Abstract
Skin, the biggest organ in mammals, protects the body from environmental hazards and prevents dehydration. Embryonic skin morphogenesis and homeostasis of adult skin require an accurately controlled gene expression in a spatiotemporally specific manner. Recently, the identification of microRNAs (miRNAs) in skin has added a new dimension in the regulatory network and attracted significant interest in this novel layer of gene regulation. Mammalian skin with its easy accessibility, well-defined lineages and established genetic tools offers an ideal system to unravel the functions of miRNAs in mammalian development and stem cells. In the past few years, significant progress has been made in determining the expression patterns of miRNAs, exploring their functions in skin morphogenesis and differentiation, as well as probing their functions in human skin diseases, for example, skin cancer. In this review, we summarized current progress in the study of miRNA in mammalian skin, provided insights gained from recent studies and offered our views for remaining challenges.
Keywords: microRNAs, mammalian skin, Dicer, DGCR8, miR-203
Mammalian skin and its appendages function as the outermost barrier of the body to protect inner organs from environmental hazards and keep essential fluid within. During embryonic development, a single layer of epidermal progenitors gives rise to the epidermis, hair follicle (HF) and sebaceous gland, a process orchestrated by an array of regulatory pathways1,2 (Figure 1). In adult, homeostasis of each of the three skin lineages is maintained for a long term through self-renewal and differentiation of distinct skin stem cells. Through extensive investigation, much has been learned about the regulatory networks that control skin morphogenesis during embryonic development and self-renewal and differentiation of adult skin stem/progenitor cells.1–4 Recently, however, a novel layer of regulation mediated by microRNAs (miRNAs) has been implicated in animal development.5–8
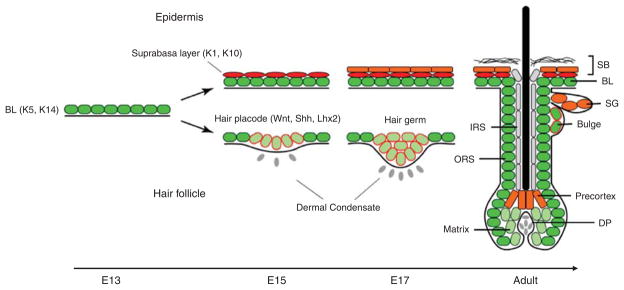
Figure 1.
Schematic illustration of embryonic epidermal development and hair follicle morphogenesis. Skin development starts with a single layer of multipotent embryonic skin stem cells in which basal layer (BL) markers such as keratin-5 and keratin-14 are highly expressed. Soon after embryonic day 13 (E13), these multipotent cells residing in the basal layer of the epidermis begin to stratify and give rise to a differentiated suprabasal layer (SB), in which terminal differentiation markers such as keratin-1 and keratin-10 are induced and the formation of the protective barrier begins. At the same time, some of these multipotent embryonic skin stem cells receive molecular instructions from underlying dermal condensates to adapt hair follicle fate. Hair placode is initiated under the regulation of signaling pathways such as Wnt and Shh pathways and transcription factors such as Lhx2. Through embryonic development, the epidermis and hair follicle continue to develop into a multiple lineage, fully protective barrier to protect the body. Note: ORS, outer-root sheath; IRS, inner-root sheath; SG, sebaceous gland; DP, dermal papilla. In the figure, stem/progenitor cells are marked in green; hair follicle stem cells are marked in green with a red border; differentiated cells in the epidermis, sebaceous gland and hair follicles are marked in red and orange; differentiated IRS cells are in gray; matrix cells are in light green
MicroRNAs are a family of non-coding, small RNAs (~19–24nt), expressed in a wide range of animals and plants.6,9,10 This newly discovered RNA species accounts for 1–3% of genes in mammalian genome. It is estimated that more than one-third of protein-encoding mRNAs are regulated by miRNAs.11,12 In turn, miRNA-mediated regulation is believed to have a widespread impact on both protein output of transcriptome13,14 and evolution of gene regulatory networks.15,16 MiRNAs’ potentials in globally regulating gene expression and developmental transitions during mammalian skin development have helped to accelerate interest in the function of these novel regulators in stem cell biology and developmental biology.
Profiling microRNA Expression in the Skin
To understand the functions of this novel class of regulators, it is important to define their expression patterns. Similar to protein-encoding mRNAs, the primary transcripts of most mammalian miRNAs are transcribed by RNA Polymerase II.17,18 Given the tissue specificity conferred by the transcriptional control of Pol II genes, this immediately suggests that miRNA expression can be spatiotemporally regulated. Unlike protein-encoding mRNAs, however, mature miRNAs are tiny, ~19–24 nt (nucleotides) in size, and do not have a 5′ m7G cap and a 3′ Poly (A) tail.6 To accommodate these miRNA-specific features, several methods have been developed for the profiling of their expression patterns (Table 1).
Table 1.
Summary of current profiling methods for miRNAs
| Methods for profiling miRNA expression | |
|---|---|
| Small RNA cloning and sequencing | The original small RNA discovery approach. Require 3′ and 5′ ligation and PCR amplification. With the application of next-generation sequencing and genome-wide bioinformatic annotation, this method becomes one of the most powerful tools to profile small RNA expression, including miRNAs. Suitable for large-scale high-throughput profiling. Absolute abundance of miRNAs can be referred by the cloning frequency. |
| MiRNA microarray | Similar to microarray for mRNAs, the expression of individual miRNAs can be compared across many different samples. Many commercial platforms are available with the requirement of <1 μg total RNA. Suitable for large-scale high-throughput profiling of miRNAs in many different samples such as clinical samples. |
| Real-time PCR-based profiling | Highly sensitive for the detection of changes in individual miRNA expression. Suitable for quantitative study of the expression of a small number of miRNAs. Multiplex approaches are also developed to profile all known miRNAs (similar to the microarray approach but with higher sensitivity). |
| In situ hybridization | Require LNA-modified probes for enhanced affinity and specificity to miRNAs. Accurately monitor individual miRNA expression in a spatiotemporally specific manner. Provide the most accurate profiling of a single miRNA in cell lineages or clinical samples. |
| Northern blotting | Classic method to measure the expression of individual miRNA. Often detect both mature miRNA and pre-miRNA at the same time. But require relatively large amount of total RNA input. Still a method of choice to validate the existence of novel miRNAs (small RNA cloning and sequencing can also be applied). |
Technical features and potential applications are listed in each column
Since the early days of miRNA research, small RNA cDNA cloning and sequencing has been instrumental for the discovery of novel miRNA species, because no previous knowledge of miRNA sequences is required.19–21 The process begins with size-fractioned RNA (19–30 nt) to enrich for miRNAs and then uses 3′ and 5′ ligation. The ligation is specifically tailored for a single-strand RNA containing the 3′ hydroxyl and 5′monophosphate group, which is characteristic of the cleavage product of Dicer, the RNase III enzyme required in miRNA generation.22 The cloned small RNA library is then subjected to PCR amplification and sequencing to determine the sequences and relative abundance (by sequencing frequency) of cloned small RNAs.23,24
This method was used to clone and sequence miRNAs from epidermal and HF samples prepared at embryonic day 17.5 (E17.5).25 At this stage, epidermal development is near completion, whereas hair morphogenesis is still at a rudimentary stage (Figure 1). MiRNAs cloned from these two libraries provided the first glance of the global expression of miRNAs and their differential localization in skin lineages. For example, the miR-199 family is highly expressed in the HF, whereas it is absent from the epidermis; members of the miR-200 family tend to express together while deriving from two genomic loci.25 The results also revealed many miRNAs that are widely expressed in other tissues, for example, let-7 family, miR-21 and miR-17 family, and so on, as well as a subset of miRNAs that are enriched in epithelial tissues, for example, miR-203.
When the sequence information of most, if not all, miRNAs became available by extensive small-RNA cloning efforts, microarray methods were developed to rapidly profile miRNAs in epithelial tissues.26,27 With a knowledge of the most abundantly expressed mammalian miRNAs in normal skin, researchers have turned toward addressing the manner in which this pattern changes in situations in which signaling pathways and/or lineages are genetically altered.26,27 Millar and colleagues adapted this strategy to identify HF-specific miRNAs when they utilized a previously established Dkk1 overexpression-transgenic mouse model in which hair development is largely abolished with the potent inhibition of the Wnt signaling pathway.26 Several miRNAs, for example, miR-200b and miR-196a, emerged as candidates that may be important for HF development as their expression was reduced in the Dkk1 transgenic skin.26 With recent development in the purification and isolation of distinct skin lineages, including the epidermis, hair germs, HF stem cells, outer root sheath, matrix and dermal papilla (DP),28–31 it is now possible to directly profile miRNA expression in these well-defined skin populations with miRNA microarrays. Insights gained from these expression studies will greatly facilitate future investigation for their functions.
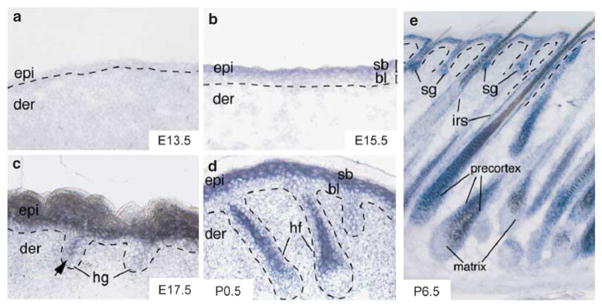
Although large-scale profiling experiments provided a panoramic view of miRNA expression, an in situ hybridization technique was adapted to determine the expression patterns of individual miRNAs.32,33 In zebrafish, whole-mount in situ hybridization can detect most, if not all, miRNAs during embryonic development.32 The elucidation of the spatiotemporal pattern of individual miRNAs unveiled tissue-specific expression of many miRNAs. In mammalian skin, miR-203 expression was examined in great detail by in situ hybridization.34 In embryonic skin development, miR-203 is induced in the suprabasal layer during epidermal differentiation, whereas it was mostly absent in basal progenitor cells. In mature skin, miR-203 is highly expressed only in differentiated cells that mostly exit cell cycle, for example, suprabasal epidermis, precortex and inner-root sheath of HFs, but not in the stem/progenitor cell compartments in which cells either possess proliferative potential or are actively dividing, for example, basal epidermis, bulge stem cells, outer-root sheath and matrix cells34 (Figure 2). Strikingly, the specific expression of miR-203 in the outermost layer of the skin is also conserved in zebrafish and humans, strongly suggesting conserved functions of this miRNA in vertebrate skin.32,34
Figure 2.
Spatiotemporally specific expression of miR-203 in the skin. (a) At E13.5, miR-203 is absent from the single layer of multipotent skin stem cells. (b) When the epidermis begins to stratify and gives rise to differentiated suprabasal layers at E15.5, miR-203 is specifically induced in the differentiated layers. (c) At E17.5, miR-203 starts to express in differentiated cells located in the hair germ (hg), whereas it continues to expand in the suprabasal epidermis. (d) With the progressive maturation of the hair follicle, the expression of miR-203 in differentiated lineages of the hair follicle becomes clearly visible at P0.5. (e) At P6.5, miR-203 is highly expressed in all differentiated skin lineages that exit the cell cycle and stop dividing while absent from all stem/progenitor skin lineages that either have proliferative potential, for example, stem cells, or actively divide, for example, matrix cells
Most recently, a next-generation deep sequencing technique was used to profile epidermal miRNAs. These results revealed a pool of ~70 miRNAs that are highly expressed in the skin (the relative expression level is more than 0.1%).35 The combination of deep coverage (more than 1 million small RNA reads from each library) and comparative study with WT, Dicer and DGCR8 knockout skin (see below) unequivocally showed that miRNAs are the major Dicer products (~19–24 nt small RNAs with the 5′ monophosphate and 3′ hydroxyl group) in the skin. With the relatively small number of highly expressed miRNAs in the skin (~70), it will soon be possible to delineate the lineage-specific expression patterns of these miRNAs by in situ hybridization. Together, the comprehensive survey of the miRNA expression firmly validated that miRNAs are differentially expressed in the skin and set the stage to further explore their biological functions.
Functional Study of microRNAs in the Skin by Targeting Critical Components in the microRNA Biogenesis Pathway
MicroRNA-mediated regulation is a novel layer of gene regulatory networks to control protein output. What is the significance of this layer of regulation in mammalian skin development? Although different miRNAs have completely different sequences, they share common biogenesis pathways wherein their precursor transcripts can fold back into a stem-loop, hairpin structure.6,36 This hairpin is first released from the long primary transcript by a Drosha-DGCR8 microprocessor complex in the nucleus37–41 and then exported to the cytoplasm by Exportin-5 in a RanGTP-dependent manner.42,43 On being transported to the cytoplasm, the hairpin is further processed by another RNase III enzyme, Dicer, to generate a dsRNA duplex, and one strand of the duplex is incorporated into an RNA-induced silencing complex to form functional miRNP.36,44 Mature miRNAs act by specifically coupling with target mRNAs at the 3′ untranslated region to impair the translation and/or mRNA stability.6,45 It is, therefore, possible to study miRNA functions by targeting critical components of the miRNA biogenesis pathways, for example, Drosha-DGCR8 or Dicer. In turn, the ablation of these components leads to the depletion of mature miRNAs and affords investigators an opportunity to examine how miRNAs may collectively function in the skin.
Constitutive Dicer knockout is embryonic lethal and the embryo is aborted around embryonic day 7.5 (E7.5),46 when the skin development is yet to start. To circumvent this problem, several groups have generated the skin conditional knockout (cKO) of Dicer.25,26 Using a floxed Dicer allele and a Cre recombinase transgene driven by a keratin-14 promoter,47 it became possible to specifically ablate Dicer from embryonic skin stem/progenitor cells starting around E13.5. As the loss of mature miRNAs is secondary to the ablation of Dicer and the relatively long half-life of miRNAs, mature miRNAs are not grossly depleted until E17.5 (R.Y. and E.F., unpublished data). The consequent depletion of miRNAs happened after the formation of the first suprabasal layer in the epidermis between E13 and E15, as well as the first wave of hair morphogenesis around E15 (see Figure 1). However, HF morphogenesis has multiple waves, with the last wave taking place between E17 and birth.1 Therefore, the consequences of a complete loss of miRNAs can be examined at this stage with the Dicer/K14-Cre cKO model.
Instead of invaginating into the dermis, Dicer cKO HFs evaginate upward.25,26 Strikingly, the evaginating HFs attract DP, which migrate with them. Thus, a continuous interaction between HF and DP is maintained, unlike β4 integrin or Shh skin cKO48,49 in which the arresting HFs are caused, at least in part, by the loss of physical interaction and communication between HF and DP. These results have suggested that the defects are likely caused by a dysregulation of gene expression on the epithelial side of the epithelial–mesenchymal communication as a consequence of ablating Dicer (miRNAs) in the skin. Furthermore, when other stratified epithelial tissues, tongue and foot pad (wherein floxed Dicer is also ablated by K14-Cre), were examined, aborted morphogenesis was also observed, suggesting that Dicer (miRNAs) is commonly required for morphogenesis of stratified epithelial tissues.25 Although it was not clear which signaling path-way(s) were specifically dysregulated in the Dicer cKO skin, we surmise that Dicer (miRNAs) is critically required for maintaining an appropriate output of the signaling pathway(s) and, in turn, the finely tuned pathway(s) are essential for the epithelial–mesenchymal communication and the morphogenesis of HFs. It is clearly of interest for future studies to determine which individual miRNA or miRNA family is responsible for these striking defects.
Although most Dicer cKO animals die neonatally, Millar and colleagues fortuitously obtained a few Dicer cKO animals that survived up to 2.5 months probably because of the mosaic deletion of Dicer.26 When examining these Dicer cKO HFs for a longer term, they found that HF stem cells in the bulge cannot be maintained and HFs are consequently degenerated.26 These results provide compelling evidence for a function of miRNAs in the maintenance of HF stem cells. Interestingly, the ablation of Dicer in the epidermis did not seem to cause the loss of epidermal stem/progenitor cells (see below). This raises the possibility that at least some miRNAs or their targets that function in this process may be specific for HF but not for epidermal stem cells. Future elucidation of which miRNAs and their targets may be involved in the regulation will enhance our understanding of the molecular basis governing the maintenance of HF stem cells.
In contrast to hypoproliferation of HFs and depletion of HF stem cells, hyperproliferation was observed in the Dicer cKO epidermis.26 In adults, the Dicer cKO epidermis is composed of multiple layers of basal and suprabasal cells, consistent with the neonatal hyperproliferative phenotype in the epidermis.26,34 These epidermal defects suggested that epidermal miRNAs control proliferation and cell cycle exit during epidermal differentiation.
Ablation of Dicer in the skin also led to mild apoptosis.25,26 Interestingly, a careful investigation revealed that apoptotic cells were enriched in the hair bulb, in which matrix cells are highly proliferative. The specific localization of apoptotic cells was suggestive of either a general requirement of Dicer (miRNAs) in rapidly dividing cells, consistent with apoptotic phenotypes observed in the Dicer cKO limb and T cells,50,51 or a specific requirement for Dicer (miRNAs) in the hair bulb.
Although studies with Dicer cKO skin provided a glimpse of how miRNAs may be functioning in the skin, two key questions arose: (i) Are the phenotypes observed in the Dicer cKO truly caused by depletion of mature miRNAs? (ii) Are miRNAs the only Dicer products in the skin? To address these questions, a recent study was conducted involving a parallel generation and examination of the skin cKO of Dicer with the equivalent cKO of DGCR8, an essential cofactor for miRNA processing in the nucleus.38–41 Although Dicer has been reported to be required for the processing of several classes of small RNAs including miRNAs and mRNA-derived siRNAs,52–54 DGCR8 has been shown to be specific for stereotypical miRNAs.35,52 When comparing the number of deep sequencing reads that can be mapped to different classes of small RNAs, stereotypical miRNAs emerged as the most abundant species in the skin. In comparing the reads in the two cKO skins, the production of stereotypical miRNAs was shown to be dependent on both Dicer and DGCR8, and the production of a few hairpin miRNAs and mRNA-derived siRNAs is dependent only on Dicer but not on DGCR8.35 More importantly, both Dicer and DGCR8 skin cKO animals showed indistinguishable phenotypes including evaginating HFs, enriched apoptosis in hair bulbs, rough and dehydrated skin and neonatal lethality.25,26,35 These results confirmed that the previously reported Dicer cKO phenotypes were indeed bona fide consequences of the loss of miRNAs in the skin and firmly established that stereotypical miRNAs are the most important Dicer products in the skin.
Exploring the Functions of Individual microRNAs in the Skin and Human Diseases
By investigating Dicer and DGCR8 skin cKOs, the global importance of miRNAs in skin development was revealed and miRNA functions were implicated in skin morphogenesis, maintenance of HF stem cells, epidermal proliferation and apoptosis. With this information at hand, the next step was to begin to dissect the functions of individual miRNAs in the skin.
Owing to its highly spatiotemporally specific expression pattern, miR-203 was the first miRNA investigated in the skin.34,55–57 Given miR-203’s abundance and its location at the transition between the dividing and differentiating layers of the epidermis,35 it was tempting to speculate that the main epidermal phenotype observed in the Dicer cKO skin,25,26,34 namely, hyperproliferation, might be attributable specifically to loss of miR-203. Indeed, a functional analysis showed a potent inhibition of the proliferative potential to skin stem/progenitor cells by miR-203 both in vivo and in vitro. Gain-of-function of the miR-203 expression in epidermal stem/progenitor cells in vivo and in cultured keratinocytes in vitro led to cell cycle exit and significantly reduced colony formation capacity, characteristic of differentiated daughter cells of these stem/progenitor cells.34,56 Loss-of-function of miR-203 expression in differentiated epidermal cells by the treatment of antagomir-203, a specific inhibitor to miR-203, increased the number of actively dividing cells in the suprabasal epidermis in vivo.34 Consistent with its function in inhibiting cell proliferation in mammalian skin, miR-203 was also shown to control fin regeneration in zebrafish, where it potently inhibits the growth of repairing fin by targeting Lef1 expression when overexpressed.58
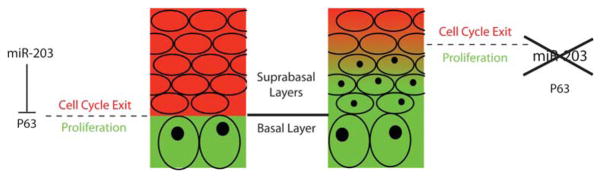
To probe the mechanism of the miR-203 function, it is critical to identify its physiological targets. Although systematic and unbiased target identification approaches are yet to be established, a few targets for miR-203 during skin development have now been documented.34,56 The best characterized is the transcription factor p63 (also known as p73-like in humans). Over the last decade, p63 has been extensively investigated for its essential function in the maintenance of ‘stemness’ in the skin and other stratified epithelial tissues.59–61 Intriguingly, p63 and miR-203 have mutually exclusive expression patterns, opposite functions and evolutionarily conserved regulatory relationships34 (see Figure 3). These observations provide a compelling case for the evolutionary conservation of the critical functions of miR-203 throughout the vertebrate kingdom.
Figure 3.
MiR-203 is specifically induced in suprabasal layers to sharpen the basal–suprabasal transition by repressing the expression of p63. In the proliferative basal layer (green), p63 is highly expressed to promote proliferation. Once the basal cells become suprabasal, miR-203 is induced to repress the residual expression of p63 mRNAs and, as a result, promote cell cycle exit (red). When miR-203 is lost, p63 is continuously expressed in otherwise differentiated suprabasal cells and stimulates their division (cells with black dot)
Owing to the potent inhibition of miR-203 to cell proliferation, miR-203 might be expected to function as a tumor suppressor. Indeed, for example, miR-203 was induced in a head and neck squamous-cell carcinoma cell line exposed to UVC irradiation, a treatment that results in cell death and cell cycle exit.56 Consistent with this notion, an elegant study in human hematopoietic tumors showed that frequent silencing of miR-203 is either genetically or epigenetically correlated with T-cell lymphomas.62 The forced expression of miR-203 directly downregulated an oncogene, BCR-ABL1, and inhibited cancer cell proliferation.62 Oddly, however, and in contrast to the epidermis, miR-203 is not detected in normal T-cell lineages.63–65 Even more paradoxical are the findings that miR-203 is upregulated in psoriasis, a common human hyperproliferative skin disease involving a proinflammatory response, and in a few epithelial tumors.55,66–69 Although further studies will be necessary to reconcile these differences, one possibility is that the expression of miR-203 can be independently induced under stress conditions, for example, cancer, psoriasis or other diseases, perhaps as a negative and, in some cases, futile feedback mechanism to suppress the proliferative state.
Clues for the functions of other skin-expressed miRNAs have also come from studies of their function in human diseases, especially cancer. MiR-200 miRNAs and miR-205 are both highly expressed in normal skin, and have been shown to specifically target the mRNA of the transcriptional repressor of E-Cadherin, ZEB1 and ZEB2.70–73 By doing so, they promote the expression of E-Cadherin. Consequently, downregulation of the miR-200 family and miR-205 leads to the inhibition of E-Cadherin, promoting an epithelial–mesenchymal transition.70–72 Consistent with these studies, miR-205 is often significantly downregulated in human epithelial tumors when compared with normal tissues.74,75 However, in other profiling studies in human epithelial cancer, the miR-200 family and/or miR-205 were found upregulated.68,76,77 These apparently contrasting results suggest that miRNAs may have different functions in different cells in which the local mRNA content may be an important determinant for their functions. Future studies are required to dissect the molecular circuit that underlies these observations.
Derived from the neural crest, melanocyte is an integral component of the skin. Moreover, melanoma is the deadliest form of skin cancer. Recent studies have begun to show miRNA expression and functions in the development and progression of melanoma.78–81 Interestingly, members of the Let-7 family were implicated in the suppression melanoma development,78,79 reminiscent of their functions in other human cancers.
Closing Remarks and Future Challenges
Although still in infancy, studies of miRNAs in mammalian skin have already provided significant insights into this novel layer of regulation. The similar and yet severe defects observed in Dicer and DGCR8 skin cKOs have firmly established the critical functions of miRNA in mammalian skin development. Unraveling the functions of individual miRNAs and their targets will be exciting and should offer important insights into this fascinating level of complex regulation of developmental biology. MiRNA-mediated regulation is particularly intriguing in that, rather than completely blocking the expression of its targets, a single miRNA can simultaneously target hundreds of mRNAs at modest levels (≤two fold) to finely tune the protein output of the transcriptome.13,14 In turn, a modestly increased expression of a thousand genes, when miRNAs are lost, leads to dramatic changes in tissue morphogenesis. An emerging scheme from our studies on miR-203 is that miRNAs may not be required to specify cell fate but rather to make an accurate transition between developmental stages. During the transition, miRNAs are upregulated to repress the expression of mRNAs inherited from the previous stage and, in turn, sharpen the boundary between two neighboring developmental lineages (Figure 3).
Given their critical functions in cancers and other human diseases, it is conceivable that miRNAs possess great potential as therapeutical targets. Owing to their small size (~19–24 nt), short oligonucleotides complementary to miR-NA sequences, for example, antagomir, can effectively block their functions in animals,34,82,83 whereas miRNA mimics can enhance miRNA functions.84 In addition, in vivo delivery strategies that have been developed for siRNAs85 can be conveniently adapted for antagomirs and for miRNA mimics. These advances have paved the way for the development of miRNA-based drugs for human diseases in the near future. In particular, because of the easy accessibility of the skin, we anticipate that skin-related diseases will be among the first to be treated with miRNA-based therapies.
Although just recently discovered, miRNAs are an integral component of gene regulatory networks throughout animal evolution.10 With the initial characterization of miRNA functions in mammalian skin, we start to appreciate the significance of an accurately regulated protein output not only in normal skin development, but also in human skin diseases. Now the stage is set to understand individual miRNA function and how critical biological events are controlled by this class of small RNA molecules.
Abbreviations
- cKO
conditional knockout
- DP
dermal papilla
- HF
hair follicle
- miRNAs
microRNAs
References
- 1.Schmidt-Ullrich R, Paus R. Molecular principles of hair follicle induction and morphogenesis. Bioessays. 2005;27:247–261. doi: 10.1002/bies.20184. [DOI] [PubMed] [Google Scholar]
- 2.Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones PH, Simons BD, Watt FM. Sic transit gloria: farewell to the epidermal transit amplifying cell? Cell Stem Cell. 2007;1:371–381. doi: 10.1016/j.stem.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs E, Horsley V. More than one way to skin. Genes Dev. 2008;22:976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 10.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 15.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 16.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 20.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 21.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 22.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21 and 22 nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeffer S, Lagos-Quintana M, Tuschl T. Cloning of small RNA molecules. Curr Protoc Mol Biol. 2005;Chapter 26(Unit 264) doi: 10.1002/0471142727.mb2604s72. [DOI] [PubMed] [Google Scholar]
- 24.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi R, O’Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 26.Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, et al. The miRNA-processing enzyme Dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 28.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 33.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 34.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 37.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 39.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 40.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 44.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 45.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 46.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 47.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dowling J, Yu QC, Fuchs E. Beta4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St-Jacques B, Dassule HR, Karavanova I, Botchkarev VA, Li J, Danielian PS, et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 50.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 55.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lena AM, Shalom-Feuerstein R, di Val Cervo PR, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 57.Aberdam D, Candi E, Knight RA, Melino G. miRNAs, ‘stemness’ and skin. Trends Biochem Sci. 2008;33:583–591. doi: 10.1016/j.tibs.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 58.Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci USA. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 60.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 61.Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 62.Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 63.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, et al. miRNA profiling of naive, effector and memory CD8T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 68.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 69.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 70.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 71.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–1178. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260. doi: 10.1016/j.jtcvs.2007.08.055. discussion 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O’Brien C, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 77.Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci USA. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muller DW, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27:6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 79.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 80.Mueller DW, Rehli M, Bosserhoff AK. MiRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 81.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 83.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 84.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]