Abstract
Antigen receptor loci poised for V(D)J rearrangement undergo germline transcription (GT) of unrearranged genes, and the accessible gene segments are associated with post-translational modifications (PTMs) on histones. Here, we performed a comprehensive analysis of the dynamic changes of 4 PTMs throughout B and T cell differentiation in freshly isolated ex vivo cells. Methylation of lysines 4 and 79 of histone H3, and acetylation of H3, demonstrated stage- and lineage-specificity, and were most pronounced at the J segments of loci poised for, or undergoing, rearrangement, except for dimethylation of H3K4, which was more equally distributed on V, D and J genes. Focusing on the IgL loci, we demonstrated there are no active PTMs in the absence of pre-BCR signaling. Kappa locus GT and PTMs on Jκ genes are rapidly induced following pre-BCR signaling in large pre-B cells. In contrast, the λ locus shows greatly delayed onset of GT and PTMs, which do not reach high levels until the immature B cell compartment, the stage at which receptor editing is initiated. Analysis of MiEκ−/− mice shows that this enhancer plays a key role in inducing not only GT but PTMs. Using an inducible pre-B cell line, we demonstrate that active PTMs on Jκ genes occur after GT is initiated, indicating that histone PTMs do not make the Jκ region accessible, but conversely, GT may play a role in adding PTMs. Our data indicate that the epigenetic profile of IgL genes is dramatically modulated by pre-BCR signaling and B cell differentiation status.
Keywords: Gene rearrangement, gene regulation, B cells
Introduction
The vast diversity of the B and T lymphocyte antigen receptors is generated by precisely orchestrated assembly of V, D and J gene segments in developing B and T lymphocytes. In pro-B cells, the first rearrangement is that of immunoglobulin heavy chain (IgH) DH to JH. When that step is completed, VH to D-JH rearrangement begins. Once a productive IgH rearrangement is made, the H chain binds to surrogate light chain genes, creating a pre-BCR (1). As a result of pre-BCR signaling, the cell subsequently proliferates and proceeds into the pre-B stage. After this transition, heavy chain rearrangement and proliferation stops and light chain rearrangement begins. Igκ light chain rearrangement begins before Igλ light chain rearrangement (2–4). T cells also undergo precisely ordered rearrangement in a similar way. Lineage specificity is shown in that T cell receptor (TCR) genes are only assembled in T cells, and Ig genes are only completely recombined in B cells, although DH to JH joining occurs in both B and T lineage cells.
The accessibility hypothesis proposes that this precise regulation of V(D)J recombination is due to controlled accessibility of the various parts of the loci to RAG-mediated rearrangement (5). In support of this hypothesis, it was observed that germline transcription (GT) of unrearranged genes precedes the developmental activation of recombination at each locus (6). This initial observation has been substantiated in numerous reports, although there have been a few rare exceptions to this general phenomenon documented in cell lines or transgenic miniloci (7–9). Transcriptional control elements within the Ig or TCR loci, including enhancers and promoters, not only play an essential role in the transcription of rearranged receptor genes, but as accessibility control elements, they are necessary for GT and determine the locus and gene segment-specific accessibility of the recombination signal sequences to RAG (10).
Research in the last several years has revealed epigenetic changes that are likely to play an important role in the control of accessibility of V(D)J rearrangement in both B and T cells. Antigen receptor loci poised for V(D)J rearrangement are typically marked by acetylated histone H3 and H4, as well as methylated H3K4 (11–19). Accessibility control elements can recruit transcriptional factors complexes which including chromatin-modifying enzymes to alter the conformation of local nucleosomes and initiate GT (10, 20). An important role of GT process, we hypothesize, may be to provide a platform for the histone methytransferases and acetyltransferases which are known to travel with the elongation of RNA polymerase II complex during elongation. In this way, some histone PTMs could be added to the chromatin during the germline transcription process (21, 22). The modified histone tails can then specifically recruit complexes containing proteins with bromodomains, chromodomains or plant homeodomains (PHD), which can further modify the chromatin structure by adding additional PTMs or by chromatin remodeling (23, 24). A particularly relevant modification in this regard is H3K4me3, which is bound by the PHD domain of RAG2 (25, 26). This recruits RAG2 to the receptor loci bearing that modification, resulting in effective recombination (25).
Systematic studies of the dynamic patterns of histone modifications during the sequential stages of B lymphocyte development at all Ig loci have not been performed. Here we show the dynamic pattern of 4 PTMs (AcH3, H3K4me2, H3K4me3, and H3K79me2) that are associated with the presence of poised V, D and J genes during lymphocyte development. A time course assay using a temperature sensitive inducible Abelson Murine Leukemia Virus (A-MuLV) pre-B cell line revealed that these PTMs occur prior to light chain rearrangement, but shortly after GT is initiated. In vivo, we observed that the induction of κ GT and induction of PTM on Jκ genes did not occur before pre-BCR signaling, but they were rapidly induced during the proliferating large pre-B cell stage. In contrast, GT and PTM in λ genes only began during the small pre-B cell stage, and were greatly increased during the immature B cell stage, the stage at which receptor editing is initiated. We show that the intronic enhancer plays an important role not only in the induction of κ GT and rearrangement, but also in the induction of PTM on the Jκ genes. Therefore, our data suggest that the epigenetic profile of IgL genes is strongly influenced by pre-BCR signaling and by the differentiative stage of the progenitor lymphocytes.
Materials and methods
Mice
Mice were bred and maintained in animal facilities at The Scripps Research Institute (TSRI) and the studies were approved by the TSRI IACUC. MiEκ −/− mice were provided by Dr. Yang Xu (27). Mb1−/− mice were provided by Dr. Michael Reth (28).
Cell culture
103/bcl2/4 inducible cells were maintained in RPMI medium supplemented with 5% FBS at 34 °C. To induce light chain GT, PTMs and rearrangements, cells were shifted to culture at 39 °C.
Purification of lineage negative cells
Bone marrow cells were isolated from 4 to 6 week old BALB/c mice by crushing the bones with a mortar and pestle. The cell suspension was filtered through nylon mesh and cotton plug to remove debris. Lineage negative cells were obtained using the Lineage Cell Depletion kit (Miltenyi Biotec) according to Miltenyi’s protocol.
Purification of pro-B, pre-B and immature B cells from bone marrow
Bone marrow cells were isolated from 4 to 6 week old BALB/c mice as described above. B220+ cells were enriched on MACS anti-PE beads (Miltenyi Biotec) after staining with B220-PE antibody. Following 4 color stain (anti B220-PE, CD43-FITC, CD19-Pacific blue and IgM-APC) of the B220+ cells, we sorted on a BD FACSAria for: pro-B=Fractions B and C (B220+CD43+CD19+IgM−), pre-B=Fraction D (B220+CD43−CD19+IgM−) and immature B=Fraction E (B220modCD43−CD19+IgM+), as defined by Hardy and colleagues (29). Pro-B cells from μMT and mb1−/− mice were purified with MACS anti-mouse CD19 microbeads (Miltenyi Biotech). Pre-B cells from μ+ RAG−/− mice were obtained by sorting for B220+CD43− cells. Pre-B cells from MiEκ−/− mice were obtained by cell sorting for B220+CD43−CD19+IgM− cells.
Purification of double negative (DN) and double positive (DP) thymocytes
DN (CD4−CD8−) thymocytes were purified from 6 week old wild-type mice using MACS anti-CD4 (L3T4) microbeads (Miltenyi Biotec, collecting the negative fraction), followed by MACS CD8a (Ly-2) MicroBeads (Miltenyi Biotec, collecting the negative fraction). For DP T cells, we sorted CD4+CD8+ cells from 4–6 week old mice by staining thymocytes with anti-CD4-APC and CD8α-PE.
Purification of large pre-B and small pre-B cells from bone marrow
Bone marrow cells were isolated from 4–6 week old C57BL/6 mice. CD19+ cells were enriched on CD19 MACS microbeads and were stained with the following antibodies: CD19-FITC, IgM-APC and CD25-PE. The CD19+IgM−CD25+ pre-B cells were identified as large or small based on forward scatter, and the large and small pre-B cells were sorted as previously described (4).
Chromatin Immunoprecipitation and real-time PCR
Formaldehyde crosslinking and ChIP were performed as described previously (19) with the exception of using QIAquick PCR Purification Kit (QIAGEN) to purify ChIP DNA after reversal of the crosslinks. We used antibodies against H3K4me2 (Upstate 07-030), H3K4me3 (Upstate 07-473), H3K9me2 (Upstate 07-441), H3K79me2 (Upstate 05-835) and AcH3 (Upstate 06-599). Real-time PCR was performed using the Quantitec SYBR PCR Kit (Qiagen) and the ABI Prism 7900 Sequence Detection System (Applied Biosystems). Data is presented relative to GAPDH (AcH3), or to CAD (H3K4me2, H3K4me3 and H3K79me2), or to neuregulin (H3K9me2). qPCR were repeated 2–3 times for each primer set for each ChIP sample, and were averaged. At least three ChIPs were done on independent samples for each cell type analyzed, and data is presented as the mean of the ChIPs ± SD. The primers are available in Supplementary Table 1.
Quantitative PCR analysis of germline transcription and VLJL rearrangements
RNA and DNA were harvested from sorted cells using AllPrep DNA/RNA Mini Kit (QIAGEN). cDNA was synthesized using the QuantiTect Reverse Transcription kit (QIAGEN). mRNA levels were assayed by real-time PCR and normalized to the value for GAPDH gene in each sample. Real time PCR for VLJL rearrangement was performed on genomic DNA and normalized to the value for GAPDH promoter region in each sample. For κ rearrangement, a Vκ family specific primer and a Jκ1 specific primer were used to amplify rearranged genomic DNA. For λ rearrangement, a primer which amplifies both Vλ1 and Vλ2 and a Jλ2-specific primer were used. Most experiments were performed more than three times on biologically independent samples of cells, with 2–3 repeats of each of the individual real time PCRs within each experiment. The primer sequences are available in Supplementary Table 1.
Results
Dynamic pattern of H3K4me2, H3K79me2, H3K4me3, and AcH3 on antigen receptor loci throughout lymphocyte differentiation
Six methylation sites on histone lysine residues, including lysines 4, 9, 27, 36, and 79 on histone H3, and lysine 20 on histone 4, have been recognized as important modifications linked to both transcriptional activation and repression (30). To systematically examine the epigenetic basis for the lineage-specific and precisely ordered -regulation of V(D)J recombination at the various loci, we characterized the dynamic pattern of histone PTMs of V, D and J genes from the IgH, κ, λ and TCRβ loci by chromatin immunoprecipitation (ChIP) on lineage negative bone marrow cells, pro-B cells, pre-B cells, immature B cells and thymocytes from BALB/c mice, and pro-B and pro-T cells from RAG1−/− mice. BALB/c cells are actively undergoing rearrangement, whereas the cells from RAG−/− mice are poised to undergo rearrangement, but, due to the absence of the recombinase, have a fully unrearranged locus. Genes that we assayed in ChIP included a variety of V, D and J genes of Ig and TCRβ covering a range from each locus. We used primer pairs flanking the RSS, so only unrearranged gene segments are detected. By screening all the sites of histone methylation, in addition to the more common studied histone acetylation, we found that several active PTMs which are correlated with the presence of poised or actively rearranging V(D)J genes during the appropriate stages of lymphocyte development.
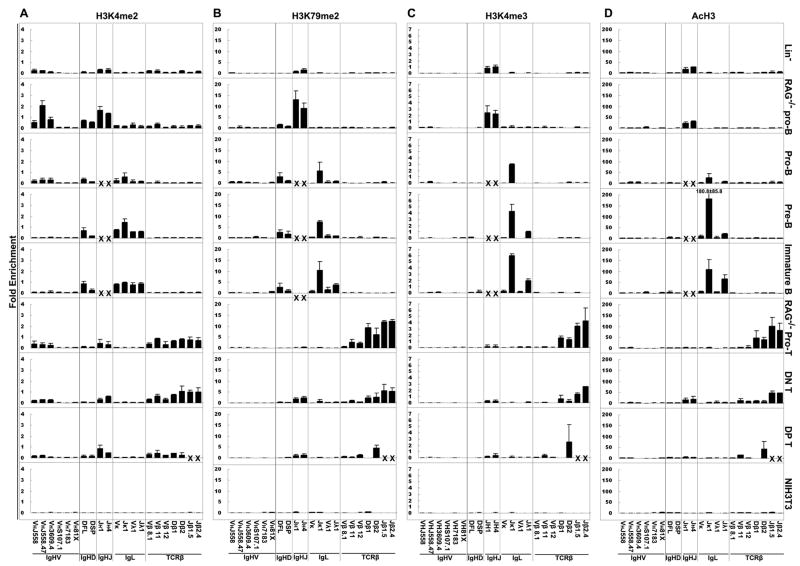
H3K4me2 is observed on V, D and J genes which are poised to undergo rearrangement, or are actively undergoing rearrangement, as previously observed in cell lines (15) (Fig. 1A). In pro-B cell fractions from RAG−/− mice, we find enrichment of this PTM with DH and JH genes, as well as on distal VH genes. The decrease in this PTM on VH genes in BALB/c pro-B cells as compared to RAG−/− pro-B cells may be a result of the ongoing VH to DJH rearrangement in the BALB pro-B cells, in which rearrangement may preferentially be targeted to the VH genes with active histone modifications. We also observed a strong association of H3K4me2 with the light chain genes in the pre-B and immature B cells. In B lineage cells, this PTM is absent on all TCRβ genes assayed, and in T lineage cells, the extent of this PTM is high on TCRβ genes. In thymocytes, it is also present on JH genes, which is consistent with the fact that thymocytes can undergo DHJH rearrangement. The very low level on distal VH genes in thymocytes and lineage negative cells was unexpected, however. In NIH3T3 fibroblast cells, this PTM is absent on both Ig and TCRβ genes.
Figure 1. The dynamic pattern of histone modifications during lymphocyte development.
A. ChIP assays were performed using antibodies reactive with H3K4me2 in pro-B and pro-T cells from 4–6 week old RAG1−/− mice; lin−, pro-B, pre-B and immature B cells from 4–6 week old BALB/c mice; and NIH3T3 cells. The name of the gene(s) assayed is shown at the bottom of the graphs. Analysis was performed by real-time PCR. JH1 cannot be detected in WT B lineage cells by real-time PCR since most DH-proximal RSS of JH1 alleles were deleted by D-J rearrangement. JH4 and Jβ genes in WT B lineage or DP T cells, respectively, were not shown because they might be in the rearranged transcribed region. Data is presented as relative to the positive control of CAD gene. Results represent the mean ± SD. B. ChIP assays were performed using antibodies reactive with H3K79me2. Data is presented as relative to the positive control of CAD gene. C. ChIP assays were performed using antibodies reactive with H3K4me3. Data is presented as relative to the positive control of CAD gene. D. ChIP assays were performed using antibodies reactive with AcH3. Data is presented as relative to the positive control of GAPDH gene.
H3K79me2 is also associated with active genes (30). We found it was highly correlated with the poised-for-rearrangement status of genes from D and J loci. In the B lineage cells, H3K79me2 was completely absent from all TCR genes, and was highest on J genes from all 3 Ig loci. In T lineage cells, H3K79me2 was enriched on both Dβ and Jβ genes. Also, low level enrichment was observed on JH genes, which do undergo limited rearrangement in thymocytes. Unlike H3K4me2, V genes do not show association with H3K79me2, in agreement with previous observations in cell lines (16) (Fig. 1B).
The two active PTMs H3K4me3 and AcH3 have very similar pattern, and are highly enriched in J genes poised for V(D)J rearrangement (Fig. 1C, D). Compared to the great extent of enrichment on the J genes, the level on D and V genes is minimal other than the Dβ genes in RAG−/− pro-T cells. H3K4me3 and AcH3 are associated with Jκ genes in BALB/c pro-B cells, and progresses to be highly enriched in pre-B and immature B cells. H3K4me3 and AcH3 on Jλ lag behind, and only begin to appear in pre-B cells, and the extent of both of those PTMs increase at the immature B cell stage. Also, in B lineage cells, there were no H3K4me3 and AcH3 modifications on any of the TCRβ genes, and in T lineage cells, we did not observe the enrichment of these two PTMs on Ig genes, other than a very low level on JH genes. In lineage negative and NIH3T3 cells, these PTMs were absent in both Ig and TCRβ genes.
Time course of induction of GT and PTM in an inducible cell line
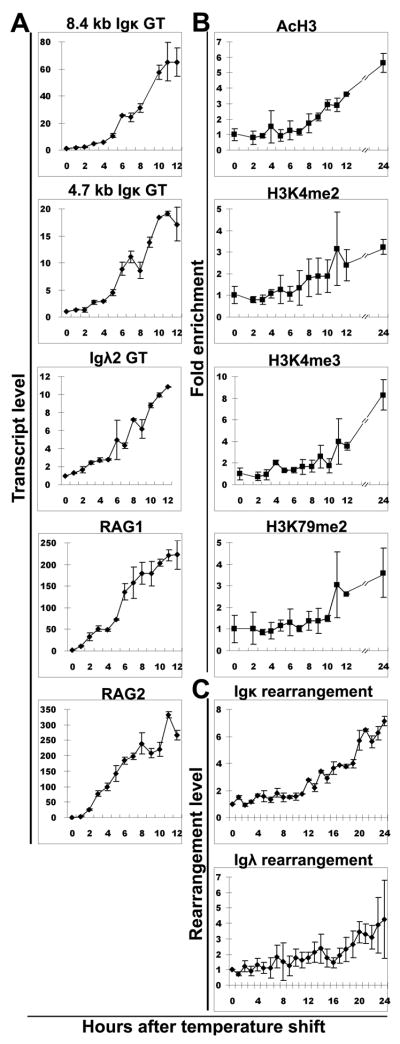
We hypothesize that an important role of GT is to recruit histone methytransferases or acetyltransferases, some of which are known to travel with the RNA polymerase II complex, so that some histone modifications can be added to the chromatin during transcription process. If this hypothesis were correct, we would predict that the GT would precede the induction of the histone modifications, and both would precede gene rearrangement. In one approach to test this hypothesis, we used the 103/bcl2/4 cell line in which light chain rearrangement can be induced. This pre-B cell line was created with a temperature sensitive version of A-MuLV (31). When the cell line is maintained at the permissive temperature of 34°C, there is no light chain rearrangement, no GT, and little RAG expression. Upon shift to the non-permissive temperature of 39 °C, v-Abl is inactivated, and RAG mRNA, GT and H3K4 methylation are induced (17, 31). We first performed a time course for the induction of the two κ GT and RAG expression. As seen in Fig. 2A, induction of GT is initiated within 4 hours after temperature shift, in agreement with published data (17, 31, 32) (Fig. 2A). We also performed a ChIP time course assay for 4 of the PTMs which we found to be associated with rearrangement status in our analysis of B cell subpopulations (Fig. 1). We observed that all four active PTMs (H3K4me2, H3K4me3, H3K79me2 and AcH3) begin to be induced on Jκ1 at 6–8 hours after temperature shift (Fig. 2B), thus lagging somewhat behind the increase in GT. The level of PTM remained low on Vκ genes as has been previously observed (32). Kappa rearrangement is much slower than PTMs, not beginning until at least 12 hours after temperature shift. Low levels of λ rearrangement can be observed in this inducible system, but it lagged behind κ rearrangement, just as λ rearrangement follows κ rearrangement in vivo (Fig. 2C).
Figure 2. Time course of germline transcription, PTMs and gene rearrangement in 103/bcl2/4 cell line after temperature shift to 39 °C.
A. Induction of expression of GT (8.4 kb and 4.7 kb Jκ, Jλ2), RAG1 and RAG2 after temperature shift. B. Induction of AcH3, H3K4me2, H3K4me3 and H3K79me2 at Jκ1 after temperature shift. C. Induction of κ and λ rearrangement after temperature shift.
Thus, in this system, GT is induced slightly earlier than the active PTMs that we measured, suggesting that the active PTMs did not make the region accessible for GT, but rather that GT preceded the induction of these active PTMs. This is consistent with the hypothesis that an important role of GT is to have histone methyltransferases and acetyltransferases travel with the RNA pol II complex and add histone PTMs, which can later play a role in facilitating V(D)J recombination.
Epigenetic status of Igκ and Igλ genes in vivo
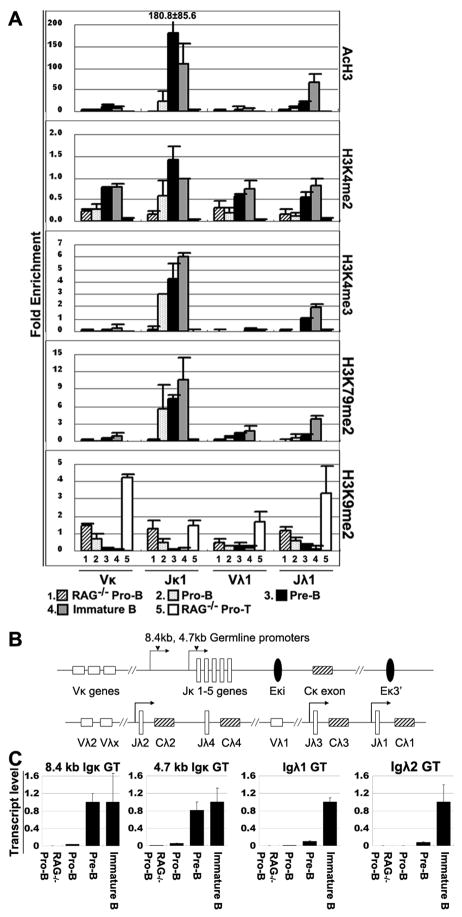
Although κ genes rearrange before λ genes in vivo, the mechanism by which the λ genes are made accessible later in B cell development than κ genes is unresolved (4). We hypothesized that there was an epigenetic basis for this phenomenon. To clearly display the dynamic patterns of the PTMs associated with light chain genes throughout B cell differentiation, we plotted the PTMs from Fig. 1 as they change throughout B cell development (Fig. 3A). The level of all the PTM other than H3K4me2 remained relatively low on V genes throughout differentiation. All active PTMs begin to appear on Jκ1 in BALB/c pro-B cells, and are very high in pre-B cells, where κ rearrangements occur. The active PTMs on Jλ are much lower than those on Jκ in pro- and pre-B stage, and rise significantly as cells progress from the pre-B compartment into the immature B compartment. The immature B cell compartment is the stage where receptor editing is initiated, and λ rearrangements are a hallmark of receptor editing. Thus the patterns of these active PTMs on Jκ and Jλ correlate well with the sequential pattern of light chain rearrangement, and therefore is a very promising one for controlling accessibility.
Figure 3. The patterns of PTMs and germline transcripts on Ig light chain genes during B cell development.
A. ChIP data from Fig. 1 on IgL genes in pro-B and pro-T cells from 4–6 week old RAG1−/− mice; pro-B, pre-B and immature B cells from 4–6 week old BALB/c mice. B. Genomic organization of the murine Igκ and Igλ locus. Top: relative positions of the Jκ gene segments and Cκ exon, and two enhancer regions: the intronic (MiEκ) and 3′Eκ enhancers. Bottom: relative positions of the Vλ and Jλ gene segments and Cλ exons. Arrows denote the positions of the 8.4 kb and 4.7 kb germline transcript promoters. C. GT levels of 8.4 kb and 4.7 kb Jκ, and Jλ2 loci. The level of each GT is plotted relative to the level in immature B cells.
We determined the level of κ and λ GT in the same B cell subpopulations as we analyzed for PTM (Fig. 3C). The κ GT are present at proximately equal level in pre-B and immature B cells, whereas λ GT rise > 10-fold between the pre-B and immature B cell compartments.
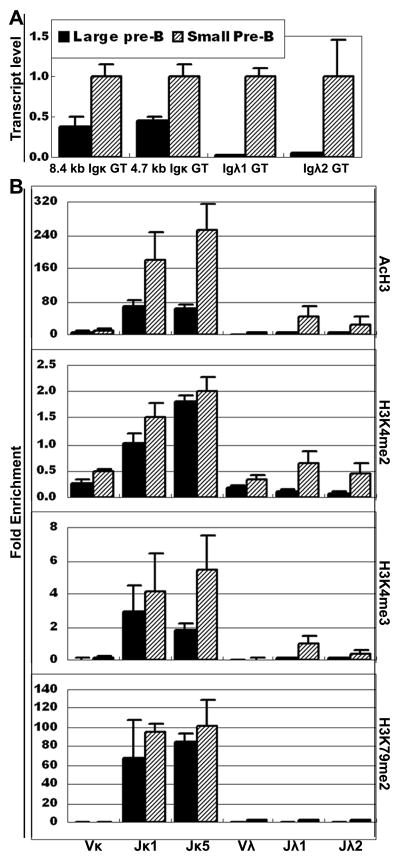
There is a noticeable difference between RAG−/− pro-B cells and BALB/c pro-B cells in both the extent of PTM and in the level of GT. All 4 active PTM are absent from RAG−/− pro-B cells except for a low level of H3K4me2. In contrast, the level of AcH3, H3K4me3 and K3K79me2 are higher on BALB/c pro-B cells than on RAG−/− pro-B cells, as is the level of κ and λ GT (Fig. 3A, 3C). The RAG−/− pro-B cells have not undergone any heavy chain rearrangement, due to the absence of the RAG recombinase. The BALB/c pro-B cells were isolated by the expression of CD43, and so also contain the more differentiated Fr. C′ cells (29), which are cells that have completed heavy chain rearrangement and are undergoing proliferation as a result of pre-BCR signaling. Thus we hypothesized that the differences between the RAG−/− pro-B and BALB/c pro-B cells were likely to be a reflection of the later developmental stages included in the BALB/c pro-B cell population. We therefore isolated pre-B cells using a different strategy by sorting for CD19+CD25+IgM− cells, and fractionating them into the large pre-B compartment (equivalent to Fr. C′ cells) and the small pre-B compartment (33) based on light scatter (4). We demonstrated that the large pre-B cells already have activated the κ locus, as measured by GT (Fig. 4A), whereas the level of λ GT is essentially undetectable at the large pre-B cell stage. This observation is consistent with data of Engel et al. and clearly demonstrates that κ GT precedes λ GT (4). Similarly, we also found that the extent of all three methylation PTMs on Jκ genes in the large pre-B cells are already almost at the level observed in small pre-B cells. In contrast, all 4 PTMs are virtually absent on Jλ genes at the large pre-B cell stage, other than low level of H3K4me2 (Fig. 4B). Even at the small pre-B cell stage, the level of PTM on Jλ genes is low, and they mainly rise on Jλ genes at the immature B cell stage (Fig. 3A). These data indicate that the delayed initiation of rearrangement of λ genes compared to κ genes could be epigenetically controlled, with delayed onset of GT and greatly delayed induction of active PTMs in λ genes.
Figure 4. Jκ and Jλ germline transcription and PTMs in large and small pre-B cells.
A. GT levels of 8.4 kb and 4.7 kb Jκ, Jλ1 and Jλ2 loci in large pre-B and small pre-B cells from 4–6 week old B6 mice. The level of each GT in large pre-B cells is plotted relative to the level in small pre-B cells. B. PTMs patterns on Ig κ and λ genes in large pre-B and small pre-B cells.
The active PTMs on Jκ genes only appear after pre-BCR signaling
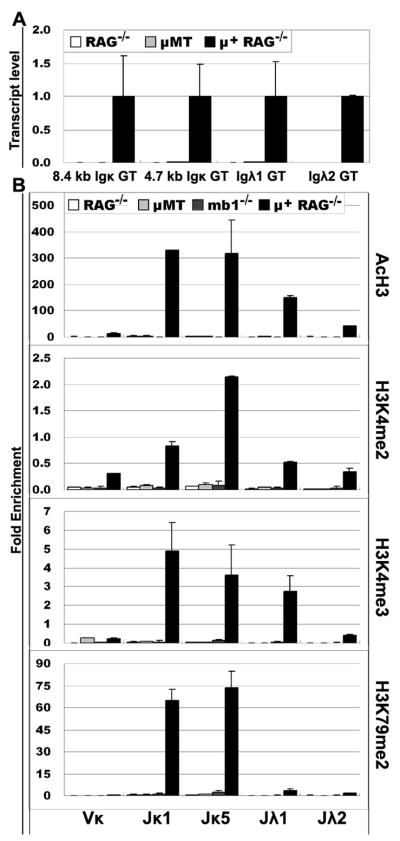
Since the proliferation in the large pre-B compartment is triggered by pre-BCR signaling, we hypothesized that pre-BCR signaling may be required for the induction of these epigenetic changes resulting in increased accessibility at the light chain loci. To test this hypothesis, we compared pro-B cells from μMT mice and from mb1−/− mice to pre-B cells from RAG−/− mice bearing a rearranged μ gene (μ+RAG−/− mice). Mb1 encodes Igα, one of the two signal transducing subunits of the pre-BCR complex (28). μMT mice have a germline disruption of the membrane exon of the μH gene, resulting in no pre-BCR signaling and hence a complete block of B cell development at the pro-B cell stage (34). In contrast to these two strains of mice, the presence of a rearranged μH gene in μ+RAG−/− mice allows pre-BCR signaling and progression of developmentally arrested RAG−/− pro-B cells to the small pre-B cell stage (35). Thus, in all cases, the cells will have a heavy chain protein, but only the cells from the μ+RAG−/− mice are capable of transmitting a pre-BCR signal. Consistent with our hypothesis, we observed that the κ locus had no active PTM in κ genes in μMT or mb1−/− pro-B cells, nor did they have any κ GT. In contrast, we observed that κ GT are dramatically induced in progenitors from μ+RAG−/− mice and we observed a large induction of the four active PTMs in Jκ light chain genes compared with pro-B cells from RAG−/− or μMT mice (Fig. 5). These results demonstrate that pre-BCR signals are necessary for both the induction of GT of light chain genes and also active PTMs on light chain J genes which are poised for rearrangement during pre-B stage.
Figure 5. Jκ and Jλ germline transcription and PTMs in μ+ RAG−/− cells, RAG−/−, mb1−/− and μMT pro-B cells.
A. Transcript levels of Jκ and Jλ loci in pre-B cells from μ+RAG−/− mice or in pro-B cells from RAG−/− or μMT mice. The level of each GT is plotted relative to the level in μ+RAG−/− pre-B cells. B. PTMs patterns on Ig κ and λ genes in pre-B cells from μ+RAG−/− mice or in pro-B cells from RAG−/−, μMT or mb1−/− mice.
Reduction of active PTMs in Jκ genes from MiEκ−/− mice
The κ locus has two enhancers, κ intronic (MiEκ) and 3′ (3′Eκ). In MiEκ−/− or 3′Eκ−/− mutant B cells, κ GT is greatly reduced and κ rearrangement is defective, and in mice deficient in both κ enhancers, κ rearrangement is abolished (27, 36, 37). To study the role of the enhancer in regulating PTMs in vivo, we isolated the pre-B cells from MiEκ −/− mice. We found 8~10-fold decreases of the two Jκ GT and of Igκ rearrangement in MiEκ −/− pre-B cells (Fig. 6A, B). In contrast Jλ GT and Igλ rearrangement were not affected in MiEκ−/− mice. ChIP analysis revealed the dramatic reduction of the four active PTMs on Jκ genes in MiEκ −/− pre-B cells, but no reduction on Jλ genes (Fig. 6C). PTMs on Vκ were unchanged. These in vivo data suggested that MiEκ, which is necessary for high levels of κ GT, also plays an important role in adding active PTMs to the Jκ genes.
Figure 6. Germline transcription, rearrangement and PTMs in MiEκ −/− pre-B cells.
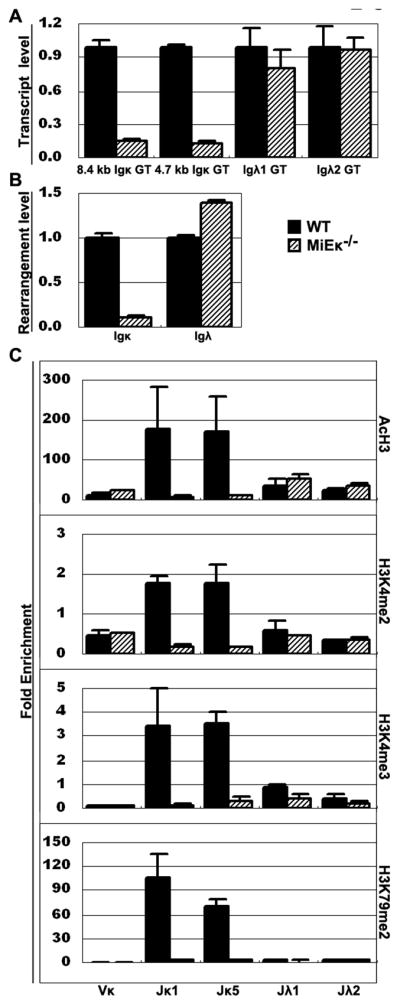
A. Transcript levels of Jκ and Jλ loci in pre-B cells from MiEκ −/− or WT mice. The level of each GT in MiEκ −/− cells is plotted relative to the level in WT pre-B cells. B. IgL rearrangement levels in pre-B cells from MiEκ−/− or WT mice. C. PTMs patterns on Igκ and Igλ genes in pre-B cells from MiEκ−/− or WT mice.
Discussion
In this study, we have characterized the pattern of histone PTMs of V, D and J genes from IgH, κ, λ and TCRβ loci during lymphocyte development. The pattern of PTMs throughout differentiation, and among lineages, is consistent with the hypothesis that chromatin changes correlate with the accessibility of genes for rearrangement. However, we observed significant differences in the extent of modifications on the different parts of the loci. Of the 4 active PTMs which we analyzed in this study, only H3K4me2 showed approximately equal modification on V, D and J genes poised for rearrangement, but also showed less stringent lineage-specificity. At the other extreme, the level of H3K4me3 and AcH3 is far higher on J genes than on D or V genes, although the level on Dβ genes is considerable. These two PTMs also demonstrate highly specific T or B lineage restriction. Many labs, including ours, have demonstrated that acetylated H3 is associated with V genes, and that the extent of acetylation can vary from gene to gene (11, 14, 19, 38). However, we show here that the magnitude of the enrichment of these 2 PTMs on J genes is far greater than on V genes, which is in agreement with studies using pro-B cell lines (15). Although the steady state level of GT from proximal VH genes is very low (6, 39), the level of GT from the distal VHJ558 genes is high. Thus, it is somewhat surprising that the distal VH genes show such low levels of H3K4me3 and AcH3 compared to J genes, suggesting that GT alone is not sufficient to result in significant levels of trimethylation of H3K4 or acetylation. H3K79me2 is present on poised D and J genes, but little is observed on poised V genes, and, like AcH3 and H3K4me3, shows very strict T vs. B lineage specificity. Enrichment of H3K4me3 on JH genes rather than VH or DH genes in pro-B cells could be responsible for the fact that DH to JH rearrangement precedes VH to DJH rearrangement since the RAG2 PHD finger, which binds to H3K4me3, has been shown to bind to JH, but not DH, genes in a pro-B cell line (25), and we would predict that RAG2 would also not bind to VH genes. Our findings on pro-B, pre-B and immature B cells suggest that all of the active PTMs appear first on heavy chain and later on light chain, and first in κ genes and later on λ genes, supporting the idea that the order and lineage specificity of V(D)J rearrangement is epigenetically controlled.
We analyzed the induction of these 4 PTM and of GT at the two light chain loci. We demonstrated that cells that cannot receive a pre-BCR signal have no PTMs on the κ genes and little GT. RAG−/− mice bearing a rearranged heavy chain gene show high levels of PTMs and normal pre-B cell levels of GT. Thus, this data, especially the analysis of the mb1−/− mice which cannot transduce the pre-BCR signal, strongly supports the hypothesis that the pre-BCR signal is required to activate the κ locus. We further showed that the Jκ region has almost full levels of histone PTMs during the proliferating large pre-B cell stage, while the λ locus is still essentially silent. Even at the small pre-B cell stage, the level of PTMs on the λ genes is quite low compared to the level on the immature B cells. Thus, the delayed onset of λ rearrangement is preceded by delayed onset of GT and delayed acquisition of active PTM in λ genes. It has been demonstrated that half of the small pre-B cells have both λ5 alleles associated with pericentric heterochromatin (40) and the VpreB2 gene shows similar association with heterochromatin (J. Skok, personal communication). λ5 is 2.2 Mb away from the λ light chain locus, and VpreB2 is 1 Mb away. If the λ locus were also associated with pericentric heterochromatin, this could help enforce the absence of active PTM until the locus was released.
The accessibility of Ig and TCR genes is strongly correlated with GT of unrearranged genes, which occurs prior to recombination (6, 41). However, it is not clear if the act of transcription makes the genes accessible, or if chromatin modifications precede GT and if the PTMs make the region accessible for TF binding and subsequent transcription. Abarrategui and Krangel demonstrated that blocking GT can block TCRα rearrangement by inserting a transcriptional terminator into the midst of the Jα locus (42). The terminator blocked transcription of the few Jα elements downstream of the terminator, and blocked rearrangement of those J genes, demonstrating a requirement of GT for rearrangement of these genes. Interestingly, the level of H3K4 methylation was significantly reduced on those same Jα genes immediately downstream of terminator, although the level of histone acetylation was not changed. The reduction in H3K4me3 by the transcriptional terminator is likely to be directly related to the block in rearrangement, since the PHD domain in RAG2 binds H3K4me3 (25, 26). Abarretegui and Krangel also targeted the same transcriptional terminator to a site just downstream of TEA, and again blocked GT for the next few Jα genes, and completely blocked their rearrangement (42). In this case, however, the induction of all histone PTMs was completely blocked including acetylation. Here we performed studies to determine if these active PTMs are added to chromatin before or after GT is induced in the inducible 103/bcl2/4 cell line, and found GT preceded the induction of these four active PTMs. We also showed the important role of the MiEκ in the induction of histone methylation on κ genes through the analysis of MiEκ −/− mice. We hypothesize that MiEκ may indirectly affect PTM levels through the GT process.
This work demonstrates that 4 PTM that are associated with active or poised genes show dynamic patterns of association with Ig and TCR gene segments throughout lymphocyte differentiation, with 3 of the 4 PTM demonstrating far higher levels on J genes than on D or V genes. As the dominant GT takes place through the J and C regions, this is consistent with the hypothesis that GT is playing an important role in the acquisition of the PTM. Analysis of mutant mice unable to signal through the pre-BCR receptor shows the key role of this process in the induction of light chain gene accessibility. Together, our data indicates that the epigenetic profile of Ig light chain genes, which is necessary for gene rearrangement, is strongly influenced by GT and by pre-BCR signaling.
Supplementary Material
Acknowledgments
We gratefully acknowledge the excellent technical support of Timothy Wong. We thank Dr. Yang Xu and Olga Gaidarenko for the MiEκ−/− mice, and Dr. Michael Reth for the mb1−/− mice. We thank Dr. Kenneth Zaret for careful reading and comments on this manuscript.
Abbreviations
- PTMs
post-translational modifications
- GT
Germline transcription
- PHD
Plant homeodomains
- ChIP
Chromatin immunoprecipitation
- A-MuLV
Abelson Murine Leukemia Virus
Footnotes
This work was supported by NIH grants R01 A129672, R01 A152313 and R01 AI61167 to A.J.F.
References
- 1.Martensson IL, Keenan RA, Licence S. The pre-B-cell receptor. Curr Opin Immunol. 2007;19:137–142. doi: 10.1016/j.coi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Durdik J, Moore MW, Selsing E. Novel kappa light-chain gene rearrangements in mouse lambda light chain-producing B lymphocytes. Nature. 1984;307:749–752. doi: 10.1038/307749a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Trounstine M, Kurahara C, Young F, Kuo CC, Xu Y, Loring JF, Alt FW, Huszar D. B cell development in mice that lack one or both immunoglobulin kappa light chain genes. EMBO J. 1993;12:821–830. doi: 10.1002/j.1460-2075.1993.tb05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel H, Rolink A, Weiss S. B cells are programmed to activate kappa and lambda for rearrangement at consecutive developmental stages. Eur J Immunol. 1999;29:2167–2176. doi: 10.1002/(SICI)1521-4141(199907)29:07<2167::AID-IMMU2167>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Yancopoulos GD, Alt FW. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]
- 6.Yancopoulos GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 7.Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc Natl Acad Sci U S A. 2002;99:12309–12314. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angelin-Duclos C, Calame K. Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment. Mol Cell Biol. 1998;18:6253–6264. doi: 10.1128/mcb.18.11.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada A, Mendelsohn M, Alt F. Differential activation of transcription versus recombination of transgenic T cell receptor beta variable region gene segments in B and T lineage cells. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 11.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol Cell Biol. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci U S A. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkins EJ, Kee BL, Ramsden DA. Histone 3 lysine 4 methylation during the pre-B to immature B-cell transition. Nucleic Acids Res. 2004;32:1942–1947. doi: 10.1093/nar/gkh523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 19.Espinoza CR, Feeney AJ. The extent of histone acetylation correlates with the differential rearrangement frequency of individual VH genes in pro-B cells. J Immunol. 2005;175:6668–6675. doi: 10.4049/jimmunol.175.10.6668. [DOI] [PubMed] [Google Scholar]
- 20.Osipovich O, Cobb RM, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat Immunol. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- 21.Svejstrup JQ. Chromatin elongation factors. Curr Opin Genet Dev. 2002;12:156–161. doi: 10.1016/s0959-437x(02)00281-2. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 24.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Ig kappa light chain intronic enhancer/matrix attachment region impairs but does not abolish V kappa J kappa rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 28.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 31.Chen YY, Wang LC, Huang MS, Rosenberg N. An active v-abl protein tyrosine kinase blocks immunoglobulin light-chain gene rearrangement. Genes Dev. 1994;8:688–697. doi: 10.1101/gad.8.6.688. [DOI] [PubMed] [Google Scholar]
- 32.Fitzsimmons SP, Bernstein RM, Max EE, Skok JA, Shapiro MA. Dynamic changes in accessibility, nuclear positioning, recombination, and transcription at the Ig kappa locus. J Immunol. 2007;179:5264–5273. doi: 10.4049/jimmunol.179.8.5264. [DOI] [PubMed] [Google Scholar]
- 33.Rolink A, Grawunder U, Winkler TH, Karasuyama H, Melchers F. IL-2 receptor alpha chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int Immunol. 1994;6:1257–1264. doi: 10.1093/intimm/6.8.1257. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 35.Spanopoulou E, Roman CA, Corcoran LM, Schlissel MS, Silver DP, Nemazee D, Nussenzweig MC, Shinton SA, Hardy RR, Baltimore D. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 36.Gorman JR, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt FW. The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 37.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 38.Hawwari A, Krangel MS. Regulation of TCR delta and alpha repertoires by local and long-distance control of variable gene segment chromatin structure. J Exp Med. 2005;202:467–472. doi: 10.1084/jem.20050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love VA, Lugo G, Merz D, Feeney AJ. Individual V(H) promoters vary in strength, but the frequency of rearrangement of those V(H) genes does not correlate with promoter strength nor enhancer-independence. Mol Immunol. 2000;37:29–39. doi: 10.1016/s0161-5890(00)00023-7. [DOI] [PubMed] [Google Scholar]
- 40.Parker MJ, Licence S, Erlandsson L, Galler GR, Chakalova L, Osborne CS, Morgan G, Fraser P, Jumaa H, Winkler TH, Skok J, Martensson IL. The pre-B-cell receptor induces silencing of VpreB and lambda5 transcription. EMBO J. 2005;24:3895–3905. doi: 10.1038/sj.emboj.7600850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Ness BG, Weigert M, Coleclough C, Mather EL, Kelley DE, Perry RP. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 1981;27:593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- 42.Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat Immunol. 2006;7:1109–1115. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.