Abstract
Crystallographic analysis revealed that the 17-member polyketide antibiotic lankacidin produced by Streptomyces rochei binds at the peptidyl transferase center of the eubacterial large ribosomal subunit. Biochemical and functional studies verified this finding and showed interference with peptide bond formation. Chemical probing indicated that the macrolide lankamycin, a second antibiotic produced by the same species, binds at a neighboring site, at the ribosome exit tunnel. These two antibiotics can bind to the ribosome simultaneously and display synergy in inhibiting bacterial growth. The binding site of lankacidin and lankamycin partially overlap with the binding site of another pair of synergistic antibiotics, the streptogramins. Thus, at least two pairs of structurally dissimilar compounds have been selected in the course of evolution to act synergistically by targeting neighboring sites in the ribosome. These results underscore the importance of the corresponding ribosomal sites for development of clinically relevant synergistic antibiotics and demonstrate the utility of structural analysis for providing new directions for drug discovery.
Keywords: lankamycin, ribosomes, synergism, resistance, rRNA
Biochemical, genetic and functional evidence indicate that a great variety of antibiotics inhibit protein synthesis by binding to ribosomal functional regions. Crystallographic studies performed over the last decade revealed the exact binding sites of a variety of such drugs (see refs.1 and 2 for review). Many natural antibiotics, as well as their clinically relevant semisynthetic derivatives, bind at the peptidyl transferase center (PTC) in the large ribosomal subunit (3–14). Most of these compounds inhibit cell growth by interfering with peptide bond formation (15). The second major antibiotic binding site in the large ribosomal subunit is located at the upper segment of the nascent peptide exit tunnel (NPET), adjacent to the PTC, and is used by macrolides and type B streptogramins (3, 7, 16–21). Binding to this site impedes progression of the nascent proteins toward the tunnel exit. Thus, compounds binding to the PTC and NPET inhibit successive steps in protein synthesis: formation of the nascent chains and their export from the ribosome.
Simultaneous inhibition of successive steps of a specific biochemical pathway often results in a synergistic action of the inhibitors (22). Nature has not ignored this opportunity when evolving ribosomal antibiotics. For example, streptogramin antibiotics, produced by several Streptomyces species, are secreted as a combination of two structurally distinct compounds that inhibit cell growth by acting upon the PTC and NPET (23, 24). Streptogramin A (SA) compounds are cyclic poly-unsaturated macrolactones that bind in the PTC, whereas type B streptogramins (SB) compounds are cyclic depsipeptides that bind in the NPET (6, 7, 25). Each of the individual streptogramin components is a fairly weak antibiotic on its own, but in combination they exhibit strong inhibitory action. The synergistic antibiotic effect of streptogramins is medically relevant—the semisynthetic formulation Synercid composed of dalfopristin (the SA derivative) and quinupristin (the SB derivative) is widely used for treatment of complicated Gram-positive infections (26).
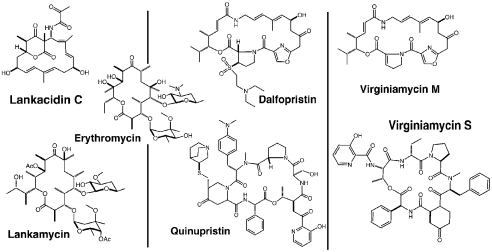
Lankacidin C (LC) and lankamycin (LM) are two inhibitory compounds produced by Streptomyces rochei 7434AN4 (27, 28). The structures of LC and LM are chemically distinct and rather different from those of streptogramins (Fig. 1). LC is a macrocyclic compound composed of a 17-membered carbocyclic ring, bridged by a 6-membered lactone. LM is a macrolide whose 14-member lactone ring is decorated with 4-acetyl-L-arcanose and D-chalcose sugars (29, 30) resembling erythromycin (ERY) (Fig. 1). LC, which is used in veterinary medicine, inhibits growth of Gram-positive bacteria by interfering with protein synthesis but has little effect on eukaryotic cell-free translation (31–34). LC and its derivatives also exhibit antitumor activity, although it is unclear whether this phenomenon is related to the drug’s effect upon protein synthesis (35). Lankamycin exhibits a weak antibiotic activity against several Gram-positive bacteria and low toxicity in animal models (36, 37).
Fig. 1.
Chemical structures of antibiotics relevant to this study. Three pairs are shown, in each the compounds that bind to the PTC are in the Upper panel, and their mates that bind to the NPET are in the Lower panel immediately below them. Erythromycin is inserted for size and structure comparisons.
LC competition with chloramphenicol for binding to the ribosome reveals the large ribosomal subunit as a likely target of its action, in accord with its classification as a protein synthesis inhibitor (32, 38). Though little data is available about activity of LM, the similarity of its structure to that of ERY indicates that it may act as a typical macrolide. Furthermore, similar to ERY and other macrolides, desacetyl-LM was reported to be capable of activating expression of inducible macrolide-resistance genes (37). Coregulation of production of LC and LM (39, 40) suggests that these drugs have been evolutionary optimized to work together. Nevertheless, although LC and LM are coproduced by the Streptomyces strain, there has been no information about their sites of action, nor any evidence of functional interaction between these two antibiotic compounds.
Here, we investigated the sites of binding and the modes of action of LC and LM by crystallographic and biochemical analyses. We show that these two compounds bind at neighboring sites in the large ribosomal subunit, which partially overlap with the binding sites of two components of streptogramin antibiotics. We present evidence that LC can bind simultaneously with LM, and that the two drugs inhibit bacterial growth synergistically, suggesting that the structures of LC and LM have been optimized in the course of evolution to allow for their simultaneous cooperative action. Based on our structural results we also suggest means for enhancing their synergetic inhibitory effect. This study presents the cases in which crystallographic analysis provided functional insights that, in turn, stimulated advanced biochemical and genetic studies, which yielded further clinical implications.
Results
Lankacidin Binding Site.
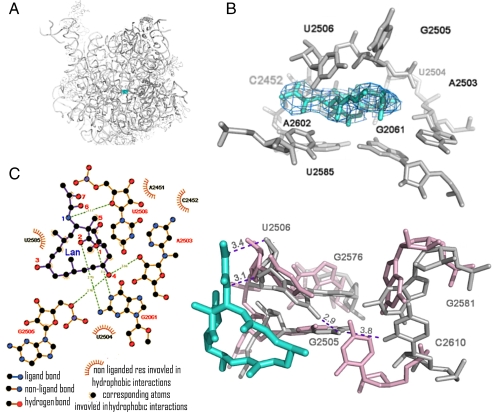
The 3.5 Å resolution (Table S1) difference electron density map calculated between the structure amplitudes of the large ribosomal subunits from Deinococcus radiodurans (D50S) in complex with LC (D50S-LC) and of the D50S native structure (41) allowed the unambiguous determination of the location (Fig. 2A) and conformation of LC in the PTC (Fig. 2B).
Fig. 2.
Interaction of lankacidin with D50S PTC. (A) LC (cyan) binding site at the PTC of D50S. (B) Modeling of LC structure in the (Fo-Fc) difference electron density map, contoured at 1.0 σ. (C) Chemical structure diagram of interactions of LC with the PTC nucleotides. (D) A network of induced interactions (in gray) between five PTC nucleotide residues that stabilize LC binding. The wild-type conformations are shown in pink.
LC binding pocket is composed of nucleotides A2602, C2452, A2503, U2504, G2505, U2585, G2061, and U2506 (E. coli numbering throughout), and the bound LC is involved in an extensive network of hydrophobic interactions with most of these nucleotides. Additionally, LC is positioned within hydrogen bond distance to N1 and N2 of G2061, the 2′ ribose hydroxyls of A2503, and O5′ of G2505 (Fig. 2B–D). It partially occupies the location of the amino acid attached to the 3′ end of A site tRNA (Fig. 3A) and barely reaches the macrolide binding site (Fig. 3C). The repositioning of the rRNA residues that occurs as a consequence of LC binding creates a unique network of interactions between five reoriented nucleotides: U2506, G2505, G2581, C2610, and G2576. Within this network G2576 stacks upon G2505, C2610 stacks upon G2581, and the exocyclic amine of G2505 is at a hydrogen bonding distance from O2 of C2610. Also, the amino group of G2505 is within a hydrogen bond reach of O4 of U2506, which shifts toward the LC’s 1,3 dicarbonyl system. These newly established contacts stabilize the placement of G2505 and U2506 in a conformation that favors binding of LC. Additionally, similar to pleuromutilins that utilize a network of remote interactions, LC binding is influenced by the second-shell nucleotides, specifically G2576, A2062, C2530, U2531, C2507, U2584, G2581, C2610, and A2059. Thus, LC exploits the PTC’s inherent flexibility for achieving high binding affinity (9, 10).
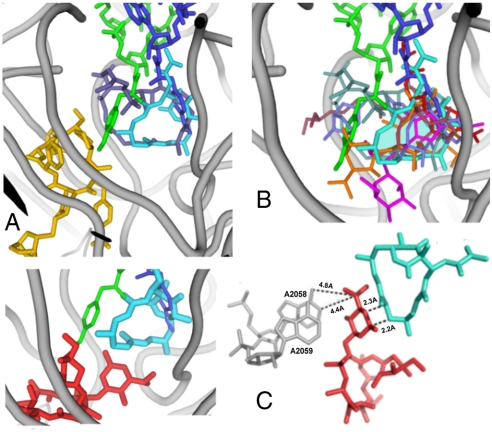
Fig. 3.
Antibiotics in PTC and NPET of the ribosome. In all the panels, LC is shown in cyan and the amino acid-esterified 3′ ends of A- and P- sites tRNAs are in blue and green, respectively (42). Ribosomal RNA backbone is colored in gray. (A) Superposition of LC and an SA compound dalfopristin (metallic blue) in PTC (6). The dalfopristin’s mate SB component, quinupristin, in the NPET (6) is shown in gold. (B) Superposition of LC and several other PTC-targeting antibiotics in their D50S binding sites: A section through the volume occupied by LC is shown in transparent cyan; the drugs shown are: clindamycin (3) in magenta, tiamulin (5) in purple; retapamulin (9) in slate, chloramphenicol (3) in red, dalfopristin (6) in metallic blue and methymycin (14) in orange and quinupristin (6) in gold. (C) Left: The relative positions of LC and ERY (red, from ref. 3) in D50S; right: The marked short distance between LC and ERY indicates a potential steric clash, thus explaining the competition of the two drugs for binding to the ribosome.
In its binding site, LC macrolactone ring fits in the shallow depression in the wall of the PTC A site, by forming van der Waals interactions with U2504, G2505 and U2506. The importance of these interactions to the drug binding is manifested by structure-activity studies that showed that hydrogenation of the macrocyclic ring alters its ring conformation and reduces the inhibitory activity of LC (43). The 2-methyl group at the lactone edge of the macrocyclic ring inserts in the opening of the hydrophobic crevice formed by the splayed out bases of A2451 and C2452. This cleft also hosts the aminoacyl moiety of A site-bound aminoacyl-tRNA and is involved in binding of other PTC-targeting antibiotics, including SA antibiotic compounds such as dalfopristin and virginiamycin M (3, 13, 44). The overlap of the LC and chloramphenicol binding sites (Fig. 3B) provides the structural basis for their competition for binding to the ribosome (32).
Despite significant size differences (Fig. 1), the position of LC closely resembles those of dalfopristin (6) and virginiamycin M (4). However, substantial differences were observed in the interactions of these compounds with the ribosome. In the D50S-Synercid complex (6) dalfopristin ring extends significantly farther toward the P site (Fig. 3A). As LC binds in the PTC center, it causes the flexible base of A2602, which plays a major role in tRNA translocation (42, 45), to undergo a 45° rotation compared to its placement in native D50S or in Synercid-bound complex. U2585, the second flexible nucleotide that also seems to play a role in A-tRNA translocation, undergoes only a minor alteration in D50S-LC complex, while it is rotated by 180° in D50S-Synercid complex. This rotation seems to occur because of steric hindrance of the dalfopristin large macrocyclic ring and to the occupation of the SB site by quinupristin, the SB component of Synercid. Other important details distinguish binding of LC and dalfopristin. While both drugs are hydrogen-bonded to the G2061 exocyclic amine, an additional H bond links dalfopristin with G2061 2′ hydroxyl (6). Both LC and Synercid induce a conformational change of C2610. However, in the D50S-LC complex C2610 stacks upon G2581 and is H-bonded to G2505, whereas in the D50S-Synercid complex it is flipped away because of steric hindrance caused by quinupristin. Additionally, while both LC and dalfopristin reorient the U2506 base, the shift of this base toward LC is unique.
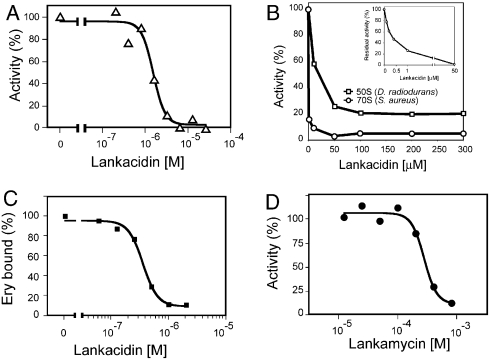
Consistent with its binding to the functionally critical PTC, LC inhibited bacterial (E. coli) cell-free transcription-translation system, with a respectable IC50 of 1.5 ± 0.1 μM (Fig. 4A). Furthermore, confirming previous observation (32, 38), we found that the drug readily interfered with the peptide bond formation inhibiting the puromycin reaction catalyzed by either Staphylococcus aureus 70S ribosomes (IC500.32 ± 0.02 μM) or isolated large ribosomal subunits of D. radiodurans (IC5010.0 ± 6.0 μM) (Fig. 4B). This result affirmed LC as an effective PTC inhibitor.
Fig. 4.
Effect of LC on (A) protein synthesis in the E. coli cell-free system and (B) peptide bond formation catalyzed by S. aureus 70S ribosomes (circles) or D. radiodurans large ribosomal subunits (squares). (C) Competition of LC with 14C-ERY for binding to D. radiodurans 50S subunits. (D) Inhibition of cell-free translation (E. coli) by LM
Lankacidin and Lankamycin Can Simultaneously Bind to the Ribosome.
S. rochei secretes two antibiotics, a 17-member ring macrocyclic LC and a 14-member ring macrolide LM. LM is structurally similar to ERY (Fig. 1) and thus we assumed that it is likely to bind to the ribosome at the site and orientation similar to ERY, namely at the NPET in immediate proximity to the PTC, the LC binding site. However, the comparison of the position of ERY in D50S (3) with the crystal structure of D50S-LC complex (Fig. 3C) revealed that the desosamine sugar of ERY approaches the macrocyclic ring of LC too close for simultaneous binding of both drugs. In agreement with this notion, competition experiments showed that LC displaced 14C-ERY from D50S with IC50 of 355 ± 26 nM (Fig. 4C). These observations raised the question whether similar to ERY, LM will also compete with LC for ribosome binding. If so, why would an antibiotic-producing microorganism synthesize two competing drugs?
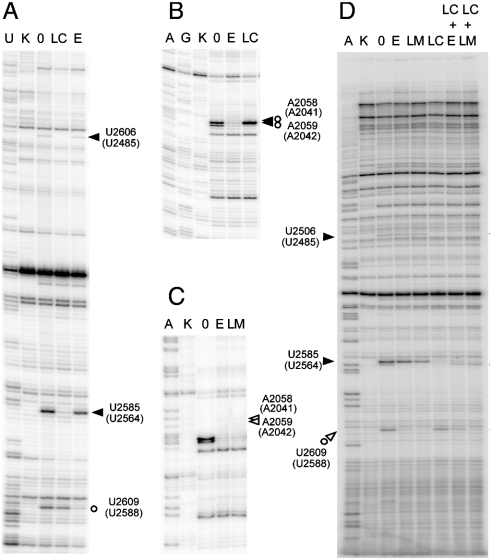
Since the lack of radiolabeled LC and LM precluded direct measurement of their binding to the ribosome, we applied an alternative technique to address this puzzle. We first verified that LM binds to the ribosome and inhibits protein synthesis. Indeed, in the E. coli cell-free system, LM inhibited translation (IC50 of 275 ± 36 μM) (Fig. 4D), arguing that the antibiotic does bind to the ribosome, albeit with only moderate affinity. RNA probing was then utilized to follow the binding of LC and LM to the ribosome. For the consistency of structural data, we carried out RNA probing experiments using D. radiodurans large ribosomal subunits. Binding of LC and LM was analyzed using chemical modifying reagents 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT) and dimethyl sulfate (DMS) (46).
In accord with crystallographic data, association of LC with D50S results in a strong protection of the PTC nucleotide residues U2506 and U2585 from CMCT modification (Fig. 5A). We also noted that upon LC binding, A2059 became partially protected from DMS, whereas modification of A2058 was increased (Fig. 5B). This effect correlated with the crystallographic structure of the LC-D50S complex where upon LC binding A2059 is stacked upon A2503, and such stacking may partly expose the surface of A2058, yielding access for DMS. This result indicated that LC-induced restructuring of the D50S nucleotide residues in the PTC can propagate allosterically to the proximal segment of the NPET.
Fig. 5.
Chemical probing of interactions of LC (filled arrowheads), LM (open arrowheads) and ERY (open circles) with the ribosome. Protection of 23S rRNA residues from (A) CMCT and (B) DMS modification by LC. (C) Protection of 23S rRNA residues from CMCT modification by LM. (D) Protections from CMCT afforded by LC, LM, and ERY present alone or in combination. Small open circles indicate bands that appeared because of slight nuclease degradation, which were not reproducible between the repeated experiments.
Similar to ERY and other 14-member ring macrolides, binding of LM results in protection of U2609 from CMCT modification and of A2058 and A2059 from DMS modification (Fig. 5C and D). We then exploited the idiosyncratic protections of 23S rRNA residues against CMCT modification, afforded by LC and LM, to interrogate their simultaneous interaction with the ribosome. At 50 μM, LC shields U2506 and U2585 from alterations. However, the accessibility of these residues to CMCT is not affected by LM or ERY (Fig. 5). Conversely, LM (at 500 μM) or ERY (at 50 μM) strongly protect U2609, while LC has no effect on this nucleotide. Thus, protection of U2506 and U2585 indicates LC binding, whereas protection of U2609 reveals LM or ERY binding. When LC and LM are present together, all the three residues (U2506, U2585, and U2609) are protected, indicating that LC and LM are simultaneously bound to the ribosome. In contrast, and in agreement with the binding experiments, the LC-mediated protection of U2506 and U2585 is partially relieved upon addition of ERY (Fig. 5D) and ERY-dependent protection of U2609 is partly reversed when LC is present. Thus, while LC and ERY compete for the binding to the ribosome, LC and LM can bind simultaneously to their respective targets in the PTC and NPET.
LC and LM Act Synergistically Upon Bacterial Cells.
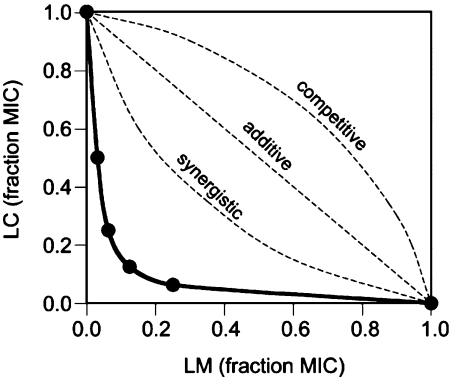
Because the binding site of LC and LM partially overlaps with that of SA and SB antibiotics, and because SA and SB act synergistically, we anticipated that despite the difference in their size and chemical properties from streptogramins, LC and LM may also exhibit synergy. To address this issue we performed in vivo and in vitro experiments, using whole cell bacteria as well as a bacterial cell-free system. Synergism was observed by an in vivo assay that utilizes a susceptible strain of Gram-positive S. aureus. We analyzed the minimal inhibitory concentrations (MIC) in a checkerboard fashion and plotted the results as a fraction of MIC (FIC) of individual compounds (Fig. 6). In this assay, antibiotics are considered synergistic if the curve has a concave shape, whereas a linear plot reflects additive action of the drugs, and a convex graph shows antagonistic interaction (47). The experimental MIC plot (solid line in Fig. 6) had a well-pronounced concave character revealing synergy in action of LC and LM. These findings were further verified in an E. coli cell-free transcription-translation system. Thus, similar to streptogramins, the two antibacterial compounds produced by S. rochei bind simultaneously to the neighboring sites in the ribosome and synergistically inhibit sensitive bacteria.
Fig. 6.
Synergistic inhibitory activity of LC and LM upon S. aureus. The plots represent changes in MIC of individual compounds when both drugs are present in combination. The general shapes of the hypothetical curves corresponding to the additive, synergistic, or antagonistic mode of the drug combinations are shown by broken lines. The experimental curve is shown as a solid line with the MIC values (shown as a fraction of MIC of LM or LC acting alone) indicated by filled circles.
Discussion
The crystallographic and biochemical data presented here firmly established the PTC as the site of LC action. The inhibitory effect of the antibiotic upon peptide bond formation is likely achieved by preventing the binding or the proper placement of aminoacyl moiety of aminoacyl-tRNA in the A site (Fig. 3A). As LC trespasses the P site, it may also affect the exact positioning of the peptidyl tRNA C terminus. Although LC is chemically distinct and is less bulky than most SA-type drugs, its binding site partially overlaps with that of SA compounds (4, 6). Furthermore, both, LC and SA drugs and U2506 that is involved in PTC functions (42, 48–50).
As can be concluded from the similarity of chemical structures of LM and ERY and from the overlap of the set of nucleotides protected by these two compounds, LM binds to the NPET macrolide binding site in a fashion similar, albeit not identical, to the other macrolides (Fig. 5). The same NPET region accommodates the B components of streptogramin antibiotics, but owing to differences in chemical nature (Fig. 1), the SB compounds and macrolides exploit a different set of interactions (3, 4, 6, 7).
Because the LC and SA binding sites in the PTC (Fig. 3C) are adjacent to the macrolides and SB binding sites, it is conceivable that compounds acting upon these two sites can either compete or cooperate in binding to the ribosome. Streptogramins are known for their synergistic action (24, 51). Such cooperativity makes evolutionary sense: The same microorganism produces both SA and SB components, and their mutually enhanced action should be highly beneficial for the antibiotic producer. This notion initially did not seem to hold true for the macrocyclic LC and a macrolide LM. Our experiments showed that LC and ERY do not cooperate, but rather compete for binding to the D. radiodurans ribosome. Hindrance may result from a direct clash between ERY desosamine sugar and the LC macrocyclic ring (Fig. 3C) or by allosteric modulation of the binding site of one compound by binding of its counterpart. The altered DMS reactivity of A2058 and A2059, the nucleotides located in the heart of the macrolide binding site (Fig. 5A), upon the LC binding might be a reflection of such allostery. However, the competition between LC and ERY is not so surprising: These drugs are produced by different microorganisms and were not “designed” to work together. In contrast, LM is coproduced with LC. Both drugs appear to be coregulated, and thus similar to the streptogramins case, seem to be intended to work together (39, 40). In agreement with this hypothesis, our results show their simultaneous binding to the ribosome. The small structural differences between ERY and LM (Fig. 1) are likely to cause the variation in their binding properties. One of the important distinctions between LM and ERY is the nature of the C5-linked sugar residue (D-desosamine in ERY vs. D-chalcose in LM). Even small modifications of desosamine in ERY can dramatically alter the antibiotic’s activity (52, 53). Replacement of 3′ dimethyl amine of ERY with a methoxy group in LM may facilitate accommodation of the C5-sugar residue in a narrow space left between the A2058/A2059 ridge and the bound LC molecule (see Fig. 3C). Similarly, minute modifications in the structure of the antibiotics or in their binding sites may have significant effects on their binding and properties (20, 37, 54, 55). LM on its own is a less potent protein synthesis inhibitor than ERY. However, LM reduced activity is compensated by its ability to act synergistically with LC. Thus, it appears that the LM synthetic pathway has been evolutionary optimized to generate a 14-member ring macrolide capable of simultaneous binding and synergistic action with LC. It is hardly a coincidence that two different combinations of synergistic protein synthesis inhibitors, the SA and SB components of streptogramins or LC and LM, utilize the same two adjacent sites in the large ribosomal subunit. These sites might be best suited within the ribosome for accommodating pairs of compounds that would be able to tightly bind and inhibit protein synthesis in a synergistic fashion. Previously, significant resources went into development of streptogramins into a clinical drug. This effort resulted in a useful and successful streptogramin antibiotic, Synercid. Yet, to the best of our knowledge, no focused attempt was dedicated to optimizing the combination of LC/LM or, for that matter, LC with any other macrolide. Our studies demonstrate the validity of LC/LM synergism and provide a structural basis for chemical modifications of either of the two components, which could lead to the improvement of the inhibitory action of the LC/LM pair and its clinical relevance. Notably, the SA compounds (for example quinupristin or virginiamycin S) are significantly larger than LC (Figs. 1 and 3A) and therefore have more chemical entities facilitating their binding to the ribosome pocket and forming a stable network of interactions. It is likely, therefore, that decorating LC with additional groups capable of forming new interaction with rRNA in its binding site may increase its affinity. Modulating the properties of the LM sugar residues may further add to the potency of the LC/LM pair.
Conclusions
We showed that two antibiotics, LC and LM, produced by S. rochei are protein synthesis inhibitors that act upon neighboring sites in the large ribosomal subunit and that their simultaneous action synergistically inhibits growth of sensitive bacteria. Our structural and biochemical data imply that optimization of this drug pair leading to high affinity concurrent binding to the ribosome may be reached by minor chemical alterations. Furthermore, our results suggest that certain combinations of PTC inhibitors with the NPET-bound compounds in their natural or chemically modified versions might exhibit synergy. Exploring such drug combinations by co-optimizing their structure or by linking them together into a single molecule may pave the way for the development of advanced antibiotics targeting the ribosome.
Materials and Methods
Crystals of D50S, grown as in ref. 41, were soaked in a solution containing lankacidin. Crystallographic data were collected with highly collimated synchrotron x-ray beam and processed with HKL2000 (56) and CCP4 (57) using the available crystal structure of lankacidin C (29). After map tracing and refinement by COOT (58) and CNS (59, 60) ribosome-antibiotic interactions were identified by LigPlot (61) and LPC (62), and images were generated by PyMol (63). Coordinates were deposited in the protein data bank (PDB) with accession code 3JQ4.
Antibiotic binding and inhibition of cell-free translation were determined as described in ref. 64. Inhibition of the peptidyl transferase reaction by LC was performed and analyzed as described in refs. 65 and 66. The data were analyzed using Prism 4 (GraphPad Software). RNA probing was carried out as described in ref. 16 and 46 using D50S. Additional details can be found in SI Text.
Supplementary Material
Table S1.
Crystallographic data for the D50S-lankacidin complex
| Parameters | |
| Space group | I222 |
| Resolution (Å) | 40-3.5 (3.63-3.5) |
| Rsym (%) | 16.3 (82.7) |
| Completeness (%) | 92.4 (91.8) |
| Redundancy | 5.3 (4.6) |
| I/σ(I) | 7.7 (1.5) |
| Unit Cell (Å) | a = 169.8 b = 410.3 c = 694.4 |
| R/Rfree (%) | 26.8/32.4 |
| Bond length (Å) | 0.006 |
| Bond angles (degrees) | 1.185 |
Acknowledgments.
We thank the ribosome group at the Weizmann Institute for participating in the experiments reported here and Dr. Llano-Sotelo (University of Illinois at Chicago) for helping with the analysis of the inhibitory and competition data. We thank Dr. Weisblum (University of Wisconsin) for providing initial samples of lankamycin and Pfizer, Inc. for lankacidin. Crystallographic data were collected at station 19ID at the Advanced Photon Source (Argonne National Laboratory) and ID23-2 at the European Synchrotron Radiation Facility, and we thank the staff of both facilities for excellent assistance. This work was supported by National Institutes of Health grants GM34360 (to A.Y.) and U19 AI56575 (to A.S.M.) and by the Kimmelman Center for Macromolecular Assemblies. CD is supported by the Adams Fellowship Program of the Israel Academy of Sciences and Humanities. A.Y. holds the Martin and Helen Kimmel Professorial Chair.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914100107/DCSupplemental.
References
- 1.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3(11):870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 2.Bottger EC. The ribosome as a drug target. Trends Biotechnol. 2006;24(4):145–147. doi: 10.1016/j.tibtech.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Schluenzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase center in eubacteria. Nature. 2001;413(6858):814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 4.Hansen JL, et al. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330(5):1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 5.Schluenzen F, et al. Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol Microbiol. 2004;54(5):1287–1294. doi: 10.1111/j.1365-2958.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- 6.Harms J, et al. Alterations at the peptidyl transferase center of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2004;2(1):4;1–10. doi: 10.1186/1741-7007-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tu D, et al. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Leach KL, et al. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell. 2007;26(3):393–402. doi: 10.1016/j.molcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Davidovich C, et al. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci USA. 2007;104(11):4291–4296. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidovich C, et al. Structural basis for cross-resistance to ribosomal PTC antibiotics. Proc Natl Acad Sci USA. 2008;105(52):20665–20670. doi: 10.1073/pnas.0810826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ippolito JA, et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. 2008;51(12):3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 12.Wilson DN, et al. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA. 2008;105(26):13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurel G, et al. U2504 determines the species specificity of the A-site cleft antibiotics the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol. 2009;389(1):146–156. doi: 10.1016/j.jmb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auerbach T, et al. Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin. Biotechnolog. 2009;84:24–35. [Google Scholar]
- 15.Polacek N, Mankin AS. The ribosomal peptidyl transferase center: Structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol. 2005;40(5):285–311. doi: 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- 16.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 17.Xiong L, et al. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase center. Mol Microbiol. 1999;31(2):633–639. doi: 10.1046/j.1365-2958.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 18.Schluenzen F, et al. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11(3):329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 19.Berisio R, et al. Structural insight into the antibiotic action of telithromycin against resistant mutants. J Bacteriol. 2003;185(14):4276–4279. doi: 10.1128/JB.185.14.4276-4279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berisio R, et al. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat Struct Biol. 2003;10(5):366–370. doi: 10.1038/nsb915. [DOI] [PubMed] [Google Scholar]
- 21.Liu M, Douthwaite S. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc Natl Acad Sci USA. 2002;99(23):14658–14663. doi: 10.1073/pnas.232580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acar JF. Antibiotic synergy and antagonism. Med Clin N Am. 2000;84(6):1391–1406. doi: 10.1016/s0025-7125(05)70294-7. [DOI] [PubMed] [Google Scholar]
- 23.Vazquez D. Mechanism of action of antimicrobial and antitumor agents. In: Corcoran JW, Hahn FE, editors. In Antibiotics III. 1975. Springer-Verlag; pp. 521–534. [Google Scholar]
- 24.Cocito C, et al. Inhibition of protein synthesis by streptogramins and related antibiotics. J Antimicrob Chemother. 1997;39(Suppl A):7–13. doi: 10.1093/jac/39.suppl_1.7. [DOI] [PubMed] [Google Scholar]
- 25.Canu A, et al. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2002;46(1):125–131. doi: 10.1128/AAC.46.1.125-131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delgado G, Jr, et al. Quinupristin-dalfopristin: An overview. Pharmacotherapy. 2000;20(12):1469–1485. doi: 10.1592/phco.20.19.1469.34858. [DOI] [PubMed] [Google Scholar]
- 27.Gaumann E, et al. Stoffwechselprodukte von Actinomyceten. 21. Mitteilung. Lankamycin und Lankacidin. Helv Chim Acta. 1960;43(2):601–606. [Google Scholar]
- 28.Kinashi H, et al. Isolation and characterization of linear plasmids from lankacidin-producing Streptomyces species. J Antibiot. 1994;47(12):1447–1455. doi: 10.7164/antibiotics.47.1447. [DOI] [PubMed] [Google Scholar]
- 29.Uramoto M, et al. The structures of Bundlin A (lankacidin) and Bundlin B. Tetrahedron Lett. 1969;27:2249–2254. doi: 10.1016/s0040-4039(01)88133-8. [DOI] [PubMed] [Google Scholar]
- 30.Harada S, et al. Isolation and structures of T-2636 antibiotics. Tetrahedron Lett. 1969;27:2239–2244. doi: 10.1016/s0040-4039(01)88131-4. [DOI] [PubMed] [Google Scholar]
- 31.Harada S, et al. Studies on lankacidin-group (T-2636) antibiotics. VII. Structure-activity relationships of lankacidin-group antibiotics. J Antibiot. 1973;26(11):647–657. doi: 10.7164/antibiotics.26.647. [DOI] [PubMed] [Google Scholar]
- 32.Retsema JA, et al. In: New Macrolides, Azalides, and Streptogramins in Clinical Practice. Neu HC, Lowell SY, Zinner SH, Acar JF, editors. Marcel; 1995. pp. 191–195. [Google Scholar]
- 33.McFarland JW, et al. Side chain modifications in lankacidin group antibiotics. Antimicrob Agents Chemother. 1984;25(2):226–233. doi: 10.1128/aac.25.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mochizuki S, et al. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol Microbiol. 2003;48(6):1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- 35.Oostu K, et al. Antitumor and immunosuppressive activities of lankacidin-group antibiotics: Structure-activity relationships. Cancer Chemother Rep. 1975;59(5):919–928. [PubMed] [Google Scholar]
- 36.Martin JR, et al. 3′-de-o-methyl-2′, 3′-anhydro-lankamycin, a new macrolide antibiotic from Streptomyces violaceoniger. Helv Chim Acta. 1976;59(5):1886–1894. doi: 10.1002/hlca.19760590545. [DOI] [PubMed] [Google Scholar]
- 37.Omura S, et al. Studies on the antibiotics from Streptomyces spinichromogenes var. kujimyceticus. II. Isolation and characterization of kujimycins A and B. J Antibiot. 1969;22(10):500–505. doi: 10.7164/antibiotics.22.500. [DOI] [PubMed] [Google Scholar]
- 38.Retsema J, Fu W. Macrolides: Structures and microbial targets. Int J Antimicrob Ag. 2001;18(Suppl 1):3–10. doi: 10.1016/s0924-8579(01)00401-0. [DOI] [PubMed] [Google Scholar]
- 39.Arakawa K, et al. gamma-Butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology+ 2007;153(Pt 6):1817–1827. doi: 10.1099/mic.0.2006/002170-0. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto S, et al. Gamma-butyrolactone-dependent expression of the Streptomyces antibiotic regulatory protein gene srrY plays a central role in the regulatory cascade leading to lankacidin and lankamycin production in Streptomyces rochei. J Bacteriol. 2008;190(4):1308–1316. doi: 10.1128/JB.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 42.Kamiya K, et al. x-ray analysis of an antibiotic, T-2636 A (Bundlin B) Tetrahedron Lett. 1969;27:2245–2248. doi: 10.1016/s0040-4039(01)88132-6. [DOI] [PubMed] [Google Scholar]
- 43.Hansen JL, et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10(1):117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 44.Bashan A, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 45.Agmon I, et al. Ribosomal crystallography: a flexible nucleotide anchoring tRNA translocation, facilitates peptide-bond formation, chirality discrimination and antibiotics synergism. FEBS Lett. 2004;567(1):20–26. doi: 10.1016/j.febslet.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 46.Merryman C, Noller HF. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith CWJ, editor. RNA:Protein Interactions. Oxford Univ Press, UK; 1998. pp. 237–253. [Google Scholar]
- 47.Hardman JG, Limbird LE, Molinoff PB, Ruddon RW. McGraw-Hill; 1996. Goodman & Gilman’s The Pharmacological Basis of Therapeutics; pp. 1046–1049. [Google Scholar]
- 48.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 49.Zarivach R, et al. Functional aspects of ribosomal architecture: Symmetry, chirality and regulation. J Phys Org Chem. 2004;17:901–912. [Google Scholar]
- 50.Schmeing TM, et al. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438(7067):520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 51.Porse BT, Garrett RA. Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23 S rRNA and the synergism of their inhibitory mechanisms. J Mol Biol. 1999;286(2):375–387. doi: 10.1006/jmbi.1998.2509. [DOI] [PubMed] [Google Scholar]
- 52.Wu YJ, Su WG. Recent developments on ketolides and macrolides. Curr Med Chem. 2001;8(14):1727–1758. doi: 10.2174/0929867013371671. [DOI] [PubMed] [Google Scholar]
- 53.Yonath A. Antibiotics targeting ribosomes: Resistance, selectivity, synergism, and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 54.Vazquez-Laslop N, et al. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30(2):190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 55.Pfister P, et al. 23S rRNA base pair 2057-2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A -- > G. Proc Natl Acad Sci USA. 2005;102(14):5180–5185. doi: 10.1073/pnas.0501598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Carter JCW, Sweet RM, editors. Processing of x-ray diffraction data collected in oscillation mode, methods in enzymology. Macromolecular Crystallography. 1997;Vol. 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.CCP4. Collaborative Computational Project, Number 4, The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 60.Brunger AT. The crystallography and NMR system. Nat Protoc. 2007;2(11):2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 61.Wallace AC, et al. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 62.Sobolev V, et al. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15(4):327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 63.DeLano WL. Palo Alto, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 64.Xiong L, et al. Binding Site of the Bridged Macrolides in the Escherichia coli Ribosome. Antimicrob Agents Chemother. 2005;49(1):281–288. doi: 10.1128/AAC.49.1.281-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maguire BA, et al. A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Mol Cell. 2005;20(3):427–436. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 66.Bartetzko A, Nierhaus KH. Mg2 + /NH4 + /polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol. 1988;164:650–658. doi: 10.1016/s0076-6879(88)64075-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.