Abstract
Thioredoxin reductases (TrxRs) regulate the intracellular redox environment by using NADPH to provide reducing equivalents for thioredoxins (Trxs). Here we present the cloning and biochemical characterization of a putative TrxR (Ta0984) and a putative Trx (Ta0866) from Thermoplasma acidophilum. Our data identifies Ta0866 as a Trx through its capacity to reduce insulin, and be reduced by E. coli TrxR in a NADPH dependent manner. Our data also establish Ta0984 as a TrxR due to its ability to reduce T. acidophilum Trx (taTrx), although not in a NADPH or NADH dependent manner. To explore the apparent inability of taTrxR to use NADPH or NADH as a reductant, we carried out a complete electrochemical characterization, which suggests that redox potential is not the source of this non-reactivity (accompanying paper). Turning to crystallographic analysis, a 2.35 Å resolution structure of taTrxR, also presented here, shows that despite the overall structural similarity to the well-characterized TrxR from E. coli (R.M.S.D. 1.30 Å2 for chain A), the “NADPH binding pocket” is not conserved. E. coli TrxR residues implicated in NADPH binding, H175, R176, R177, and R181 have been substituted with E185, M186, Y187, and M191 in the ta protein. Thus, we have identified a Trx and TrxR protein system from T. acidophilum for which the TrxR shares overall structural and redox properties with other TrxRs, but lacks the appropriate binding motif to use the standard NADPH reductant. Our discovery of a TrxR that does not use NADPH provides a new twist in redox regulation.
Tight regulation of the cellular redox environment is crucial for cell survival. Cellular viability relies on the reduction of metabolites for energy production, on the oxidation of small molecules to assemble structural scaffolds, and on the maintenance of the proper disulfide state in the extracellular or intracellular environments. An imbalance in redox regulation can trigger a cascade of genetic responses, the most extreme of which can lead to cell death (Reviewed in (1-4).
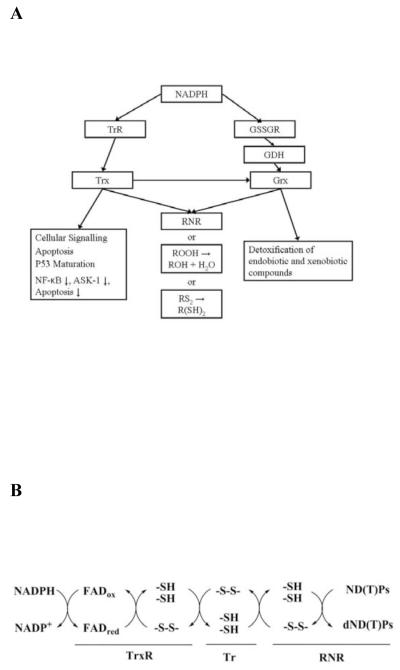
The principal protein systems that act as reductants are NADPH-dependent thioredoxin reductase (TrxR) / thioredoxin (Trx), and glutathione reductase (GSSGR) / glutathione (GDH) / glutaredoxin (Grx) (Figure 1A) (1, 2, 4-6). NADPH reduces the FAD of TrxRs, which in turn reduces Trs through disulfide exchange (Figure 1B). The TrxR / Trx system is responsible for providing reducing equivalents for many cellular processes including the reduction of ribonucleotide reductase (RNR) during the conversion of nucleotides to deoxynucleotides (7). This reaction is essential for maintaining the appropriate levels of deoxynucleotides in the cell for DNA repair, and for cellular replication (8, 9). The significance of the redox regulation of these and other biochemical pathways has lead to renewed interest in anti-bacterial or anti-tumorgenic therapies that are targeted to disrupt redox regulation (1, 10-13).
Figure 1.
NADPH participation in cellular redox regulation. (A) The NADPH-dependent redox regulatory pathways. (B) Mechanism of NADPH-dependent cellular reduction by the thioredoxin reductase system.
Here we study putative TrxR and Trx from Thermoplasma acidophilum, an organism first isolated from a self-heating coal refuse pile at the Friar Tuck mine in southwest Indiana. This organism lacks a cell wall and optimal growth occurs at 59 °C and at pH 1-2 (14). Despite the organism’s ability to grow at low pH, the cytoplasmic pH of T. acidophilum was reported to be between 5.5 and 6.9 (15, 16). The 1.56 Mb genomic sequence of T. acidophilum encoding 1,509 open reading frames (ORFs) has been reported (17), allowing us to identify a putative TrxR (Ta0984) and Trx (Ta0866).
We report the cloning, expression, and purification of gene products Ta0984 and Ta0866, and show that Ta0866 exhibits thioredoxin-like activity, and that Ta0984 is able to reduce T. acidophilum Trx (taTrx), establishing its identity as a thioredoxin reductase. An accompanying paper describes the full electrochemical characterization of T. acidophilum TrxR (taTrxR). While the redox potential is within the expected range for TrxRs, we find that the standard TrxR reducing agents NADPH or NADH do not reduce the taTrxR flavin. The X-ray structure of taTrxR to 2.35 Å resolution presented here reveals a major modification of the classic TrxR NADPH binding site, suggesting that T. acidophilum may utilize a novel reductant.
EXPERIMENTAL PROCEDURES
Materials
The organism Thermoplasma acidophilum (ATCC number 27656) was obtained from the American Tissue Culture Center. TOPO and TOPO Zero Blunt PCR cloning kits were obtained from Invitrogen (Carlsbad, CA). NovaBlue competent cells, Rosetta™ (DE3) pLysS competent cells, and pET28a plasmid were purchased from Novagen (Madison, WI, USA). pfuTurbo DNA polymerase and a PCR Optimization Kit were obtained from Stratagene (Cedar Creek, TX, USA). Primers were obtained from Integrated DNA Technologies (Coralville, IA, USA). Nickel nitrilotriacetic acid (Ni-NTA) was purchased from Qiagen (Valencia, CA, USA). Restriction enzymes were purchased from New England BioLabs (Beverly, MA, USA). Kanamycin, chloroamphenicol, TRIS-HCl, ethylenediaminetetraacetic acid (EDTA), and phenylmethylsulfonylfluoride (PMSF) were obtained from Sigma-Aldrich (St. Louis, MA, USA).
Genomic analysis of Thermoplasma acidophilum genome
The genome of Thermoplasma acidophilum DSM1728 (RefSeq NC_002578, GenBank AL139299) was analyzed using the web interface at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) for putative thioredoxin reductase (TrxR) and thioredoxin (Trx) open reading frames (ORFs). An ORF encoding a 319 amino acid protein, Ta0984, and an ORF encoding for a 113 amino acid protein, Ta0866, were identified by sequence homology to known TrxRs and Trxs using PSI-BLAST (18). Protein sequences were aligned using the program ClustalW (19) and the output was rendered using the web interface ESPript 2.2 (20) (Figure S1 and S2).
Cloning of taTrxR and taTrx
The taTrxR and taTrx genes were amplified by the polymerase chain reaction (PCR) using genomic DNA from a cell suspension and the primers 5′- A TGC CAT ATG GCA TAT GGA ATT TAA CCT GCA TGC AGT A-3′ and 5′-TCA GCC AAG CTT CAT TTT TTG GAT ATA GAA TCA-3′ for taTrxR and 5′ A TGC CAT ATG GCA TAT GAA GAA TTA TAT GGG CTG CG-3′ and 5′-TAG CGG CCG CTC AGA CGG AAT CGC AGG GA-3′ for taTrx (restriction sites are underlined). PCR products were cloned into the expression vector pET28a using NdeI and HindIII restriction enzymes for taTrxR and NdeI and NotI for taTrx. The sequence of the pET28a-taTrxR and pET28a-taTrx cDNA was confirmed at the MIT Biopolymers Sequencing Facility (Figures S3 and S4). After confirmation of the correct sequences, the resulting constructs were transformed into Rosetta™ (DE3)pLysS cells for facile protein expression.
Purification of recombinant taTrxR and taTrx
Rosetta™ (DE3)pLysS transformed with either pET28ataTrxR or pET28ataTrx constructs were used to inoculate TB media supplemented with 20 μg ml−1 kanamycin and 34 μg ml−1 chloroamphenicol. The culture was grown at 37°C to an OD600 of 0.8 and induced with 0.8 mM isopropylthio-ß-D-galactoside (IPTG) for 4 hrs. The cells were harvested by centrifugation, resuspended in lysis buffer (10 mM TRIS-HCl pH 8.0, 150 mM NaCl, 10 mM imidazole, 1 mM PMSF), and disrupted by sonication using a Digital Sonifier 250 (Branson, CN, USA). Insoluble material was removed by ultracentrifugation at 40,000 x g using a Beckman Avanti J-25 centrifuge (Beckman, PA, CA, USA). The supernatant was subjected to heating at 70°C for 45 min, and then centrifuged at 40,000 x g. The clarified supernatant was loaded onto a Ni-NTA column, washed with high salt buffer, and eluted with buffer containing 10 mM TRIS-HCl pH 8.0, 150 mM NaCl, 200 mM imidazole. Eluted fractions were analyzed by an SDS-PAGE 4-20% gel and those fractions containing proteins with comparable size to taTrxR or taTrx were concentrated using an Amicon (Millipore, MA, USA) stirred ultrafiltration cell.
The eluted protein was dialyzed overnight in 20 mM HEPES, pH 7.5, 20 mM NaCl, 0.5 mM EDTA, 1 mM 1,4-dithio-DL-threitol (DTT) with thrombin (0.05 unit ml−1) to remove the N-terminal poly-His-tag. The digested protein fraction was loaded onto an SEC-30 high-prep size exclusion column from Amersham Biosciences (Piscataway, NJ, USA), which was pre-equilibrated with storage buffer (10 mM HEPES, pH 7.0, 20 mM NaCl). Fractions containing protein of either MW ~34 kDa (taTrxR) or MW ~13 kDa (taTrx) were concentrated using Centriprep YM-3 filter from Amicon (Millipore, Bedford, MA, USA). Protein purity was analyzed by SDS/PAGE 4-20% gel (Figure S5) and protein concentration was determined from the theoretical extinction coefficient at 280 nm (23,040 M−1 cm−1 for taTrxR and 16,500 M−1cm−1 for taTrx) (ProtParam, ExPaSy web server). A Cary 300 spectrophotometer (Varian, CA, USA) was used to analyze the purified taTrxR for flavin incorporation.
Temperature stability of NADPH and NADH
NADPH (0.22 μM) and NADH (0.26 μM) were analyzed for temperature dependent degradation in 50 mM TRIS-HCl, pH 7.5, 150 mM NaCl (Figure S6). Initial UV / visible spectra were collected at 20° C from 800 nm to 240 nm. The temperature was increased to 80° C and the reaction was allowed to equilibrate for 5 min before full spectra were collected. The reaction was cooled to 20° C and then another full spectrum was collected. To measure time dependent degradation, NADPH or NADH was heated to the desired temperature (20° C, 60° C, and 80° C) and a reading was collected at 340 nm in 5 min increments for 45 min. Degradation of NADPH or NADH was inferred from the decrease in absorbance at 340 nm normalized to 100 % of the absorbance for the sample at time zero. Experiments were performed in triplicate.
Activity of taTrx measured by insulin and DTT assays
Purified taTrx was assayed for disulfide reductase activity by the insulin reduction method (21). The standard assay mixture contained 1.25 mM bovine insulin, 100 mM potassium phosphate, pH 7.0, 2 mM EDTA and taTrx (43 μM, 66 μM, or 70 μM). The reaction was initiated by the addition of 1 mM DTT. An increase in absorbance at 650 nm was monitored at 25 °C.
Purified oxidized taTrx was also assayed for its ability to be reduced by ecTrxR using the DTNB method (22). Briefly, this colorimetric assay measures the Trx dependant reduction of DTNB to 5-thio-2-nitrobenzoic acid (TNB), which absorbs strongly at 405-414 nm, using TrxR and NADPH as electron donors. The reaction was performed using 3.8 μM ecTrxR, in 50 mM TRIS-HCl, pH 8.0, 2 mM EDTA, 0.5 mM NADPH, and 0.6 mM DTNB. Each assay was initiated by the addition of taTrx (5 – 60 μM) to reaction mixture and the conversion of DTNB to TNB was monitored by the increase in absorbance at 412 nm.
Titration of taTrxR and ecTrxR with NADPH and NADH
taTrxR (43.3 μM) was titrated with NADPH or with NADH (0.5 to 5.0 mole NAD(P)H / mole FAD) under anaerobic conditions at 59° C in 50 mM TRIS, pH 7.5, 150 mM NaCl. Spectra were collected from 800 nm to 240 nm to monitor the reduction of the enzyme bound flavin by NADPH or NADH. ecTrxR (40.5 μM) was titrated with NADPH or with NADH (0.5 to 5.0 mol NADPH or NADH / mol FAD) under anaerobic conditions at 25 °C in 50 mM TRIS, pH 7.5, 150 mM NaCl. Spectra for NADPH reduction of ecTrxR were recorded as described above, and were corrected for dilution. The reduction of bound FAD was monitored by observing the change in absorbance at 457 nm over 45 minutes. taTrxR (43.3 μM) and ecTrxR (40.5 μM) were titrated with 2.5 mM NADPH or NADH for 45 minutes. For taTrxR, spectra were collected every minute and for ecTrxR spectra were collected every 0.1 minute for initial 5 minutes and then every 5 minutes thereafter.
Ability of taTrxR to reduce taTrx
To verify that the reduced taTrxR was capable of passing electrons to taTrx, the xanthine/xanthine oxidase reduction system, described by Massey and co-workers, was used (23). After complete reduction of a sample of taTrxR, the re-oxidation of taTrxR (4 μM) was monitored spectroscopically in the visible region following the addition of taTrx or ecTrx (8 μM). The taTrxR reduction system was as follows: in a 1 cm path length cuvette stirred continuously and held at 25° C in an MBraun Labmaster glove box, a solution of 100mM phosphate buffer (pH 7.5) with 1 μM benzyl viologen, 250 μM xanthine and 4 μM taTrxR, an aliquot of xanthine oxidase was added to a final concentration of ~ 0.25 nM, to initiate the reaction. This solution was monitored in the visible region of the spectrum, using an SI Photonics spectrophotometer contained within the glovebox, over the course of 1.5 hours, upon which complete reduction of the taTrxR was achieved. Oxidized Trx was then added to a final concentration of 8 μM, and the spectral features were monitored.
Analysis of T. acidophilum genome for alternative redox regulation pathways
To investigate if T. acidophilum uses alternative characterized reduction pathways, the T. acidophilum genome was mined for known pathways by using NCBI BLAST (18) to compare known proteins involved in the formation of glutathione or of coenzyme F420 to the ORFs of T. acidophilum. Pathway members were identified using ExPASy PATHWAY service (http://ca.expasy.org/cgi-bin/lists?pathway.txt).
Crystallization and data collection
taTrxR was crystallized by incubating 2.0 μL of protein solution (65 mg mL−1 taTrxR in 10 mM HEPES, pH 7.5 and 20 mM NaCl) and 2.0 μL of precipitant solution (0.28 M MgCl2, 0.1 M Bis-Tris, pH 5.5, 25% (w/v) PEG 3350) in a sitting drop at room temperature. Crystals had rhombohedral geometry, and their bright yellow color confirmed the incorporation of flavin adenine dinucleotide (FAD) by the enzyme. Crystals appeared cubic and grew to approximately 100 μM per side in 3 to 4 days. These crystals were soaked for 30 seconds in a cryo-protectant solution of 0.28 M MgCl2, 0.1 M Bis-Tris, pH 5.5, 25% (w/v) PEG 3350, 25% PEG 400, before cryo-cooling in liquid nitrogen.
All diffraction data were collected at 100 K at the Advanced Light Source (Berkeley, CA), beam line 5.0.2. Data were processed and scaled using DENZO and SCALEPACK from the HKL (24) program suite (See Table 1). The crystals belong to the I23 (I-centered cubic) space group (a = b = c = 165.95 Å), with one homodimer per asymmetric unit.
Table 1.
Data and refinement statistics for or taTrxR structure
| Dataset | Native |
|---|---|
| Wavelength (Å) | 1.2710 |
| Beamline | ALS 5.0.2 |
| Space Group | I23 |
| a=b=c (Å) | 165.95 |
| Resolution (Å) | 50. - 2.35 |
| Rsym (%) | 6.9 (52.1)a |
| No. of Observations | 434,549 |
| Unique Observation | 31,586 |
| I/σI | 32.9 (7.6) |
| Completeness (%) | 99.9 (100.0) |
| Refinement Statistics | |
| Resolution Limits (Å) | 50. - 2.35 |
| # Unique Reflections | 31,128 |
| # Reflections - Test Set | 1,809 |
| Rwork (%)b | 22.5 |
| Rfree (%)c | 26.0 |
| Wilson B factor (Å) | 68.3 |
| B factor Chain A (Å) | 55.4 |
| B factor Chain B (Å) | 62.1 |
| B factor FAD 1 (Å) | 49.2 |
| B factor FAD 2 (Å) | 59.4 |
| B factor Waters (Å) | 68.0 |
| Solvent Content (%) | 56.4 |
| # protein atoms | 2272 (2285)d |
| # water atoms | 111 |
| # non-protein atoms (FAD) | 106 |
| Bond length deviation (Å) | 0.0073 |
| Bond angle deviation (°) | 1.2 |
| Ramachandran Plot | |
| Residues in most favored regions (%) | 93.6 |
| Res in allowed region (%) | 6.2 |
| Res in disallowed region (%) | 0.2 |
Statistics for highest resolution shell (2.35 Å to 2.43 Å) are in parentheses
Rwork = Σ∥Fobs| - |Fcalc∥ / Σ|Fobs|
Rfree calculated using 5% of the total reflections, which were not used in refinement
Number of atoms in chain A and chain B
Molecular replacement solution and refinement
The crystal structure of the thioredoxin reductase was solved by the method of molecular replacement. Initial phases were obtained from a full length poly-alanine model of thioredoxin reductase from Escherichia coli (PDB code 1TDE) (25). The solution was found using PHASER (26) with two search models consisting of the flavin binding domain and the NADPH-binding domain of the E. coli structure, utilizing data from a resolution range of 4.0 – 12.0 Å. PHASER was first used to find a solution for the FAD-binding domain of taTrxR (residues 1 – 126 and 250 – 319). This solution was fixed and a second round of molecular replacement was performed to find the NADPH-binding domain (residues 127 – 249).
Refinement was carried out in CNS (27) with rigid body refinement followed by simulated annealing. Manual fitting of amino acid residues was done using XFIT (28). Subsequent rounds of refinement included simulated annealing, energy minimization, and B-factor refinement without sigma cutoff. Topology and parameter files for the FAD moiety were obtained from HIC-UP (29) (PDB code 1N1P). Water molecules were added with CNS and were manually checked against 2Fo-Fc and Fo-Fc electron density maps. A composite omit map was prepared in CNS and used to confirm the assignment of model atoms. The final model contains a biologically relevant dimer in the asymmetric unit, which consists of residues 14-319 for molecule ‘A’ of 319 and residues 13-319 of 319 for molecule ‘B’, one flavin molecule per monomer and 114 water molecules.
RESULTS
Cloning, purification, and characterization of taTrxR and taTrx
Using the T. acidophilum genome database, we identified two ORFs (Accession no. Ta0984 and Ta0866): one encoding a putative thioredoxin reductase and the other one encoding a thioredoxin homologue. The Ta0984 gene encodes a protein of 319 amino acids with a predicted molecular mass of 34116 Da (Figure S3). The deduced amino acid sequence contains 28% identity (48% similar) to the ecTrxR (Figure S1). The Ta0866 gene encodes a 133 amino acid protein (Figure S4) with a CxxC disulfide-reducing motif and displays 34% identity (58% similarity) to ecTrx(Figure S2). These ORFs have been amplified by PCR, cloned, sequenced, and compared to the sequence in the T. acidophilum genome database.
The purified taTrxR yields visible absorption spectra typical for flavoproteins with absorbance maxima at 380 and 460 nm. Freshly purified protein has an A280 / A460 ratio of between 7.5 and 8.0, in agreement with the full incorporation of FAD as previously observed for ecTrxR (30). Expression and subsequent purification yields ~18.5 mg from a 1 liter culture. The molecular mass of the protein is estimated to be approximately 34 kDa by SDS-PAGE (Figure S5a). This molecular mass is in agreement with values predicted by the gene analysis. The native molecular mass is determined to be about 70 kDa by gel-filtration using a Sephacryl SEC-75 HR 16/60 column. These results suggest that the native state of taTrxR is a homodimer, similar to ecTrxR (31). Expression of taTrx yields a protein of 13.2 kDa as analyzed by SDS PAGE (Figure S5b). Subsequent gel-filtration using a Sephacryl SEC-30 HR 16/60 column reveals a native molecular mass of ~13 kDa, suggesting that the taTrx is a monomer in its native state.
Lack of activity of taTrxR with NADPH or NADH
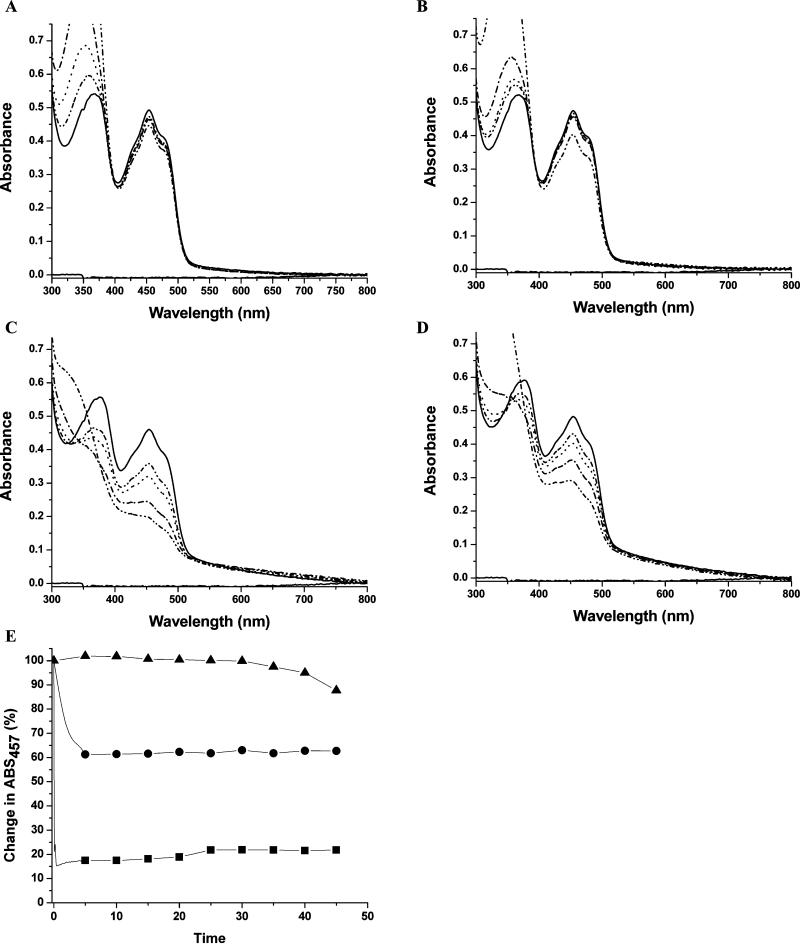
All previously characterized TrxRs are reduced by NADPH (Reviewed in (2, 32, 33)). For ecTrxR, reaction of NADPH with the FAD cofactor occurs rapidly (2,000 turnovers FAD−1 min−1) with a Km for NADPH of 1.2 μM (34). While there is no measurement of Km for NADH with ecTrxR, it has been estimated to be 400 fold higher than that of NADPH (31). Therefore, it is surprising that titration of taTrxR with concentrations of up to 5 fold molar excess NADPH does not reduce its FAD cofactor to any appreciable extent, showing no significant spectral change in the visible part of the spectra (Figure 2A). For comparison, titration of ecTrxR with NADPH results in a rapid reduction of the bound FAD, decreasing the absorbance at 460 nm (Figure 2C). Titration of taTrxR with NADH shows minimal reduction of FAD as indicated by the small change in the shoulder at 460 nm (Figure 2B). Although NADPH is the preferred substrate for ecTrxR, NADH does reduce the flavin to some extent in this protein (Figure 2D). The reduction is, in fact, greater than that observed for taTrxR. Compared to the rapid reduction of ecTrxR by NADPH (turnover rate is 2,000 FAD−1 min−1) (34), the changes observed for taTrxR appear too slow and too small to be physiologically relevant (Figure 2E). The inability of NADPH or NADH to rapidly reduce taTrxR is not due to instability of these components at 59 °C, the physiological temperature of T. acidophilum. Analysis of the time-dependent degradation of NADPH and NADH at 59 °C shows that there is negligible degradation at 5 min under assay conditions, not enough to explain this lack of activity (Figure S6). At the higher temperature of 80°C, degradation is increased, but even then less than 10% of NADPH or NADH is degraded during the first five minutes of the assay, and only 30% is degraded by 45 minutes (Figure S6).
Figure 2.
Anaerobic titration of taTrxR and ecTrxR with NADPH and NADH. (A) Visible spectra of taTrxR (43.3 μM) titrated with NADPH at 59 °C. Solid line is without NADPH, dashed line is with 0.5 moles NADPH / mole FAD, dotted line is with 1.0 mole NADPH, dash dot line is with 2.0 mole NADPH, dash dot dot line is with 5 mole NADPH. (B) Visible spectra of taTrxR titrated with NADH at 59 °C. (C) Visible spectra of ecTrxR (40.5 μM) titrated with NADPH at 25 °C. (D) Visible spectra of ecTrxR (40.5 μM) titrated with NADH at 25 °C. Spectra (B), (C), and (D) were recorded as in (A). (E) Time dependent reduction of taTrxR and NADPH at 59 °C (closed triangles), taTrxR and NADH at 59 °C (closed circles), and ecTrxR at 25 °C with NADPH (closed squares).
Disulfide reductase activity of taTrx
The lack of measurable activity between taTrxR and NADPH prevented the use of a coupled assay for taTrx activity, requiring us to turn to other methods to characterize the taTrx. The reduction of insulin disulfides can be monitored by following the increase in turbidity at 650 nm due to the precipitation of the free B-chain (21). The recombinant taTrx reduced insulin in a protein concentration-dependent manner (Figure S7). To investigate whether taTrx is able be reduced by ecTrxR we performed the DTNB assay using NADPH, ecTrxR, and taTrx. taTrx is reduced by ecTrxR, with an estimate of the apparent Km of 14.4 (±7.0) μM for the ecTrxR (Figure S8). This Km value is higher than for the ecTrxR / ecTrx system (Km of 2.8 μM) (31), which is not surprising in that the two proteins (ecTrxR and taTrx) did not evolve to interact. These experiments confirm that the taTrx is a thioredoxin-like protein capable of performing disulfide exchange.
Thioredoxin reductase activity of taTrxR
To verify that taTrxR can catalyze a disulfide exchange with taTrx, we pre-reduced taTrxR using a modified xanthine / xanthine oxidase system (see methods) at 25° C; after complete reduction of the taTrxR flavin, the reduced enzyme was treated with oxidized taTrx. Upon the introduction of two equivalents of taTrx, the taTrxR was rapidly oxidized, achieving complete re-oxidation of the FAD within one min (Figure 3). This experiment indicates that taTrxR is a biologically relevant TrxR, which does not use NADPH as a reductant.
Figure 3.
Demonstration of taTrxR/Trx reactivity. Reduction of oxidized taTrxR (4 μM) was achieved by use of a modified xanthine/xanthine oxidase system, as described in the Materials and Methods section. Oxidized taTrxR (solid line) was reduced with xanthine as the source of electrons, over the course of 90 minutes, in order to build up a stable reduced form of the enzyme (dashed line). Upon introduction of 2 equivalents of the taTrx, the sample was completely reoxidized within 1 minute (dotted line).
Overall Structure
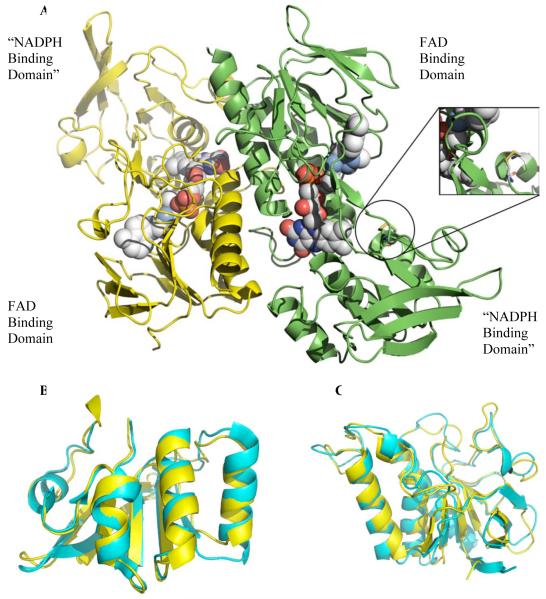
The structure of taTrxR was solved to 2.35 Å resolution with the physiological dimer in the asymmetric unit and an R and Rfree of 22.6% and 26.8%, respectively (See Table 1 for statistics). Each taTrxR monomer consists of an FAD-binding domain (residues 14-125 and 251-319) and an “NADPH binding domain” (residues 126-250) (Figure 4A). Each domain of taTrxR is quite similar to the equivalent domain of ecTrxR, with an r.m.s.d. of 1.23 Å for the 166 Cα of the FAD-binding domain, and an r.m.s.d of 1.17 Å for 121 Cα of the “NADPH-binding domains” (Figure 4B and 4C). The taTrxR structure has one FAD molecule bound in a similar conformation as the ecTrxR structure (Figure S9), and with similar protein-FAD interactions (Figure S10).
Figure 4.
Ribbon diagram structure of taTrxR dimer. (A) Monomers of taTrxR are colored by chain. The flavin molecule (1 per monomer) is represented as space filled spheres and colored by atom (C in light grey, N in blue, O in red, P in orange). Circle highlights C145/C148 disulfide. (B) Superposition of the taTrxR NADPH binding domain with ecTrxR NADPH binding domain. (C) Superposition of the taTrxR FAD binding domain in yellow with ecTrxR FAD binding domain in cyan.
ecTrxR is known to undergo a large conformational change during catalysis, rotating between a “flavin oxidizing” form (FO) and a “flavin reducing” form (FR) (25, 35, 36). In the FR form, the FAD domain is positioned such that NADPH can reduce the FAD by hydride transfer (Figure 4A). In the FO form, the FAD domain is positioned adjacent to the disulfide such that the FADH2 can be oxidized by the disulfide. Structures of both conformations are available for the E. coli enzyme: the structure of wild type ecTrxR represents the FO state, and a structure of a complex between ecTrxR and ecTrxrepresents the FR state. Interestingly, while all other structures of uncomplexed TrxRs are in the FO state, the taTrxR structure is of the FR form. The taTrxR FR conformation is very similar to that of ecTrxR in the FR state with an r.m.s.d. of 1.30 Å for chain A and 1.73 Å for chain B when all the Cα atoms are superimposed. As with the ecTrxR FR structure, the taTrxR FAD domain is positioned near the putative NADPH binding site, and away from the catalytic cysteines (Cys145 and Cys148), which are oxidized with an S-S distance of 2.04 Å. A discussion of the conformational dynamics of TrxRs is available in the accompanying paper.
Major Differences in the Domain that Typically Binds NADPH
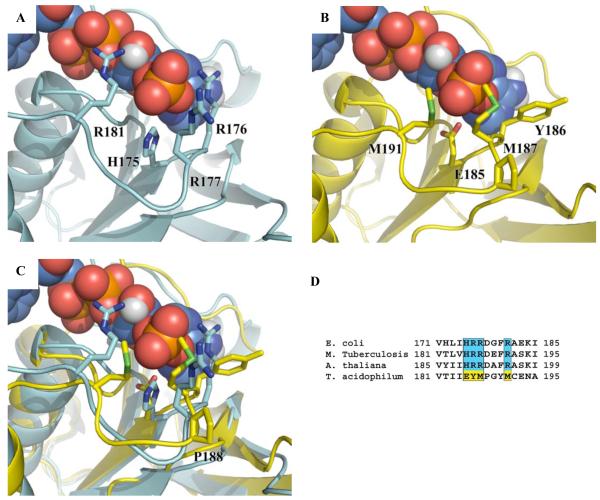
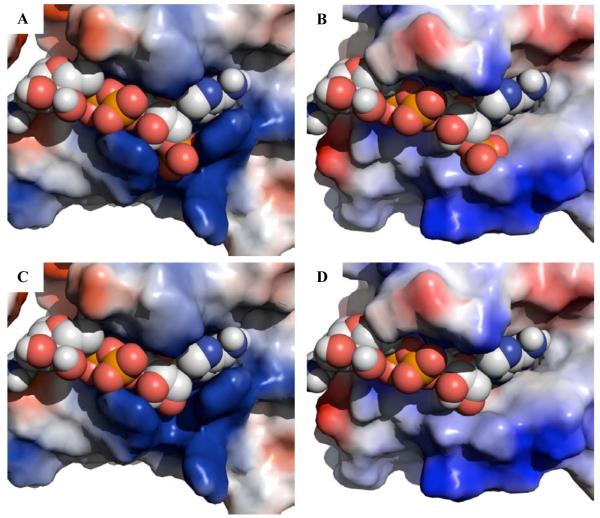
Structural superposition of the “NADPH binding domains” from ecTrxR and taTrxR reveal that the backbone of residues 184 - 191 of the taTrxR structure is constrained into a slightly different conformation than is found in ecTrxR. This different loop conformation appears to result from the presence of a proline (P188) in the taTrxR sequence (Figure 5C), and not the presence or absence of nucleotide in the structure (Figure S12). This variation in loop conformation is subtle and is unlikely to explain the lack of affinity of NADPH for the ta enzyme. In contrast, while the “NADPH-binding domains” for taTrxR and ecTrxR display an overall similar fold, close examination reveals several highly conserved residues in all bacterial TrxRs that are not conserved in the taTrxR sequence. The ecTrxR structure, which was co-crystallized with NADP+, shows that the NADPH 2′ phosphate is surrounded by a histidine (H175) and three arginines (R176, R177, and R181) creating a positively charged pocket that complements the negatively charged phosphate (Figures 5A, 6A, and S11A). The lower activity (400-fold) for NADH compared to NADPH (31) is presumably due to the loss of interactions between the cofactor and residues that form this positively charged pocket (Figure 6C). In taTrxR, none of the residues that form this pocket are conserved. The classic NADPH binding motif ‘VxxxHRRDxxRA’ is replaced with ‘VxxxEYMPxxMC’ in taTrxR (Figure 5D). ecTrxR’s histidine 175, which points directly toward the 2′ phosphate of the NADPH, is substituted with a glutamic acid (E185) in taTrxR, and the three arginine residues (R176, R177, and R181) are replaced by a tyrosine (Y186) and two methionines (M187 and M191) (Figures 5B and 5C). These substitutions dramatically change the electrostatics of this protein surface (Figures 6 and S11). In a model of NADH bound to taTrxR, an unfavorable interaction between E185 and the 2′ phosphate of NADPH is relieved (Figure 6D), but neither NADPH nor NADH would be expected to bind with any kind of reasonable affinity, consistent with the lack of rapid reduction of FAD by these molecules. Analysis of all known bacterial and archaeal genomes shows that only one other organism, the close cousin to T. acidophilum, Thermoplasma volcanicum, contains an identical ‘VxxxEYMPxxMC’ sequence. All others use a variant of the Arg-rich ‘VxxxHRRDxxRA’ motif.
Figure 5.
Ribbon representation of the ecTrxR NADPH binding domain (cyan) (PDB code 1TDF) and taTrxR “NADPH binding domain” (yellow). (A) ecTrxR with NADPH binding amino acids, H175, R176, R177, and R181, represented in stick form. (B) taTrxR structure with corresponding structurally aligned amino acids, E185, Y186, M187 and M191 represented in stick form with modeled NADPH. (C) Superposition of ecTrxR and taTrxR NADPH binding domains with NADPH from ecTrxR structure (D) Sequence alignment of the NADPH binding domain regions.
Figure 6.
Surface representation of NADPH binding pocket with blue representing basic residues, red acidic residues, and white neutral residues. (A) ecTrxR structure with NADPH bound. (B) taTrxR structure with modeled NADPH. (C) ecTrxR structure with NADH modeled in the NADPH binding site. (D) taTrxR structure with NADH modeled in the NADPH binding site. The orientations shown in Figure 6 are similar to those shown in Figure 5, with the panels in Figure 6 rotated slightly to the left so that the NAD(P)H adenine rings are more clearly visible. Surface calculations were made using the APBS module in Pymol (45).
DISCUSSION
The protein derived from Ta0984 is a thioredoxin reductase. Our structural and biochemical studies reveal a protein with an FAD cofactor and an overall similar fold to the ecTrxR. However, unlike all other characterized TrxRs, taTrxR is not reduced by NADPH. This lack of reactivity is unlikely to be due to redox potential differences between TrxRs (See accompanying paper). The redox potential of taTrxR is within 35 mV of ecTrxR (−305 vs. −270 mV) (37), and when reduced with a xanthine / xanthine oxidase system, taTrxR is able to react stoichiometrically with taTrx.
Examination of the taTrxR structure suggests that the lack of activity with NADPH is due to amino acid substitutions as compared to the E. coli enzyme. The “NADPH-binding site” of taTrxR does not appear to be designed to bind NADPH, with neutral and negatively charged amino acids replacing the canonical Arg-rich motif. The “NADPH-binding site” is also structurally constrained by P188. Sequence analysis shows that only one other organism, the closely related Thermoplasma volcanicum, shares taTrxR’s unusual sequence.
The lack of a NADPH-dependent TrxR / Trx reduction system in T. acidophilum led us to investigate whether this organism might rely heavily on GSSGR / Grx systems instead of on thioredoxins. Analysis of the T. acidophilum genome, however, revealed no GSSGR / Grx system, nor enzymes for the biosynthesis of glutaredoxin (38). Our extensive data mining and sequence alignments have not produced any near or distantly related GSSGR / Grx proteins from this organism.
We also considered whether T. acidophilum uses NADH or NADPH in other cellular processes. Our genomic analysis reveals a putative NAD kinase, the enzyme responsible for the conversion of NAD to NADP, and several putative NADH and NADPH utilizing enzymes, which suggests that T. acidophilium does use these cofactors. To date, there are only a few characterized enzymes in T. acidophilum that use NADH (aldohexose dehydrogenase) (39) or NADPH (glyderaldehyde dehydrogenase) (40) or both NADH and NADPH as substrates (41). However, the use of NADPH or NADH by any T. acidophilum enzyme, in addition to the presence of the NAD kinase gene, indicates that availability of these cofactors is not the reason for the observed variation in the TrxR protein.
The above data challenges the paradigm shown in Figure 1A. Our data blocks the arrow to the left, and the lack of GSSGR / Grx blocks the arrow to the right. Given this, we revisited the T. acidophilum genome to search again for other putative TrxR sequences and found none. Unless there is a TrxR with a very different sequence, we have cloned the only one in T. acidophilum.
If NADPH does not reduce this taTrxR, and this TrxR is the only TrxR in T. acidophilum, and there are no obvious GSSGR / Grx candidates, there must be some small molecule that can provide reducing equivalents or the cytoplasmic environment would quickly oxidize and the organism would expire. One such molecule, coenzyme F420, has been shown to be a physiological reductant in methane reducing thermophilic organisms (Reviewed in (42-44)). However, genomic analysis of T. acidophilum does not reveal any of the enzymes involved in the biosynthesis of F420. The lack of molecular pathways for the biosynthesis of coenzyme F420, along with the lack of other F420-dependent enzymes in T. acidophilum, seems to indicate that coenzyme F420 is not a reductant of taTrxR. Of course, we cannot rule out the possibility that F420 is generated by a different set of enzymes in this organism.
Conclusions
To date, there is no well characterized example of an organism that uses a reductant other than NADPH to maintain its intracellular redox environment. Here we present evidence that one organism, T. acidophilum, which does not contain a Grx system, uses an alternative method to reduce the bound FAD of the TrxR system. This work lays the foundation for a biochemical search for the molecular redox partners of T. acidophilum TrxR.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to thank Dr. Scott Mulrooney and Prof. Charles Williams, Jr. for the generous donation of the ecTrxR over-expression system. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Abbreviations
- Trx
thioredoxin
- TrxR
thioredoxin reductase
- Grx
glutaredoxin
- GDH
glutathione
- GSSGR
glutathione reductase
- RNR
ribonucleotide reductase
- ORF
open reading frame
- TRIS-HCl
tris(hydroxymethyl)-aminomethane hydrochloride
- PMSF
phenyl methyl sulfonyl fluoride
- PCR
polymerase chain reaction
- DTT
1,4-dithio-DL-theritol
- HEPES
N-2-hydroxyethyl-piperazine-N’-2-ethanesulfonic acid
- Ec
Escherichia coli
- EDTA
ethylenediaminetetraacetic acid
- FAD
flavin adenine dinucleotide
- FADH2
flavin adenine dinucleotide, reduced form
- FO
flavin oxidizing conformation
- FR
flavin reducing conformation
- NADH
nicotinamide adenine dinucleotide, reduced form
- NADPH
nicotinamide adenine dinucleotide phosphate, reduced form
- SafO
safranine O
- Ta
Thermoplasma acidophilum
- XO
xanthine oxidase
Footnotes
This work was supported by the NIGMS Ruth L. Kirschstein pre-doctoral fellowship GM73569 (H.H.H.), the National Institutes of Health Grant GM65337 (C.L.D.), and the Richard Allan Barry Fund of the Boston Foundation (S.J.E).
The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (www.rcsb.org) as entry 3CTY.
SUPPORTING INFORMATION AVAILABLE
Supporting information contains sequence alignments of TrxRs and Trxs, sequences of taTrxR and taTrx, a figure showing gel analysis of purified protein, figures showing the temperature dependent degradation of NADH and NADPH, results of insulin assays, results of taTrx and ecTrxR reduction of DTNB, a figure of FAD in composite omit map density, schematic representation interactions between FAD and taTrxR, and structural comparison figures. This material is available free of charge via the Internet at http//:pubs.acs.org.
REFERENCES
- 1.Jaeger T, Flohe L. The thiol-based redox networks of pathogens: unexploited targets in the search for new drugs. Biofactors. 2006;27(14):109–120. doi: 10.1002/biof.5520270110. [DOI] [PubMed] [Google Scholar]
- 2.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267(20):6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 3.Masutani H, Ueda S, Yodoi J. The thioredoxin system in retroviral infection and apoptosis. Cell Death Differ. 2005;12(Suppl 1):991–998. doi: 10.1038/sj.cdd.4401625. [DOI] [PubMed] [Google Scholar]
- 4.Gromer S, Urig S, Becker K. The thioredoxin system--from science to clinic. Med Res Rev. 2004;24(1):40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 5.Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000;346(Pt 1):1–8. [PMC free article] [PubMed] [Google Scholar]
- 6.Stroher E, Dietz KJ. Concepts and approaches towards understanding the cellular redox proteome. Plant Biol (Stuttg) 2006;8(4):407–418. doi: 10.1055/s-2006-923961. [DOI] [PubMed] [Google Scholar]
- 7.Toledano MB, Kumar C, Le Moan N, Spector D, Tacnet F. The system biology of thiol redox system in Escherichia coli and yeast: differential functions in oxidative stress, iron metabolism and DNA synthesis. FEBS Lett. 2007;581(19):3598–3607. doi: 10.1016/j.febslet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Herrick J, Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 2007;63(1):22–34. doi: 10.1111/j.1365-2958.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 9.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 10.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6(5):409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 11.Hashemy SI, Ungerstedt JS, Zahedi Avval F, Holmgren A. Motexafin gadolinium, a tumor-selective drug targeting thioredoxin reductase and ribonucleotide reductase. J Biol Chem. 2006;281(16):10691–10697. doi: 10.1074/jbc.M511373200. [DOI] [PubMed] [Google Scholar]
- 12.Urig S, Becker K. On the potential of thioredoxin reductase inhibitors for cancer therapy. Semin Cancer Biol. 2006;16(6):452–465. doi: 10.1016/j.semcancer.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Becker K, Gromer S, Schirmer RH, Muller S. Thioredoxin reductase as a pathophysiological factor and drug target. Eur J Biochem. 2000;267(20):6118–6125. doi: 10.1046/j.1432-1327.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 14.Darland G, Brock TD, Samsonoff W, Conti SF. A thermophilic, acidophilic mycoplasma isolated from a coal refuse pile. Science. 1970;170(965):1416–1418. doi: 10.1126/science.170.3965.1416. [DOI] [PubMed] [Google Scholar]
- 15.Searcy DG. Thermoplasma acidophilum: intracellular pH and potassium concentration. Biochim Biophys Acta. 1976;451(1):278–286. doi: 10.1016/0304-4165(76)90278-6. [DOI] [PubMed] [Google Scholar]
- 16.Hsung JC, Haug A. Intracellular pH of Thermoplasma acidophila. Biochim Biophys Acta. 1975;389(3):477–482. doi: 10.1016/0005-2736(75)90158-3. [DOI] [PubMed] [Google Scholar]
- 17.Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407(6803):508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins D, Gibson T.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressivemultiple sequence alignment through sequence weighting,position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;(22):4673–4680. doi: 10.1093/nar/22.22.4673. T. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 21.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254(19):9627–9632. [PubMed] [Google Scholar]
- 22.Arner ES, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 23.Massey V, Brumby PE, Komai H. Studies on milk xanthine oxidase. Some spectral and kinetic properties. J Biol Chem. 1969;244(7):1682–1691. [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Waksman G, Krishna TS, Williams CH, Jr., Kuriyan J. Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis. J Mol Biol. 1994;236(3):800–816. [PubMed] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125(23):156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 29.Kleywegt GJ, Jones TA. Databases in protein crystallography. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 6 Pt 1):1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 30.Williams CH, Jr., Zanetti G, Arscott LD, McAllister JK. Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and thioredoxin. J Biol Chem. 1967;242(22):5226–5231. [PubMed] [Google Scholar]
- 31.Thelander L. Thioredoxin reductase. Characterization of a homogenous preparation from Escherichia coli B. J Biol Chem. 1967;242(5):852–859. [PubMed] [Google Scholar]
- 32.Williams CH, Arscott LD, Muller S, Lennon BW, Ludwig ML, Wang PF, Veine DM, Becker K, Schirmer RH. Thioredoxin reductase two modes of catalysis have evolved. Eur J Biochem. 2000;267(20):6110–6117. doi: 10.1046/j.1432-1327.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 33.Biaglow JE, Miller RA. The thioredoxin reductase/thioredoxin system: novel redox targets for cancer therapy. Cancer Biol Ther. 2005;4(1):6–13. doi: 10.4161/cbt.4.1.1434. [DOI] [PubMed] [Google Scholar]
- 34.Williams CH., Jr. Flavin-Containing Dehydrogenases. In: Boyer PD, editor. The Enzymes. Third Edition ed. Vol. 1. Academic Press; New York: 1976. pp. 89–173. [Google Scholar]
- 35.Lennon BW, Williams CH, Jr., Ludwig ML. Crystal structure of reduced thioredoxin reductase from Escherichia coli: structural flexibility in the isoalloxazine ring of the flavin adenine dinucleotide cofactor. Protein Sci. 1999;8(11):2366–2379. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang PF, Veine DM, Ahn SH, Williams CH., Jr. A stable mixed disulfide between thioredoxin reductase and its substrate, thioredoxin: preparation and characterization. Biochemistry. 1996;35(15):4812–4819. doi: 10.1021/bi9526793. [DOI] [PubMed] [Google Scholar]
- 37.O’Donnell ME, Williams CH., Jr. Proton stoichiometry in the reduction of the FAD and disulfide of Escherichia coli thioredoxin reductase. Evidence for a base at the active site. J Biol Chem. 1983;258(22):13795–13805. [PubMed] [Google Scholar]
- 38.Copley SD, Dhillon JK. Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 2002;3(5):1–16. doi: 10.1186/gb-2002-3-5-research0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reher M, Schonheit P. Glyceraldehyde dehydrogenases from the thermoacidophilic euryarchaeota Picrophilus torridus and Thermoplasma acidophilum, key enzymes of the non-phosphorylative Entner-Doudoroff pathway, constitute a novel enzyme family within the aldehyde dehydrogenase superfamily. FEBS Lett. 2006;580(5):1198–1204. doi: 10.1016/j.febslet.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 40.Nishiya Y, Tamura N, Tamura T. Analysis of bacterial glucose dehydrogenase homologs from thermoacidophilic archaeon Thermoplasma acidophilum: finding and characterization of aldohexose dehydrogenase. Biosci Biotechnol Biochem. 2004;68(12):2451–2456. doi: 10.1271/bbb.68.2451. [DOI] [PubMed] [Google Scholar]
- 41.Smith LD, Budgen N, Bungard SJ, Danson MJ, Hough DW. Purification and characterization of glucose dehydrogenase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Biochem J. 1989;261(3):973–977. doi: 10.1042/bj2610973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer M, Bacher A. Biosynthesis of flavocoenzymes. Nat Prod Rep. 2005;22(3):324–350. doi: 10.1039/b210142b. [DOI] [PubMed] [Google Scholar]
- 43.Graupner M, Xu H, White RH. The pyrimidine nucleotide reductase step in riboflavin and F(420) biosynthesis in archaea proceeds by the eukaryotic route to riboflavin. J Bacteriol. 2002;84(7):1952–1957. doi: 10.1128/JB.184.7.1952-1957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mack M, Grill S. Riboflavin analogs and inhibitors of riboflavin biosynthesis. Appl Microbiol Biotechnol. 2006;71(3):265–275. doi: 10.1007/s00253-006-0421-7. [DOI] [PubMed] [Google Scholar]
- 45.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.