Abstract
Any system, natural or human-made, is better understood if we analyze both its history and its structure. Here we combine structural analyses with a “Reconstructed Evolutionary Adaptive Path” (REAP) analysis that used the evolutionary and functional history of DNA polymerases to replace amino acids to enable polymerases to accept a new class of triphosphate substrates, those having their 3′-OH ends blocked as a 3′-ONH2 group (dNTP-ONH2). Analogous to widely used 2′,3′-dideoxynucleoside triphosphates (ddNTPs), dNTP-ONH2s terminate primer extension. Unlike ddNTPs, however, primer extension can be resumed by cleaving an O-N bond to restore an -OH group to the 3′-end of the primer. REAP combined with crystallographic analyses identified 35 sites where replacements might improve the ability of Taq to accept dNTP-ONH2s. A library of 93 Taq variants, each having replacements at three or four of these sites, held eight variants having improved ability to accept dNTP-ONH2 substrates. Two of these (A597T, L616A, F667Y, E745H, and E520G, K540I, L616A) performed notably well. The second variant incorporated both dNTP-ONH2sand ddNTPs faithfully and efficiently, supporting extension-cleavage-extension cycles applicable in parallel sequencing and in SNP detection through competition between reversible and irreversible terminators. Dissecting these results showed that one replacement (L616A), not previously identified, allows Taq to incorporate both reversible and irreversible terminators. Modeling showed how L616A might open space behind Phe-667, allowing it to move to accommodate the larger 3′-substituent. This work provides polymerases for DNA analyses and shows how evolutionary analyses help explore relationships between structure and function in proteins.
Keywords: DNA polymerases, evolutionary analysis, sequencing technology, single nucleotide polymorphisms, synthetic biology
As illustrated by the Human Genome Project (1), we can now sequence the complete genome of any species on Earth, given sufficient financial resources. Traditional Sanger sequencing cannot, however, broadly meet this opportunity because of its cost (2). Accordingly, many seek “next generation” sequencing platforms, in particular, those that can sequence millions of oligonucleotides in parallel (3). Chemistry that supports such sequencing might also help meet other goals in the analytical chemistry of DNA, including multiplexed detection of single-nucleotide polymorphisms (SNPs) in complex biological environments, discovery of genetic variation distinguishing diseased and nondiseased tissues from a single patient, and point-of-care diagnostics that personalize treatment based on multiplexed detection of genetic markers.
Some architectures for next generation sequencing use “reversible terminators” to better identify homopolymeric runs that are difficult to read using pyrosequencing. Exemplified by 2′,3′-dideoxynucleoside triphosphates (ddNTPs), these are nucleoside triphosphates that initially terminate primer extension after being incorporated into a growing DNA strand (4–10). Unlike 2′,3′-ddNTPs, however, reversible terminators are chemically structured so that appropriate treatment restores a 3′-extendable primer, allowing extension to resume. Proposed 3′-O-blocking groups include 3′-O-acyl (4), 3′-O-allyl (5, 6), 3′-O-methoxymethyl, 3′-O-nitrobenzyl (7), and 3′-O-azidomethylene (8, 9), the last reportedly used on the most recent Solexa-Illumina instrument. Metzker has suggested that reversible termination can also be achieved by groups appended not to the 3′-O atom but rather to the nucleobases (10).
However, application of reversible terminators in architectures for DNA analysis requires polymerases that accept appropriately modified nucleoside triphosphate analogs efficiently and faithfully. Finding such polymerases is especially difficult when the unnatural triphosphates are modified at their 2′- and 3′-positions. Because polymerases have evolved for billions of years to discriminate between ribonucleoside triphosphates and 2′-deoxyribonucleoside triphosphates, they have evolved to inspect the 2′- and 3′- positions of their substrates closely (11).
We report here a previously unreported reversible terminating group that replaces the 3′-OH group on the triphosphates with an -ONH2 group and conditions that cleave the 3′-blocking group to restore a 3′-OH group. To obtain polymerases that accept them, we developed a strategy that combined evolutionary analyses with structural analyses to identify replacements at sites in the polymerase. Called “Reconstructing Evolutionary Adaptive Paths” (REAP) (12), this strategy examines patterns of variation and conservation within a family of enzymes to identify sites that are likely to alter the behavior of an enzyme without destroying its core catalytic activity.
Results
Experiments with reversible terminators have yet to generate a 3′-O modification that is efficiently and faithfully incorporated and cleaved with sufficient ease to support architectures for DNA sequencing that involve long (> 30) read lengths; the 3′-O-azidomethylene blocking group may be the most successful so far (13). Some time ago, experiments with this and other 3′-O blocking groups suggested a “rule”: Smaller 3′-appendages are more likely to be accepted by polymerases than larger modifications (14).
Applying this rule, we hypothesized that a 3′-ONH2 blocking group (Fig. 1) might serve as a reversible terminator on a nucleoside triphosphate (15). Inspection of the periodic table in light of interatomic bonding patterns suggested that this would be the smallest 3′-O-blocking group sufficiently stable to be practically useful and able to be removed under conditions sufficiently mild to leave DNA intact.
Fig. 1.
Reversible terminator having its 3′-OH group blocked with a 3′-ONH2 moiety.
A series of reagents and conditions were then explored to identify cleavage conditions that might convert a 3′-ONH2 to a 3′-OH. These included treatment with oxidants, reductants, and electrophiles. Of these, treatment with buffered (pH 5.5) sodium nitrite was preferred. Cleavage restoring the native 3′-OH of a primer is over 99% complete in less than 2 min. Controls lasting 72 h with sodium nitrite under these conditions suggested that fewer than one guanine per 10,000 in a single stranded template would be deaminated in 2 min under these conditions; adenine and cytosine were found to be still more stable (SI Appendix).
With the cleavability of a 3′-ONH2 terminating group under mild conditions established, we sought polymerases that would accept 3′-ONH2 blocked nucleoside triphosphates. Screening commercially available polymerases found that only a few accepted triphosphates with a 3′-ONH2 blocking group. Further, those that did either did so poorly or had other problems that made them ill-suited in DNA analysis architectures. Therefore, we decided to search for mutants of the DNA polymerase from Thermus aquaticus (Taq) that might accept these triphosphates.
To search for replacements, we implemented the REAP (12) strategy to analyze the evolutionary history of Family A polymerases. REAP recognizes that the evolutionary history of a protein family, including the sequences of ancestral proteins represented by nodes in an evolutionary tree, can be inferred from the sequences of the descendent proteins (16). After inferring those ancestral sequences, REAP examines patterns of change in that history, recognizing that when that history differs from the history expected from simple Markov models for protein sequence divergence, that difference contains information about the relation between sequence and behavior in the protein (17).
REAP analysis of the polymerase family began by identifying amino acid replacements in the history of polymerase evolution that might have been responsible for functional divergence between viral and nonviral polymerases. Sequence alignments and a phylogenetic tree (Fig S1) were constructed for 719 Family A polymerases sequences extracted from the PFAM database (PF00476) (18). These were clustered in two branches in the evolutionary tree. Probabilistic amino acid sequences were inferred for ancestral polymerases throughout the tree.
Because viral polymerases are empirically known to be more likely to accept modified nucleotides than nonviral polymerase (19), the sites identified by evolutionary analysis as changing during the evolutionary episode when viral and nonviral polymerases were diverging were hypothesized to be candidate sites that, in nonviral polymerases, could undergo amino acid replacements to generate new polymerases that might accept modified nucleoside triphosphates without substantial loss of catalytic activity or fidelity. This approach differs from the in clinico selection approach that we previously used to identify polymerases that incorporate nonstandard nucleoside triphosphates (20).
These candidate sites, identified by a historical analysis, were considered within the context of a high-resolution structure of the Taq polymerase (PDB number 2ktq). Sites identified by REAP were preferred if they lay near the active center and if they had been identified by Henry and Romesberg as “sites of interest” based on their review of interactions between polymerases and nucleoside triphosphate analogs (21).
This combined historical-structural analysis identified 35 sites (Fig. S2) of special interest. These held 57 possible amino acid replacements (when comparing viral polymerases with nonviral polymerases). A set of 93 Taq polymerase variants was then designed that replaced three or four amino acids at the sites identified (Table S1). The amino acid replacements were combinatorially distributed such that each replacement was present in exactly six variants (22). Genes for the variants were synthesized with codons optimized for expression in Escherichia coli; wild-type polymerase was called Co-Taq. The genes were then cloned and expressed.
The 93 variant Taq polymerases were then screened for their ability to incorporate nucleoside triphosphates carrying the 3′-ONH2 unit (Fig. S3A). Co-Taq polymerase and the Taq polymerase from New England Biolabs (NEB Taq, catalog number M0267) were also tested in parallel as controls. NEB Taq generated at most a trace of the N + 1 product upon incubation at 72 °C for 2 min in the presence of a template that called for the incorporation of dTTP-ONH2. In contrast, 30 of the 93 polymerase variants (approximately 32% of the REAP library) were catalytically active with natural triphosphates (despite the number of replacements near the active site) and were able to incorporate dTTP-ONH2 opposite dA in a synthetic template better than NEB Taq. Of these variants, eight extended the primer by more than 50% to give N + 1 products, thereby also performing better than Co-Taq. In particular, REAP-42 (A597T, L616A, F667Y, E745H; numbering is from the crystal structure 2ktq) and REAP-58 (E520G, K540I, L616A) showed the best incorporation of dTTP-ONH2. The absence of an N + 2 band in Fig. S3A indicated that the 3′-ONH2 group, once appended to the primer, successfully terminated primer extension.
The results were similar when dCTP-ONH2 was used as a substrate with a template that placed a dG at the first two positions to be copied (Fig. S4A). A scatter plot of the amount of N + 1 product formed with dTTP-ONH2 versus the amount of N + 1 product formed with dCTP-ONH2 (Fig. S4B) suggested that the incorporation of cytosine opposite guanine was generally easier with almost all of the 93 polymerases in the REAP library than the incorporation of thymine opposite adenine. This is most simply explained by the increased number of hydrogen bonds joining the C:G pair compared with just two joining the T:A pair.
We then explored the potential of these variant polymerases to incorporate the corresponding 2′,3′-dideoxynucleoside triphosphates, the irreversible terminators. Polymerase variants were challenged to incorporate ddTTP opposite template dA (Fig. S3B). Of the 93 variants, 27 were found to incorporate ddTTP to a detectable extent. Three of these converted more than 50% of the primer to N + 1 product using ddTTP (REAP-38, REAP-45, and REAP-70).
Interestingly, the ability of a polymerase variant to accept the 3′-ONH2 reversible terminator correlated only weakly (r2 ≈ 0.17) with its ability to accept 2′,3′-dideoxynucleoside triphosphates (Fig. S3C). The low r2 value arose primarily because variants with replacements at site 667 lay off the trend line. If variants having a F667Y replacement at site 667 were removed from the analysis, the correlation coefficient (r2) rose to 0.64 (Fig. S3D).
The exceptionality of site 667 was recognized a decade ago by Tabor and Richardson in their effort to find Taq variants that accept 2′,3′-dideoxynucleoside triphosphates (23). Tabor and Richardson suggested that a Taq polymerase having a single F667Y replacement better accepted ddNTPs because introduction of a hydroxyl group via the Tyr-for-Phe replacement compensated for the loss of the hydroxyl group at the 3′-position of ddNTPs. This rationale would not apply to 3′-ONH2 replacement, of course, accounting for the position of variants holding the F667Y replacement above the trend line.
Although structural analysis originally drove the identification of the F667Y replacement, it was also identified by the REAP analysis. Phe-667 is conserved in bacterial polymerases, but not in viral, mycobacteria and mitochondrial polymerases, all of which are “functional outgroups” in the REAP analysis. About 20% of the viral polymerases have Tyr at this position (24).
In general, polymerase variants in the REAP library prefer 3′-ONH2 deoxynucleoside triphosphates over the 2′,3′-dideoxynucleoside triphosphates (Fig. S3A). This suggests that to REAP variants, a 3′-ONH2 group resembles more closely a 3′-OH group than a 3′-H group.
Deoxythymidine triphosphate was also tested with REAP variants (Fig. S5). As expected, variants that do not incorporate this natural triphosphate also do not incorporate nonstandard triphosphates. Most REAP variants with enhanced ability to accept reversible terminators showed lower fidelity in incorporation of dTTP.
To determine whether single amino acid replacements at single sites identified by the combined historical-structural analyses could account for the observations made with reversible and irreversible terminators, seven Taq variants were prepared, examined, and tested for their ability to incorporate dTTP-ONH2, each carrying just one amino acid replacement chosen from those found in the eight REAP variants that extended a primer with dTTP-ONH2 more than 50% in 2 min (Fig. 2A). The variant with the L616A replacement alone was best able to incorporate dTTP-ONH2; this ability was only modestly lower than with REAP-58 itself. Variants with the I614G replacement alone and the F667Y replacement alone (the Tabor–Richardson replacement) were second and third best, respectively.
Fig. 2.
Incorporation of dTTP-ONH2 (A) and ddTTP (B) by seven Taq variants with only one amino acid replacement. In a 10-μL reaction volume, gamma-32P-labeled primer (2.5 pmol), cold primer (22.5 pmol) and the template (30 pmol) (see below) were annealed. Cell-free extracts (4 μL) containing Taq were then added to reactions and the mixtures were incubated at 72 °C for 2 min. Reactions were initiated by the addition of dTTP-ONH2 or ddTTP (final concentration 100 μM). The resulting reaction mixtures were separated on 14% PAGE and visualized by autoradiography. The primer and templates are as follows:
Primer (24 nt): 5′-GCGTAATACGACTCACTATGGACG-3′.
Temp A (36 nt): 3′-CGCATTATGCTGAGTGATACCTGCAATGTGCTTCTG-5′.
We then examined the fidelity of REAP-42 and REAP-58. The former had the Tabor–Richardson F667Y replacement; the latter did not. Very little infidelity was observed with both (Fig. 3A) when they were challenged to misincorporate reversibly terminating triphosphates. This was evident by the essentially complete absence of all misincorporation products with REAP-58 at position N + 1 when the “wrong” dNTP-ONH2 was presented to the polymerase. Trace amounts of products were seen by PAGE to indicate a mismatch only of incoming dCTP-ONH2 opposite template dC and incoming dATP-ONH2 opposite template dA, but only with REAP-42.
Fig. 3.
Fidelity of incorporation of reversible and irreversible terminators by REAP-42 and REAP-58 using the following primer and templates (polymerases were purified by Qiagen Ni-NTA Spin Kit and quantified according to Bradford assay):
Primer (24 nt): 5′-GCGTAATACGACTCACTATGGACG-3′.
Temp A (36 nt): 3′-CGCATTATGCTGAGTGATACCTGCAATGTGCTTCTG-5′.
Temp G (36 nt): 3′-CGCATTATGCTGAGTGATACCTGCGGTGTGCTTCTG-5′.
Temp T (36 nt): 3′-CGCATTATGCTGAGTGATACCTGCTTTGTGCTTCTG-5′.
Temp C (36 nt): 3′-CGCATTATGCTGAGTGATACCTGCCCTGTGCTTCTG-5′.
Interestingly, whereas the fidelity of REAP-58 and REAP-42 with the reversible terminator both are high, this was not so for 2′,3′-dideoxy irreversibly terminating triphosphates and REAP-42 (Fig. 3B). Whereas the REAP-58 variant displayed high fidelity with ddNTPs, REAP-42 yielded detectable mismatches in almost every mismatch extension experiment with ddNTPs.
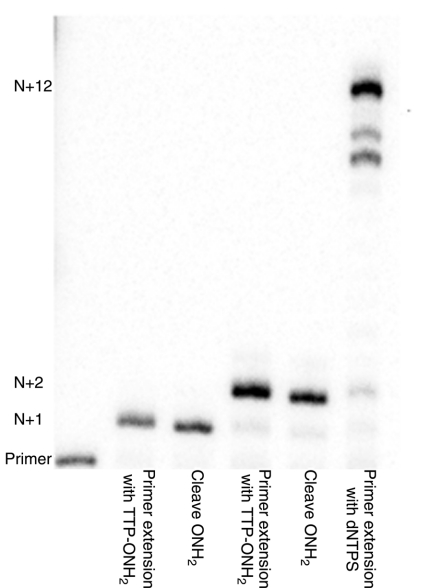
We then demonstrated that these variant polymerases could support cyclic reversible termination with 3′-ONH2 terminators. A 5′-32P-labeled primer was annealed to a biotinylated template calling for the addition of two consecutive Ts. The duplex was immobilized on magnetic beads with streptavidin. The beads were then incubated with REAP-58 polymerase in the presence of T-ONH2 triphosphate (100 μM, 2 min, 72 °C). After extension, the beads were treated with buffered sodium nitrite to cleave the -ONH2 group. After washing, a second round of primer extension was achieved with REAP-58 polymerase and dTTP-ONH2. Following cleavage of the second terminating 3′-ONH2 group, full length product was obtained by adding a third batch of REAP-58 polymerase and all dNTPs.
After each step, the products were released from the bead and resolved by gel electrophoresis. The results (Fig. 4) showed that oligonucleotides modified with a 3′-ONH2 unit move slightly slower in electrophoresis than unmodified oligonucleotides. This decreased mobility is interpreted as a consequence of either a slightly larger molecule arising from the replacement of -OH by -ONH2 or of a slightly less anionic product arising from a partly cationic  group (pKa ∼ 6). Regardless, this mobility difference allowed us to conclude that each step went essentially to completion. Pausing bands observed late in the last extension to give full length product were attributed to the presence of a biotin at the 5′-end of the template; biotin labeling is often observed to hinder complete extension of primers to give full length products.
group (pKa ∼ 6). Regardless, this mobility difference allowed us to conclude that each step went essentially to completion. Pausing bands observed late in the last extension to give full length product were attributed to the presence of a biotin at the 5′-end of the template; biotin labeling is often observed to hinder complete extension of primers to give full length products.
Fig. 4.
Addition of the 3′-ONH2 reversible terminator followed by cleavage and continued extension. The template is immobilized to the bead; the 5′-radiolabeled primer is observed (position indicated by “Primer” marking) after each cycle. The template calls for incorporation of consecutive Ts. Full length product is indicated by “N + 12”.
Lane 1: Primer. Lane 2: Products obtained after incubation with REAP-58 (1 μL diluted) show complete conversion of the primer (N) to an extended product (N + 1), with the 3′-ONH2 completely preventing formation of N + 2 product. Lane 3: N + 1 product after treatment with buffered sodium nitrite. Lane 4: Product following second incubation with the TONH2 triphosphate. Full conversion of the band at N + 1 to a band at N + 2 shows the completion of the cleavage reaction. Lane 5: Product after deblocking with a second cycle with buffered sodium nitrite. Lane 6: Product following deblocking and incubation with all four standard triphosphates, showing the formation of the full length product (N + 12).
5′-32P-GCGTAATACGACTCACTATGGACG.
3′-CGCATTATGCTGAGTGATACCTGCAATGTGCTTCTG-Biotin-5′.
Reversible terminators have uses in the analysis of DNA in addition to sequencing, including to identify SNPs. Several of these architectures require irreversible terminators and reversible terminators to compete in the same assay. For example, SNPs might be detected with low noise if an irreversibly terminating dideoxynucleotide were incorporated opposite the standard base in a standard template, with an ONH2-terminated nucleoside incorporated opposite an SNP base in an SNP template. The irreversible terminator allows the polymerase to not have to make a choice between doing nothing or misincorporating a mismatch with the nonmutated template, which is often in large excess in clinical samples. Only for the SNP product can the extension be resumed for sequencing or cloning.
To demonstrate the use of these variant polymerases to support SNP detection architectures combining reversibly and irreversibly terminating triphosphates, a model template with T at an “SNP site,” and three SNP templates containing A, G, or C, were tested singly. After being annealed with a primer that queried the SNP site, these were incubated with one irreversible terminator (ddATP) and three reversible terminators (dTTP-ONH2, dCTP-ONH2, dGTP-ONH2) and REAP-58. After 5 min, the reaction was quenched and the mixture treated with oxidized polyethylene glycol (PEG) having an aldehyde moiety at one end. This forms an oxime with the 3′-ONH2 tagged extension product (DNA-O-N = CH-O(CH2CH2-O)2-H), but not the dideoxy-terminated product. The tagged product migrates more slowly upon electrophoresis than the untagged species.
As expected, the primer encountering the natural template was irreversibly capped by a ddATP (S6), whereas the primer that encountered an SNP template was reversibly capped and ran more slowly than the primer capped with the irreversible terminator (S6Left). Capture with PEG aldehyde caused the SNP product, but not the non-SNP product, to electrophorese still more slowly (S6Right). The presence of the irreversibly terminating ddATP in the assay mixture provided the polymerase the opportunity to introduce a match, diminishing the chance that it would incorporate a mismatched nucleotide opposite template T rather than doing nothing at all.
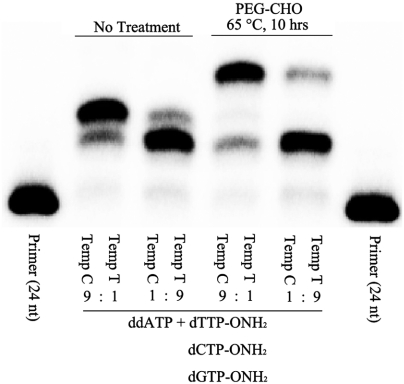
To further explore the use of competition between reversible and irreversible inhibitors in SNP detection, standard and SNP sequences were combined in 1∶9 and 9∶1 ratios (Fig. 5). The capture products had, to within 5%, the ratios of the standard and SNP sequences presented to the assay, as verified by PEG aldehyde capture.
Fig. 5.
Gel shift showing the PEG-aldehyde capture 3′-ONH2 reversible terminator extended on SNP templates. 14% PAGE of primer extension reactions containing Templates with a T or C at position N + 1 were incubated with ddATP and dGTP-ONH2. Aliquots of the samples were treated with PEG aldehyde at 65 °C for 10 h. The samples were resolved on a 14% PAGE gel for analysis.
Primer (24 nt): 5′-GCGTAATACGACTCACTATGGACG-3′.
Standard Temp T (36 nt): 3′-CGCATTATGCTGAGTGATACCTGCTATGTGCTTCTG-5.
SNP Temp C (36 nt): 3′-CGCATTATGCTGAGTGATACCTGCCATGTGCTTCTG-5′.
Discussion
This research both answers scientific questions and meets technological goals. From a scientific perspective, this work provides a working example where a REAP analysis was applied to construct mutant enzymes. REAP identified two replacements (L616A and I614G) that have previously not been suggested by structure-based analysis, although it is interesting to note that Loeb′s laboratory used in vitro selection to identify 614 as a site where alteration might influence fidelity and deoxy/ribo selectivity in polymerases (21, 25). It also identified the F667Y replacement that had been previously suggested by structural analyses.
These replacements generate polymerases that incorporate unnatural nucleoside triphosphates carrying a 3′-ONH2 reversible terminator. This result suggests a utility for the REAP approach, which is presumably successful because it diminishes the size of libraries that must be examined by focusing on sites where replacement does not destroy core fold and function. We anticipate that REAP will help guide the engineering of other types of proteins, as well as provide further insights into the interaction between sequence, function, and evolution in natural proteins.
From a technological perspective, the polymerases provided here have a useful ability to incorporate a reversible terminator with a small 3′-ONH2 substituent, both directly and in competition with irreversible terminators. These are able to support extension-cleavage-extension-cleavage cycles as well as architectures that might detect SNPs.
Returning to science and the discovery of useful mutations through evolutionary analyses, these experiments show that a single L616A replacement allows a polymerase to incorporate well both 3′-ONH2 and 2′-3′-dideoxynucleoside triphosphates (Fig. 2 and Fig. S7). This is an intriguing result, especially in the context of the reasoning used by Tabor and Richardson when they proposed the F667Y replacement. That reasoning focused on a presumed need to add an -OH unit to an amino acid in the active site to compensate for the loss of the 3′-OH in 2′,3′-dideoxynucleoside triphosphate substrates.
Replacing the Leu 616 by Ala does not, of course, compensate for a missing 3′-OH group in the same way. Therefore, this replacement cannot create a polymerase that accepts a 2′,3′-dideoxynucleoside triphosphate under a “replacement of a functional group” model. This indicates that the L616A replacement must work for another reason.
To better understand our observations, we modeled the active site of Taq DNA polymerase using the crystal structure defining the closed conformation (PDB file 3ktq, Fig. 6) (26). The structure was subjected to 500 rounds of gradient energy minimization to optimize its stereochemistry (rmsd from 3ktq = 0.59 Å) using PHENIX. The L616A mutation was modeled on this structure, and the model subjected to the same energy minimization procedure (rmsd from 2ktq = 0.59 Å2) to correct for any stereochemical or geometrical strain introduced by the replacement.
Fig. 6.
L616A creates additional space for bulky 3′-substituents behind the phenyl ring of F667. Close-up view of the DNA binding cleft with an idealized model for the Large Fragment of DNA Pol I from Taq (pdb 3ktq) in green. The cavity (free space) behind L616 (A) or A616 (B) in the “fingers domain” is depicted as a transparent green surface. Nucleotides are shown as cyan sticks; Mg++ ions are gold spheres. The 3′- position of bound ddCTP is highlighted. Key residues in the binding pocket are highlighted showing how L616 supports the positioning of F667 in the pocket.
This modeling (Fig. 6) provides a view of the impact of the L616A replacement on the geometry of the active site. L616 is in close van der Waals contact with the phenyl ring of F667 (3.5 Å), which is on helix O at the edge of the dNTP and DNA binding cleft. In turn, the phenyl ring of Phe-667 in the 3ktq structure is in close proximity on its opposite side to the 2′- and 3′-atoms of the ribose ring of the incoming nucleoside triphosphate. The aromatic ring is perpendicular to the ribose ring and approximately bisected by its 2′- and 3′-positions. Thus, this phenyl ring forms a “wall” in the active site, separating a hydrophobic region in the “fingers” domain of the polymerase from the incoming triphosphate. The side chain of Leu- 616 directly supports positioning of aromatic ring of Phe-667 residue. The interaction of Phe-667 and the triphosphate in the closed active site is modeled to be stabilizing (ddG = -2.24 kcal/mol, CUPSAT).
The most obvious impact of the L616A replacement is to open space behind F677 relative to the bound ddCTP. This space might allow the ring to move away from the 2′- and 3′-atoms of the ribose when such motion is needed to accommodate a larger substrate. In addition to creating a void behind the phenyl ring, the L616A replacement also reduces contact surface between the mobile O helix and the P helix, generating greater conformational freedom for Phe-667.
According to this model, the 3′-ONH2 substituent in the incoming triphosphate would sterically clash with the phenyl ring of Phe-667 as it is positioned in the wild-type polymerase. From a pure geometric complementarity perspective, the movement of that phenyl ring away from the ribose allowed by the L616A replacement should allow the active site to accommodate a 3′-ONH2 substituent on the ribose. This mutation may not lower fidelity or activity, as geometric complementation may still support the buttressing of the incoming ribose through close contact with the 2′- and 3′ atoms. A similar argument might apply to the I614G replacement, as Gly is also much smaller than Ile (Figs. 2 and 6).
However, this does not account for the ability of the L616 replacement to accept ddNTPs well with respectable fidelity; clearly, a ddNTP does not need and cannot use the extra space. Considering this, we speculated that solvent may play a role. The space opened by the L616A replacement is sufficient to allow a water molecule to enter the active site. Whereas a water molecule should not enter the hydrophobic pocket between the phenyl ring and the side chain of the amino acid at site 616, it might enter between the more hydrophilic ribose and the phenyl ring.
With ddNTP substrates, we speculate that such a water molecule might compensate for the oxygen missing from the 2′,3′-dideoxynucleoside substrate. However, when a dNTP-ONH2 is the substrate, this water molecule would be displaced by the 3′-ONH2 amino group. This would account for the ability of the L616A polymerase variant to accept both the 2′,3′-dideoxy and the 3′-ONH2 substrates with reasonable fidelity. The I614G replacement may perform similarly for these reasons as well.
This model, which involves replacements of residues not in direct contact with the substrate, but in the “second shell” contacting residues that do make direct contact, is consistent with various models that emphasize flexibility over rigidity in the active sites of enzyme active sites (27). Although to our knowledge no systematic study has been done, it appears that amino acid replacements in the second shell may also be frequent in natural evolution seeking to adapt natural proteins to changes in the structure of natural substrates.
Methods
For details, see SI Appendix.
Supplementary Material
Acknowledgments.
We are indebted to the National Institutes of Health (R41HG004589, 2R42HG003668-02A, R01HG004831, and 1R41GM074433), and a contract from Nucleic Acids Licensing LLC for support of this research.
Footnotes
Conflict of interest statement: S.A.B., E.A.G., D.H., and the Foundation for Applied Molecular Evolution hold patents and patent applications for various of the compounds and enzymes described here.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908463107/DCSupplemental.
References
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.França LT, Carrilho E, Kist TB. A review of DNA sequencing techniques. Q Rev Biophys. 2002;35:169–200. doi: 10.1017/s0033583502003797. [DOI] [PubMed] [Google Scholar]
- 3.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92(5):255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Hiatt AC. Compositions for enzyme catalyzed template-independent creation of phosphodiester bonds using protected nucleotides. 6232465. US Patent. 2001
- 5.Ruparel H, et al. Design and synthesis of a 3′-O-allyl photocleavable fluorescent nucleotide as a reversible terminator for DNA sequencing by synthesis. Proc Natl Acad Sci USA. 2005;102:5932–5937. doi: 10.1073/pnas.0501962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju J, et al. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc Natl Acad Sci USA. 2006;103:19635–196340. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, et al. 3′-O-modified nucleotides as reversible terminators for pyrosequencing. Proc Natl Acad Sci USA. 2007;104(42):16462–16467. doi: 10.1073/pnas.0707495104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J, et al. Four-color DNA sequencing with 3′-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides. Proc Natl Acad Sci USA. 2008;105:9145–9150. doi: 10.1073/pnas.0804023105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ju J, et al. DNA sequencing with non-fluorescent nucleotide reversible terminators and cleavable label modified nucleotide terminators. 2008. PCT Int. Appl. Publ. WO2009054922.
- 10.Wu W, et al. Termination of DNA synthesis by N6-alkylated, not 3′-O-alkylated, photocleavable 2′-deoxyadenosine triphosphates. Nucleic Acids Res. 2007;35:6339–6449. doi: 10.1093/nar/gkm689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benner SA. Understanding nucleic acids using synthetic chemistry. Acc Chem Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 12.Gaucher EA. In: Ancestral Sequence Reconstruction. Liberles David A., editor. New York: Oxford Univ Press; 2007. pp. 20–33. [Google Scholar]
- 13.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canard B, Cardona B, Sarfati RS. Catalytic editing properties of DNA polymerases. Proc Natl Acad Sci USA. 1995;92:10859–10863. doi: 10.1073/pnas.92.24.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benner SA. Method for sequencing DNA and RNA by synthesis. 7544794. US Patent. 2009
- 16.Pauling L, Zuckerkandl E. Chemical paleogenetics molecular restoration studies of extinct forms of life. Acta Chem Scand. 1963;17:S9–S16. [Google Scholar]
- 17.Benner SA. Interpretive proteomics. Finding biological meaning in genome and proteome databases. Adv Enzyme Regul. 2003;43:271–359. doi: 10.1016/s0065-2571(02)00024-9. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horlacher J, et al. Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with nonstandard hydrogen bonding patterns. Proc Natl Acad Sci USA. 1995;92:6329–6333. doi: 10.1073/pnas.92.14.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sismour AM, et al. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from human immunodeficiency virus-1. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry AA, Romesberg FE. The evolution of DNA polymerases with novel activities. Curr Opin Biotechnol. 2005;16:370–377. doi: 10.1016/j.copbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Liao J, et al. Engineering proteinase K using machine learning and synthetic genes. BMC Biotechnol. 2007;7:16. doi: 10.1186/1472-6750-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabor S, Richardson CC. A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel P, Loeb LA. Multiple Amino Acid Substitutions Allow DNA Polymerases to Synthesize RNA. J Biol Chem. 2000;275:40266–40272. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsou CL. Conformational flexibility of enzyme active sites. Science. 1993;262:380–1. doi: 10.1126/science.8211158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.