilotinib, and dasatinib in biochemical assays with purified, dephosphorylated, native ABL and ABLT315I. All inhibitors diminished the enzymatic activity of native ABL, but only AP24534 was effective against the ABLT315I mutant (Figure 2). Similar potent inhibition by AP24534 was observed for additional imatinib-resistant ABL mutants tested, including ABLG250E, ABLY253F, and ABLE255K (not shown), establishing that AP24534 directly targets native and mutant ABL kinase, including ABLT315I.
Figure 2. AP24534 Inhibits the Autophosphorylation of ABL and ABLT315I.
In vitro [γ-32P]-ATP autophosphorylation of full-length (A) ABL or (B) ABLT315I kinase treated with AP24534, imatinib, nilotinib, or dasatinib. After incubation of tyrosine-dephosphorylated enzyme with the indicated inhibitor in the presence of [γ-32P]-ATP and separation by SDS-PAGE, signal intensity was measured by autoradiography.
Kinase Selectivity Profile of AP24534
The in vitro potency and selectivity of AP24534 was assessed in kinase assays with multiple recombinant kinase domains and peptide substrates (Table 1 and Table S1). AP24534 potently inhibited native ABL (IC50: 0.37 nM), ABLT315I (IC50: 2.0 nM), and other clinically important ABL kinase domain mutants (IC50: 0.30–0.44 nM) (Table 1). AP24534 also inhibited SRC (IC50: 5.4 nM) and members of the VEGFR, FGFR, and PDGFR families of receptor tyrosine kinases (Table 1 and Table S1). AP24534 did not inhibit Aurora kinase family members, nor did it inhibit insulin receptor or cyclin-dependent kinase 2 (CDK2)/Cyclin E (IC50 > 1000-fold relative to native ABL).
Table 1.
Kinase Inibition Profile of AP24534 for Native ABL, ABLT315I, and Selected Kinases
| Kinase | IC50 (nM) |
|---|---|
| ABL | 0.37 |
| ABLT315I | 2.0 |
| ABLQ252H | 0.44 |
| ABLY253 F | 0.30 |
| ABLM351T | 0.30 |
| ABLH396P | 0.34 |
| c-SRC | 5.4 |
| LYN | 0.24 |
| c-KIT | 12.5 |
| VEGFR2 | 1.5 |
| FGFR1 | 2.2 |
| PDGFRα | 1.1 |
| IR | >1000 |
| IGF-1R | >1000 |
| Aurora A | >1000 |
| CDK2/Cyclin E | >1000 |
AP24534 Inhibits the Growth of Cells Expressing Native or Mutant BCR-ABL
Cellular proliferation assays were performed with parental Ba/F3 cells and Ba/F3 cells expressing native BCR-ABL or BCR-ABL with a range of single mutations in the kinase domain. AP24534 potently inhibited proliferation of Ba/F3 cells expressing native BCR-ABL (IC50: 0.5 nM). All BCR-ABL mutants tested remained sensitive to AP24534 (IC50: 0.5–36 nM; Table 2) including BCR-ABLT315I (IC50: 11 nM). Annexin V staining confirmed that inhibition of proliferation by AP24534 correlated with induction of apoptosis (not shown). Growth of parental Ba/F3 cells was inhibited only at significantly higher IC50 (1713 nM), indicating a substantial differential selectivity for inhibition of BCR-ABL-positive cells. Ba/F3 BCR-ABLT315I cells grown in the presence of IL-3 exhibited an IC50 (1804 nM) similar to that of parental Ba/F3 cells.
Table 2.
AP24534 IC50 Values for Ba/F3 Cellular Proliferation Assays and CML and Non-CML Cellular Proliferation Assays
| Cell lines | AP24534 IC50 (nM) |
|---|---|
| Ba/F3 cells | |
| Native BCR-ABL | 0.5 |
| M244V | 2.2 |
| G250E | 4.1 |
| Q252H | 2.2 |
| Y253F | 2.8 |
| Y253H | 6.2 |
| E255K | 14 |
| E255V | 36 |
| T315A | 1.6 |
| T315I | 11 |
| F317L | 1.1 |
| F317V | 10 |
| M351T | 1.5 |
| F359V | 10 |
| H396P | 1.1 |
| Parental | 1713 |
| T315I + IL-3 | 1804 |
| CML leukemia cells | |
| K562 | 3.9 |
| KY01 | 0.4 |
| LAMA | 0.3 |
| Non-CML leukemia cells | |
| Marimo | 2215 |
| HEL | 2522 |
| 1CMK | 1652 |
We also tested AP24534 against BCR-ABL-positive and -negative cell lines derived from leukemic patients. While we observed potent growth inhibition of K562, KY01, and LAMA cells (derived from CML patients in blast crisis), there was no significant activity against three BCR-ABL-negative leukemia cell lines (Table 2).
AP24534 Inhibits BCR-ABL-Mediated Signaling in Cells Expressing BCR-ABLT315I
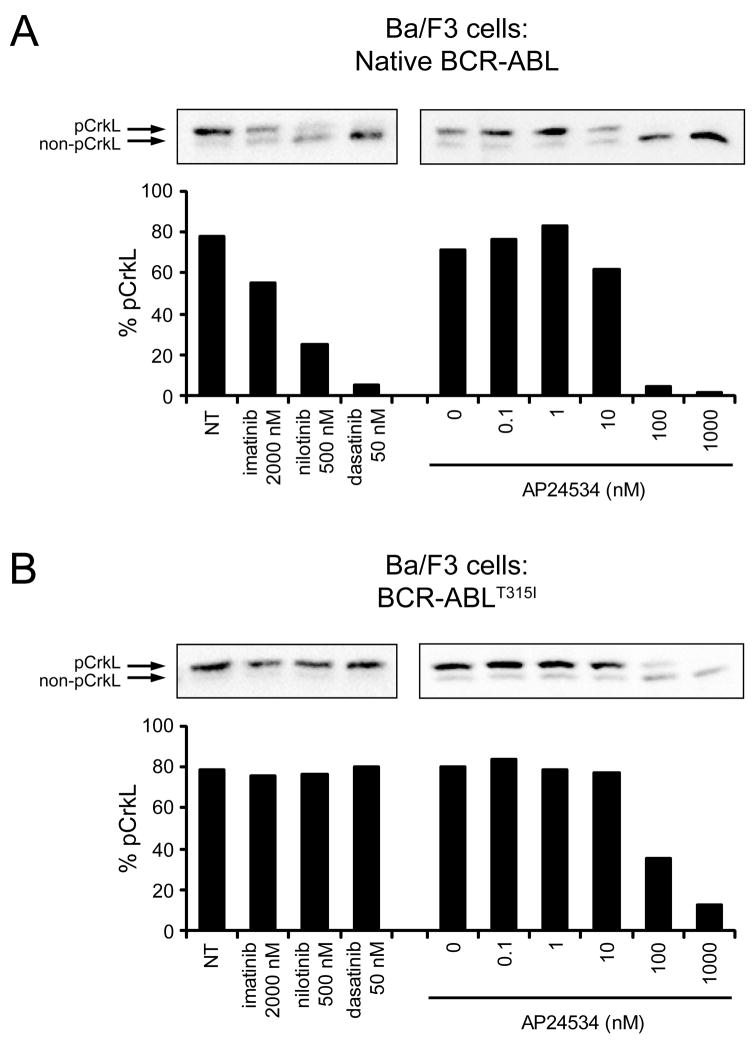
To confirm target inhibition in Ba/F3 cells expressing native BCR-ABL or BCR-ABLT315I, we examined the effect of AP24534 on the tyrosine phosphorylation status of BCR-ABL (Figure S1) and the direct BCR-ABL substrate CrkL (Figure 3), with the three approved ABL inhibitors included for comparison. Monitoring CrkL tyrosine phosphorylation status as a surrogate for BCR-ABL kinase activity has been the preferred pharmacodynamic assay in clinical trials of BCR-ABL inhibitors (Druker et al., 2001; Talpaz et al., 2006). In the CrkL gel shift assay, the percentage of tyrosine-phosphorylated CrkL (upper band) decreases in response to inhibition of BCR-ABL. While all tested inhibitors were effective against Ba/F3 cells expressing native BCR-ABL (Figure 3A), only AP24534 demonstrated activity against the T315I mutant (Figure 3B). Inhibition of BCR-ABL phosphorylation was observed in parallel experiments (Figure S1).
Figure 3. AP24534 Inhibits BCR-ABL Signaling in CML Cell Lines Expressing Native BCR-ABL or BCR-ABLT315I.
(A) Immunoblot analysis of CrkL phosphorylation in Ba/F3 cells expressing native BCR-ABL treated with imatinib, nilotinib, dasatinib, or AP24534. Cells were cultured for 4 hr with inhibitors, harvested, lysed, and analyzed by immunoblot using an antibody for CrkL, a substrate of BCR-ABL whose phosphorylation is an established clinical marker of BCR-ABL kinase activity. Both the phosphorylated and non-phosphorylated forms are resolved by electrophoretic mobility, and bands are quantitated by densitometry and expressed as a % phosphorylated CrkL. (B) Immunoblot analysis of CrkL phosphorylation in Ba/F3 BCR-ABLT315I cells treated with imatinib, nilotinib, dasatinib, or AP24534. Assays and analysis were carried out as described above in panel (A). Abbreviations: NT, no treatment.
Treatment of CML Primary Cells with AP24534 Inhibits Cellular Proliferation
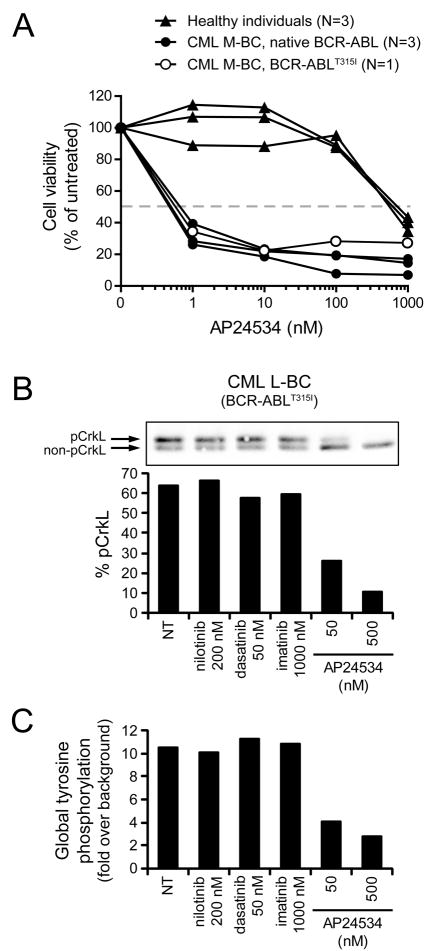
To assess the efficacy of AP24534 on primary cells from patients with BCR-ABL-driven leukemia, we exposed mononuclear cells derived from blood or bone marrow from CML myeloid blast crisis patients harboring native BCR-ABL or BCR-ABLT315I and from healthy individuals to graded concentrations of AP24534 and assayed viable cells after 72 hr. Consistent with biochemical and cell line viability data, AP24534 induced a selective reduction of viable cell numbers in primary CML cells, with IC50 values approximately 500-fold lower than those observed with normal cells (Figure 4A). Neither imatinib nor dasatinib reached an IC50 (highest concentration: 1000 nM) in primary CML BCR-ABLT315I cells (data not shown).
Figure 4. Ex Vivo Treatment of CML Primary Cells with AP24534 Inhibits Cellular Proliferation and BCR-ABL-Mediated Signaling.
(A) Cellular proliferation assays for ex vivo AP24534-treated mononuclear cells from CML myeloid blast crisis (M-BC) patients harboring native BCR-ABL (N=3) or BCR-ABLT315I (N=1) and from healthy individuals (N=3). For reference, the dashed line indicates 50% cell viability relative to untreated cells.
(B) Immunoblot analysis of CrkL phosphorylation in mononuclear cells from a CML lymphoid blast crisis (L-BC) patient harboring BCR-ABLT315I following ex vivo exposure to AP24534, imatinib, nilotinib, or dasatinib. Cells were cultured overnight with inhibitors, harvested, lysed, and analyzed by CrkL immunoblot. Both the phosphorylated and non-phosphorylated forms are resolved by electrophoretic mobility, and bands are quantitated by densitometry and expressed as a % phosphorylated CrkL.
(C) FACS analysis of global tyrosine phosphorylation in mononuclear cells from the CML L-BC BCR-ABLT315I patient in panel (B). After overnight culture with inhibitors, cells were fixed and permeabilized, incubated with a FITC-labeled antibody for phosphorylated tyrosine, and analyzed by FACS. Values reported are as fold increase in mean fluorescence intensity relative to unstained controls. Abbreviations: NT, no treatment.
AP24534 Inhibits BCR-ABLT315I Kinase Activity and Colony Formation in Primary CML Cells
To monitor target inhibition following ex vivo exposure to AP24534 of mononuclear cells obtained from a CML T315I lymphoid blast crisis patient, we carried out an assay similar to that described for Ba/F3 cell lines, wherein cells were incubated with inhibitors and then analyzed for CrkL phosphorylation by immunoblot. Exposure to AP24534 resulted in a reduction in phosphorylated CrkL signal while none of the other ABL inhibitors had an effect (Figure 4B); similar results were obtained upon analysis for global tyrosine phosphorylation by flow cytometry (Figure 4C).
We also evaluated the efficacy of AP24534 in myeloid colony formation assays using mononuclear cells from a CML T315I accelerated phase patient and from a healthy individual. Whereas neither nilotinib nor dasatinib showed an effect against patient-derived T315I cells, AP24534 inhibited the formation of colonies in a concentration-dependent manner (Figure S2A) and exhibited no toxicity to normal hematopoietic cells at concentrations below 500 nM (Figure S2B), consistent with cellular proliferation assay data obtained using normal cells (Figure 4A).
Oral AP24534 Prolongs Survival and Reduces Tumor Burden in Mice with BCR-ABLT315I-Dependent Disease
To examine the pharmacologic properties of AP24534, mice were administered a single oral dose and plasma concentrations were then measured at multiple time points. In mice administered a dose of 2.5 mg/kg, mean plasma levels of 90, 58, and 2 nM were achieved at 2, 6, and 24 hr post-dose, respectively. At a dose of 30 mg/kg, mean plasma levels reached 782, 561, and 8 nM at the same time points. These results demonstrate that plasma levels exceeding the in vitro IC50 values for all tested BCR-ABL mutants can be sustained in mice for >6 hr at oral doses that are well tolerated, indicating that adequate target inhibition for a therapeutic effect should be reached (Shah et al., 2008).
We next evaluated the efficacy of AP24534 in a survival model in which Ba/F3 cells expressing native BCR-ABL were injected intravenously. As shown in Figure 5A (left), the median survival time for vehicle-treated mice was 19 days. Daily oral treatment with 2.5 or 5 mg/kg AP24534 for 19 days prolonged median survival to 27.5 and 30 days, respectively (p<0.01 for both dose levels). These results were comparable to those achieved following daily oral administration of 5 mg/kg dasatinib (a regimen previously reported to be efficacious (Lombardo et al., 2004)), in which median survival was 27 days (p<0.01).
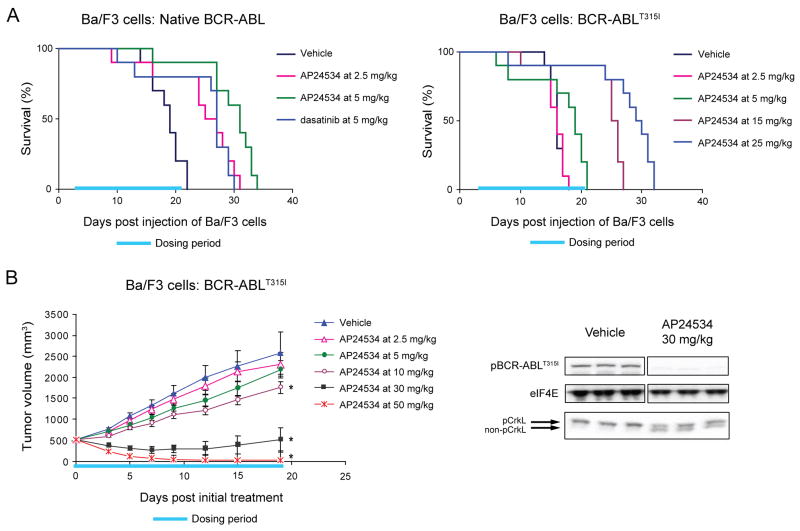
Figure 5. AP24534 is Effective in Mouse Xenograft Models of BCR-ABL-Driven and BCR-ABLT315I-Driven Tumor Growth.
(A) Effect of AP24534 on survival of SCID mice after intravenous injection of Ba/F3 cells expressing native BCR-ABL (left) or BCR-ABLT315I (right). Ba/F3 cells expressing native BCR-ABL or BCR-ABLT315I were injected into the tail vein of SCID mice, and animals were treated once daily by oral gavage with vehicle, AP24534, or dasatinib for the indicated dosing period (days 3–21).
(B) In vivo efficacy and BCR-ABL signaling suppression by AP24534 in a subcutaneous xenograft model using Ba/F3 BCR-ABLT315I cells. Tumor-bearing animals were treated once daily by oral gavage with vehicle or the indicated doses of AP24534 for 19 consecutive days (dosing period indicated) with mean tumor volume plotted (error bars represent S.E.M.). Each AP24354 treatment group was compared to the vehicle group using Dunnett’s test, with statistical significance (p<0.05) indicated by an asterisk. BCR-ABL and CrkL phosphorylation were evaluated by immunoblot in animals treated with a single oral dose of vehicle or 30 mg/kg AP24534 (N=3 per group).
In a survival model in which mice were instead injected with Ba/F3 BCR-ABLT315I cells, administration of dasatinib at doses as high as 300 mg/kg had no effect on survival time, as expected (Figure S3). By contrast, treatment with AP24534 prolonged survival in a dose-dependent manner (Figure 5A, right). AP24534 dosed orally for 19 days at 5, 15, and 25 mg/kg prolonged median survival to 19.5, 26, and 30 days, respectively compared to 16 days for vehicle-treated mice (p<0.01 for all three dose levels).
The anti-tumor activity of AP24534 was further assessed in a xenograft model in which Ba/F3 BCR-ABLT315I cells were injected subcutaneously into mice. Tumor growth was inhibited by AP24534 in a dose-dependent manner (Figure 5B, left) compared to vehicle-treated mice, with significant suppression of tumor growth upon daily oral dosing at 10 and 30 mg/kg (%T/C = 68% and 20%, respectively; p<0.01 for both dose levels). Daily oral dosing of 50 mg/kg AP24534 caused significant tumor regression (%T/C = 0.9%, p<0.01), with a 96% reduction in mean tumor volume at the final measurement compared to the start of treatment. AP24534 was well tolerated at all efficacious dose levels for the duration of the study; maximal decreases in body weight were <5%, <5%, and <12% for the 10, 30, and 50 mg/kg dose groups, respectively, with no signs of overt toxicity.
To confirm target inhibition, levels of phosphorylated BCR-ABLT315I and phosphorylated CrkL were assessed in tumors from mice harvested 6 hr after one-time dosing with vehicle or AP24534. As shown in Figure 5B (right), a single oral dose of 30 mg/kg markedly decreased levels of phosphorylated BCR-ABL and phosphorylated CrkL.
Single-Agent AP24534 Completely Suppresses Outgrowth of Resistant Clones
To survey for potential sites of vulnerability to resistance, we tested AP24534 in our established accelerated mutagenesis assay. This assay has previously been used to characterize the resistance profile of imatinib, nilotinib, and dasatinib, and has proved to be predictive of clinical experience with these inhibitors (Bradeen et al., 2006). In this screen, a BCR-ABL-driven cell line is exposed to mutagen, and then plated into tissue culture wells with graded concentrations of inhibitor. Outgrowth of cells reflects the emergence of resistant subclones, which are sequenced to identify BCR-ABL mutations.
Initially, we performed mutagenesis experiments using Ba/F3 cells expressing native BCR-ABL at several concentrations of AP24534 (5–40 nM) and found a concentration-dependent reduction in both the percentage of wells with outgrowth and in the scope of mutations observed (Figure 6A). At 5 nM AP24534, all wells (576/576) exhibited outgrowth and 90% of the sequenced representative subclones expressed native BCR-ABL (Table S2). Raising the concentration of AP24534 to 10 nM resulted in both a marked reduction in outgrowth (168/1440 wells; 11.7%) and an increased frequency of mutated subclones (33.1%; Table S2). Mutations recovered included occurrences at several P-loop residues (G250, Q252, Y253, and E255), a cluster at the C-helix (K285, E292, and L298), and T315 (T315I), as well as F317, V339, F359, L387, and S438. Among the recovered mutations, nearly all have been previously encountered in resistance to imatinib, nilotinib, and/or dasatinib (reviewed by (O’Hare et al., 2007)). No mutations were encountered that were specific for AP24534 only.
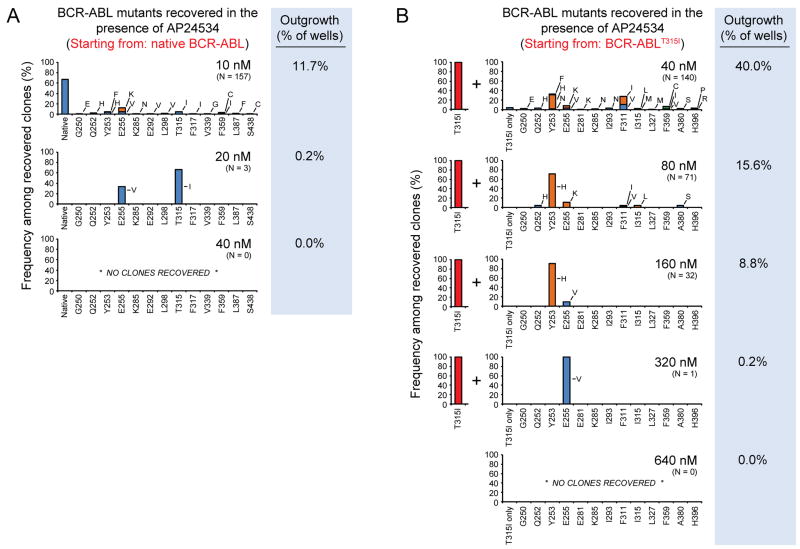
Figure 6. Single-Agent AP24534 Completely Suppresses Resistant Outgrowth in Cell-Based Mutagenesis Screens.
(A) Resistant clones recovered from ENU-treated Ba/F3 cells starting from native BCR-ABL cultured with graded concentrations of AP24534 (10, 20, 40 nM). Each bar represents the relative percentage of the indicated BCR-ABL kinase domain mutant among recovered subclones. Since the percentage of surviving resistant subclones and the concentration of AP24534 are inversely related, a different number of sequenced subclones are represented in the graph for each concentration of AP24534 (Table S2). The percent of wells surveyed that contained outgrowth is indicated to the right of each graph.
(B) ENU-treated Ba/F3 BCR-ABLT315I cells were cultured with graded concentrations of AP24534 (40, 80, 160, 320, 640 nM). Experiments and data analysis were performed as described in panel (A). All recovered subclones contain T315I in addition to the secondary mutation indicated on each graph.
We next investigated 20 nM AP24534 and found that outgrowth was sharply curtailed (3/1440 wells; 0.2%), with only two mutations, E255V and T315I, persisting (Figure 6A and Table S2). Thus, within our extensive survey, no previously undiscovered mutations capable of conferring high-level resistance to AP24534 were identified. At 40 nM AP24534, which is 43-fold lower than the IC50 for parental BaF/3 cells, complete suppression of in vitro resistance was achieved. This absence of resistant outgrowth was further confirmed at higher concentrations of AP24534 (80–320 nM; not shown).
Effects of AP24534 on Compound Mutants
Having identified a limited resistance susceptibility profile for AP24534 at the level of single mutations, we wanted to investigate the vulnerability to compound mutations, defined as two kinase domain mutations in the same allele, which have been detected in some treatment failures (Khorashad et al., 2008; Shah et al., 2007; Stagno et al., 2008). To simulate the situation in which AP24534 is used to treat a patient with a predominant T315I subclone, we repeated the accelerated mutagenesis assay, this time starting with an existing T315I mutation (Figure 6B and Table S3). We found that there was still a concentration-dependent hierarchy and that AP24534 at a concentration of 160 nM or lower overcame all compound mutants involving T315I except Y253H/T315I and E255V/T315I. At 320 nM, the only remaining compound mutant was E255V/T315I, which couples the two most resistant single mutants, and outgrowth was completely suppressed at the highest concentration (640 nM) tested, still ~3-fold below the IC50 for parental Ba/F3 cell line inhibition. This resistance profile was confirmed in a subsequent screen starting from a background of BCR-ABLE255V, the most resistant single BCR-ABL kinase domain mutation to AP24534, with the E255V/T315I compound mutant again persisting to 320 nM and being eliminated at 640 nM (Table S4).
DISCUSSION
AP24534 is a next-generation ABL kinase inhibitor optimized using structure-based drug design to bind to the inactive, DFG-out conformation of ABL and ABLT315I. The key structural feature of the molecule is a carbon-carbon triple bond linkage that makes productive hydrophobic contact with the side chain of I315, allowing inhibition of the T315I mutant. The triple bond also acts as an inflexible connector that enforces correct positioning of the two binding segments of AP24534 into their established binding pockets. AP24534 maintains an extensive hydrogen-bonding network and occupies a region of the kinase that overlaps significantly with the imatinib binding site.
A key design feature of AP24534 underlying its pan-BCR-ABL inhibitor profile is incorporation of multiple contact points to confer very high potency and to balance and distribute the overall binding affinity. While each of the hydrogen-bonding and contact residue interactions contribute substantially to the inhibitor’s affinity for its target, mutation-based disruption of one element of the binding network or distortion of a sub-region within the binding pocket results in only a slight reduction in affinity. As a consequence, AP24534 also retains potency against other imatinib-resistant ABL mutants in addition to ABLT315I. While mutations that destabilize the inactive conformation of ABL to which AP24534 binds, including T315I and E255V, result in modest reductions in binding affinity, substantial reductions would be expected to require at least two changes at non-proximal residues – a prediction consistent with findings from our mutagenesis screen.
Kinase selectivity studies showed that AP24534 does not inhibit Aurora kinases, clearly distinguishing it from other T315I inhibitors in development. These studies also revealed inhibition of SRC, LYN, PDGFRα, and c-KIT with <10-fold selectivity compared to ABLT315I. Several of these kinases are important clinical targets of imatinib, nilotinib and/or dasatinib, although only dasatinib has been reported to inhibit SRC family kinases. Although assay differences preclude direct comparison of the kinase profiles of AP24534 and dasatinib, a comprehensive kinase interaction map for dasatinib was recently reported (Karaman et al., 2008). In general, the linearity of the triple bond in AP24534 is predicted to minimize steric clash between the inhibitor and hydrophobic gatekeeper residues. This feature probably contributes to the relatively broad kinase specificity profile of AP24534, which includes VEGFR and FGFR family kinases, receptors not inhibited by the three currently approved BCR-ABL drugs. As SRC, VEGFR, FGFR, and PDGFR family kinases are potential targets in a variety of other malignancies, this supports the potential testing of AP24534 in a wider range of cancers.
Evaluation of AP24534 in cellular proliferation assays confirmed its potent pan-BCR-ABL inhibition against cells expressing native or mutant BCR-ABL, including BCR-ABLT315I, while retaining a high degree of selectivity (>1000-fold) for Ph-positive cells. Among the BCR-ABL mutants tested, the E255V mutant, which confers high-level resistance to imatinib and intermediate-level resistance to nilotinib and dasatinib (O’Hare et al., 2007), was most resistant to AP24534. Notably, AP24534 potently inhibited mutants at residues Y253 and F359 (which have been reported in patients failing nilotinib (Cortes et al., 2007; Kantarjian et al., 2007)), as well as F317 (implicated in clinical resistance to dasatinib (Burgess et al., 2005; Cortes et al., 2007; Khorashad et al., 2008; Shah et al., 2007; Talpaz et al., 2006)). While clinically achievable and effective doses will need to be determined, the sizeable selectivity for BCR-ABL-expressing cells (regardless of mutational status) over normal cells suggests the potential for efficacy with minimal toxicity.
In clinical studies of BCR-ABL inhibitors, pharmacodynamic evaluation of target inhibition is an important component of dose optimization. In the preclinical studies reported here we monitored phosphorylation of CrkL, a direct substrate of native and mutant BCR-ABL, by immunoblot analysis. In both Ba/F3 cells and primary CML BCR-ABLT315I cells, treatment with AP24534 resulted in a marked reduction in phosphorylated CrkL, while imatinib, dasatinib, and nilotinib had no effect. This assay was recently used to monitor BCR-ABL activity in patients treated with nilotinib; values of percent phosphorylated CrkL from serially collected peripheral blood samples were consistent with BCR-ABL kinase domain mutation status and matched closely with other measures of response, including BCR-ABL transcript levels and white cell counts (La Rosee et al., 2008). Given its extensive validation in the clinic, this assay is being employed to monitor the pharmacodynamic effects of AP24534 in its phase 1 evaluation.
The oral bioavailability of AP24534 was confirmed in mouse pharmacology studies, where concentrations above the IC50s for all tested mutants could be safely sustained following daily oral dosing. AP24534 demonstrated potent activity after daily oral administration in a series of mouse models of CML driven by native BCR-ABL or BCR-ABLT315I. In a survival model using Ba/F3 cells expressing native BCR-ABL, AP24534 significantly prolonged survival at low doses of 2.5 and 5 mg/kg and demonstrated similar efficacy to dasatinib. In an analogous model using BCR-ABLT315I cells, AP24534 significantly extended survival whereas dasatinib, as expected, was inactive. AP24534 was also active in a subcutaneous BCR-ABLT315I tumor model, where tumor stasis or regression occurred at doses of 30 and 50 mg/kg, and suppression of BCR-ABL signaling was demonstrated using the shift CrkL phosphorylation assay. AP24534 was well tolerated at all dose levels used in these studies. Thus, AP24534 is orally bioavailable, inhibits its molecular target, and has a wide therapeutic range (5–50 mg/kg) in BCR-ABLT315I dependent CML animal models.
Mutation-mediated resistance to clinical ABL inhibitors is the main route of BCR-ABL signaling reactivation, particularly in chronic phase disease. As AP24534 advances into clinical evaluation, anticipating potential resistance liabilities, especially compared to those of nilotinib and dasatinib, will be important for prospective treatment decisions. Several mutations have been reported in association with clinical resistance to nilotinib (L248V, Y253F/H, E255K/V, T315I, F359C/V; (Cortes et al., 2007; Kantarjian et al., 2007)) or dasatinib (V299L, T315A/I, F317I/L; (Cortes et al., 2007; Khorashad et al., 2008; Shah et al., 2007)) which are largely consistent with our in vitro profiling (Bradeen et al., 2006). In our accelerated mutagenesis screens for AP24534, we found a concentration-dependent reduction in both the percentage of wells with outgrowth and in the range of mutations observed. Although at 10 nM AP24534 we observed 16 different substitutions across 13 different residues, the only resistant subclones recovered at 20 nM harbored either a T315I or E255V mutation, and at 40 nM AP24534 and above complete suppression of outgrowth was observed. Depending on achievable plasma levels, our data suggest that AP24534 may have the potential to overcome single-mutation-based resistance in the clinical setting. This result has been previously achieved in this assay only with combinations of nilotinib or dasatinib and a pre-clinical T315I inhibitor (O’Hare et al., 2008). To our knowledge no other ABL kinase inhibitor has been shown to have this potential as a single agent.
As compound (multiple) mutations of BCR-ABL represent a rare but challenging scenario clinically, we carried out additional accelerated mutagenesis screens starting with cells expressing either of the two individually most resistant mutants, BCR-ABLT315I or BCR-ABLE255V. This predictive assay implicated certain compound mutations, especially those involving any two of Y253H, E255V, and T315I in moderate to high-level resistance to AP24534. Among these, Y253H/T315I and E255V/T315I are predicted to be the most resistant pairings, although high concentrations of AP24534 still prevented these mutations emerging. Thus, AP24534 has the capability to eliminate compound mutations involving T315I and E255V predicted to be highly resistant to all other inhibitors. Currently, the number of clinically documented compound mutations within the kinase domain of BCR-ABL associated with treatment failure is low (Table S5). Nonetheless, they represent a formidable problem for those patients harboring them, and incidence may increase with the prolonged survival of CML patients and with more patients undergoing sequential ABL kinase inhibitor treatment (Shah et al., 2007). Overall, although no mutagenesis screen can be completely exhaustive, our data indicate AP24534 has the potential to address this currently unmet clinical issue.
Our pre-clinical profiling indicates that AP24534 has potential as an important option for controlling resistance in CML. The combined results of our biochemical, cell-based, and in vivo studies suggest that AP24534 exhibits sufficient activity against native BCR-ABL and all tested BCR-ABL mutants to warrant consideration for single-agent use as a pan-BCR-ABL inhibitor. Moreover, our results indicate that AP24534 holds promise for controlling compound mutants involving T315I, while raising awareness that it is advantageous to eliminate resistant subclones at the single-mutation stage. In the longer term, this may advocate for the potential future use of a pan-BCR-ABL inhibitor such as AP24534 in a first-line therapeutic capacity.
Clinical use of a pan-BCR-ABL inhibitor active against T315I could make long-term remissions an achievable goal at least for some patients with advanced CML. A phase 1 clinical trial evaluating oral AP24534 in patients with refractory CML and other hematologic malignancies is ongoing (NCT00660920, www.clinicaltrials.gov).
EXPERIMENTAL PROCEDURES
Inhibitors
AP24534, 3-(imidazo[1,2b]pyridazin-3-ylethynyl)-4-methyl-N-(4-((4-methylpiperazin-1-yl)methyl)-3-(trifluoromethyl)phenyl)benzamide was synthesized at ARIAD Pharmaceuticals. Imatinib, dasatinib, and nilotinib were purchased from the Oregon Health & Science University (OHSU) pharmacy or made at ARIAD. All inhibitors were prepared as 10.0 mM stock solutions and stored at −20°C. Serial dilutions of 10.0 mM stock solutions were carried out just prior to use in each experiment.
Crystallization and Structural Determination of ABLT315I:AP24534 Complex
The kinase domain of murine ABLT315I (residues 229–515) was co-expressed with YopH protein tyrosine phosphatase in E. coli as described (Zhou et al., 2007) and purified in the presence of AP24534 to near homogeneity (> 95%) using metal affinity, Mono Q, and size exclusion chromatography. The typical yield of purified ABLT315I bound with AP24534 was about 1 mg/L.
Co-crystals of ABLT315I and AP24534 were grown by the hanging drop vapor diffusion method at 4oC by mixing equal volumes of the AP24534:ABLT315I complex (25 mg/mL) and well solution (30% w/v polyethylene 4000, 0.2 M sodium acetate, 0.1 M Tris, pH 8.5). After 1–2 days, crystals reached a typical size of 50 × 50 × 300 μm3 and were harvested in mother liquor supplemented with 30% v/v glycerol as cyroprotectant. X-ray diffraction data were collected at 100K at beamline 19 BM (Advanced Photon Service, Argonne, IL). The data were indexed and scaled in space group P21 by using the HKL2000 package (Otwinowski and Minor, 1997).
The structure of AP24534 in complex with ABLT315I was determined by molecular replacement by AMoRe (Navaza, 1994) using the structure of native ABL bound with imatinib (PDB code: 1IEP). There were two ABLT315I molecules in the asymmetric unit. The structure was refined with CNX combined with manual rebuilding in Quanta (Accelrys Inc., San Diego, CA), and AP24534 was built into the density after several cycles of refinement and model building, which then continued until convergence was reached. The final model, refined to 1.95 Å, consists of residues 228 through 511, with 386–397 in the activation loop disordered. The electron density for bound AP24534 as well as the side chain of I315 was well resolved in both complexes, leaving no ambiguities for the binding mode of the inhibitor.
Autophosphorylation Assays For ABLT315I
Kinase autophosphorylation assays with full length, tyrosine-dephosphorylated ABL, ABLG250E, ABLY253F, ABLE255K and ABLT315I (Invitrogen; San Diego, CA) were performed in the presence of imatinib, nilotinib, dasatinib, or AP24534 (0–1000 nM) as per (O’Hare et al., 2004).
Kinase Selectivity Profile of AP24534
AP24534 was profiled against >100 kinases by Reaction Biology Corporation (Malvern, PA) using the Kinase Hotspot assay, which utilizes 10 μM [33P]-ATP, recombinant kinase domain, peptide substrate, and a range of 10 concentrations of inhibitor to establish an IC50 value (http://www.reactionbiology.com/pages/kinase.htm).
Collection of Patient Samples
Clinical samples were obtained with informed consent and under the approval of the OHSU Institutional Review Board. Blood or bone marrow from patients or healthy individuals was separated on a Ficoll gradient (GE Healthcare) for isolation of mononuclear cells.
Cell Lines
Ba/F3 transfectants (expressing native BCR-ABL or BCR-ABL with a single kinase domain mutation) were maintained in RPMI 1640 supplemented with 10% FCS, 1 unit/mL penicillin G, and 1 mg/mL streptomycin (complete media) at 37°C and 5% CO2. The Ba/F3 BCR-ABLT315A cell line was a kind gift of Dr. Neil Shah, UCSF. Parental Ba/F3 cells (and as a control, Ba/F3 BCR-ABLT315I cells) were supplemented with IL-3 provided by WEHI-conditioned media. Prior to cell proliferation assays, RNA was isolated from each Ba/F3 cell line, and kinase domain mutations were confirmed by RT-PCR followed by DNA sequence analysis using Mutation Surveyor software (SoftGenetics, State College, PA).
Cell Proliferation Assays
Ba/F3 cell lines were distributed in 96-well plates (4 × 103 cells/well) and incubated with escalating concentrations of AP24534 for 72 hr. The inhibitor ranges used were: 0–625 nM for cells expressing BCR-ABL and 0–10,000 nM for BCR-ABL negative cells. Proliferation was measured using a methanethiosulfonate (MTS)-based viability assay (CellTiter96 Aqueous One Solution; Promega). IC50 values are reported as the mean of three independent experiments performed in quadruplicate. For cell proliferation experiments with CML or normal primary cells, mononuclear cells were plated in 96-well plates (5 × 104 cells/well) over graded concentrations of AP24534 (0–1000 nM) in RPMI supplemented with 10% FBS, L-glutamine, penicillin/streptomycin, and 100 μM β-mercaptoethanol. Following a 72 hr incubation, cell viability was assessed by subjecting cells to an MTS assay. All values were normalized to the control wells with no drug.
CrkL Phosphorylation in Ba/F3 Cell Lines
Ba/F3 cells expressing native BCR-ABL or BCR-ABLT315I (5 × 106 per well) were cultured 4 hr in complete media alone or with imatinib (2000 nM), dasatinib (50 nM), nilotinib (500 nM), or AP24534 (0.1–1000 nM). Lysates produced by boiling cells in SDS-PAGE loading buffer supplemented with protease and phosphatase inhibitors. Lysates were subjected to SDS-PAGE and immunoblotted with anti-CrkL antibody C-20 (Santa Cruz). Phosphorylated and non-phosphorylated CrkL signals were distinguished based on differential band migration, quantified by densitometry on a Lumi Imager (Roche) and expressed as a % phosphorylated CrkL.
Ex Vivo Exposure of BCR-ABLT315I Patient Samples to AP24534
Peripheral blood mononuclear cells from a patient with CML in lymphoid blast crisis (CML L-BC) with a BCR-ABLT315I mutation were isolated by Ficoll centrifugation. RT-PCR and sequencing analysis confirmed that the sample predominantly contained the BCR-ABLT315I mutant. Mononuclear cells (5 × 106 cells/well) were cultured overnight in serum-free IMDM media (Invitrogen) supplemented with 20% BIT (StemCell), 40 μg/mL human low-density lipoprotein, and 100 μM β-mercaptoethanol alone or with imatinib (1000 nM), dasatinib (50 nM), nilotinib (200 nM), or AP24534 (50 nM, 500 nM). Cells were lysed directly into boiling SDS-PAGE loading buffer supplemented with protease and phosphatase inhibitors. Lysates were subjected to SDS-PAGE and immunoblotted with anti-CrkL antibody C-20 (Santa Cruz). Phosphorylated and non-phosphorylated CrkL were distinguished based on differential band migration. Band signal intensities were quantified by densitometry on a Lumi Imager (Roche).
Global Tyrosine Phosphorylation by FACS
Mononuclear cells (2 × 105) were cultured overnight in serum-free media alone or with imatinib (1000 nM), dasatinib (50 nM), nilotinib (200 nM), or graded concentrations of AP24534 (50, 500 nM). Cells were fixed and permeabilized according to the manufacturer’s instructions (Caltag), incubated with 2 μg of anti-phosphotyrosine 4G10-FITC antibody (BD Biosciences) for 1 hr, washed twice with PBS supplemented with 1% BSA and 0.1% sodium azide, and fixed in 1% formaldehyde. FITC signal intensity was analyzed on a FACSAria instrument (BD) and mean fluorescence intensity (MFI) was calculated. Values are reported as fold increase in MFI relative to unstained controls.
Hematopoietic Colony Forming Assays of Primary CML Cells and Normal Bone Marrow
To assess the effect of AP24534 against primary CML cells harboring BCR-ABLT315I and normal hematopoietic progenitors, bone marrow mononuclear cells isolated by Ficoll density centrifugation were cultured with graded concentrations of AP24534 (CML patient: 0–50 nM; healthy individual: 0–1000 nM). Cells were plated in triplicate (5×104 cells/plate) in 1 mL of IMDM:methylcellulose media (1:9 v/v) containing 50 ng/mL SCF, 10 ng/mL GM-CSF, and 10 ng/mL IL-3 (Methocult GF H4534; Stem Cell Technologies) to assess granulocyte/macrophage colony formation (CFU-GM). After culturing at 37°C for 14–18 days, colonies (>50 cells) were counted and results reported as the percentage of colonies relative to untreated control ± SEM.
Pharmacokinetics
All animal experiments were approved by ARIAD’s IACUC and conformed to relevant regulatory standards. The pharmacokinetic profile of AP24534 (in citrate buffer, pH 2.74) was assessed in CD-1 female mice after a single dose by oral gavage. Blood samples were collected at various time points and AP24534 concentrations in plasma determined by an internal standard LC/MS/MS method using protein precipitation and calibration standards prepared in blank mouse plasma. Reported concentrations are average values from 3-mice/time point/dose group.
Ba/F3 survival model
Ba/F3 cells expressing native BCR-ABL or BCR-ABLT315I were injected into the tail vein of female SCID mice (100 μL of a 1×107 cells/mL suspension in serum-free medium). Beginning 72 hr later mice were treated once daily by oral gavage with vehicle (25 mM citrate buffer, pH 2.75), AP24534, or dasatinib for up to 19 consecutive days. Moribund animals were sacrificed as per IACUC guidelines. On necropsy, mice had marked splenomegaly due to tumor cell infiltration. Survival data were analyzed using Kaplan-Meier method, and statistical significance was evaluated with a Log-rank test (GraphPad PRISM) comparing the survival time of each treatment group with the vehicle group.
Ba/F3 Tumor Model
Ba/F3 BCR-ABLT315I cells were implanted subcutaneously into the right flank of female nude mice (100 μL of a 1×107 cells/mL cell suspension in serum-free medium). Mice were randomized to treatment groups when the average tumor volume reached approximately 500 mm3. Mice were treated once daily by oral gavage with vehicle (25 mM citrate buffer, pH 2.75) or AP24534 for up to 19 consecutive days. Tumor volume (mm3) was calculated using the following formula: tumor volume = L×W2×0.5. To determine tumor growth inhibition when the treatment period was finished, mean tumor volume for treatment group/mean tumor volume for control group (%T/C) was calculated at the final measurement. The mean tumor volume from the last measurement of all groups was compared using a one-way ANOVA test (GraphPad PRISM) and each treatment group was further compared to that of vehicle-treated mice for statistical significance using Dunnett’s test.
For analysis of tyrosine-phosphorylated BCR-ABL and CrkL levels, tumor-bearing animals were treated with a single dose of vehicle or 30 mg/kg AP24534 by oral gavage. Six hr after dosing, animals (N=3/group) were sacrificed and tumor samples collected for immunoblot analysis with antibodies against pBCR-ABL and eIF4E (Cell Signaling Technology) and total CrkL (C-20; Santa Cruz).
Accelerated cell-based mutagenesis screen: Single-agent AP24534
Ba/F3 cells expressing native BCR-ABL were treated overnight with N-ethyl-N-nitrosourea (ENU; 50 μg/mL), pelleted, resuspended in fresh media, and distributed into 96-well plates at a density of 1×105 cells/well in 200 μL complete media supplemented with graded concentrations of AP24534. The wells were observed for cell growth under an inverted microscope and media color change every two days throughout the 28-day experiment. The contents of wells exhibiting cell outgrowth were transferred to a 24-well plate containing 2 mL complete media supplemented with AP24534 at the same concentration as in the initial 96-well plate. If growth was simultaneously observed in all wells of a given condition, 24 representative wells were expanded for further analysis. At confluency, cells in 24-well plates were collected by centrifugation. DNA was extracted from the cell pellets using a DNEasy Tissue kit (QIAGEN). The BCR-ABL kinase domain was amplified using primers B2A (5′ TTCAGAAGCTTCTCCCTGACAT 3′) and ABL4317R (5′ AGCTCTCCTGGAGGTCCTC 3′), PCR products were bi-directionally sequenced by a commercial contractor (Agencourt Bioscience) using primers ABL3335F (5′-ACCACGCTCCATTATCCAGCC-3′) and ABL4275R (5′-CCTGCAGCAAGGTAGTCA-3′), and the chromatograms were analyzed for mutations using Mutation Surveyor software (SoftGenetics). Results from this screen are reported as the cumulative data from three independent experiments (see Table S2). The mutagenesis screen was also conducted as described above for single-agent AP24534 starting with Ba/F3 cells expressing BCR-ABLT315I (see Table S3) or BCR-ABLE255V (see Table S4) in single independent experiments.
Crystallographic Coordinates
Crystallographic coordinates for the AP24534:ABLT315I complex have been deposited at the RCSB Protein Data Bank. The PDB accession number is 3IK3.
Supplementary Material
Figure S1: Inhibition of BCR-ABL phosphorylation in Ba/F3 cells expressing native BCR-ABL or BCR-ABLT315I. BCR-ABL phosphorylation was evaluated in Ba/F3 cells expressing either (A) native BCR-ABL or (B) BCR-ABLT315I treated for 4 hr with imatinib, nilotinib, dasatinib, or AP24534. Samples were analyzed by immunoblot analysis with antibodies against pBCR-ABL and eIF4E (loading control). The phosphorylation status of CrkL in these same lysates was determined by immunoblot analysis as described in Figure 3.
Figure S2: Colony Formation Assays for CML T315I Patient and Normal Primary Cells Against AP24534. Mononuclear cells from a CML accelerated phase (AP) patient harboring BCR-ABLT315I and from a healthy individual were plated in methylcellulose containing nilotinib, dasatinib, or AP24534 and cultured for 14–18 days. Colonies were counted under an inverted microscope, and results were expressed as the mean of three replicates (error bars represent S.E.M.).
(A) Colony formation assays in the presence of AP24534 using mononuclear cells from a CML AP patient harboring BCR-ABLT315I.
(B) Colony formation assays in the presence of AP24534 using mononuclear cells from a healthy individual.
Figure S3: Effect of dasatinib in mouse models using Ba/F3 cells expressing BCR-ABLT315I. Survival curves are shown for mice injected intravenously with Ba/F3 cells expressing BCR-ABLT315I treated during the indicated dosing period with vehicle or dasatinib by oral gavage. Median survival was calculated using the Kaplan-Meier method, and statistical significance was evaluated with a Log-rank test (GraphPad PRISM) by comparing the survival time of each treatment group with the vehicle group.
Table S1: AP24534 kinase panel screening data
Table S2: Tabulated AP24534 single-agent mutagenesis data (starting from Ba/F3 native BCR-ABL cells)
Table S3: Tabulated AP24534 single-agent mutagenesis data (starting from Ba/F3 BCR-ABLT315I cells)
Table S4: Tabulated AP24534 single-agent mutagenesis data (starting from Ba/F3 BCR-ABLE255V cells)
Table S5: BCR-ABL compound mutations involving T315I or E255V conferring moderate to high level resistance to AP24534
Acknowledgments
The authors thank D. Wen, I. Chen, G. Banda, L. Cai, J. Romero, S. Das, S. Lentini, and S. Liu for chemical synthesis, S. Lamore for biological experiments, K. Russian, M. Broudy, and N. Narasimhan for pharmacokinetic analyses and discussions, J. Iuliucci for discussions pertaining to pharmacology, and M. Wong for enzymatic assays. W.C.S., X.Z., V.M.R, F.W., T.Z., W.H., S.W., Y.N., J.A.K., Y.W., M.T., L.C., D.C.D. and T.C. are employees of ARIAD Pharmaceuticals, Inc. M.W.N.D. serves as a consultant for Novartis and Bristol Myers Squibb and receives research support from Calistoga Pharmaceuticals and Genzyme. OHSU and B.J.D. have a financial interest in MolecularMD. Technology used in this research has been licensed to MolecularMD. This potential conflict of interest has been reviewed and managed by the OHSU Conflict of Interest in Research Committee and the Integrity Program Oversight Council. OHSU has clinical trial contracts with ARIAD, Novartis and Bristol-Myers-Squibb to pay for patient costs, nurse and data manager salaries, and institutional overhead. B.J.D. and M.W.N.D. do not derive salary, nor do their laboratories receive funds from these contracts. The research was supported in part by funding from Howard Hughes Medical Institute and The Leukemia & Lymphoma Society 7393-06.
Footnotes
SIGNIFICANCE
The ABL kinase inhibitor imatinib is the current standard first-line therapy for patients with BCR-ABL-positive leukemia. The second-line ABL kinase inhibitors nilotinib and dasatinib are effective salvage therapies for patients who relapse on imatinib. BCR-ABLT315I, a mutant resistant to all approved ABL kinase inhibitors, is emerging as a common pathway of failure on all three inhibitors. AP24534 is an orally active multi-targeted kinase inhibitor that potently inhibits BCR-ABLT315I. It may represent a treatment option for patients with this mutation and complement the current clinically employed inhibitors. Longer term, a pan-BCR-ABL inhibitor such as AP24534 may offer important strategic advantages in a first-line capacity by minimizing BCR-ABL kinase domain mutation-based drug resistance.
SUPPLEMENTAL DATA
The Supplemental Data include three supplemental figures (Figure S1: AP24534 effectively inhibits BCR-ABL-mediated signaling in CML cell lines expressing native or T315I-mutant BCR-ABL; Figure S2: AP25434 potently inhibits colony formation of primary BCR-ABLT315I cells with minimal toxicity to normal cells; Figure S3: Effect of dasatinib in mouse models using Ba/F3 BCR-ABLT315I cells) and five supplemental tables (Table S1: AP24534 kinase panel screening data; Table S2: AP24534 cell-based mutagenesis assay (starting from native BCR-ABL); Table S3: AP24534 cell-based mutagenesis assay (starting from BCR-ABLT315I); Table S4: AP24534 cell-based mutagenesis assay (starting from BCR-ABLE255V); Table S5: BCR-ABL compound mutations involving T315I or E255V conferring moderate to high level resistance to AP24534), and can be found online with this article at (insert location).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bradeen HA, Eide CA, O’Hare T, Johnson KJ, Willis SG, Lee FY, Druker BJ, Deininger MW. Comparison of imatinib, dasatinib (BMS-354825), and nilotinib (AMN107) in an n-ethyl-n-nitrosourea (ENU)-based mutagenesis screen: high efficacy of drug combinations. Blood. 2006;108:2332–2338. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A. 2005;102:3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J, Jabbour E, Kantarjian H, Yin CC, Shan J, O’Brien S, Garcia-Manero G, Giles F, Breeden M, Reeves N, et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood. 2007;110:4005–4011. doi: 10.1182/blood-2007-03-080838. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Giles FJ, Cortes J, Jones D, Bergstrom D, Kantarjian H, Freedman SJ. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- Huang WS, Zhu X, Wang Y, Azam M, Wen D, Sundaramoorthi R, Thomas RM, Liu S, Banda G, Lentini SP, et al. 9-(Arenethenyl)purines as Dual Src/Abl Kinase Inhibitors Targeting the Inactive Conformation: Design, Synthesis, and Biological Evaluation. J Med Chem. 2009;52:4743–4756. doi: 10.1021/jm900166t. [DOI] [PubMed] [Google Scholar]
- Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, Baccarani M, Cortes J, Cross NC, Druker BJ, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour E, Cortes J, Kantarjian H. Treatment selection after imatinib resistance in chronic myeloid leukemia. Target Oncol. 2009;4:3–10. doi: 10.1007/s11523-008-0100-y. [DOI] [PubMed] [Google Scholar]
- Kantarjian HM, Giles F, Gattermann N, Bhalla K, Alimena G, Palandri F, Ossenkoppele GJ, Nicolini FE, O’Brien SG, Litzow M, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110:3540–3546. doi: 10.1182/blood-2007-03-080689. [DOI] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Khorashad JS, Milojkovic D, Mehta P, Anand M, Ghorashian S, Reid AG, De Melo V, Babb A, de Lavallade H, Olavarria E, et al. In vivo kinetics of kinase domain mutations in CML patients treated with dasatinib after failing imatinib. Blood. 2008;111:2378–2381. doi: 10.1182/blood-2007-06-096396. [DOI] [PubMed] [Google Scholar]
- La Rosee P, Holm-Eriksen S, Konig H, Hartel N, Ernst T, Debatin J, Mueller MC, Erben P, Binckebanck A, Wunderle L, et al. Phospho-CRKL monitoring for the assessment of BCR-ABL activity in imatinib-resistant chronic myeloid leukemia or Ph+ acute lymphoblastic leukemia patients treated with nilotinib. Haematologica. 2008;93:765–769. doi: 10.3324/haematol.12186. [DOI] [PubMed] [Google Scholar]
- Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- Navaza J. A MoRe. Acta Crystallogr A. 1994;A50:157–163. [Google Scholar]
- O’Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–2249. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- O’Hare T, Eide CA, Tyner JW, Corbin AS, Wong MJ, Buchanan S, Holme K, Jessen KA, Tang C, Lewis HA, et al. SGX393 inhibits the CML mutant Bcr-AblT315I and preempts in vitro resistance when combined with nilotinib or dasatinib. Proc Natl Acad Sci U S A. 2008;105:5507–5512. doi: 10.1073/pnas.0800587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hare T, Pollock R, Stoffregen EP, Keats JA, Abdullah OM, Moseson EM, Rivera VM, Tang H, Metcalf CA, 3rd, Bohacek RS, et al. Inhibition of wild-type and mutant Bcr-Abl by AP23464, a potent ATP-based oncogenic protein kinase inhibitor: implications for CML. Blood. 2004;104:2532–2539. doi: 10.1182/blood-2004-05-1851. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A, Cortes J. Therapeutic options against BCR-ABL1 T315I-positive chronic myelogenous leukemia. Clin Cancer Res. 2008;14:4392–4399. doi: 10.1158/1078-0432.CCR-08-0117. [DOI] [PubMed] [Google Scholar]
- Shah NP, Kasap C, Weier C, Balbas M, Nicoll JM, Bleickardt E, Nicaise C, Sawyers CL. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008;14:485–493. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Shah NP, Skaggs BJ, Branford S, Hughes TP, Nicoll JM, Paquette RL, Sawyers CL. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- Stagno F, Stella S, Berretta S, Massimino M, Antolino A, Giustolisi R, Messina A, Di Raimondo F, Vigneri P. Sequential mutations causing resistance to both Imatinib Mesylate and Dasatinib in a chronic myeloid leukaemia patient progressing to lymphoid blast crisis. Leuk Res. 2008;32:673–674. doi: 10.1016/j.leukres.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O’Brien S, Nicaise C, Bleickardt E, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- Tokarski JS, Newitt JA, Chang CY, Cheng JD, Wittekind M, Kiefer SE, Kish K, Lee FY, Borzillerri R, Lombardo LJ, et al. The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shakespeare WC, Huang WS, Sundaramoorthi R, Lentini S, Das S, Liu S, Banda G, Wen D, Zhu X, et al. Novel N9-arenethenyl purines as potent dual Src/Abl tyrosine kinase inhibitors. Bioorg Med Chem Lett. 2008;18:4907–4912. doi: 10.1016/j.bmcl.2008.06.042. [DOI] [PubMed] [Google Scholar]
- Weisberg E, Manley PW, Breitenstein W, Brüggen J, Ray A, Cowan-Jacob SW, Fabbro D, Fendrich G, Hall-Meyers E, Huntly BJ, et al. Characterization of AMN107, a selective inhibitor of wild-type and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Zhou T, Parillon L, Li F, Wang Y, Keats J, Lamore S, Xu Q, Shakespeare W, Dalgarno D, Zhu X. Crystal structure of the T315I mutant of Abl kinase. Chem Biol Drug Des. 2007;70:171–181. doi: 10.1111/j.1747-0285.2007.00556.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Inhibition of BCR-ABL phosphorylation in Ba/F3 cells expressing native BCR-ABL or BCR-ABLT315I. BCR-ABL phosphorylation was evaluated in Ba/F3 cells expressing either (A) native BCR-ABL or (B) BCR-ABLT315I treated for 4 hr with imatinib, nilotinib, dasatinib, or AP24534. Samples were analyzed by immunoblot analysis with antibodies against pBCR-ABL and eIF4E (loading control). The phosphorylation status of CrkL in these same lysates was determined by immunoblot analysis as described in Figure 3.
Figure S2: Colony Formation Assays for CML T315I Patient and Normal Primary Cells Against AP24534. Mononuclear cells from a CML accelerated phase (AP) patient harboring BCR-ABLT315I and from a healthy individual were plated in methylcellulose containing nilotinib, dasatinib, or AP24534 and cultured for 14–18 days. Colonies were counted under an inverted microscope, and results were expressed as the mean of three replicates (error bars represent S.E.M.).
(A) Colony formation assays in the presence of AP24534 using mononuclear cells from a CML AP patient harboring BCR-ABLT315I.
(B) Colony formation assays in the presence of AP24534 using mononuclear cells from a healthy individual.
Figure S3: Effect of dasatinib in mouse models using Ba/F3 cells expressing BCR-ABLT315I. Survival curves are shown for mice injected intravenously with Ba/F3 cells expressing BCR-ABLT315I treated during the indicated dosing period with vehicle or dasatinib by oral gavage. Median survival was calculated using the Kaplan-Meier method, and statistical significance was evaluated with a Log-rank test (GraphPad PRISM) by comparing the survival time of each treatment group with the vehicle group.
Table S1: AP24534 kinase panel screening data
Table S2: Tabulated AP24534 single-agent mutagenesis data (starting from Ba/F3 native BCR-ABL cells)
Table S3: Tabulated AP24534 single-agent mutagenesis data (starting from Ba/F3 BCR-ABLT315I cells)
Table S4: Tabulated AP24534 single-agent mutagenesis data (starting from Ba/F3 BCR-ABLE255V cells)
Table S5: BCR-ABL compound mutations involving T315I or E255V conferring moderate to high level resistance to AP24534