Abstract
Transplantation and pregnancy, in which two diploid genomes reside in one body, can each lead to diseases in which immune cells from one individual target antigens encoded in the other’s genome. One such disease, graft-versus-host disease (GVHD) after hematopoetic stem cell transplantation (HSCT, or bone marrow transplant), is common even after transplants between HLA-identical siblings, indicating that cryptic histocompatibility loci exist outside the HLA locus. The immune system of an individual whose genome is homozygous for a gene deletion can recognize epitopes encoded by that gene as alloantigens. Analyzing common gene deletions in three HSCT cohorts (1,345 HLA-identical sibling donor-recipient pairs), we found that risk of acute GVHD was greater (OR = 2.5 [95%CI, 1.4–4.6]) when donor and recipient were mismatched for homozygous deletion of UGT2B17, a gene expressed in GVHD-affected tissues and giving rise to multiple histocompatibility antigens. Human genome structural variation merits investigation as a potential mechanism in diseases of alloimmunity.
GVHD is a serious, common complication of allogeneic hematopoietic stem cell transplantation (HSCT, or bone marrow transplant) in which immune responses by donor-derived lymphocytes target alloantigens in the host. GVHD rarely if ever occurs after transplants between monozygotic twins 1,2 but frequently occurs after transplants between HLA-identical siblings, indicating that additional histocompatibility loci must exist outside the HLA locus.
One clear example of a non-HLA compatibility locus is the Y chromosome, as there is increased risk of GVHD when HSCT involves a female donor and a male recipient 3. This effect arises from immune recognition (by donor-derived lymphocytes and antibodies) of antigens encoded by a few Y-linked genes that are expressed in the soma 4–9; these genes collectively differ in sequence from their X-linked paralogs at only a few hundred amino acids 10. This observation demonstrates that changes in an individual’s antigen repertoire of hundreds of amino acids – the size of many individual autosomal genes – can increase risk of GVHD.
The human genome is increasingly recognized to have extensive structural polymorphism 11,12, including deletions of entire autosomal genes 13,14. Some of these gene deletion alleles are sufficiently common that individuals inherit them from both parents and therefore completely lack a protein-coding gene that is expressed in other individuals 13. Because the immune system of an individual with a homozygous gene deletion presumably has not learned to tolerate the protein encoded by that gene, immune recognition of that protein as an alloantigen 15–17 by immune cells or antibodies from that individual could in principle contribute to risk of alloimmune disease.
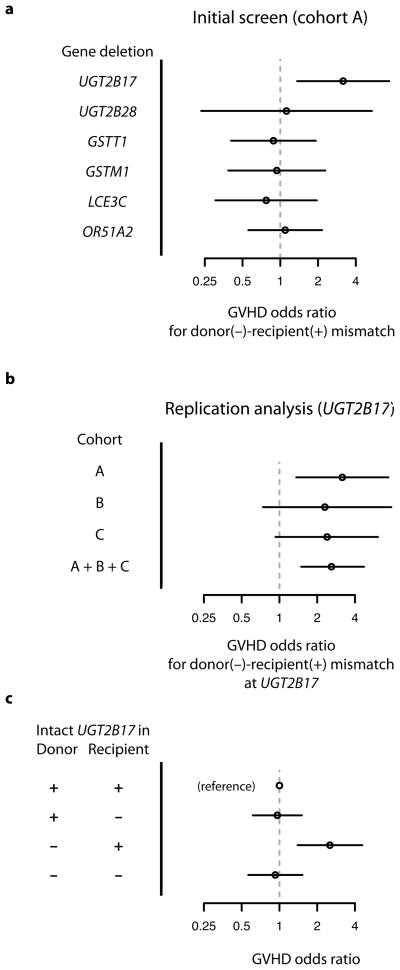
To assess whether donor-recipient mismatch for homozygous gene deletions increases risk of GVHD after transplantation, we first typed a set of common gene deletions in 400 HSCT patients and their HLA-identical sibling donors (Cohort A; Methods, Supplementary Table 1). This screen involved gene-deletion polymorphisms that we identified from a genome-wide survey of copy-number variation 18 as satisfying the following criteria: a deletion allele that (1) removes the gene protein-coding sequence and (2) segregates in the population with allele frequency greater than 10%; (3) expression of the gene in one or more of the tissues commonly involved in acute GVHD (liver, intestine, skin); and (4) significant sequence difference (tens to hundreds of amino acids) from any paralogous gene encoded elsewhere in the human genome (Supplementary Table 2). The following genes were identified from this screen: UGT2B17, UGT2B28, GSTM1, GSTT1, LCE3C, OR51A2. We assessed association with GVHD risk by first typing each deletion in HSCT patients and their sibling donors, so as to determine which transplants involved homozygous gene deletion in the donor but not the recipient. At each locus, we then assessed whether such mismatches were associated with increased risk of acute GVHD (Fig. 1a).
Figure 1.
(a) Initial screen for association of acute GVHD with donor-recipient mismatch for common gene deletions, in Cohort A. Six common gene-deletion polymorphisms were screened by typing in donors and recipients; data represent association of donor-recipient mismatch (donor −, recipient +) with the development of GVHD after transplantation. Odds ratios and 95% confidence intervals are shown. (b) Analysis in additional patient cohorts of the association of donor-recipient mismatch at UGT2B17 with acute GVHD. (c) Association of UGT2B17 deletion in donor and recipient with GVHD risk. The group of transplants in which both donor and patient were UGT2B17-positive is used as the reference group for analysis. Odds ratios and confidence intervals were calculated using the Cochran-Mantel-Haenszel test to combine data from the 1,345 donor-recipient pairs from Cohorts A, B, and C.
In this initial screen, donor-recipient mismatch for homozygous deletion of one of these genes, UGT2B17, showed a promising potential association with GVHD (Fig. 1a; OR = 3.0 [1.3–6.9], nominal p = 0.006, by Cochran-Mantel-Haenszel test; 0.03 after Bonferroni correction). UGT2B17 encodes a 530-amino-acid cell-surface protein that is highly expressed in the same tissues – liver, intestine, and skin – that are affected by clinically apparent GVHD and targeted by donor-derived lymphocytes. For the other five gene deletions tested, we observed no evidence of association of donor-recipient mismatch with acute GVHD (Fig. 1a).
We further assessed the contribution of UGT2B17 mismatches to GVHD in two additional patient cohorts (Cohorts B and C, Supplementary Table 1). Outcomes in Cohorts B and C also involved an increased risk in transplants involving donor-recipient mismatch at UGT2B17 (OR = 2.4 [95%CI, 1.1–5.1], p=0.02, by Cochran-Mantel-Haenszel test), strengthening the overall evidence for association (Fig. 1b) (OR = 2.5, p = 5 × 10−4, by Cochran-Mantel-Haenszel test).
We evaluated alternative models for the association of GVHD with donor-recipient mismatch at UGT2B17. In particular, the observed association might in principle be due to donor genotype or patient genotype independent of donor-recipient mismatch, particularly since variation at UGT2B17 associates to other clinical phenotypes 19–21. We therefore evaluated the data from all 1,345 donor-recipient pairs in Cohorts A, B, and C (Cochran-Mantel-Haenszel test) to assess the risk of GVHD for each combination of donor and recipient UGT2B17 status, relative to a reference group in which donor and recipient were both UGT2B17 (+) (Fig. 1c). Increased risk was confined to the group of transplants for which donors were UGT2B17 (−) and recipients were UGT2B17 (+) (Fig. 1c). In particular, the UGT2B17 status of the HSC donor was not associated with GVHD when HSC recipients were UGT2B17 (−), and the UGT2B17 status of the HSC recipient was not associated with GVHD when HSC donors were UGT2B17 (+) (Fig. 1c).
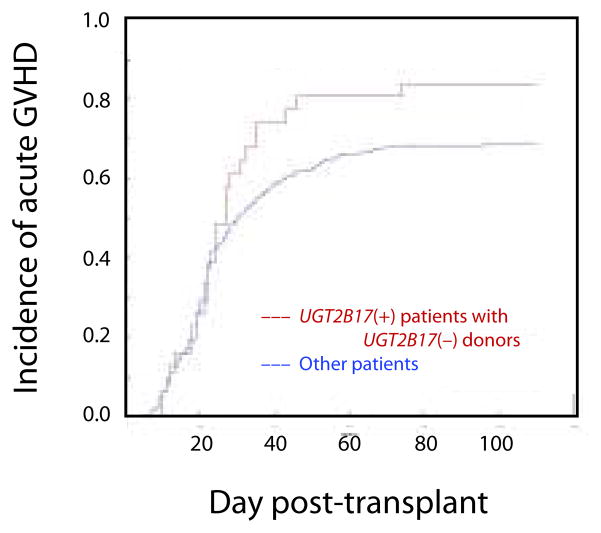
To assess the time course of GVHD incidence in patients with UGT2B17-mismatched donors, a cumulative incidence analysis was performed (Fig. 2). UGT2B17(+) patients with UGT2B17(−) donors showed an unremarkable incidence of acute GVHD during the first 20 days after transplantation; GVHD then began to increase beyond the level observed in other patients after day 20, a pattern similar to that observed in male patients who receive transplants from female donors.
Figure 2.
Cumulative incidence of acute GVHD during the first 100 days after HSCT (Cohort C). In this analysis, death and recurring malignancy before the onset of grades II – IV GVHD were treated as competing risks.
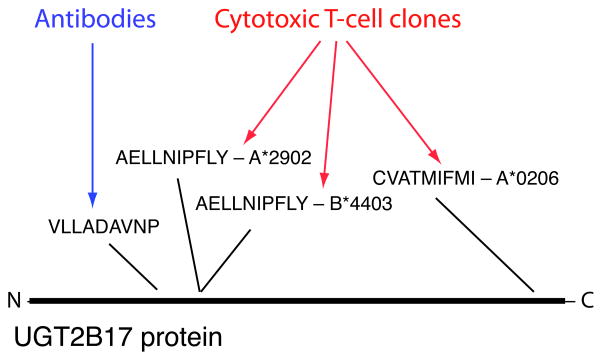
A corollary of the model in which common gene deletions contribute to GVHD risk is that immune responses to multiple antigens derived from UGT2B17 would be present in patients. Several years ago, a cytotoxic T-cell clone derived from a patient with acute GVHD of the intestine was used to screen an expression library to identify the antigen recognized by the T-cell; the antigen was determined to be AELLNIPFLY, a peptide derived from UGT2B17 and presented by HLA-A*2902 in this patient 17. AELLNIPFLY was subsequently found to be presented also by HLA-B*4403 and recognized in this form by a distinct T-cell clone from the same patient 22. Screening of a cytotoxic T-cell clone from a different patient recently identified CVATMIFMI, a different UGT2B17-derived peptide, as the antigen recognized by the clone, when presented by the common HLA allele HLA-A*0206 23. Given evidence that antibody responses can also contribute to GVHD 24–27, and the possibility that UGT2B17’s predicted localization to the cell surface would facilitate such responses, we screened sera from 26 GVHD patients (including ten with UGT2B17-mismatched donors) against an array of overlapping peptides designed to span the UGT2B17 protein sequence. Serum from one patient (a UGT2B17-positive patient whose donor was UGT2B17-negative) showed a robust antibody response to the peptide LQESKFDVLLADAVNPCGEL (UGT2B17 141-160); we fine-mapped this response to the epitope VLLADAVNP (UGT2B17 148-156). Antibodies from this patient distinguished this epitope from paralogous peptide sequences encoded by all other UGT2B genes in the human genome. These data collectively indicate that UGT2B17 gives rise to multiple histocompatibility antigens (Fig. 3), offering a candidate molecular and cellular mechanism for genetic association of UGT2B17 mismatch with GVHD.
Figure 3.
Multiple histocompatibility antigens derived from UGT2B17. An antibody response to the UGT2B17-derived peptide VLLADAVNP was detected in the serum of a UGT2B17-positive GVHD patient whose donor was UGT2B17-negative. Responses of T-cell clones against multiple antigens derived from UGT2B17 and presented by three distinct HLA alleles have been detected in other GVHD patients 17,22,23.
Several features of the UGT2B17 protein may make UGT2B17 a more-potent histocompatibility locus than other gene deletions: (1) UGT2B17 is a large protein (530 amino acids), increasing the likelihood that it contains multiple antigenic epitopes; (2) UGT2B17 is abundant in liver, intestine, and skin, the tissues in which pre-HSCT conditioning elicits the strongest inflammation and in which immune surveillance for alloantigens may therefore be strongest; (3) UGT2B17 is expressed on the cell surface, well-positioned to contribute to antibody-mediated as well as cell-mediated immune responses; and (4) UGT2B17 is also abundant in blood, skin, semen, and placenta, tissues that give rise to inter-individual immune exposures that may pre-expose and immunize UGT2B17 (−) individuals against UGT2B17, a phenomenon that has been observed in healthy female donors for some of the antigens encoded on the Y chromosome 24.
While an estimate of effect size for donor-recipient UGT2B17 mismatches in GVHD based on the cohorts analyzed here (OR = 2.5) is comparable to the established effect of sex mismatch (female donor, male recipient), UGT2B17 mismatches cannot explain a comparable fraction of GVHD incidence, due to the lower frequency at which UGT2B17 mismatches arise between siblings. This sibling mismatch frequency varies among populations due to population variation in frequency of the UGT2B17 deletion allele (19%–85%, an unusually high level of variation that has been attributed to adaptive evolution of UGT2B17 copy number 28); as a result, the expected frequency of sibling mismatches ranges from 2% in African Americans to 5% in most European populations to 9% in Gujarati Indians but does not approach the frequency of female-to-male sex mismatch (25%) in any population (Supplementary Table 4). We also caution that the association observed here (like all such associations in genetics) should be considered preliminary until confirmed by independent, multicenter investigations, and that it may not extend to transplants involving unrelated donors 20, for which the potent effects of donor-recipient mismatches at untyped HLA loci 29 may dominate the effects of mismatches outside the HLA.
The gene deletion polymorphisms analyzed here could in principle be a leading edge of a larger class of polymorphisms that have multifold effects on antigen repertoire. Such polymorphisms might include not only deletion alleles, but SNPs that introduce early stop codons and frame-shifts into protein-coding sequences, alleles that alter transcript splicing, and null regulatory alleles. As large-scale sequence data increasingly make it possible to catalog all the single-nucleotide and structural polymorphisms that segregate in human populations, it will be important to identify the polymorphisms that have multifold effects on antigen repertoire.
Human genome structural variation has been proposed to affect phenotypes by altering gene dosage and by affecting the regulation of nearby genes. Structural polymorphism may also influence disease by a very-different mechanism – by giving rise to multifold differences between individuals’ antigen repertoires that arise from specific genomic loci. The generality of such relationships – in other transplantation settings and in pregnancy – will be an important subject of investigation.
Online Methods
Patients
The first study population (Cohort A) consisted of 424 allogeneic HSCT recipients and their HLA-identical sibling donors transplanted at Helsinki University Central Hospital (HUCH) and the Dana-Farber Cancer Institute (DFCI) for the treatment of a hematological disease (Supplementary Table 1). Analysis was retrospective. Recipients had myeloablative conditioning and received a graft from an HLA-identical sibling between 1993 and 2005. All aspects of human subjects research adhered to protocols approved by the institutional review boards of HUCH and DFCI (Protocol 01-206).
A second study cohort (Cohort B, Supplementary Table 1) consisted of 336 bone marrow recipients and their HLA-identical sibling donors. This cohort has been described previously 30 and consists of sibling donor-recipient pairs who had allogeneic bone marrow transplantation from 1985 through 1993 at several U.S. institutions. Samples were obtained from patients with grades III or IV acute GVHD and from patients without acute GVHD. Patients with grades I or II acute GVHD were not included in this cohort.
A third study cohort (Cohort C, Supplementary Table 1) consisted of 595 bone marrow recipients and their HLA-identical sibling donors. These cases were included in a whole genome scanning study of approximately 1,500 unrelated donor/recipient and HLA-identical sibling pairs randomly selected from patients who had HSCT with myeloablative conditioning regimens at Fred Hutchinson Cancer Research Center (FHCRC) from 1992 through 2004 for treatment of a hematological malignancy or myelodysplasia. Lack of available DNA was the only reason for exclusion from the study cohort. This human subjects research was performed according to protocols approved by the institutional review board of the FHCRC.
Grading of GVHD
Acute GVHD was diagnosed and graded according to standard criteria employed at the time of documented patient care. For the purpose of the current study, affected individuals were defined as those with grades II–IV acute GVHD; unaffected individuals were those with grades 0-I acute GVHD. Diagnostic sensitivity in the grading of GVHD can differ between clinical institutions 31 though it is generally well-harmonized within institutions. Note that the criteria for selection of patients (which were defined prior to genetic analysis, and which for Cohorts B and C reflected the design of other studies) differed from institution to institution; for example, Cohort C utilized a cohort design, while Cohort B utilized a case/control design and over-sampled patients with severe (grades III–IV) acute GVHD.
Genotyping of deletion polymorphisms
Quantitative PCR assays were developed for typing each deletion polymorphism (Supplementary Note). Each internally-controlled, two-color-fluorescence assay allowed the individuals in a cohort to be assigned to three clear genotype classes, consisting of individuals with 0, 1, or 2 gene copies. To ensure the quality of gene-deletion genotypes, we verified that (i) membership in the three genotype classes (corresponding to 0, 1, 2 copies) exhibited Hardy-Weinberg equilibrium; and (ii) regression of patient genotypes against the genotypes of their sibling donors yielded a regression coefficient that was not significantly different from the expected value of 0.5. The accuracy of these assays was further evaluated by using them to genotype the gene deletions in the 270 HapMap samples; these results showed 99.7% concordance with results from an independent experimental approach (use of the Affymetrix SNP 6.0 array 18, with analysis by the Canary algorithm in Birdsuite 32). These assays were used for the initial screen in Cohort A and the replication analysis in Cohort B.
Genotyping of the UGT2B17 gene deletion in Cohort C utilized data from the Affymetrix SNP 5.0 array 18, which is being used for an ongoing genome-wide association study of transplant outcomes at the Fred Hutchinson Cancer Research Center. Copy-number probes spanning the deleted segment containing UGT2B17 were identified 18, and the intensity measurements from these probes were summarized into a single measurement for each deletion polymorphism in each patient (Supplementary Note). These measurements exhibited a trimodal distribution identifying individuals with 0, 1 or 2 copies of each locus (Supplementary Note).
Determination of mismatches
Transplants were determined to involve a donor-recipient mismatch for a gene deletion if the donor had a homozygous deletion for that gene (0 copies) and the recipient had 1 or 2 gene copies.
Statistical analysis
The association of donor-recipient mismatch with case/control status within cohort A was evaluated with the use of a Cochran-Mantel-Haenszel test to combine data from the two subcohorts (Supplementary Table 1). Separate analyses of cohort B and of cohort C utilized a chi-square test. All analyses of multiple study cohorts utilized a Cochran-Mantel-Haenszel test. The hypothesis tested was directional in two important dimensions: “mismatch” was defined in the direction prescribed by the antigenicity model (homozygous deletion of a gene in the HSC donor but not the recipient); the direction of effect was also prescribed by this model (mismatch associated with increased risk). The directionality of this hypothesis is not typical of genome-wide association studies. Nonetheless, given the typical use of two-sided hypothesis tests in genome-wide association studies, we report the results of both one- and two-sided statistical tests in Supplementary Table 3.
ELISA
ELISA utilized an approach similar to earlier studies of alloantibodies to Y antigens 25,26. Overlapping 20mer peptides (10-amino-acid overlaps) across UGT2B17 and UGT2B28 were synthesized. To prepare each ELISA plate, 0.5ug of each peptide was dissolved in coating buffer (BioFX), added to pre-treated ELISA plates (Evergreen), and incubated overnight at 4°C. Wells were blocked with BSA blocking buffer (BioFX) for 3 hours at 37°C. Stored serum samples from patients diagnosed with GVHD (generally obtained 6–18 months after transplantation) were used. Patient serum (1:1000) was incubated in each well (1 hour at 37°C). To assay patient sera for antibodies, goat anti-human IgG conjugated to alkaline phosphatase (Abcam) (1:1000) was incubated for 1 hour at 37°C. Plates were washed with PBS wash solution (BioFX) and pNPP(Chemicon) was added and allowed to develop at room temperature for 20 min. Plates were read at 410nm. Each serum sample was used to screen the peptide library on its own 96-well plate. (Several highly charged peptides yielded signal in all patient sera and were discarded.) A peptide was scored positive if the test result exceeded the median of the signals for all other peptides by at least six standard deviations. Positive results were re-tested in three replicates and considered confirmed only if they scored positive in re-tests both (i) relative to other peptides for the same serum sample, and (ii) relative to other serum samples for the same peptide.
Supplementary Material
Table 1.
Characteristics of the clinical populations studied. Acute GVHD was diagnosed and graded according to standard criteria employed at the time of documented patient care. Note that the ratio of affected to unaffected individuals is not uniform across cohorts, primarily because the criteria for patient selection (which were defined prior to genetic analysis, and which for Cohorts B and C reflected the design of earlier studies) differed from institution to institution; for example, Cohort C utilized a cohort design, while Cohort B utilized a case/control design and over-sampled patients with severe acute GVHD to maximize power. Diagnostic sensitivity in the grading of GVHD can also differ between clinical institutions 33 though it is generally well-harmonized within institutions.
| Cohort A (initial screen) | Cohort B | Cohort C | ||
|---|---|---|---|---|
| A1 | A2 | |||
| Hospital | Helsinki University Central Hospital | Dana-Farber Cancer Institute | Various | Fred Hutchinson Cancer Research Center |
| Pre-transplant conditioning | Myeloablative | Myeloablative | Myeloablative | Myeloablative |
| Transplant year | 1993–2005 | 1995–2005 | 1985–1993 | 1992–2004 |
| Donor type | HLA-identical sibling | HLA-identical sibling | HLA-identical sibling | HLA-identical sibling |
| Donor/recipient pairs | 232 | 182 | 336 | 595 |
| Graft type | ||||
| Bone marrow | 77 | 36 | 336 | 265 |
| Peripheral blood stem cells | 155 | 146 | 0 | 226 |
| Both | 0 | 0 | 0 | 4 |
| Donor/patient sex | ||||
| Female/male | 50 | 45 | 74 | 164 |
| Female/female | 57 | 46 | 86 | 115 |
| Male/male | 66 | 51 | 90 | 203 |
| Male/female | 59 | 40 | 86 | 113 |
| Acute GVHD (grade II–IV) | ||||
| Yes | 39 | 54 | 243 | 410 |
| No | 193 | 128 | 93 | 180 |
| Not gradable | 0 | 0 | 0 | 5 |
| Disease | ||||
| Non-malignant disease | 7 | 8 | 0 | 0 |
| Malignant hematological disease | 225 | 174 | 0 | 595 |
| N/A | 0 | 0 | 336 | 0 |
Table 2.
Characteristics of the gene deletions studied.
| Gene | Peptide length | Genomic location | Sites of expression | Deletion size (kb) | Deletion extent |
|---|---|---|---|---|---|
| UGT2B17 | 530 | chr4: 69.2 | Liver, intestine, skin, prostate, lymphocytes | 129 | all exons |
| UGT2B28 | 529 | chr4: 70.3 | Liver, mammary gland, kidney, salivary gland | 104 | all exons |
| GSTT1 | 240 | chr22: 22.7 | All tissues | 114 | all exons |
| GSTM1 | 218 | chr1: 109.9 | All tissues | 19 | all exons |
| LCE3C | 95 | chr1: 149.4 | Connective epithelia | 30 | all exons |
| OR51A2 | 313 | chr11: 4.9 | Olfactory and intestinal epithelia | 8 | all exons |
Acknowledgments
The authors wish to thank the patients, donors, and clinical care teams at Helsinki University Central Hospital, the Dana-Farber Cancer Institute, and the Fred Hutchinson Cancer Research Center. We also wish to thank Doreen Sese and colleagues at the Brigham and Women’s Hospital Tissue Typing Laboratory; the Ted and Eileen Pasquarello Tissue Bank; Qiheng Yang and colleagues at the Quality Assurance Office for Clinical Trials at Dana-Farber Cancer Institute; Eric Lander, Winfred Williams, Yishay Ofran, Jason Chien, and Mark Daly, for helpful conversations on the project and data; and Bruce Blazar, Carolyn Keever-Taylor, and David Senitzer, for providing patient samples for the Nichols et al. cohort further studied here. This work was supported by the Broad Institute of MIT and Harvard (DA, SM), the Academy of Finland (LV, HT, JP), the Helsinki University Central Hospital Research Rund (LV), the Fred Hutchinson Cancer Research Center, a Lilly Life Sciences Research fellowship (SM), the S. Juselius Foundation (JP), the Center of Excellence Program of the Finnish Academy (AP), and the National Institutes of Health (HA070149 to JA; AI29530 to JR; AI33484, CA18029, and HL087690 to JH).
Footnotes
Author contributions. SM, JB and DA developed an initial study plan with insights from RS, JR, AP, EW and PM. Patient collections including DNA samples were established and/or further developed by LV, HT, TR and JP in Helsinki; JB, SL, JA, JR and RS in Boston; DG in Michigan; and PM, SL, BS, and JH in Seattle. Analyses of patient clinical data were led, performed, and/or further analyzed by LV and TR at Helsinki University Central Hospital; SL, JB, JA, RS and JR at Dana-Farber Cancer Institute; and BS, PM, SL, EW and JH at the Fred Hutchinson Cancer Research Center. Deletion polymorphisms were genotyped by SM, HT and SC, using molecular assays developed by SM. BZ and LZ analyzed array-based data to genotype deletions and analyze association and time course in the FHCRC cohort. SM performed statistical analyses of genotype-phenotype correlation with feedback from other authors especially PM, AP and DA. SC and HT performed ELISA experiments. SM wrote the manuscript with extensive input and feedback from coauthors.
References
- 1.Gale RP, Champlin RE. How does bone-marrow transplantation cure leukaemia? Lancet. 1984;2:28–30. doi: 10.1016/s0140-6736(84)92009-9. [DOI] [PubMed] [Google Scholar]
- 2.Gale RP, et al. Identical-twin bone marrow transplants for leukemia. Ann Intern Med. 1994;120:646–52. doi: 10.7326/0003-4819-120-8-199404150-00004. [DOI] [PubMed] [Google Scholar]
- 3.Randolph SS, Gooley TA, Warren EH, Appelbaum FR, Riddell SR. Female donors contribute to a selective graft-versus-leukemia effect in male recipients of HLA-matched, related hematopoietic stem cell transplants. Blood. 2004;103:347–52. doi: 10.1182/blood-2003-07-2603. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, et al. Human H-Y: a male-specific histocompatibility antigen derived from the SMCY protein. Science. 1995;269:1588–90. doi: 10.1126/science.7667640. [DOI] [PubMed] [Google Scholar]
- 5.Meadows L, et al. The HLA-A*0201-restricted H-Y antigen contains a posttranslationally modified cysteine that significantly affects T cell recognition. Immunity. 1997;6:273–81. doi: 10.1016/s1074-7613(00)80330-1. [DOI] [PubMed] [Google Scholar]
- 6.Warren EH, et al. The human UTY gene encodes a novel HLA-B8-restricted H-Y antigen. J Immunol. 2000;164:2807–14. doi: 10.4049/jimmunol.164.5.2807. [DOI] [PubMed] [Google Scholar]
- 7.Vogt MH, et al. The DBY gene codes for an HLA-DQ5-restricted human male-specific minor histocompatibility antigen involved in graft-versus-host disease. Blood. 2002;99:3027–32. doi: 10.1182/blood.v99.8.3027. [DOI] [PubMed] [Google Scholar]
- 8.Spierings E, et al. Identification of HLA class II-restricted H-Y-specific T-helper epitope evoking CD4+ T-helper cells in H-Y-mismatched transplantation. Lancet. 2003;362:610–5. doi: 10.1016/S0140-6736(03)14191-8. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov R, et al. Identification of a 40S ribosomal protein S4-derived H-Y epitope able to elicit a lymphoblast-specific cytotoxic T lymphocyte response. Clin Cancer Res. 2005;11:1694–703. doi: 10.1158/1078-0432.CCR-04-1772. [DOI] [PubMed] [Google Scholar]
- 10.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 11.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 12.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 13.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 14.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 15.Colin Y, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–52. [PubMed] [Google Scholar]
- 16.Aguilera I, et al. Antibodies against glutathione S-transferase T1 (GSTT1) in patients with de novo immune hepatitis following liver transplantation. Clin Exp Immunol. 2001;126:535–9. doi: 10.1046/j.1365-2249.2001.01682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata M, Warren EH, Riddell SR. A human minor histocompatibility antigen resulting from differential expression due to a gene deletion. J Exp Med. 2003;197:1279–89. doi: 10.1084/jem.20030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 19.Schulze JJ, et al. Doping test results dependent on genotype of uridine diphospho-glucuronosyl transferase 2B17, the major enzyme for testosterone glucuronidation. J Clin Endocrinol Metab. 2008;93:2500–6. doi: 10.1210/jc.2008-0218. [DOI] [PubMed] [Google Scholar]
- 20.Terakura S, et al. A UGT2B17-positive donor is a risk factor for higher transplant-related mortality and lower survival after bone marrow transplantation. Br J Haematol. 2005;129:221–8. doi: 10.1111/j.1365-2141.2005.05427.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang TL, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet. 2008;83:663–74. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terakura S, et al. A single minor histocompatibility antigen encoded by UGT2B17 and presented by human leukocyte antigen-A*2902 and -B*4403. Transplantation. 2007;83:1242–8. doi: 10.1097/01.tp.0000259931.72622.d1. [DOI] [PubMed] [Google Scholar]
- 23.Kamei M, et al. HapMap scanning of novel human minor histocompatibility antigens. Blood. 2008 doi: 10.1182/blood-2008-07-171678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miklos DB, et al. Antibody response to DBY minor histocompatibility antigen is induced after allogeneic stem cell transplantation and in healthy female donors. Blood. 2004;103:353–9. doi: 10.1182/blood-2003-03-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miklos DB, et al. Antibody responses to H-Y minor histocompatibility antigens correlate with chronic graft-versus-host disease and disease remission. Blood. 2005;105:2973–8. doi: 10.1182/blood-2004-09-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zorn E, et al. Minor histocompatibility antigen DBY elicits a coordinated B and T cell response after allogeneic stem cell transplantation. J Exp Med. 2004;199:1133–42. doi: 10.1084/jem.20031560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutler C, et al. Rituximab for steroid-refractory chronic graft-versus-host disease. Blood. 2006;108:756–62. doi: 10.1182/blood-2006-01-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue Y, et al. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet. 2008;83:337–46. doi: 10.1016/j.ajhg.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4:e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols WC, et al. Polymorphism of adhesion molecule CD31 is not a significant risk factor for graft-versus-host disease. Blood. 1996;88:4429–34. [PubMed] [Google Scholar]
- 31.Martin PJ, et al. Increasingly frequent diagnosis of acute gastrointestinal graft-versus-host disease after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2004;10:320–7. doi: 10.1016/j.bbmt.2003.12.304. [DOI] [PubMed] [Google Scholar]
- 32.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–60. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.