Abstract
Farnesyltransferase inhibitors (FTIs) represent a new class of anticancer drugs that show promise in blocking the growth of tumors. Here, we report that FTIs are capable of inducing apoptosis of transformed but not untransformed cells. Treatment of v-K-ras-transformed normal rat kidney (KNRK) cells with FTIs leads to the induction of apoptotic cell morphology, chromatin condensation and DNA fragmentation. In addition, fluorescence-activated cell sorter analysis of FTI-treated KNRK cells shows a sub-G1 apoptotic peak (chromosome content of <2 N). This FTI-induced apoptosis is evident only when the cells are grown in low serum conditions (0.1% fetal calf serum) and is observed selectively with transformed KNRK cells and not with untransformed NRK cells. Further analysis of the mechanism underlying this apoptosis has shown that FTI treatment of KNRK cells results in the activation of caspase 3 but not caspase 1. Moreover, the addition of Z-DEVD-fmk, an agent that interferes with caspase 3 activity, can inhibit FTI-induced apoptosis in a dose-dependent manner. Introduction of the CASP-3 gene into MCF7 cells, which lack caspase 3 activity, results in a significant increase of FTI-induced apoptosis. Furthermore, FTI induces the release of cytochrome c into the cytosol. This release is an important feature of caspase 3-mediated apoptosis. These results suggest that FTIs induce apoptosis through the release of cytochrome c from the mitochondria resulting in caspase 3 activation.
Recent studies on protein farnesylation have led to the development of a new class of anticancer drugs, known as farnesyltransferase inhibitors (FTIs), which show promise in blocking the growth of tumors with minimal toxicity to normal cells (1–3). These compounds inhibit protein farnesyltransferase (4), an enzyme that catalyzes the farnesylation of a number of proteins including members of the Ras superfamily of small G proteins. These farnesylated proteins terminate in a C-terminal CAAX motif (C is cysteine, A is an aliphatic amino acid, and X is usually serine, cysteine, glutamine, methionine, or alanine) that is recognized by protein farnesyltransferase. A variety of animal studies have demonstrated the ability of FTIs to block or even regress growth of tumor cells (5–7). FTIs have been tested on a collection of human tumor cell lines, and have been shown to block anchorage-independent growth in >70% of cell lines tested, suggesting that FTIs inhibit the growth of a wide range of human tumors (8). FTIs are currently being assessed in clinical trials (9).
We previously reported that FTIs cause dramatic morphological changes of v-K-ras-transformed normal rat kidney (KNRK) cells (10). We now report that FTI is also able to induce apoptosis in KNRK cells as well as other transformed cell lines. This observation is significant since the induction of apoptosis by FTI suggests that protein farnesylation plays a critical role in apoptosis. Furthermore, FTI-induced apoptosis could be exploited as a treatment to eliminate cancer cells. For these reasons, we have undertaken a systematic study to further characterize FTI-induced apoptosis. Previous reports (11–13) have appeared that support the idea that FTI induces apoptosis. However, these results are fragmentary and sometimes conflicting. Lebowitz et al. (11) observed FTI induced apoptosis in H-ras-transformed Rat1 cells when these cells were detached from the substratum. On the other hand, Hung and Chuang (12) reported that FTI alone was sufficient to induce apoptosis of human ovarian cancer cells. Therefore, we first sought to establish FTI’s capability to induce apoptosis. We also addressed a number of issues including whether induction of apoptosis occurs with different classes of FTI compounds and whether this induction is specific to transformed cells. Furthermore, we investigated whether FTI induces apoptosis through the caspase cascade.
Caspases are a family of cysteinyl aspartate-specific proteinases that play key roles in apoptosis (14, 15). To date, 13 caspases have been reported (14). Among these caspases, caspase 3 is considered to be a crucial enzyme since it is commonly activated by a number of apoptotic stimuli and has a variety of substrates that include poly(ADP-ribose) polymerase, nuclear lamins, and focal adhesion kinase (14, 15). Furthermore, caspase-3 knock-out mice exhibit defects in apoptosis of neuronal cells and brain development (16). One mechanism of caspase 3 activation involves cytochrome c, which is released from the mitochondria into the cytosol during apoptosis (17–19). The release of cytochrome c activates the apoptotic protease activating factor, Apaf-1, which then initiates a protease cascade resulting in the activation of caspase 3. Therefore, we investigated the possible roles of caspase 3 and cytochrome c in FTI-induced apoptosis.
In this paper, we report that a number of different FTI compounds are capable of inducing apoptosis in KNRK as well as other tumor cell lines. This FTI-induced apoptosis is more specific to transformed cells, because apoptosis is observed in KNRK cells but not in NRK cells. In addition, we demonstrate that caspase 3 is activated by FTI and that this activation is critical for FTI-induced apoptosis. We further show that FTI induces cytochrome c release from the mitochondria, an event that has been shown to trigger a cascade leading to caspase 3 activation.
MATERIALS AND METHODS
Materials.
Four different FTI compounds were used in this study. SCH56582, a tricyclic inhibitor of protein farnesyltransferase and a derivative of SCH44362 (20, 21), was provided by W. R. Bishop (Schering-Plough). The peptidomimetic inhibitors, BMS191563 (22) and B1088 (7), were provided by V. Manne (Bristol-Myers Squibb) and A.M. Garcia (Eisai Research Institute, Andover, MA), respectively. Another peptidomimetic inhibitor, FTI-277 (23), was provided by S. Sebti (University of South Florida) or purchased from Calbiochem. The caspase-3 fluorogenic substrate, Ac-DEVD-AMC, and caspase-1 fluorogenic substrate, Ac-YVAD-AMC, were purchased from PharMingen (AMC, 7-amino-4-methylcoumarin). Z-DEVD-fmk was purchased from Enzyme Systems Products. 4,6-Diamidino-2-phenylindole (DAPI) was purchased from Sigma. A plasmid containing the caspase 3 gene (CASP-3) was provided by K. Ueno (Kagoshima University, Japan). PD98059 was purchased from NEB, and wortmannin was purchased from Calbiochem.
Cells.
KNRK (CRL-1569), NRK (CRL-1570), and MCF7 cells were obtained from American Type Culture Collection (Manassas, VA). KNRK cells are NRK cells transformed with v-K-ras (24). MCF7 cells are human breast carcinoma cells that lack caspase 3 activity owing to a 47-bp deletion in exon 3 of the CASP-3 gene (25). Spon 8 cells are tumorigenic, metastatic mouse lung cancer cells (26) and were provided by M. You (Ohio State University). KNRK cells and NRK cells were grown in DMEM containing 10% or 0.1% fetal calf serum (FCS), while MCF7 cells and Spon 8 cells were grown in RPMI 1640 medium containing 10% or 0.1% FCS (26).
Preparation of Cytosolic and Mitochondrial Fractions and Western Blot Analysis.
Cytosolic and mitochondrial fractions from FTI-treated KNRK or NRK cells were prepared essentially as described (27). Briefly, cells treated with FTI were collected and resuspended in 300 μl of buffer A (20 mM Hepes-KOH, pH 7.5/10 mM MgCl2/1 mM EDTA/1 mM EGTA/1 mM DTT) containing 250 mM sucrose and 1× protease inhibitor cocktail, Complete (Boehringer Mannheim). After homogenization, unbroken cells, large plasma membrane pieces, and nuclei were removed by centrifugation at 1,000 × g for 10 min. The supernatant was subjected to centrifugation at 10,000 × g for 20 min. The pellet fraction containing mitochondria was resuspended in 50 μl of TNC buffer (10 mM Tris-acetate, pH 8.0/0.5% Nonidet P-40/5 mM CaCl2). The supernatant was further centrifuged at 50,000 g for 2 h to generate cytosol. For detection of cytochrome c, cytosolic and the mitochondrial fractions were analyzed by Western blotting as described (22) using a monoclonal anti-cytochrome c antibody (PharMingen). A monoclonal anti-actin antibody (Sigma) was used to detect actin. Proteins were visualized using enhanced chemiluminescence (ECL; Amersham) after incubation with the appropriate peroxidase-conjugated secondary antibody (Sigma).
DAPI Staining and DNA Fragmentation.
Cells were harvested, washed in 1× PBS, and fixed with 4% paraformaldehyde. The cells were then permeabilized by 0.5% Triton X-100 in 1× PBS. After permeabilization, cells were stained for 30 min with DAPI (1 μg/ml) and analyzed via fluorescence microscopy to assess chromatin condensation and segregation. To detect DNA fragmentation, cellular DNA was prepared using the Blood and Cell Culture Mini DNA kit (Qiagen, Chatsworth, CA). Purified DNA was then incubated for 2 h at 37°C with 0.2 mg/ml RNase A and analyzed on 1.7% agarose gel. DNA was visualized by ethidium bromide staining.
Detection of Cell Death.
Cells treated with FTIs were analyzed by propidium iodide (PI) staining and flow cytometry with a FACScan flow cytometer (Becton Dickinson Immunocytometry Systems) to assess apoptosis. 10,000 events were collected to maximize the statistical validity of the compartmental analysis. Cell death was also evaluated by trypan blue dye exclusion. Thirty microliters of 0.4% trypan blue solution was added to 30 μl of cell suspension (5 × 104 cells/ml). Both viable (unstained) and nonviable (stained) cells were counted using a hemocytometer. A minimum of 100 cells was counted for each data point in a total of nine microscopic fields. Cell death was calculated as the percent of stained cells out of the total number of cells counted.
Caspase Activity Assay.
Caspase 3 activity in KNRK cells treated with FTI, PD98059 (28) or FTI plus Z-DEVD-fmk (29) were analyzed by using the substrate Ac-DEVD-AMC (PharMingen) essentially as described by Hughes et al. (30). Active caspase 3 cleaves the substrate Ac-DEVD-AMC after the aspartic acid (D) residue and before the AMC group. The released AMC becomes fluorescent, and the fluorescence is quantified by flow cytometry. At least 10,000 cells were examined for statistical validity. The caspase 1 assay was carried out similarly using the substrate Ac-YVAD-AMC (PharMingen).
Stable CASP-3 (Caspase 3) Transformants of MCF7 Cells.
The gene encoding wild type caspase 3 was inserted into the EcoRI-XbaI sites of pEGFP-C1 vector (CLONTECH). The mammalian pEGFP-C1 vector carries the green fluorescent protein gene under the control of the cytomegalovirus (CMV) promoter. The resulting pEGFP-C1-CASP-3 plasmid and the pEGFP-C1 vector were transfected into MCF7 cells using Lipofectamine (GIBCO). Stable MCF7 transformants expressing GFP-casapase-3 (MCF7-CASP-3) or expressing GFP (MCF7-vector) were established by selecting for resistance to 500 μg/ml G418.
RESULTS
FTI Induces Apoptosis in Low Serum Conditions.
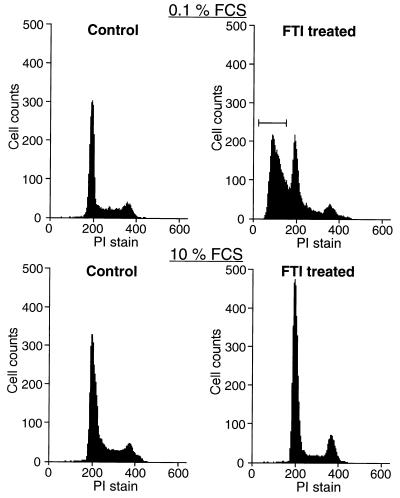
We previously demonstrated that KNRK cells (v-K-ras-transformed NRK cells) revert to a flattened, elongated phenotype resembling untransformed NRK cells when treated with the tricyclic FTI, SCH56582 (10). In these experiments, which were performed in 10% FCS medium, no cell death or toxicity was observed (10). However, we found that treating KNRK cells with SCH56582 in low serum medium (0.1% FCS) resulted in significant cell death. Under these conditions, apoptotic KNRK cells were evident 12–24 h after the addition of FTI. These cells showed morphology typical of apoptotic cells. Fig. 1 Upper shows the fluorescence-activated cell sorter (FACS) analysis of KNRK cells in 0.1% FCS medium. The addition of SCH56582 led to the appearance of a significant fraction of cells in the region corresponding to cells with a DNA content less than 2 N, a characteristic typical of apoptotic cells. Apoptotic cells were not detected when KNRK cells were treated with dimethyl sulfoxide (DMSO) instead of FTI (Control). When KNRK cells were grown in media containing 10% FCS, apoptotic cells were not detected even after FTI treatment (Fig. 1, Lower). In this case, as reported (10), an increased percentage of G1 phase cells and decreased percentage of S phase cells were observed.
Figure 1.
Flow cytometric analysis of apoptosis in KNRK cells. KNRK cells were plated at a density of 5 × 105 cells/60-mm dish in 10% FCS medium. After 24 h, the medium was changed to either 0.1% FCS medium or 10% FCS medium and the cells were treated with DMSO (Control) or with 20 μM SCH56582 (FTI treated). The cells were collected, stained with PI, and analyzed for DNA content by flow cytometry. Apoptotic cells with a DNA content of <2 N are marked by brackets. At least two separate experiments were carried out with results similar to those shown here.
FTI-induced apoptosis is not unique to KNRK cells. In the mouse lung cancer cell line, Spon 8, 66.3% of SCH56582-treated cells appeared as apoptotic cells (<2 N DNA content) by FACS analysis (Table 1), while only 4.7% of DMSO-treated cells had <2 N DNA content. In addition, the ovarian cancer cell line PA-1 also underwent apoptosis; 58.5% of SCH56582-treated cells were apoptotic by FACS analysis. In a control experiment using DMSO instead of FTI, only 7.6% of PA-1 cells were apoptotic.
Table 1.
Induction of apoptosis by a variety of FTI in Spon 8 cells
| Compound | Cell line | % cells with <2 N DNA content |
|---|---|---|
| DMSO (control) | Spon 8 | 4.7 |
| SCH56582 (FTI) | Spon 8 | 66.3 |
| BMS191563 (FTI) | Spon 8 | 62.4 |
| B1088 (FTI) | Spon 8 | 35.5 |
| FTI-277 (FTI) | Spon 8 | 27.6 |
Spon 8 cells were incubated in media containing 0.1% FCS and treated with DMSO, 20 μM SCH56582, 100 μM BMS191563, 80 μM B1088, or 80 μM FTI-277 for 36 h. The cells were analyzed by propidium iodide staining and flow cytometry as described in Materials and Methods. 10,000 cells were examined in each experiment. Apoptotic cell populations were analyzed by using CELL-Quest software (Becton Dickinson Immunocytometry Systems). At least two separate experiments were carried out with results similar to those shown here.
A Variety of FTI Compounds, but Not MEK Inhibitor or Wortmannin, Induce Apoptosis in KNRK cells.
We also investigated whether FTI compounds other than the tricyclic inhibitor SCH56582 are capable of inducing apoptosis. The compounds BMS191563, B1088, and FTI-277 were also tested. These compounds are peptidomimetic inhibitors derived from CAAX tetrapeptides (1–3). All of these compounds induced apoptosis of Spon 8 cells as evidenced by the appearance of cells with <2 N DNA content (Table 1). Though the efficiencies of the induction differed somewhat among these compounds, these results indicate that the induction of apoptosis is a property common to a variety of different FTI compounds.
We then investigated whether or not FTIs are unique among signal transduction inhibitors in their ability to efficiently induce apoptosis in transformed cells. We tested two widely used compounds, PD98059 (MEK inhibitor), which inhibits MEK activation of Erk1 and Erk2 (28), and wortmannin KY12420, which inhibits phosphatidylinositol 3-kinase (31). These inhibitors are of particular interest because they have been shown to block activation of the mitogen-activated protein kinase pathway and the phosphatidylinositol 3-kinase/Akt pathway, two pathways that function downstream of Ras (32, 33). In addition, these two pathways are reported to modulate apoptotic signals (32, 33). The effects of these compounds on KNRK cells grown in 0.1% FCS are shown in Table 2. Addition of PD98059 did not result in the appearance of apoptotic cells. The concentration of PD98059 used in these experiments was sufficient to inhibit activation of Erk in KNRK cells as determined by the use of an antiphosphorylated Erk antibody (data not shown). Addition of wortmannin, even at concentrations greater than that required to inhibit phosphatidylinositol 3-kinase (31), also did not result in the appearance of apoptotic cells. These results suggest that the pathway that is affected by FTI is distinct from the mitogen-activated protein kinase and the phosphatidylinositol 3-kinase/Akt pathways.
Table 2.
Induction of apoptosis by FTI but not inhibitors of Ras signaling in KNRK cells
| Compound | Cell line | % cells with <2 N DNA content |
|---|---|---|
| DMSO (control) | KNRK | 0.9 |
| SCH56582 (FTI) | KNRK | 56.3 |
| Wortmannin | KNRK | 0.9 |
| PD98059 | KNRK | 3.3 |
KNRK cells were treated with DMSO, 20 μM SCH56582, 1 μM wortmannin KY12420 (phosphatidylinositol 3-kinase inhibitor), or 20 μM PD98059 (MEK inhibitor) for 24 h in 0.1% FCS medium. The cells were analyzed by propidium iodide staining and flow cytometry as described in Materials and Methods; 10,000 cells were examined in each experiment. Apoptotic cell populations were analyzed by using CELL-Quest software (Becton Dickinson Immunocytometry Systems). At least two separate experiments were carried out with results similar to those shown here.
FTI Induces Apoptosis Preferentially in Transformed Cells.
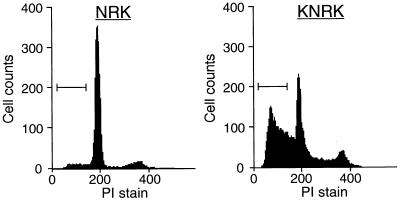
In contrast to transformed cells, FTI was less efficient in inducing apoptosis in untransformed cells. NRK and KNRK cells were grown in 0.1% FCS with the addition of FTI, and the induction of apoptosis was examined by FACS analysis. As can be seen in Fig. 2, apoptosis was efficiently induced in KNRK cells treated with FTI. In contrast, no significant apoptosis was induced in untransformed NRK cells. These results provide the first demonstration that FTI can preferentially induce apoptosis in transformed cells. It also suggests that there is a difference in apoptotic signaling processes between transformed and untransformed cells.
Figure 2.
FTI induces apoptosis in KNRK cells and not in NRK cells. NRK and KNRK cells were plated at a density of 5 × 105 cells/60-mm dish in 10% FCS medium. After 24 h, the medium was changed to 0.1% FCS medium and the cells were treated with DMSO (Control) or with 15 μM SCH56582 (FTI treated). The cells were collected, stained with PI and analyzed for DNA content by flow cytometry. Apoptotic cells with a DNA content of <2 N are marked by brackets. At least two separate experiments were carried out with results similar to those shown here.
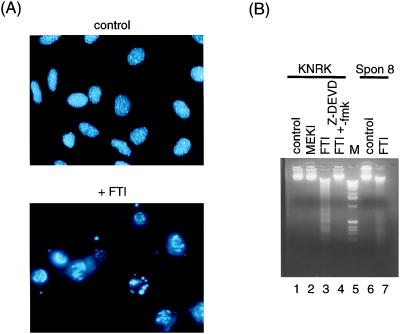
FTI Induces Chromatin Condensation and the Generation of Internucleosomal DNA Fragmentation.
Cells undergoing apoptosis show a characteristic condensation of their chromatin as well as the appearance of internucleosomal DNA ladders (34, 35). These features were examined in FTI treated cells to further characterize the FTI-induced apoptosis. FTI-treated KNRK cells stained with DAPI and visualized by immunofluorescence microscopy showed chromatin condensation and segregation characteristic of apoptotic cells (Fig. 3A). In addition, treatment of KNRK and Spon 8 cells with FTI in low serum medium resulted in the appearance of an internucleosomal DNA ladder (Fig. 3B, lanes 3 and 7). DNA fragmentation was not observed in either KNRK or Spon 8 cells treated with DMSO (Fig. 3B, lanes 1 and 6). In contrast to FTI, the MEK inhibitor PD98059 did not induce DNA fragmentation (Fig. 3B, lane 2). Furthermore, the addition of Z-DEVD-fmk, a caspase 3 inhibitor, almost completely blocked the FTI-induced DNA fragmentation (Fig. 3B, lane 4). This inhibition suggests that caspase 3 is involved in the apoptosis induced by FTI.
Figure 3.
FTI induces chromatin condensation and DNA fragmentation. (A) Chromatin condensation and segregation. KNRK cells were plated at a density of 5 × 104 cells/well in a four-well chamber slide (LAB-TEK) in 10% FCS medium. After 24 h, the medium was changed to 0.1% FCS and the cells were treated with DMSO (Control) or with 20 μM SCH56582 (+ FTI). After 24 h, cells were washed and fixed in 4% paraformaldehyde and stained with DAPI to check chromatin condensation and segregation as described in Materials and Methods. At least three separate experiments were carried out with results similar to those shown here. (B) Internucleosomal DNA fragmentation. KNRK and Spon 8 cells were seeded at 5 × 105 cells/60-mm dish in 10% FCS medium. After 24 h, the medium was changed to 0.1% FCS medium supplemented with DMSO (lanes 1 and 6), 20 μM SCH56582 (lanes 3 and 7), 20 μM PD98059 (lane 2), or 20 μM SCH56582 plus 50 μM Z-DEVD-fmk (lane 4). After 24 h, genomic DNA was prepared and analyzed with a 1.7% agarose gel as described in Materials and Methods. Lane 5 shows DNA size markers (1 kb DNA ladder, GIBCO/BRL).
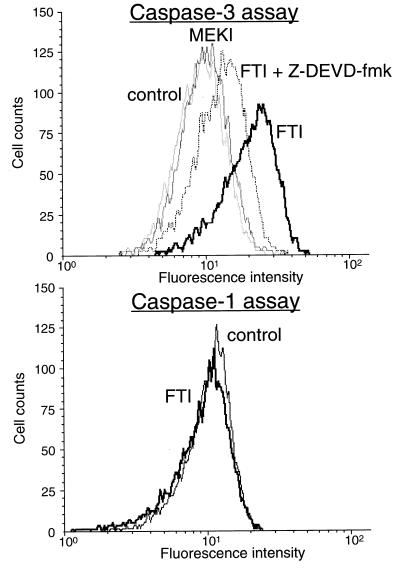
FTI Treatment Induces Activation of Caspase 3 but not Caspase 1.
To gain insights into the mechanism by which FTI induces apoptosis, we investigated effects of FTI on caspases. Of the many caspases involved in apoptosis, we focused on caspase 3, which is activated by a number of apoptotic signals (14, 15). The activity of caspase 3 was measured using the fluorogenic substrate, Ac-DEVD-AMC. Active caspase 3 cleaves Ac-DEVD-AMC between the second aspartic acid (D) and AMC, resulting in the release of the fluorescent AMC. This causes an increase in fluorescence intensity that can be quantified by FACS (excitation 380 nm, emission 450 nm). As shown in the upper panel of Fig. 4, FTI treatment resulted in a marked increase in fluorescence intensity indicating that caspase 3 was activated. Furthermore, the increase in fluorescence was blocked significantly by Z-DEVD-fmk, a caspase 3 inhibitor. FTI’s ability to activate caspase 1 was also tested. As seen in Fig. 4 Lower, FTI treatment did not result in an increase in fluorescence intensity that would be expected from the cleavage of the caspase 1 substrate, Ac-YVAD-AMC. This result indicates that FTI treatment does not activate caspase 1.
Figure 4.
FTI induces activation of caspase 3 but not caspase 1. KNRK cells were treated with DMSO (control), 20 μM PD98059 (MEK inhibitor), 20 μM SCH56582 (FTI), or 20 μM SCH56582 plus 50 mM Z-DEVD-fmk (FTI + Z-DEVD-fmk) in 0.1% FCS medium. Cells were collected after 24 h, and caspase 3 and caspase 1 activities were measured as described in Materials and Methods by using the substrates Ac-DEVD-AMC and Ac-YVAD-AMC, respectively. At least 10,000 cells were examined in each assay.
Caspase 3 Inhibitors Block FTI-Induced Apoptosis.
The idea that caspase 3 plays an important role in FTI-induced apoptosis was further investigated by the use of the caspase 3 peptide inhibitor, Z-DEVD-fmk (29). Z-DEVD-fmk blocks FTI-induced caspase 3 activation as well as FTI-induced DNA fragmentation (Fig. 3 and Fig. 4). To quantitate the amount of apoptosis, percent of cells that underwent apoptosis was determined using trypan blue dye exclusion. As shown in Fig. 5A, a dose-dependent inhibition of apoptosis was observed, with the addition of 50 μM Z-DEVD-fmk resulting in a >70% inhibition of apoptosis. The inhibition was also confirmed by FACS analysis, which showed a drastic reduction in the sub-G1 apoptotic peak in both KNRK and Spon 8 cells (data not shown). These results suggest that caspase 3 activation is required for FTI-induced apoptosis.
Figure 5.
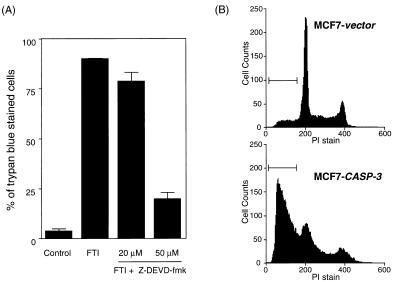
(A) Z-DEVD-fmk blocks FTI-induced apoptotic death in KNRK cells. KNRK cells were treated with DMSO (Control), 20 μM SCH56582 (FTI), 20 μM SCH56582 plus 20 μM Z-DEVD-fmk, or 20 μM SCH56582 plus 50 μM Z-DEVD-fmk in 0.1% FCS medium. The number of dead cells was determined 24 h after the treatment by counting the number of trypan blue-stained cells in 100–200 randomly selected cells. Bars = SD. (B) Introduction of the CASP-3 gene restores FTI-induced apoptosis to MCF7 cells. MCF7-vector (stable MCF7 transformants that express GFP) and MCF7-CASP-3 (stable MCF7 transformants that express GFP fused with the wild-type human caspase 3) cells were treated with 150 μM BMS191563 (FTI) for 24 h in 0.1% FCS medium. The cells were analyzed by PI staining and flow cytometry as described in Materials and Methods. Apoptotic cells with a DNA content of <2 N are marked by brackets. At least two separate experiments were carried out with results similar to those shown here.
Introduction of the CASP-3 Gene Restores FTI Sensitivity to MCF7 Cells.
To further confirm that caspase 3 is critical for the induction of apoptosis by FTI, we took advantage of the breast cancer cell line, MCF7. These cells have no detectable caspase 3 activity due to a deletion in exon 3 of the CASP-3 gene, which encodes procaspase 3 (25). We speculated that MCF7 cells would be resistant to FTI induced apoptosis, and that introduction of the CASP-3 gene into MCF7 cells would restore sensitivity to FTI. To assess this idea, we prepared stable transformants of MCF7 cells expressing caspase 3 (MCF7-CASP-3). A fusion with the green fluorescent protein (GFP) gene was used to confirm expression of CASP-3. MCF7 cells stably transformed with a GFP vector (MCF7-vector) were used as a control. FTI sensitivity of MCF7-vector and MCF7-CASP-3 cells was compared by FACS analysis. As shown in Fig. 5B, FTI treatment induced significant apoptosis in MCF7-CASP-3 cells as is evidenced by the appearance of apoptotic cells containing <2 N DNA content (61% cells detected as apoptotic). In contrast, only a small percentage (9%) of FTI-treated MCF7-vector cells appeared as apoptotic cells. Treatment of both MCF7-vector cells and MCF7-CASP-3 cells with DMSO resulted in ≈5% apoptotic cells. These results provide a convincing demonstration that caspase 3 is critical for FTI-induced apoptosis.
Cytochrome c Is Released by FTI Treatment.
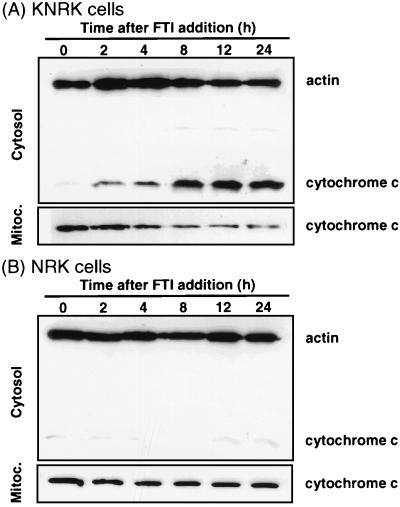
Recent work on apoptosis revealed that cytochrome c is released from the mitochondria during apoptosis (17–19). The released cytochrome c binds Apaf-1, which then activates caspase 9 that, in turn, activates caspase 3. To examine whether this pathway is activated by FTI, we investigated whether FTI induces cytochrome c release. FTI-treated KNRK and NRK cells grown in 0.1% FCS were collected and fractionated into cytosolic and mitochondrial fractions. The presence of cytochrome c in these fractions was detected by Western blot analysis using an anti-cytochrome c monoclonal antibody. As shown in Fig. 6A, increasing amount of cytochrome c was detected in the cytosol in a time-dependent manner after the addition of FTI to KNRK cells with a concomitant decrease of cytochrome c in the mitochondrial fraction. The time course of cytochrome c release into the cytosol correlates well with the appearance of apoptotic cells and the activation of caspase 3. In contrast, no significant release of cytochrome c into the cytosol or change in the amount of cytochrome c in the mitochondrial fraction was detected with FTI-treated NRK cells (Fig. 6B). The amount of actin present in each preparation was used to standardize differences in loading. These results suggest that FTI induces release of cytochrome c into the cytosol preferentially in KNRK cells.
Figure 6.
Cytochrome c release from mitochondria into cytosol of FTI-treated KNRK but not NRK cells. KNRK (A) or NRK (B) cells were treated with 100 μM BMS191563 in 0.1% FCS medium for the time intervals indicated. Cells were harvested and cytosolic and mitochondrial fractions were prepared. The levels of cytochrome c and actin were determined by Western blot analysis as described in Materials and Methods with the use of monoclonal anti-cytochrome c and anti-actin antibodies, respectively. At least two separate experiments were carried out with results similar to those shown here.
DISCUSSION
Our work clearly establishes that FTIs are capable of inducing apoptosis in transformed cells. In this paper, we have shown that FTIs induce apoptosis in four different transformed cell lines, KNRK, Spon 8 mouse metastatic lung cancer cells, PA-1 human ovarian cancer cells, and MCF7 cells expressing CASP-3 gene. We recently studied a variety of human cancer cell lines and found that FTIs are capable of inducing apoptosis of a number of human leukemic cell lines as well as human lung and colon adenocarcinoma cell lines (T. Nemoto and F.T., unpublished observations). Thus, FTIs induce apoptosis of a variety of cancer cell lines. Furthermore, the induction of apoptosis appears to be a general property of FTIs, because apoptosis is induced by the four different FTI compounds studied. These compounds differ in their structures with one being a tricyclic compound while the others are CAAX peptidomimetics. In addition, Hung and Chuang (12, 36) showed that another type of FTI, a competitor of farnesyl diphosphate, also induces apoptosis.
An important feature of FTI-induced apoptosis is its specificity for transformed cells. In this paper, we used transformed KNRK and untransformed NRK cells to demonstrate that FTI induces apoptosis specifically in transformed cells. A similar preferential induction of apoptosis is observed in NIH 3T3 cells as FTIs induce apoptosis in H-ras-transformed NIH 3T3 cells but not untransformed NIH 3T3 cells (N.S. and F.T., unpublished observations). The preferential effect FTIs have on transformed cells is further supported by our observation that FTIs induce the release of cytochrome c in KNRK cells but not in NRK cells. These results suggest that significant differences exist in the apoptotic pathways of transformed vs. untransformed cells.
Our analysis of the apoptotic pathways affected by FTIs provides insights into understanding the mechanism by which FTIs induce apoptosis. We have shown that caspase 3 activation is critical for FTI-induced apoptosis by using a caspase 3 inhibitor as well as MCF7 cells. Furthermore, we have shown that FTIs induce the release of cytochrome c from the mitochondria. The time course of cytochrome c release corresponds with caspase 3 activation and the appearance of apoptotic cells; these events become evident 12–24 h after the addition of FTIs. Cytochrome c release initiates a cascade that leads to the activation of caspase 3 through Apaf-1 and caspase 9 (17, 18). Our results support the idea that FTIs induce apoptosis through this cytochrome c pathway. FTI-induced apoptosis does not appear to involve the pathway initiated by tumor necrosis factor signals (37), since FTIs do not induce apoptosis in MCF7 cells even though they still retain partial sensitivity to tumor necrosis factor-mediated apoptosis (25).
It is important to emphasize that FTIs do not induce apoptosis when cells are grown in 10% FCS. Thus, FTIs themselves are not sufficient to induce apoptosis. A second signal is required to induce apoptosis and is provided by low serum conditions. To gain insights into how low serum conditions affect FTI-induced apoptosis, we compared the level of caspase 3 activation induced by FTI in both 10% and 0.1% FCS. Similar levels of caspase 3 activation were detected in both cases (N.S. and F.T., unpublished observations). Thus, exposure to 0.1% FCS is not necessary for the activation of caspase 3 by FTI. These results suggest that the apoptotic signal, which is triggered by low serum conditions, converges at a step downstream of caspase 3 activation. The nature of this signal remains to be defined.
How does FTI induce the release of cytochrome c? Cytochrome c normally resides in the inter-membrane space of the mitochondria and is involved in energy transfer (38). However, a population of cytochrome c is released into the cytosol during apoptosis. This cytochrome c release appears to involve the Bcl-2 family of proteins, which regulate apoptosis (39–41). The Bcl-2 family of proteins may form channels on the mitochondrial membrane to facilitate the release of cytochrome c (41–43). Bcl-xL, Bcl-2, and Bax are capable of forming channels on artificial membranes in vitro (41, 42), and the Bcl-xL protein contains structural similarity to channel forming proteins such as diphteria toxins and colicins (43). Therefore, one possible mechanism for the FTI-induced cytochrome c release is that FTI affects the channel forming activities of the Bcl-2 family proteins. Alternatively, FTI may affect the function of an as yet unidentified farnesylated protein(s) involved in antagonizing the release of cytochrome c from the mitochondria. FTI could affect the function of these proteins by inhibiting their farnesylation. Further studies are needed to determine the mechanism by which FTIs induce cytochrome c release.
In summary, our work establishes that FTI is capable of inducing apoptosis in a number of transformed cell lines. This apoptosis is accomplished by inducing the release of cytochrome c from the mitochondria and the subsequent activation of caspase 3. Our results clearly implicate protein farnesylation in apoptosis, and further work may provide valuable insights into the roles that farnesylated proteins play in programmed cell death.
Acknowledgments
We are grateful to Dr. W. Robert Bishop and Dr. Veeraswamy Manne for providing SCH56582 and BMS191563, respectively. We thank Dr. Ana Maria Garcia for providing B1088; Dr. Said Sebti for providing our initial supply of FTI-277; and Dr. John Colicelli and Dr. Genghong Cheng for critical reading of this paper. We thank Ingrid Schmid, Nathan Regimbal, and Iris Williams in the University of California, Los Angeles, Flow Cytometry Core Laboratory, for assistance on flow cytometry analysis. We also thank Dr. Tetsuo Nemoto and Nicole Robinson for helpful discussion and comments. This work is supported by National Institutes of Health Grant CA41996. J.U. is also supported by U.S. Public Health Service National Research Service Award GM07185.
ABBREVIATIONS
- FTI
farnesyltransferase inhibitor
- NRK
normal rat kidney
- KNRK
v-K-ras-transformed NRK
- AMC
7-amino-4-methylcoumarin
- DAPI
4,6-diamidino-2-phenylindole
- FCS
fetal calf serum
- PI
propidium iodide
- FACS
fluorescence-activated cell sorter
- GFP
green fluorescent protein
- DMSO
dimethyl sulfoxide
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Sattler I, Tamanoi F. In: Regulation of the RAS Signaling Network. Maruta H, Burgess A W, editors. Austin, TX: Landes; 1996. pp. 95–137. [Google Scholar]
- 2.Yang W, Del Villar K, Urano J, Mitsuzawa H, Tamanoi F. J Cell Biochem. 1997;S27:12–19. [PubMed] [Google Scholar]
- 3.Gibbs J B, Oliff A. Annu Rev Pharmacol Toxicol. 1997;37:143–166. doi: 10.1146/annurev.pharmtox.37.1.143. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F L, Casey P J. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 5.Kohl N E, Omer C A, Conner M W, Anthony N J, Davide J P, deSolms S J, Giuliani E A, Gomez R P, Graham S L, Hamilton K, et al. Nat Med. 1995;1:792–7977. doi: 10.1038/nm0895-792. [DOI] [PubMed] [Google Scholar]
- 6.Sun J, Qian Y, Hamilton A D, Sebti S M. Cancer Res. 1995;55:4243–4247. [PubMed] [Google Scholar]
- 7.Nagasu T, Yoshimatsu K, Rowell C, Lewis M D, Garcia A M. Cancer Res. 1995;55:5310–5314. [PubMed] [Google Scholar]
- 8.Sepp-Lorenzino L, Ma Z, Rands E, Kohl N E, Gibbs J B, Oliff A, Rosen N. Cancer Res. 1995;55:5302–5309. [PubMed] [Google Scholar]
- 9.Barinaga M. Science. 1997;278:1036–1039. doi: 10.1126/science.278.5340.1036. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki N, Del Villar K, Tamanoi F. Proc Natl Acad Sci USA. 1998;95:10499–10504. doi: 10.1073/pnas.95.18.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebowitz P F, Sakamuro D, Prendergast G C. Cancer Res. 1997;57:708–713. [PubMed] [Google Scholar]
- 12.Hung W C, Chuang L Y. Intl J Oncol. 1998;12:137–140. [PubMed] [Google Scholar]
- 13.Barrington R E, Subler M A, Rands E, Omer C A, Miller P J, Hundley J E, Koester S K, Troyer D A, Bearss D J, Conner M W, et al. Mol Cell Biol. 1998;18:85–92. doi: 10.1128/mcb.18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 15.Villa P, Kaufmann S H, Earnshaw W C. Trends Biochem Sci. 1997;22:388–393. doi: 10.1016/s0968-0004(97)01107-9. [DOI] [PubMed] [Google Scholar]
- 16.Kuida K, Zheng T S, Na S, Kuan C-Y, Yang D, Karasuyama H, Rakic P, Flavell R A. Nature (London) 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 17.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 18.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 19.Li F, Srinivasan, Wang Y, Armstrong R C, Tomaselli K J, Fritz L C. J Biol Chem. 1997;272:30299–30305. doi: 10.1074/jbc.272.48.30299. [DOI] [PubMed] [Google Scholar]
- 20.Bishop W R, Bond R, Petrin J, Wang L, Patton R, Doll R, Njoroge G, Catino J, Schwartz J, Windsor W, et al. J Biol Chem. 1995;270:30611–30618. doi: 10.1074/jbc.270.51.30611. [DOI] [PubMed] [Google Scholar]
- 21.Whyte D B, Kirschmeier P, Hockenberry T N, Nunez-Oliva I, James L, Catino J J, Bishop W R, Pai J K. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 22.Hunt J T, Lee V G, Leftheris K, Seizinger B, Mabus J, Ricca C, Yan N, Manne V. J Med Chem. 1996;39:353–358. doi: 10.1021/jm9507284. [DOI] [PubMed] [Google Scholar]
- 23.Vogt A, Qian Y, McGuire T, Hamilton A D, Sebti S M. Oncogene. 1996;13:1991–1999. [PubMed] [Google Scholar]
- 24.Aaronson S A, Weaver C A. J Gen Virol. 1971;13:245–252. doi: 10.1099/0022-1317-13-2-245. [DOI] [PubMed] [Google Scholar]
- 25.Janicke R U, Sprengart M L, Wati M R, Porter A G. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 26.Herzog C R, Soloff E V, McDoniels A L, Tyson F L, Malkinson A M, Haugen-Strano A, Wiseman R W, Anderson M W, You M. Oncogene. 1996;13:1885–1891. [PubMed] [Google Scholar]
- 27.Tang D G, Li L, Zhu Z, Joshi B. Biochem Biophys Res Commun. 1998;242:380–384. doi: 10.1006/bbrc.1997.7969. [DOI] [PubMed] [Google Scholar]
- 28.Servant M J, Giasson E, Meloche S. J Biol Chem. 1996;271:16047–16052. doi: 10.1074/jbc.271.27.16047. [DOI] [PubMed] [Google Scholar]
- 29.Pastorino J G, Chen S T, Tafani M, Snyder J W, Farber J L. J Biol Chem. 1998;273:7770–7775. doi: 10.1074/jbc.273.13.7770. [DOI] [PubMed] [Google Scholar]
- 30.Hughes F M, Jr, Bortner C D, Purdy G D, Cidlowski J A. J Biol Chem. 1997;272:30567–30576. doi: 10.1074/jbc.272.48.30567. [DOI] [PubMed] [Google Scholar]
- 31.Okada T, Sakuma L, Fukui Y, Hazeki O, Ui M. J Biol Chem. 1994;269:3563–3567. [PubMed] [Google Scholar]
- 32.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1330. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 33.Zundel W, Giaccia A. Genes Dev. 1998;12:1941–1946. doi: 10.1101/gad.12.13.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyllie A H. Nature (London) 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Li P, Widlak P, Zou H, Luo X, Garrard W T, Wang X. Proc Natl Acad Sci USA. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hung W-C, Chuang L-Y. Intl J Oncol. 1998;12:1339–1342. doi: 10.3892/ijo.12.6.1339. [DOI] [PubMed] [Google Scholar]
- 37.Nagata S. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 38.Cortese J D, Voglino A L, Hackenbrock C R. Biochim Biophys Acta. 1995;1228:216–228. doi: 10.1016/0005-2728(94)00178-8. [DOI] [PubMed] [Google Scholar]
- 39.Jurgensmeier J M, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed J C. Proc Natl Acad Sci USA. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosse T, Olivier R, Monney L, Rager M, Conus S, Fellay I, Jansen B, Borner C. Nature (London) 1998;391:496–499. doi: 10.1038/35160. [DOI] [PubMed] [Google Scholar]
- 41.Chao D T, Korsmeyer S J. Annu Rev Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 42.Reed J C. Nature (London) 1997;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- 43.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Chang B S, Thompson C B, Wong S, et al. Nature (London) 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]