Abstract
Cyclodextrin glucanotransferases (CGTases) are industrially important enzymes that produce cyclic α-(1,4)-linked oligosaccharides (cyclodextrins) from starch. Cyclodextrin glucanotransferases are also applied as catalysts in the synthesis of glycosylated molecules and can act as antistaling agents in the baking industry. To improve the performance of CGTases in these various applications, protein engineers are screening for CGTase variants with higher product yields, improved CD size specificity, etc. In this review, we focus on the strategies employed in obtaining CGTases with new or enhanced enzymatic capabilities by searching for new enzymes and improving existing enzymatic activities via protein engineering.
Keywords: Amylase, Biocatalysis, Directed evolution, Glycoside hydrolase, Protein engineering, Starch
Introduction
Cyclodextrin glucanotransferases (CGTases; EC 2.4.1.19) convert starch into cyclic α-1,4-glucans, called cyclodextrins (CDs). Cyclodextrins were identified in 1891 and structurally characterized in the preceding years. The main products of CGTases are α-, β-, and γ-CDs, composed of 6, 7, or 8 glucose residues, although, much larger cyclic glucans are produced in the early phases of the reaction (Terada et al. 1997; Zheng et al. 2002; Qi et al. 2007). In nature, certain bacteria and archaea presumably excrete CGTases to monopolize on the starch substrate, converting it into CDs, which cannot be utilized by competing microorganisms (Pajatsch et al. 1999; Hashimoto et al. 2001).
Cyclodextrins have numerous applications in the pharmaceutical, cosmetics, and food and textile industry, etc., as reviewed (Martin Del Valle 2009; Li et al. 2007; Biwer et al. 2002), because of their capacity to encapsulate hydrophobic molecules within their hydrophobic cavity. Encapsulation is used to solubilize hydrophobic molecules in water (CDs have a hydrophilic outside), which is particularly advantageous as many drug molecules are poorly soluble in water (Loftsson and Duchene 2007), or to protect guest molecules from light, heat, or oxidizing conditions (Astray et al. 2009). Cyclodextrins are also used to lower the volatility of odor molecules in perfumes and room refreshers for controlled release of the odor. In the chemical industry, CDs are used in the separation of enatiomers to extract toxic chemicals from waste streams (Martin Del Valle 2009) and in soil bioremediation (Fava and Ciccotosto 2002). Various other applications of CDs include the suppression of undesirable (bitter) tastes and the extraction of compounds such as cholesterol from foods (Szente and Szejtli 2004; Szejlti and Szente 2005).
CGTases
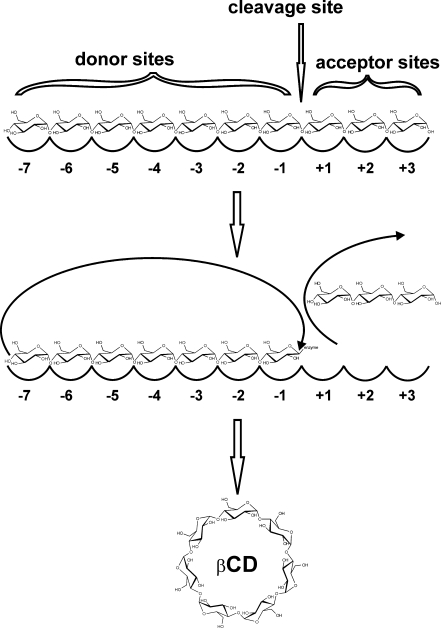
Cyclodextrin glucanotransferases are members of the largest family of glycoside hydrolases acting on starch and related α-glucans, glycoside hydrolase family 13 (Stam et al. 2006) (http://www.cazy.org). The first 3D structure of this enzyme (Klein and Schulz 1991) revealed that CGTases are five domain proteins with the active site located at the bottom of a (β/α)8-barrel in the A domain. Substrates bind across the enzyme’s surface in a long groove formed by the domains A and B that can accommodate at least 7 glucose residues at the donor subsites and 3 at the acceptor subsites (Fig. 1, labeled −7 to +3) as revealed by kinetic studies and crystal structures of substrate/inhibitor/product-CGTase complexes (Bender 1990; Kanai et al. 2001; Leemhuis et al. 2003a; Wind et al. 1998; Schmidt et al. 1998; Uitdehaag et al. 1999b). The C-terminal region of CGTases is formed by C, D, and E-domains. The function of domain D is unknown, domain C has been implied in substrate binding (Penninga et al. 1996), and domain E is a raw starch-binding domain (Penninga et al. 1996; Dalmia et al. 1995; Chang et al. 1998). The E-domain is classified as a family 20 carbohydrate binding module (CBM20) (Cantarel et al. 2009; Machovic and Janecek 2006) (http://www.cazy.org).
Fig. 1.
Schematic view of CD formation by CGTase
Cyclodextrin glucanotransferases cleave the α-1,4-glycosidic bonds between the subsites -1 and +1 in α-glucans yielding a stable covalent glycosyl-intermediate bound at the donor subsites (Fig. 1) (Uitdehaag et al. 1999a). The glycosyl-intermediate is then transferred to the 4-hydroxyl of its own nonreducing end forming a new α-1,4-glycosidic bond to yield a cyclic product (Fig. 1). Cyclodextrin glucanotransferases can also transfer the glycosyl-intermediate to a second α-glucan yielding a linear product (disproportionation) or to water (hydrolysis). In addition, CGTase can degrade CDs by opening the CD ring and transferring the linearalized CD to a sugar acceptor to yield a linear oligosaccharide (coupling). The large amount of structural information together with site-directed mutagenesis data have been used to elucidate the mechanistic functions of the residues at the catalytic center of CGTases (e.g., donor subsite -1) (Nakamura et al. 1993; Leemhuis et al. 2003c; Klein et al. 1992; Haga et al. 2003). However, as these mutations generally resulted in very low catalytic proficient CGTase mutants, they are not discussed here. Mutagenesis studies affecting reaction specificities are discussed below.
CD production
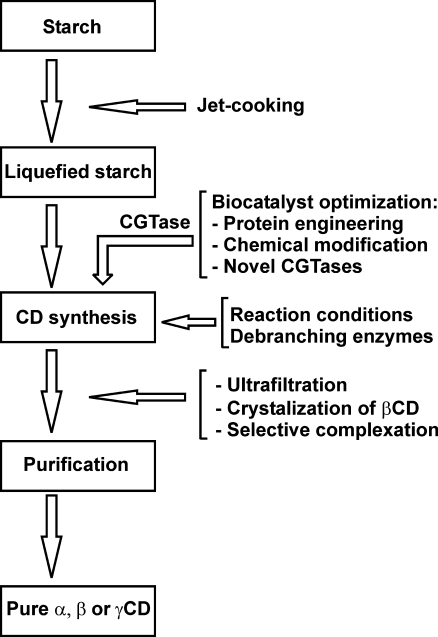
Cyclodextrins are produced in thousands of tons from starch annually by several manufactures, and demands are still rising. The starch is first liquefied, usually via an energy consuming jet-cooking step (Buchholz and Seibel 2008) (Fig. 2). Unfortunately, the total conversion of starch into CDs is closer to 50% than 100%. One of the reasons for this lack of efficiency is that CGTases have difficulty in bypassing the α-1,6-branches in amylopectin yielding CGTase limit dextrin (van der Maarel et al. 2002). The addition of isoamylase or pullulanase debranching enzymes increases the accessibility of the amylopectin fraction of starch, thus, increasing the CD yield (Rendleman 1997). Cyclodextrin yields are also limited due to enzyme product inhibition (Leemhuis et al. 2003a; Gaston et al. 2009) and breakdown of CDs by CGTases into linear oligosaccharides in the coupling reaction. The effects of both product inhibition and CD degradation are minimized by keeping the CD concentrations in the reactor low, which is generally achieved by adding complexing agents leading to the precipitation of the CDs. Moreover, the type of complexing agent used strongly influences the ratio of α-, β-, and γ-CD produced, for details see Blackwood and Bucke (2000), Biwer et al. (2002), and Zhekova et al. (2009). The breakdown of CDs is reduced further by restricting the accumulation of short oligosaccharides through the use of CGTases with low hydrolytic activity. Indeed, it has been shown that at high concentrations of saccharides, CGTases do not produce CDs from starch (Martin et al. 2001).
Fig. 2.
Flow scheme of CD production. Highlighted are the steps where protein engineers and process controllers can influence the process efficiency
The other major issue in CD production is that CGTases produce a mixture of CDs. A selective purification step is, thus, required to obtain pure α-CD, β-CD, or γ-CD, through the use of complexing agents during CD synthesis and the variation in solubility of the different CDs to allow selective precipitation (Matioli et al. 2000; Lee and Kim 1992; Son et al. 2008). The source of CGTase is the key factor in the type of CDs produced (Table 1), along with reaction parameters such as the type of starch used, the buffer composition, reaction temperature, and pH (Qi et al. 2004; Kamaruddin et al. 2005; Alves-Prado et al. 2008).
Table 1.
Characterized CGTases and their CD specificity
| Strain | Main CD produced | Reference |
|---|---|---|
| Archaea | ||
| Pyrococcus furiosus DSM 3638 | β | (Lee et al. 2007) |
| Thermococcus kodakaraensis KOD1 | β | (Rashid et al. 2002) |
| Thermococcus sp. B1001 | α | (Hashimoto et al. 2001) |
| Bacteria | ||
| Alkalophilic Bacillus agaradhaerens LS-3C | β | (Martins et al. 2003a) |
| Alkalophilic Bacillus sp. 1-1 | β | (Schmid et al. 1988) |
| Alkalophilic Bacillus sp. 17-1 | β | (Kaneko et al. 1989) |
| Alkalophilic Bacillus sp. 38-2 | β | (Hamamoto and Kaneko 1987) |
| Alkalophilic Bacillus sp. 1011 | β | (Kimura et al. 1987) |
| Alkalophilic Bacillus sp. 8SBa | β | (Atanasova et al. 2008) |
| Alkalophilic Bacillus sp. 20RFa | β | (Atanasova et al. 2008) |
| Alkalophilic Bacillus sp. A2-5a | β | (Ohdan et al. 2000) |
| Alkalophilic Bacillus sp. G-825-6 | γ | (Hirano et al. 2006) |
| Alkalophilic Bacillus sp. I-5 | β | (Shim et al. 2004) |
| Anaerobranca gottschalkii | α | (Thiemann et al. 2004) |
| Bacillus circulans 8 | β | (Nitschke et al. 1990) |
| Bacillus circulans 251 | β | (Lawson et al. 1994) |
| Bacillus circulans A11 | β | (Rimphanitchayakit et al. 2005) |
| Bacillus circulans DF 9R | α/β | (Marechal et al. 1996) |
| Bacillus clarkii 7384 | γ | (Takada et al. 2003) |
| Bacillus clausii E16a | β | (Alves-Prado et al. 2008) |
| Bacillus firmus 290-3 | β/γ | (Englbrecht et al. 1988) |
| Bacillus firmus 7Ba | β | (Moriwaki et al. 2007) |
| Bacillus firmus NCIM 5119a | β | (Gawande et al. 1999) |
| Bacillus firmus no. 37a | β | (Matioli et al. 2001) |
| Bacillus licheniformis | α/β | (Hill et al. 1990) |
| Bacillus macerans b | α | (Takano et al. 1986) |
| Bacillus megaterium a | β | (Pishtiyski et al. 2008) |
| Bacillus obhensis | β | (Sin et al. 1991) |
| Bacillus sp. B1018 | β | (Itkor et al. 1990) |
| Bacillus sp. BL-31 | β | (Go et al. 2007) |
| Bacillus sp. G1 | β | (Ong et al. 2008) |
| Bacillus sp. KC201 | β | (Kitamoto et al. 1992) |
| Bacillus sp. TS1-1 | β | (Rahman et al. 2006) |
| Bacillus stearothermophilus NO2c | α/β | (Fujiwara et al. 1992) |
| Brevibacillus brevis CD162 | β | (Kim et al. 1998) |
| Geobacillus stearothermophilus ET1 | β | (Chung et al. 1998) |
| Klebsiella pneumoniae M5a1 | α | (Binder et al. 1986) |
| Paenibacillus sp. BT01a | α/β | (Yampayont et al. 2006) |
| Paenibacillus sp. C36 | β | (Kinder 2007) |
| Paenibacillus sp. RB01d | β | (Charoensakdi et al. 2007a) |
| Paenibacillus sp. T16 | β/γb | (Charoensakdi et al. 2007b) |
| Paenibacillus campinasensis H69-3a | β | (Alves-Prado et al. 2007) |
| Paenibacillus graminis NC22.13 | α/β | (Vollu et al. 2008) |
| Paenibacillus illinoisensis ST-12 Ka | β | (Doukyu et al. 2003) |
| Paenibacillus pabuli US132 | β | (Jemli et al. 2008) |
| Thermoanaerobacter sp. 501 | α/β | (Norman and Jorgensen 1992) |
| Thermoanaerobacter sp. ATCC 53627 | β | (Jørgensen et al. 1997) |
| Thermoanaerobacter sp. P4a, d | βa | (Avci and Donmez 2009) |
| Thermoanaerobacterium thermosulfurigenes EM1 | α/β | (Wind et al. 1998) |
aCGTase purified from host organism, but the gene has not been identified and sequenced
bAlso known as Paenibacillus macerans
cAlso known as Geobacillus stearothermophilus NO2
dFormation of α- and γCD was not assayed
Searching nature for better performing CGTases
As the origin of the CGTase is the key factor in determining the ratios at which the different CDs are produced, scientists are searching nature for better performing CGTases. Moreover, novel CGTases may be better suited to the various industrial processing parameters, including substrate conversion, stability, and activity, than the currently available enzymes. There are 3 approaches to identify novel CGTases: genome mining of DNA databases, cloning environmental DNA in expression vectors and identifying recombinant clones displaying CGTase activity, or isolating bacteria/archaea expressing CGTase activity. Today CGTases of about fifty microorganisms have been (partly) characterized for CD specificity (Table 1). Fifteen of these enzymes were identified in the last 2 years, displaying a broad variation in optimal reaction pH, temperature, stability, and CD size specificity. Despite the availability of a large number of these enzymes, a CGTase that requires little or no reengineering for industrial optimization has yet to be identified. The CGTase from Bacillus clarkii, for example, was shown to produce approximately 80% γ-CDs (Takada et al. 2003). However, its overall conversion of starch into CDs was low at 14%.
Chemical modification, immobilization, and enzyme cross-linking
Before recombinant DNA technologies were developed, proteins engineers relied on chemical modification of amino acids such as lysine, cysteine, etc., to improve enzyme function and to gain insights into functionally important residues. When combined with mass spectrometry, one can determine exactly which amino acid residues are modified, as shown for Bacillus circulans DF 9R CGTase (Costa et al. 2009). Chemical modifications of amino acids can have various effects on the reaction specificity as demonstrated for Thermoanaerorbacter CGTase, where succinylation and acetylation enhanced the transglycosylation (Alcalde et al. 2001) and hydrolytic activities of the enzyme, respectively (Alcalde et al. 1999). Chemical modification is also the first step in the synthesis of cross-linked enzyme crystals, which are insoluble particles that retain catalytic activity under harsh conditions such as extreme pH, high temperature, and high solvent concentrations as demonstrated for B. macerans CGTase (Kim et al. 2003). Cross-linking can also be performed in the presence of substrate/product molecules, known as molecular imprinting, were one tries to fixate a productive conformation of an enzyme. Molecular imprinting of Paenibacillus sp. A11 and Bacillus macerans CGTase with γ-CD yielded CGTase crystals that converted over 10% of starch into CDs larger than γ-CD (Kaulpiboon et al. 2007), while the corresponding wild-type does not produce these large CDs.
Closely related to the chemical modification method is the immobilization of enzymes onto particles to facilitate the recovery of the expensive biocatalyst from product streams for reuse of the expensive biocatalyst. In addition, immobilization usually stabilizes the biocatalyst under industrial settings. A small number of reports have described the immobilization of CGTases covalently to supports, such as Eupergit C (Martin et al. 2003) and glyoxyl-agarose (Ferrarotti et al. 2006), or by entrapment in sodium alginate beads (Arya and Srivastava 2006). Covalent immobilization is, generally, more favorable as the biocatalyst is not leaking away, but unfortunately, this typically reduces the activity of CGTase to below 10% due to the inaccessibility of a large portion of the immobilized enzyme for the polymeric substrate. One report describes an alternative strategy to facilitate the recovery of biocatalyst, namely, entrapping bacterial cells that display CGTases on the surface in polyvinyl-cryogel beads (Martins et al. 2003b). Chemical modification processes can, therefore, provide a means of improving the application range of CGTases in industry.
Protein engineering for improving CD production by CGTases
The technique of choice for improving enzyme performance by today’s protein engineers is directed evolution. This approach, generally, delivers better enzymes than site-directed mutagenesis, which is a consequence of our limited understanding of structure/function relationships of enzymes. Directed evolution involves the construction of thousands to millions of variants of preexisting enzymes, using polymerase-chain-reaction techniques followed by high-throughput screening for better performing biocatalysts (Leemhuis et al. 2009; Kelly et al. 2009a). Site-directed mutagenesis is, nevertheless, highly valuable for investigating residues selected on basis of 3D structural knowledge or sequence alignments. Moreover, directed evolution and site-directed mutagenesis are frequently combined, randomizing functionally important regions of enzymes via saturation mutagenesis (Reetz et al. 2008).
Stability
The stability of a biocatalyst is an important factor in industrial applications. Cyclodextrin glucanotransferases are available from both mesophiles and extremophiles (Table 1) allowing selection of a CGTase with an appropriate thermostability. Highly stable CGTases, however, display greater hydrolytic activity on starch than their less stable counterparts (Kelly et al. 2009b). This may result in lower CD yields as hydrolytic products stimulate the degradation of CDs in the coupling reaction. The stability of enzymes with otherwise beneficial properties can be enhanced via protein engineering (Eijsink et al. 2004), but there is only one report were mutagenesis was used to improve the temperature stability of a CGTase. The stability of Bacillus circulans 251 CGTase was raised by engineering a salt bridge on the surface of the B-domain (Leemhuis et al. 2004a). Other site-directed mutagenesis and directed evolution studies have revealed that engineering of CGTases for reaction specificity, generally, delivers variants with reduced thermostability (Kelly et al. 2008a).
The alternative approach is to engineer existing highly stable CGTases towards the desired reaction specificity. The highly thermostable, but highly hydrolytic, Thermoanaerobacterium thermosulfurigenes EM1 CGTase forms large amounts of short oligosaccharides and degrades CDs in the later phases of starch conversion via the coupling reaction. Using directed evolution, a variant of this CGTase (mutant S77P) was engineered that formed almost no hydrolytic products while maintaining native CD forming activity and stability (Kelly et al. 2008b). Moreover, the coupling activity of this mutant was very low with no degradation of CDs in the later phases of the starch conversion.
CD specificity
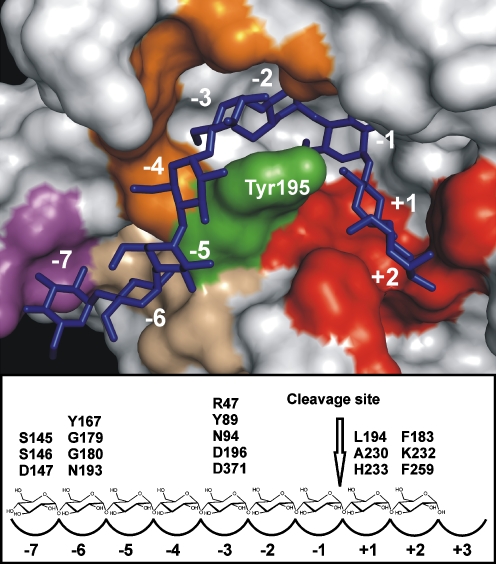
The long standing goal of CGTase engineering is the construction of variants producing a single type of CD only, which is extremely challenging as substrates of various lengths bind along the enzymes surface to allow for circularization of glucan chains. The challenge is to engineer CGTases that only permit the binding of glucan chains of a defined length at the donor subsites prior to covalent glycosyl-intermediate formation and CD formation. Such specific CGTase variants have not yet been engineered, although, CD ratios are strongly influenced by mutations at the -3/-6/-7 donor subsites. The -1/-2 donor subsites are crucial for catalytic activity. Strong enzyme-substrate interactions exist at subsite -6 while a lack of these interactions at subsites -4/-5 ensures the binding of a substrate long enough for CD formation. Engineering CD specificity by mutating conserved acceptor subsites residues of CGTases is not advisable as this typically results in highly hydrolytic CGTases (Table 2) that form small quantities of CDs (Shim et al. 2004; Kelly et al. 2007). Mutating the nonconserved residue 232 (Lys or Ala) at acceptor subsite +2 slightly altered CD ratios but also lowered CD yields (Nakagawa et al. 2006; Kelly et al. 2008a). Figure 3 shows the CGTase residues/regions important for reaction specificity.
Table 2.
Mutations affecting reaction and CD product specificity of CGTasesa
| Position (subsite) | Residues found in wild-type CGTases | Mutation | % conversion of starch into indicated CD | % ratio of indicated CD (%) | Hydrolysis (U/mg) |
|---|---|---|---|---|---|
| 195 (central)b | Y, F | Y195We | γCD: 8 → 15 | − | − |
| Y195Wf | − | γCD: 20 → 50 | − | ||
| Y195Lg | − | βCD: 64 → 86 | − | ||
| 259 (+2) | F, Y | F259Nh | − | − | 3 → 60 |
| F259Ii | − | − | 4 → 29 | ||
| F259Ej | − | − | 54 → 177 | ||
| 232 (+2) | K, A | K232Et | αCD: 6 → 1 | − | − |
| A232Ru | − | γCD: 26 → 35 | − | ||
| 194 (+1) | L | L194Tk | − | αCD: 10 → 2 | − |
| 230 (+1) | A | A230Vl | − | − | 3 → 72 |
| 47 (−3) | R, K, H, T | H47Tm | − | γCD: 10 → 39 | − |
| R47Qn | − | αCD: 17 → 8 | − | ||
| 89 (−3)c | Y, G, D, E, Q | Y89Ko | αCD: 15 → 19 | − | − |
| Y89Ro | αCD: 15 → 21 | − | − | ||
| Y89Dr | αCD: 5.6 → 6.8 | − | − | ||
| 371 (−3) | D | D371Ko | αCD: 15 → 20 | − | − |
| D371Rp | αCD: 9.8 → 1.7 | − | − | ||
| D371Rp | γCD: 4.9 → 7.5 | − | − | ||
| 167 | Y167Fq | αCD: 4.9 → 6.7 | − | − | |
| 179 | G179Lq | αCD: 4.9 → 2.7 | − | − | |
| 180 | G180Lq | αCD: 4.9 → 5.5 | − | − | |
| 193 (−6) | N | N193Gq | αCD: 4.9 → 8.2 | − | − |
| 146 (−7)c | S, E, L, F | S146Pr | αCD: 5.6 → 9.6 | − | − |
| 145–151 (−7) | Δ145–151 → Dk | − | γCD: 20 → 40 | − | |
| 146/89 (−6/−3) | S146P/Y89Dr | αCD: 5.6 → 12 | − | − | |
| 77d | S | S77Ps | − | − | 40 → 3 |
aNumbering follows that of B. circulans 251 CGTase. Only the most effective mutations in CGTase engineering are listed
bThis is the centrally located residue in the substrate binding groove (Fig. 3)
cNote that the length and conformation of this loop is variable among CGTases
dS77 is a second shell residue that is important for the orientation of the acid/base catalyst Glu257
e Bacillus ohbensis (Sin et al. 1994)
f B. circulans 8 (Parsiegla et al. 1998)
g B. circulans 251 (Penninga et al. 1995)
h B. circulans 251 (van der Veen et al. 2001)
iAlkalophilic Bacillus sp. I-5 (Shim et al. 2004)
j T. thermosulfurigenes EM1 (Leemhuis et al. 2002a)
k B. circulans 8 (Parsiegla et al. 1998)
l B. circulans 251 (Leemhuis et al. 2003d)
m Bacillus sp. G1 (Goh et al. 2009)
n B. circulans 251 (van der Veen et al. 2000a)
o P. macerans (Li et al. 2009)
p T. thermosulfurigenes EM1 (Wind et al. 1998)
q B. circulans 251 (Leemhuis et al. 2002b)
r B. circulans 251 (van der Veen et al. 2000b)
s T. thermosulfurigenes EM1 (Kelly et al. 2008b), CD forming activity unaffected
t B. circulans 251 (Kelly et al. 2008a)
u Bacillus clarkii 7364 (Nakagawa et al. 2006)
Fig. 3.
Substrate binding at the active site of CGTase. The upper panel shows the binding mode of a maltononaose substrate (blue sticks) at the active site of B. circulans 251 CGTase (crystal structure 1CXK from the protein data bank). Green—Tyr195; red—subsites +1/+2; orange—subsite -3; wheat—subsite -6; and magenta—subsite -7. Figure was created with PyMOL (DeLano 2002). The lower panel gives a schematic overview of the subsites and the residues providing the substrate interactions important for reaction specificity
Cyclodextrin glucanotransferase 3D structures indicate that in the process of CD formation, the glucan chain wraps around the Tyr/Phe195 residue (Fig. 3) suggesting that this residue has an important role in determining CD specificity. Indeed, an Y195W mutation increased the amount of γ-CD formed by Bacillus ohbensis CGTase (Table 2) and enhanced the ratio of γ-CD formed by B. circulans 8. In B. circulans 251, CGTase mutant Y195L increased the ratio of β-CD strongly (Table 2). Thus, the centrally located Tyr/Phe195 is an important target for engineering CD size specificity, but the effect of a substitution turns out to be different in various CGTases.
Engineering of subsites -3/-6/-7 has been successful in altering CD specificity of CGTases. Substituting the conserved residues of subsite -6 increased (Y167F, G180L, and N193G) the amount of α-CDs produced by B. circulans 251 CGTase (Table 2) (Leemhuis et al. 2002b). A D371R mutation at subsite -3 of T. thermosulfurigenes EM1 CGTase strongly increased γ-CD and lowered α-CD production (Table 2). In Bacillus sp. G1 a H47T mutation raised the ratio of γ-CD formed (Table 2), though the total γ-CD yield was only mildly enhanced. In Paenibacillus macerans mutations Y89R and D371K at subsite -3 enhanced the α-CD yield (Table 2). Substrate interactions at subsite -7 are provided by residues from loop 145-151 and are thought to create extra interactions with the seventh sugar residue to favor β-CD formation. Removing this loop strongly increased the ratio of γ-CD produced by B. circulans 8 (Table 2). The S146P mutation at subsite -7, in contrast, increased the conversion of starch into α-CDs (Table 2) and in combination with the Y89D substitution at subsite -3 even more α-CD was produced (van der Veen et al. 2000b).
Thus, CD ratios and amounts produced by CGTases can be engineered by mutations, but the effects have been studied mostly by incubating starch with the mutant enzymes, which is quite different from industrial process settings. We expect that most mutations will be even more effective under industrial production conditions where continuous precipitation of CDs, etc., selects more for the initial rates of the enzymes, which are generally more affected by the mutations than the final CD ratios. As described above, our current insights are restricted to knowing many “hotspot” residues for CD size specificity. However, as the polymeric starch substrates interact with at least 9 glucose moieties (Fig. 3), single mutants are not expected to change CD size specificity completely (see Table 2). Naturally, the extension of recent understanding of CGTase structure/function would fast track the targeting of specific areas for combinatorial site-saturation mutagenesis of the “hotspot” residues followed by high-throughput screening for highly active and CD size specific variants. The screening procedure is, ultimately, the constricting factor in the selection of CD specific CGTase variants. Screening requires a high-throughput HPLC platform to accurately assess the CD specificity, as the dyes methyl orange, phenolphthalein, and bromocresol green used to reveal the formation of α-, β-, and γ-CD, respectively, show cross-reactivity with other CD sizes. Moreover, as CGTases will eventually breakdown CDs in the coupling reaction, the amount of CDs produced should be measured at several time points. Thus, the protein engineer must analyze CGTase variants for CD specificity and percentage of substrate conversion over a number of time points, which should be economically feasible to most laboratories using 96-well format high-throughput HPLC systems.
Baking
Slowing down the retrogradation of the starch fraction in baked goods is the key to raise their shelf life. This can be accomplished by exo-acting enzymes, such as maltogenic amylase, that partly degrade the exterior of the amylopectin chains during baking process (De Stefanis and Turner 1981). Novamyl, a maltose forming maltogenic amylase, is used commercially as an antistaling enzyme. This enzyme shares ∼50% sequence identity with CGTases, but does not form CDs, although, a few mutations are enough to change it into a CD forming enzyme (Beier et al. 2000). The opposite experiment, changing a CGTase into a maltogenic amylase, was also successful (Leemhuis et al. 2003b). The high similarity of Novamyl with CGTases initiated research to investigate and improve the antistaling properties of CGTases. Protein engineers constructed highly hydrolytic CGTase variants with antistaling properties, using both site-directed mutagenesis (van der Veen et al. 2001; Lee et al. 2002; Leemhuis et al. 2002a) and directed evolution (Shim et al. 2004; Leemhuis et al. 2003d; Kelly et al. 2007). Kelly et al. engineered the most hydrolytic “CGTases” with virtually no CD forming activity (Kelly et al. 2007). All the hydrolytic variants mentioned carry mutations in the residues Phe183, Ala230, or Phe259 at the acceptor subsites +1/+2 and have (strongly) reduced cyclization activities. Recently, a CGTase with improved hydrolyzing activity was manipulated for bakers yeast surface display, enhancing the application of these antistaling enzymes in the baking industry (Shim et al. 2007). Increasingly, protein engineers are opening new routes in quality improvements in baking.
CGTases in carbohydrate synthesis
Oligosaccharides play key roles in living organisms, however, their chemical synthesis remains challenging, explaining the frequent use of enzymes in carbohydrate synthesis (Faijes and Planas 2007; Plou et al. 2007; Homann and Seibel 2009; Kaper et al. 2007). CGTases primarily transfer linear α-1,4-glucans to the 4-hydroxyl of glucose or longer α-glucans making a new α-1,4-glycosidic bond. Cyclodextrin glucanotransferase can, however, also transfer α-1,4-glucans to various other molecules displaying a single glucose moiety or even molecules simply displaying a hydroxyl function. An example of this reaction is the glycosylation of the low calorie sweeter stevioside, to reduce its bitter after-taste (Jung et al. 2007). Table 3 gives a list of compounds glycosylated by CGTases. Glycosylation may increase the water solubility, improve the bifidogenic characteristics, and lower cytotoxicity or improve the shelf life of these compounds. In these acceptor reactions starches, maltodextrins or cyclodextrins are used as the donor substrates and if desired, the attached α-glucans maybe trimmed by exo-glycosidases such as β-amylase or α-glucosidase. CGTases have also been applied in the synthesis of long (>10) oligosaccharides using CDs as donor and glucose as acceptor substrate (Yoon and Robyt 2002b). In addition, a donor subsite -1 mutant (H140A) CGTase has been shown to use the potent inhibitor acarbose as substrate and transfers the unnatural α-glucan compound acarviosyl (acarviosyl is a pseudo disaccharide composed of C7-cyclitol bound via an imino bridge to 4-amino-4,6-dideoxyglucose) to acceptor sugars (Leemhuis et al. 2004b). This demonstrates that mutant CGTases can be engineered to couple “α-glucan” like molecules to acceptor sugars.
Table 3.
Acceptor substrates of CGTase
| Acceptora | Reference |
|---|---|
| Acarboseb | (Yoon and Robyt 2002a) |
| Anhydro-D-fructosec | (Yoshinaga et al. 2003) |
| Arbutinc | (Sugimoto et al. 2003) |
| Ascorbic acidd | (Jun et al. 2001) |
| Benzo[h]quinazolinese | (Markosyan et al. 2009) |
| Curcumin β-D-glucosidec | (Shimoda et al. 2007) |
| Daidzein 7-O-β-D-glucopyranosidec | (Shimoda et al. 2008b) |
| Genistinc | (Li et al. 2005) |
| Glycerolf | (Nakano et al. 2003) |
| 7-Glycolylpaclitaxel 2-O-α-D-glucopyranosidec | (Shimoda et al. 2008a) |
| Hesperidinc, g | (Go et al. 2007) |
| Inositolh | (Sato et al. 1992) |
| Isomaltosec | (Vetter et al. 1992) |
| Luteoling | (Radu et al. 2006) |
| Naringinc,g | (Go et al. 2007) |
| Pentaerythritoli | (Nakano et al. 1992) |
| Phenyl β-D-glucopyranosidec | (Yoon and Robyt 2006) |
| Rutinc, g | (Go et al. 2007) |
| Salicing | (Yoon et al. 2004) |
| Saponinsc | (Kim et al. 2001) |
| Sorbitolj | (Park et al. 1998) |
| Steviosidec | (Kochikyan et al. 2006; Jung et al. 2007) |
| Sucrosec | (Martin et al. 2004) |
| Sucrose lauratec | (Okada et al. 2007) |
| Trimethylolpropanei | (Nakano et al. 1992) |
aMore information on the type of hydroxyl group used as acceptor is provided below the table
bCyclitol (2,3,4-trihydroxyl-5-(hydroxyl)-5,6-cyclohexene
cGlucose moiety
dHydroxyl of lactone ring
ePrimary hydroxyl
fAlcohol
gPhenolic
h1,2,3,4,5,6-hexahydroxylcyclo-hexane
iReduced glucose
jPolyol
Maltotriose and maltotetraose are the shortest natural donor substrates utilized by CGTases, however, CGTase can effectively use short α-fluorides of glucose and maltose as donor substrate because they carry an excellent leaving group (Mosi et al. 1997). For example, branched oligosaccharides were synthesized by glycosylation of panose at its 2 free 4-hydroxyl groups using α-fluoride maltose (with a protected 4-hydroxyl group) and CGTase as biocatalyst (Greffe et al. 2003). Cyclodextrin glucanotransferases also use α-fluoride maltose derivatives where the oxygen atom of the glycosidic linkage was substituted with a carbon or sulfur atom as substrate to synthesize linear and circular “α-glucans” (Bornaghi et al. 1997). In the near future, we may see an increase in the utilization of engineered (mutant) CGTase in carbohydrate synthesis, as they may have a broader donor and acceptor substrate specificity.
Future CGTases
Biocatalysis is, generally, regarded as an environment friendly technology. Nevertheless, CD synthesis may become even more environmental friendly if CGTase variants can be obtained, either via screening or protein engineering, that effectively convert native starch granules into specific CDs. The long standing aim of CGTase engineering is to construct variants producing a single type of CD. We feel that the current developments in the understanding of CGTase structure/function and advances in CGTase protein engineering will allow the creation of such a desirable catalyst in the coming years, employing combinatorial site-saturation mutagenesis. Moreover, new insights may lead to the design of CGTases forming high amounts of CDs consisting of 9 or more glucose monomers or CGTase variants that are capable of glycosylating, a wide variety of molecules bearing a hydroxyl function.
Acknowledgements
HL is supported by the Netherlands Organization for Scientific Research (NWO).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alcalde M, Plou FJ, Andersen C, Martin MT, Pedersen S, Ballesteros A. Chemical modification of lysine side chains of cyclodextrin glycosyltransferase from Thermoanaerobacter causes a shift from cyclodextrin glycosyltransferase to α-amylase specificity. FEBS Lett. 1999;445:333–337. doi: 10.1016/s0014-5793(99)00134-9. [DOI] [PubMed] [Google Scholar]
- Alcalde M, Plou FJ, Teresa MM, Valdes I, Mendez E, Ballesteros A. Succinylation of cyclodextrin glycosyltransferase from Thermoanaerobacter sp. 501 enhances its transferase activity using starch as donor. J Biotechnol. 2001;86:71–80. doi: 10.1016/s0168-1656(00)00422-3. [DOI] [PubMed] [Google Scholar]
- Alves-Prado HF, Gomes E, da Silva R. Purification and characterization of a cyclomaltodextrin glucanotransferase from Paenibacillus campinasensis strain H69–3. Appl Biochem Biotechnol. 2007;137–140:41–55. doi: 10.1007/s12010-007-9038-2. [DOI] [PubMed] [Google Scholar]
- Alves-Prado HF, Carneiro AA, Pavezzi FC, Gomes E, Boscolo M, Franco CM, da Silva R. Production of cyclodextrins by CGTase from Bacillus clausii using different starches as substrates. Appl Biochem Biotechnol. 2008;146:3–13. doi: 10.1007/s12010-007-8093-z. [DOI] [PubMed] [Google Scholar]
- Arya SK, Srivastava SK. Kinetics of immobilized cyclodextrin glucanotransferase produced by Bacillus macerans ATCC 8244. Enzyme Microb Technol. 2006;39:507–510. [Google Scholar]
- Astray G, Gonzalez-Barreiro C, Mejuto JC, Rial-Otero R, Simal-Gandara J. A review on the use of cyclodextrins in foods. Food Hydrocolloids. 2009;23:1631–1640. [Google Scholar]
- Atanasova N, Petrova P, Ivanova V, Yankov D, Vassileva A, Tonkova A. Isolation of novel alkaliphilic Bacillus strains for cyclodextrin glucanotransferase production. Appl Biochem Biotechnol. 2008;149:155–167. doi: 10.1007/s12010-007-8128-5. [DOI] [PubMed] [Google Scholar]
- Avci A, Donmez S. A novel thermophilic anaerobic bacteria producing cyclodextrin glycosyltransferase. Process Biochem. 2009;44:36–42. [Google Scholar]
- Beier L, Svendsen A, Andersen C, Frandsen TP, Borchert TV, Cherry JR. Conversion of the maltogenic alpha-amylase Novamyl into a CGTase. Protein Eng. 2000;13:509–513. doi: 10.1093/protein/13.7.509. [DOI] [PubMed] [Google Scholar]
- Bender H. Studies of the mechanism of the cyclisation reaction catalysed by the α-cyclodextrin glycosyltransferase from Klebsiella pneumoniae M5a1, and the β-cyclodextrin glycosyltransferase from Bacillus circulans 8. Carbohydr Res. 1990;206:257–267. doi: 10.1016/0008-6215(90)80065-b. [DOI] [PubMed] [Google Scholar]
- Binder F, Huber O, Bock A. Cyclodextrin-glycosyltransferase from Klebsiella pneumoniae M5a1: cloning, nucleotide sequence and expression. Gene. 1986;47:269–277. doi: 10.1016/0378-1119(86)90070-3. [DOI] [PubMed] [Google Scholar]
- Biwer A, Antranikian G, Heinzle E. Enzymatic production of cyclodextrins. Appl Microbiol Biotechnol. 2002;59:609–617. doi: 10.1007/s00253-002-1057-x. [DOI] [PubMed] [Google Scholar]
- Blackwood AD, Bucke C. Addition of polar organic solvents can improve the product selectivity of cyclodextrin glycosyltransferase. Solvent effects on CGTase. Enzyme Microb Technol. 2000;27:704–708. doi: 10.1016/s0141-0229(00)00270-2. [DOI] [PubMed] [Google Scholar]
- Bornaghi L, Utille JP, Rekai D, Mallet JM, Sinay P, Driguez H. Transfer reactions catalyzed by cyclodextrin glucosyltransferase using 4-thiomaltosyl and C-maltosyl fluorides as artificial donors. Carbohydr Res. 1997;305:561–568. doi: 10.1016/s0008-6215(97)00262-0. [DOI] [PubMed] [Google Scholar]
- Buchholz K, Seibel J. Industrial carbohydrate biotransformations. Carbohydr Res. 2008;343:1966–1979. doi: 10.1016/j.carres.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Irwin PM, Nikolov ZL. Effects of mutations in the starch-binding domain of Bacillus macerans cyclodextrin glycosyltransferase. J Biotechnol. 1998;65:191–202. doi: 10.1016/s0168-1656(98)00115-1. [DOI] [PubMed] [Google Scholar]
- Charoensakdi R, Iizuka M, Ito K, Rimphanitchayakit V, Limpaseni T. A recombinant cyclodextrin glycosyltransferase cloned from Paenibacillus sp. strain RB01 showed improved catalytic activity in coupling reaction between cyclodextrins and disaccharides. J Incl Phenom Macrocycl Chem. 2007;57:53–59. [Google Scholar]
- Charoensakdi R, Murakami S, Aoki K, Rimphanitchayakit V, Limpaseni T. Cloning and expression of cyclodextrin glycosyltransferase gene from Paenibacillus sp. T16 isolated from hot spring soil in northern Thailand. J Biochem Mol Biol. 2007;40:333–340. doi: 10.5483/bmbrep.2007.40.3.333. [DOI] [PubMed] [Google Scholar]
- Chung HJ, Yoon SH, Lee MJ, Kim MJ, Kweon KS, Lee I, Kim JW, Oh BH, Lee HS, Spiridonova VA, Park KH. Characterization of a thermostable cyclodextrin glucanotransferase isolated from Bacillus stearothermophilus ET1. J Agric Food Chem. 1998;46:952–959. [Google Scholar]
- Costa H, del Canto S, Ferrarotti S, de Jimenez Bonino MB. Structure-function relationship in cyclodextrin glycosyltransferase from Bacillus circulans DF 9R. Carbohydr Res. 2009;344:74–79. doi: 10.1016/j.carres.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Dalmia BK, Schutte K, Nikolov ZL. Domain E of Bacillus macerans cyclodextrin glucanotransferase: an independent starch-binding domain. Biotechnol Bioeng. 1995;47:575–584. doi: 10.1002/bit.260470510. [DOI] [PubMed] [Google Scholar]
- De Stefanis VA, Turner EW (1981) Modified enzyme system to inhibit bread firming method for preparing same and use of same in bread and other bakery products. US Pat 4299848. http://www.uspto.gov/patft/index.html
- DeLano WL (2002) The PyMOL molecular graphics system. http://www.pymol.org
- Doukyu N, Kuwahara H, Aono R. Isolation of Paenibacillus illinoisensis that produces cyclodextrin glucanotransferase resistant to organic solvents. Biosci Biotechnol Biochem. 2003;67:334–340. doi: 10.1271/bbb.67.334. [DOI] [PubMed] [Google Scholar]
- Eijsink VGH, Bjork A, Gaseidnes S, Sirevag R, Synstad B, van den Burg B, Vriend G. Rational engineering of enzyme stability. J Biotechnol. 2004;113:105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Englbrecht A, Harrer G, Lebert M. Biochemical and genetic characterization of a CGTase from an alkalophilic bacterium forming primarily gamma-cyclodextrin. In: Szejtli HO, editor. Proceedings of the fourth international symposium on cyclodextrins. Dordrecht: Kluwer; 1988. pp. 87–92. [Google Scholar]
- Faijes M, Planas A. In vitro synthesis of artificial polysaccharides by glycosidases and glycosynthases. Carbohydr Res. 2007;342:1581–1594. doi: 10.1016/j.carres.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Fava F, Ciccotosto VF. Effects of randomly methylated-beta-cyclodextrins (RAMEB) on the bioavailability and aerobic biodegradation of polychlorinated biphenyls in three pristine soils spiked with a transformer oil. Appl Microbiol Biotechnol. 2002;58:393–399. doi: 10.1007/s00253-001-0882-7. [DOI] [PubMed] [Google Scholar]
- Ferrarotti SA, Bolivar JM, Mateo C, Wilson L, Guisan JM, Fernandez-Lafuente R. Immobilization and stabilization of a cyclodextrin glycosyltransferase by covalent attachment on highly activated glyoxyl-agarose supports. Biotechnol Prog. 2006;22:1140–1145. doi: 10.1021/bp0600740. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Kakihara H, Woo KB, Lejeune A, Kanemoto M, Sakaguchi K, Imanaka T. Cyclization characteristics of cyclodextrin glucanotransferase are conferred by the NH2-terminal region of the enzyme. Appl Environ Microbiol. 1992;58:4016–4025. doi: 10.1128/aem.58.12.4016-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston JAR, Szerman N, Costa H, Krymkiewicz N, Ferrarotti S. Cyclodextrin glycosyltransferase from Bacillus circulans DF 9R: activity and kinetic studies. Enzyme Microb Technol. 2009;45:36–41. [Google Scholar]
- Gawande BN, Goel A, Patkar AY, Nene SN. Purification and properties of a novel raw starch degrading cyclomaltodextrin glucanotransferase from Bacillus firmus. Appl Microbiol Biotechnol. 1999;51:504–509. doi: 10.1007/s002530051424. [DOI] [PubMed] [Google Scholar]
- Go YH, Kim TK, Lee KW, Lee YH. Functional characteristics of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. BL-31 highly specific for intermolecular transglycosylation of bioflavonoids. J Microbiol Biotechnol. 2007;17:1550–1553. [PubMed] [Google Scholar]
- Goh KM, Mahadi NM, Hassan O, Rahman RNZRA, Illias RM. A predominant β-CGTase next term G1 engineered to elucidate the relationship between protein structure and product specificity. J Molecul Catalysis B: Enzymatic. 2009;57:270–277. [Google Scholar]
- Greffe L, Jensen MT, Bosso C, Svensson B, Driguez H. Chemoenzymatic synthesis of branched oligo- and polysaccharides as potential substrates for starch active enzymes. Chembiochem. 2003;4:1307–1311. doi: 10.1002/cbic.200300692. [DOI] [PubMed] [Google Scholar]
- Haga K, Kanai R, Sakamoto O, Aoyagi M, Harata K, Yamane K. Effects of essential carbohydrate/aromatic stacking interaction with Tyr100 and Phe259 on substrate binding of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp. 1011. J Biochem. 2003;134:881–891. doi: 10.1093/jb/mvg215. [DOI] [PubMed] [Google Scholar]
- Hamamoto T, Kaneko THK. Nucleotide sequence of the cyclomaltodextrin glucanotransferase (CGTase) gene from alkalophilic Bacillus sp. strain no. 38–2. Agric Biol Chem. 1987;51:2019–2022. doi: 10.1099/00221287-134-1-97. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Yamamoto T, Fujiwara S, Takagi M, Imanaka T. Extracellular synthesis, specific recognition, and intracellular degradation of cyclomaltodextrins by the hyperthermophilic archaeon Thermococcus sp. B1001. J Bacteriol. 2001;183:5050–5057. doi: 10.1128/JB.183.17.5050-5057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DE, Aldape R, Rozzell JD. Nucleotide sequence of a cyclodextrin glucosyltransferase gene, cgtA, from Bacillus licheniformis. Nucleic Acids Res. 1990;18:199. doi: 10.1093/nar/18.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Ishihara T, Ogasawara S, Maeda H, Abe K, Nakajima T, Yamagata Y. Molecular cloning and characterization of a novel gamma-CGTase from alkalophilic Bacillus sp. Appl Microbiol Biotechnol. 2006;70:193–201. doi: 10.1007/s00253-005-0041-7. [DOI] [PubMed] [Google Scholar]
- Homann A, Seibel J. Towards tailor-made oligosaccharides-chemo-enzymatic approaches by enzyme and substrate engineering. Appl Microbiol Biotechnol. 2009;83:209–216. doi: 10.1007/s00253-009-1989-5. [DOI] [PubMed] [Google Scholar]
- Itkor P, Tsukagoshi N, Udaka S. Nucleotide sequence of the raw starch-digesting amylase gene from Bacillus sp. B1018 and its strong homology to the cyclodextrin glucanotransferase genes. Biochem Biophys Res Commun. 1990;166:630–636. doi: 10.1016/0006-291x(90)90855-h. [DOI] [PubMed] [Google Scholar]
- Jemli S, Ben ME, Ben MS, Bejar S. The cyclodextrin glycosyltransferase of Paenibacillus pabuli US132 strain: molecular characterization and overproduction of the recombinant enzyme. J Biomed Biotechnol. 2008;2008:692573. doi: 10.1155/2008/692573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen ST, Tangney M, Starnes RL, Amemiya K, Jørgensen PL. Cloning and nucleotide sequence of a thermostable cyclodextrin glycosyltransferase gene from Thermoanaerobactersp. ATCC 53627 and its expression in Escherichia coli. Biotechnol Lett. 1997;19:1027–1031. [Google Scholar]
- Jun H-K, Bae K-M, Kim S-K. Production of 2-O-α-d-glucopyranosyl L-ascorbic acid using cyclodextrin glucanotransferase from Paenibacillus sp. Biotechnol Lett. 2001;23:1793–1797. [Google Scholar]
- Jung S-W, Kim T-K, Lee K-W, Lee Y-H. Catalytic properties of β-cyclodextrin glucanotransferase from alkalophilic Bacillus sp. BL-12 and intermolecular transglycosylation of stevioside. Biotechnol Bioprocess Eng. 2007;12:207–212. [Google Scholar]
- Kamaruddin K, Illias RM, Aziz SA, Said M, Hassan O. Effects of buffer properties on cyclodextrin glucanotransferase reactions and cyclodextrin production from raw sago (Cycas revoluta) starch. Biotechnol Appl Biochem. 2005;41:117–125. doi: 10.1042/BA20040040. [DOI] [PubMed] [Google Scholar]
- Kanai R, Haga K, Yamane K, Harata K. Crystal structure of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 complexed with 1-deoxynojirimycin at 2.0 A resolution. J Biochem. 2001;129:593–598. doi: 10.1093/oxfordjournals.jbchem.a002895. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Song K, Harmamoto T, Kudo T, Horikoshi K. Construction of a chimeric series of Bacillus cyclodextrin glucanotransferases and analysis of the thermal stability and pH optima of the enzymes. J Gen Microbiol. 1989;135:3447–3457. doi: 10.1099/00221287-135-12-3447. [DOI] [PubMed] [Google Scholar]
- Kaper T, Leemhuis H, Uitdehaag JC, van der Veen BA, Dijkstra BW, van der Maarel MJE, Dijkhuizen L. Identification of acceptor substrate binding subsites +2 and +3 in the amylomaltase from Thermus thermophilus HB8. Biochemistry. 2007;46:5261–5269. doi: 10.1021/bi602408j. [DOI] [PubMed] [Google Scholar]
- Kaulpiboon J, Pongsawasdi P, Zimmermann W. Molecular imprinting of cyclodextrin glycosyltransferases from Paenibacillus sp. A11 and Bacillus macerans with gamma-cyclodextrin. FEBS J. 2007;274:1001–1010. doi: 10.1111/j.1742-4658.2007.05649.x. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Leemhuis H, Dijkhuizen L. Conversion of a cyclodextrin glucanotransferase into an alpha-amylase: assessment of directed evolution strategies. Biochemistry. 2007;46:11216–11222. doi: 10.1021/bi701160h. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Leemhuis H, Gatjen L, Dijkhuizen L. Evolution toward small molecule inhibitor resistance affects native enzyme function and stability, generating acarbose-insensitive cyclodextrin glucanotransferase variants. J Biol Chem. 2008;283:10727–10734. doi: 10.1074/jbc.M709287200. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Leemhuis H, Rozeboom HJ, van Oosterwijk N, Dijkstra BW, Dijkhuizen L. Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution. Biochem J. 2008;413:517–525. doi: 10.1042/BJ20080353. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Dijkhuizen L, Leemhuis H. The evolution of cyclodextrin glucanotransferase product specificity. Appl Microbiol Biotechnol. 2009;84:119–133. doi: 10.1007/s00253-009-1988-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RM, Dijkhuizen L, Leemhuis H. Starch and α-glucan acting enzymes, modulating their properties by directed evolution. J Biotechnol. 2009;140:184–193. doi: 10.1016/j.jbiotec.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kim MH, Sohn CB, Oh TK. Cloning and sequencing of a cyclodextrin glycosyltransferase gene from Brevibacillus brevis CD162 and its expression in Escherichia coli. FEMS Microbiol Lett. 1998;164:411–418. doi: 10.1111/j.1574-6968.1998.tb13117.x. [DOI] [PubMed] [Google Scholar]
- Kim YH, Lee YG, Choi KJ, Uchida K, Suzuki Y. Transglycosylation to ginseng saponins by cyclomaltodextrin glucanotransferases. Biosci Biotechnol Biochem. 2001;65:875–883. doi: 10.1271/bbb.65.875. [DOI] [PubMed] [Google Scholar]
- Kim W, Chae H, Park C, Lee K. Stability and activity of cross-linking enzyme crystals of cyclodextrin glucanotransferase isolated from Bacillus macerans. J Molec Catalys B: Enzymatic. 2003;26:287–292. [Google Scholar]
- Kimura K, Kataoka S, Ishii Y, Takano T, Yamane K. Nucleotide sequence of the beta-cyclodextrin glucanotransferase gene of alkalophilic Bacillus sp. 1011 and similarity of its amino acid sequence to those of α-amylases. J Bacteriol. 1987;169:4399–4402. doi: 10.1128/jb.169.9.4399-4402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ (2007) Cloning and expression of a bacterial CGTase and impacts of transgenic plants on phytoremediation of organic pollutants. Thesis, Michigan State University
- Kitamoto N, Kimura T, Kito Y, Ohmiya K. Cloning and sequencing of the gene encoding cyclodextrin glucanotransferase from Bacillus sp. KC201. J Ferment Bioeng. 1992;74:345–351. [Google Scholar]
- Klein C, Schulz GE. Structure of cyclodextrin glycosyltransferase refined at 2.0 A resolution. J Mol Biol. 1991;217:737–750. doi: 10.1016/0022-2836(91)90530-j. [DOI] [PubMed] [Google Scholar]
- Klein C, Hollender J, Bender H, Schulz GE. Catalytic centre of cyclodextrin glycosyltransferase derived from X-ray structure analysis combined with site-directed mutagenesis. Biochemistry. 1992;31:8740–8746. doi: 10.1021/bi00152a009. [DOI] [PubMed] [Google Scholar]
- Kochikyan VT, Markosyan AA, Abelyan LA, Balayan AM, Abelyan VA. Combined enzymatic modification of stevioside and rebaudioside. Appl Biochem Microbiol. 2006;42:31–37. [PubMed] [Google Scholar]
- Lawson CL, van Montfort R, Strokopytov B, Rozeboom HJ, Kalk KH, de Vries GE, Penninga D, Dijkhuizen L, Dijkstra BW. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans 251 in a maltose-dependent crystal form. J Mol Biol. 1994;236:590–600. doi: 10.1006/jmbi.1994.1168. [DOI] [PubMed] [Google Scholar]
- Lee YD, Kim HS. Effect of organic solvents on enzymatic production of cyclodextrins from unliquefied corn starch in an attrition bioreactor. Biotechnol Bioeng. 1992;39:977–983. doi: 10.1002/bit.260391002. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kim YW, Lee S, Auh JH, Yoo SS, Kim TJ, Kim JW, Kim ST, Rho HJ, Choi JH, Kim YB, Park KH. Modulation of cyclizing activity and thermostability of cyclodextrin glucanotransferase and its application as an antistaling enzyme. J Agric Food Chem. 2002;50:1411–1415. doi: 10.1021/jf010928q. [DOI] [PubMed] [Google Scholar]
- Lee MH, Yang SJ, Kim JW, Lee HS, Kim JW, Park KH. Characterization of a thermostable cyclodextrin glucanotransferase from Pyrococcus furiosus DSM3638. Extremophiles. 2007;11:537–541. doi: 10.1007/s00792-007-0061-6. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Dijkstra BW, Dijkhuizen L. Mutations converting cyclodextrin glycosyltransferase from a transglycosylase into a starch hydrolase. FEBS Lett. 2002;514:189–192. doi: 10.1016/s0014-5793(02)02362-1. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Uitdehaag JCM, Rozeboom HJ, Dijkstra BW, Dijkhuizen L. The remote substrate binding subsite-6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism. J Biol Chem. 2002;277:1113–1119. doi: 10.1074/jbc.M106667200. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Dijkstra BW, Dijkhuizen L. Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase: mechanism and kinetics of inhibition by acarbose and cyclodextrins. Eur J Biochem. 2003;270:155–162. doi: 10.1046/j.1432-1033.2003.03376.x. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Kragh KM, Dijkstra BW, Dijkhuizen L. Engineering cyclodextrin glycosyltransferase into a starch hydrolase with a high exo-specificity. J Biotechnol. 2003;103:203–212. doi: 10.1016/s0168-1656(03)00126-3. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Rozeboom HJ, Dijkstra BW, Dijkhuizen L. The fully conserved Asp residue in conserved sequence region I of the alpha-amylase family is crucial for the catalytic site architecture and activity. FEBS Lett. 2003;541:47–51. doi: 10.1016/s0014-5793(03)00286-2. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Rozeboom HJ, Wilbrink M, Euverink GJ, Dijkstra BW, Dijkhuizen L. Conversion of cyclodextrin glycosyltransferase into a starch hydrolase by directed evolution: the role of alanine 230 in acceptor subsite +1. Biochemistry. 2003;42:7518–7526. doi: 10.1021/bi034439q. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Rozeboom HJ, Dijkstra BW, Dijkhuizen L. Improved thermostability of Bacillus circulans cyclodextrin glycosyltransferase by the introduction of a salt bridge. Proteins. 2004;54:128–134. doi: 10.1002/prot.10516. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Wehmeier UF, Dijkhuizen L. Single amino acid mutations interchange the reaction specificities of cyclodextrin glycosyltransferase and the acarbose-modifying enzyme acarviosyl transferase. Biochemistry. 2004;43:13204–13213. doi: 10.1021/bi049015q. [DOI] [PubMed] [Google Scholar]
- Leemhuis H, Kelly RM, Dijkhuizen L. Directed evolution of enzymes: library screening strategies. IUBMB Life. 2009;61:222–228. doi: 10.1002/iub.165. [DOI] [PubMed] [Google Scholar]
- Li D, Roh SA, Shim JH, Mikami B, Baik MY, Park CS, Park KH. Glycosylation of genistin into soluble inclusion complex form of cyclic glucans by enzymatic modification. J Agric Food Chem. 2005;53:6516–6524. doi: 10.1021/jf050732g. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang M, Wang F, Gu Z, Du G, Wu J, Chen J. γ-Cyclodextrin: a review on enzymatic production and applications. Appl Microbiol Biotechnol. 2007;77:245–255. doi: 10.1007/s00253-007-1166-7. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang J, Wang M, Gu Z, Du G, Li J, Wu J, Chen J. Mutations at subsite-3 in cyclodextrin glycosyltransferase from Paenibacillus macerans enhancing α-cyclodextrin specificity. Appl Microbiol Biotechnol. 2009;83:483–490. doi: 10.1007/s00253-009-1865-3. [DOI] [PubMed] [Google Scholar]
- Loftsson T, Duchene D. Cyclodextrins and their pharmaceutical applications. Int J Pharm. 2007;329:1–11. doi: 10.1016/j.ijpharm.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Machovic M, Janecek S. Starch-binding domains in the post-genome era. Cell Mol Life Sci. 2006;63:2710–2724. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal LR, Rosso AM, Marechal MA, Krymkiewicz N, Ferrarotti SA. Some properties of a cyclomaltodextrin-glucanotransferase from Bacillus circulans DF9R type. Cell Mol Biol. 1996;42:659–664. [PubMed] [Google Scholar]
- Markosyan AA, Abelyan LA, Markosyan AI, Abelyan VA. Transglycosylation of benzo[h]quinazolines. Appl Biochem Microbiol. 2009;45:130–136. [Google Scholar]
- Martin Del Valle EMM. Cyclodextrins and their uses: a review. Process Biochem. 2009;39:1033–1046. [Google Scholar]
- Martin MT, Alcalde M, Plou FJ, Dijkhuizen L, Ballesteros A. Synthesis of malto-oligosaccharides via the acceptor reaction catalyzed by cyclodextrin glycosyltransferases. Biocatal Biotransform. 2001;19:21–35. [Google Scholar]
- Martin MT, Plou FJ, Alcalde M, Ballesteros A. Immobilization on Eupergit C of cyclodextrin glucosyltransferase (CGTase) and properties of the immobilized biocatalyst. J Molecul Catalysis B: Enzymatic. 2003;21:299–308. [Google Scholar]
- Martin MT, Cruces A, Alcalde M, Plou FJ, Bernabe M, Ballesteros A. Synthesis of maltooligosyl fructofuranosides catalyzed by immobilized cyclodextrin glucosyltransferase using starch as donor. Tetrahedron. 2004;60:529–534. [Google Scholar]
- Martins RF, Delgado O, Hatti-Kaul R. Sequence analysis of cyclodextrin glycosyltransferase from the alkaliphilic Bacillus agaradhaerens. Biotechnol Lett. 2003;25:1555–1562. doi: 10.1023/a:1025430532333. [DOI] [PubMed] [Google Scholar]
- Martins RF, Plieva FM, Santos A, Hatti-Kaul R. Integrated immobilized cell reactor-adsorption system for beta-cyclodextrin production: a model study using PVA-cryogel entrapped Bacillus agaradhaerens cells. Biotechnol Lett. 2003;25:1537–1543. doi: 10.1023/a:1025408727114. [DOI] [PubMed] [Google Scholar]
- Matioli G, Zanin GM, de Moraes FF. Enhancement of selectivity for producing γ-cyclodextrin. Appl Biochem Biotechnol. 2000;84–86:955–962. [PubMed] [Google Scholar]
- Matioli G, Zanin GM, de Moraes FF. Characterization of cyclodextrin glycosyltransferase from Bacillus firmus strain no. 37. Appl Biochem Biotechnol. 2001;91–93:643–654. doi: 10.1385/abab:91-93:1-9:643. [DOI] [PubMed] [Google Scholar]
- Moriwaki C, Costa GL, Pazzetto R, Zanin GM, Moraes FF, Portilho M, Matioli G. Production and characterization of a new cyclodextrin glycosyltransferase from Bacillus firmus isolated from Brazilian soil. Process Biochem. 2007;42:1384–1390. [Google Scholar]
- Mosi R, He S, Uitdehaag J, Dijkstra BW, Withers SG. Trapping and characterization of the reaction intermediate in cyclodextrin glycosyltransferase by use of activated substrates and a mutant enzyme. Biochemistry. 1997;36:9927–9934. doi: 10.1021/bi970618u. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Takada M, Ogawa K, Hatada Y, Horikoshi K. Site-directed mutations in alanine 223 and glycine 255 in the acceptor site of γ-cyclodextrin glucanotransferase from Alkalophilic Bacillus clarkii 7364 affect cyclodextrin production. J Biochem. 2006;140:329–336. doi: 10.1093/jb/mvj158. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Haga K, Yamane K. Three histidine residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 effects of the replacement on pH dependence and transition-state stabilization. Biochemistry. 1993;32:6624–6631. doi: 10.1021/bi00077a015. [DOI] [PubMed] [Google Scholar]
- Nakano H, Kitahata S, Tominaga Y, Kiku Y, Ando K, Kawahashi Y, Takenishi S. Syntheses of glucosides with trimethylolpropane and two related polyol moieties by cyclodextrin glucanotransferase and their esterification by lipase. J Ferment Bioeng. 1992;73:237–238. [Google Scholar]
- Nakano H, Kiso T, Okamoto K, Tomita T, Manan MB, Kitahata S. Synthesis of glycosyl glycerol by cyclodextrin glucanotransferases. J Biosci Bioeng. 2003;95:583–588. [PubMed] [Google Scholar]
- Nitschke L, Heeger K, Bender H, Schulz GE. Molecular cloning, nucleotide sequence, and expression in Escherichia coli of the β-cyclodextrin glycosyltransferase gene from Bacillus circulans no. 8. Appl Microbiol Biotechnol. 1990;33:542–546. doi: 10.1007/BF00172548. [DOI] [PubMed] [Google Scholar]
- Norman BE, Jorgensen ST. Thermoanaerobacter sp. CGTase: its properties and application. Denpun Kagaku. 1992;39:101–108. [Google Scholar]
- Ohdan K, Kuriki T, Okada S. Cloning of the cyclodextrin glucanotransferase gene from alkalophilic Bacillus sp. A2–5a and analysis of the raw starch-binding domain. Appl Microbiol Biotechnol. 2000;53:430–434. doi: 10.1007/s002530051637. [DOI] [PubMed] [Google Scholar]
- Okada K, Zhao H, Izumi M, Nakajima S, Baba N. Glucosylation of sucrose laurate with cyclodextrin glucanotransferase. Biosci Biotechnol Biochem. 2007;71:826–829. doi: 10.1271/bbb.60646. [DOI] [PubMed] [Google Scholar]
- Ong RM, Goh KM, Mahadi NM, Hassan O, Rahman RN, Illias RM. Cloning, extracellular expression and characterization of a predominant beta-CGTase from Bacillus sp. G1 in E. coli. J Ind Microbiol Biotechnol. 2008;35:1705–1714. doi: 10.1007/s10295-008-0462-2. [DOI] [PubMed] [Google Scholar]
- Pajatsch M, Andersen C, Mathes A, Bock A, Benz R, Engelhardt H. Properties of a cyclodextrin-specific, unusual porin from Klebsiella oxytoca. J Biol Chem. 1999;274:25159–25166. doi: 10.1074/jbc.274.35.25159. [DOI] [PubMed] [Google Scholar]
- Park KH, Kim MJ, Lee HS, Han NS, Kim D, Robyt JF. Transglycosylation reactions of Bacillus stearothermophilus maltogenic amylase with acarbose and various acceptors. Carbohydr Res. 1998;313:235–246. doi: 10.1016/s0008-6215(98)00276-6. [DOI] [PubMed] [Google Scholar]
- Parsiegla G, Schmidt AK, Schulz GE. Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the γ-cyclodextrin production. Eur J Biochem. 1998;255:710–717. doi: 10.1046/j.1432-1327.1998.2550710.x. [DOI] [PubMed] [Google Scholar]
- Penninga D, Strokopytov B, Rozeboom HJ, Lawson CL, Dijkstra BW, Bergsma J, Dijkhuizen L. Site-directed mutations in tyrosine 195 of cyclodextrin glycosyltransferase from Bacillus circulans 251 affect activity and product specificity. Biochemistry. 1995;34:3368–3376. doi: 10.1021/bi00010a028. [DOI] [PubMed] [Google Scholar]
- Penninga D, van der Veen BA, Knegtel RM, van Hijum SA, Rozeboom HJ, Kalk KH, Dijkstra BW, Dijkhuizen L. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans 251. J Biol Chem. 1996;271:32777–32784. doi: 10.1074/jbc.271.51.32777. [DOI] [PubMed] [Google Scholar]
- Pishtiyski I, Popova V, Zhekova B. Characterization of cyclodextrin glucanotransferase produced by Bacillus megaterium. Appl Biochem Biotechnol. 2008;144:263–272. doi: 10.1007/s12010-007-8009-y. [DOI] [PubMed] [Google Scholar]
- Plou FJ, Gomez de Segura A, Ballesteros A (2007) Application of glycosidases and transglycosidases in the synthesis of oligosaccharides. In: Industrial enzymes. Springer, Netherlands, pp 141–157
- Qi Q, She X, Endo T, Zimmermann W. Effect of the reaction temperature on the transglycosylation reactions catalyzed by the cyclodextrin glucanotransferase from Bacillus macerans for the synthesis of large-ring cyclodextrins. Tetrahedron. 2004;60:799–806. [Google Scholar]
- Qi Q, Mokhtar MN, Zimmermann W. Effect of ethanol on the synthesis of large-ring cyclodextrins by cyclodextrin glucanotransferases. J Incl Phenom Macrocycl Chem. 2007;57:95–99. [Google Scholar]
- Radu OL, Armand S, Lenouvel F, Driguez H, Cimpean A, Iordachescu D. Luteolin glycosylation in organic solvents under catalytic action of cyclodextrin glycosyltransferase from Bacillus circulans. Rev Roum Chim. 2006;51:147–152. [Google Scholar]
- Rahman K, Illias RM, Hassan O, Mahmood NKN, Rashid NAA. Molecular cloning of a cyclodextrin glucanotransferase gene from alkalophilic Bacillus sp. TS1–1 and characterization of the recombinant enzyme. Enzyme Microb Technol. 2006;39:74–84. [Google Scholar]
- Rashid N, Cornista J, Ezaki S, Fukui T, Atomi H, Imanaka T. Characterization of an archaeal cyclodextrin glucanotransferase with a novel C-terminal domain. J Bacteriol. 2002;184:777–784. doi: 10.1128/JB.184.3.777-784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reetz MT, Kahakeaw D, Lohmer R. Addressing the numbers problem in directed evolution. Chembiochem. 2008;9:1797–1804. doi: 10.1002/cbic.200800298. [DOI] [PubMed] [Google Scholar]
- Rendleman JA., Jr Enhancement of cyclodextrin production through use of debranching enzymes. Biotechnol Appl Biochem. 1997;26(Pt 1):51–61. [PubMed] [Google Scholar]
- Rimphanitchayakit V, Tonozuka T, Sakano Y. Construction of chimeric cyclodextrin glucanotransferases from Bacillus circulans A11 and Paenibacillus macerans IAM1243 and analysis of their product specificity. Carbohydr Res. 2005;340:2279–2289. doi: 10.1016/j.carres.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Sato N, Nakamura K, Nagano H, Yagi Y, Koizumi K. Synthesis of glucosyl-inositol using a CGTase, isolation and characterization of the positional isomers, and assimilation profiles for intestinal bacteria. Biotechnol Lett. 1992;14:659–664. [Google Scholar]
- Schmid G, Englbrecht A, Schimd D. Cloning and nucleotide sequence of a cyclodextrin glycosyltransferase gene from the alkalophilic Bacillus sp.1–1. In: Szejtli HO, editor. Proceedings of the fourth international symposium on cyclodextrins. Dordrecht: Kluwer; 1988. pp. 77–81. [Google Scholar]
- Schmidt AK, Cottaz S, Driguez H, Schulz GE. Structure of cyclodextrin glycosyltransferase complexed with a derivative of its main product β-cyclodextrin. Biochemistry. 1998;37:5909–5915. doi: 10.1021/bi9729918. [DOI] [PubMed] [Google Scholar]
- Shim JH, Kim YW, Kim TJ, Chae HY, Park JH, Cha H, Kim JW, Kim YR, Schaefer T, Spendler T, Moon TW, Park KH. Improvement of cyclodextrin glucanotransferase as an antistaling enzyme by error-prone PCR. Protein Eng Des Sel. 2004;17:205–211. doi: 10.1093/protein/gzh035. [DOI] [PubMed] [Google Scholar]
- Shim JH, Seo NS, Roh SA, Kim JW, Cha H, Park KH. Improved bread-baking process using Saccharomyces cerevisiae displayed with engineered cyclodextrin glucanotransferase. J Agric Food Chem. 2007;55:4735–4740. doi: 10.1021/jf070217d. [DOI] [PubMed] [Google Scholar]
- Shimoda K, Hara T, Hamada H, Hamada H. Synthesis of curcumin β-maltooligosaccharides through biocatalytic glycosylation with Strophanthus gratus cell culture and cyclodextrin glucanotransferase. Tetrahedron Lett. 2007;48:4029–4032. [Google Scholar]
- Shimoda K, Hamada H, Hamada H. Chemo-enzymatic synthesis of ester-linked taxol–oligosaccharide conjugates as potential prodrugs. Tetrahedron Lett. 2008;49:601–604. [Google Scholar]
- Shimoda K, Sato N, Kobayashi T, Hamada H, Hamada H. Glycosylation of daidzein by the Eucalyptus cell cultures. Phytochemistry. 2008;69:2303–2306. doi: 10.1016/j.phytochem.2008.05.024. [DOI] [PubMed] [Google Scholar]
- Sin K, Nakamura A, Kobayashi K, Masaki H, Uozumi T. Cloning and sequencing of a cyclodextrin glucanotransferase gene from Bacillus ohbensis and its expression in Escherichia coli. Appl Microbiol Biotechnol. 1991;35:600–605. doi: 10.1007/BF00169623. [DOI] [PubMed] [Google Scholar]
- Sin KA, Nakamura A, Masaki H, Matsuura Y, Uozumi T. Replacement of an amino acid residue of cyclodextrin glucanotransferase of Bacillus ohbensis doubles the production of γ–cyclodextrin. J Biotechnol. 1994;32:283–288. doi: 10.1016/0168-1656(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Son YJ, Rha CS, Park YC, Shin SY, Lee YS, Seo JH. Production of cyclodextrins in ultrafiltration membrane reactor containing cyclodextrin glycosyltransferase from Bacillus macerans. J Microbiol Biotechnol. 2008;18:725–729. [PubMed] [Google Scholar]
- Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Nishimura T, Nomura K, Sugimoto K, Kuriki T. Syntheses of arbutin-α-glycosides and a comparison of their inhibitory effects with those of α-arbutin and arbutin on human tyrosinase. Chem Pharm Bull (Tokyo) 2003;51:798–801. doi: 10.1248/cpb.51.798. [DOI] [PubMed] [Google Scholar]
- Szejlti J, Szente L. Elimination of bitter, disgusting tastes of drugs and foods by cyclodextrins. Eur J Pharm Biopharm. 2005;61:115–125. doi: 10.1016/j.ejpb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Szente L, Szejtli J. Cyclodextrins as food ingredients. J Food Sci Technol. 2004;15:137–142. [Google Scholar]
- Takada M, Nakagawa Y, Yamamoto M. Biochemical and genetic analyses of a novel γ-cyclodextrin glucanotransferase from an alkalophilic Bacillus clarkii 7364. J Biochem. 2003;133:317–324. doi: 10.1093/jb/mvg043. [DOI] [PubMed] [Google Scholar]
- Takano T, Fukuda M, Monma M, Kobayashi S, Kainuma K, Yamane K. Molecular cloning, DNA nucleotide sequencing, and expression in Bacillus subtilis cells of the Bacillus macerans cyclodextrin glucanotransferase gene. J Bacteriol. 1986;166:1118–1122. doi: 10.1128/jb.166.3.1118-1122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, Yanase M, Takata H, Takaha T, Okada S. Cyclodextrins are not the major cyclic α-1, 4-glucans produced by the initial action of cyclodextrin glucanotransferase on amylose. J Biol Chem. 1997;272:15729–15733. doi: 10.1074/jbc.272.25.15729. [DOI] [PubMed] [Google Scholar]
- Thiemann V, Donges C, Prowe SG, Sterner R, Antranikian G. Characterisation of a thermoalkali-stable cyclodextrin glycosyltransferase from the anaerobic thermoalkaliphilic bacterium Anaerobranca gottschalkii. Arch Microbiol. 2004;182:226–235. doi: 10.1007/s00203-004-0717-x. [DOI] [PubMed] [Google Scholar]
- Uitdehaag JC, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat Struct Biol. 1999;6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- Uitdehaag JCM, Kalk KH, van der Veen BA, Dijkhuizen L, Dijkstra BW. The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a γ-cyclodextrin-CGTase complex at 1.8-A resolution. J Biol Chem. 1999;274:34868–34876. doi: 10.1074/jbc.274.49.34868. [DOI] [PubMed] [Google Scholar]
- van der Maarel MJEC, van der Veen BA, Uitdehaag JCM, Leemhuis H, Dijkhuizen L. Properties and applications of starch-converting enzymes of the alpha-amylase family. J Biotechnol. 2002;94:137–155. doi: 10.1016/s0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- van der Veen BA, Uitdehaag JC, Dijkstra BW, Dijkhuizen L. The role of arginine 47 in the cyclization and coupling reactions of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 implications for product inhibition and product specificity. Eur J Biochem. 2000;267:3432–3441. doi: 10.1046/j.1432-1327.2000.01353.x. [DOI] [PubMed] [Google Scholar]
- van der Veen BA, Uitdehaag JC, Penninga D, van Alebeek GJ, Smith LM, Dijkstra BW, Dijkhuizen L. Rational design of cyclodextrin glycosyltransferase from Bacillus circulans 251 to increase α-cyclodextrin production. J Mol Biol. 2000;296:1027–1038. doi: 10.1006/jmbi.2000.3528. [DOI] [PubMed] [Google Scholar]
- van der Veen BA, Leemhuis H, Kralj S, Uitdehaag JC, Dijkstra BW, Dijkhuizen L. Hydrophobic amino acid residues in the acceptor binding site are main determinants for reaction mechanism and specificity of cyclodextrin-glycosyltransferase. J Biol Chem. 2001;276:44557–44562. doi: 10.1074/jbc.M107533200. [DOI] [PubMed] [Google Scholar]
- Vetter D, Thorn W, Brunner H, Konig WA. Directed enzymatic synthesis of linear and branched gluco-oligosaccharides, using cyclodextrin-glucanosyltransferase. Carbohydr Res. 1992;223:61–69. doi: 10.1016/0008-6215(92)80006-m. [DOI] [PubMed] [Google Scholar]
- Vollu RE, da Mota FF, Gomes EA, Seldin L. Cyclodextrin production and genetic characterization of cyclodextrin glucanotransferase of Paenibacillus graminis. Biotechnol Lett. 2008;30:929–935. doi: 10.1007/s10529-008-9635-3. [DOI] [PubMed] [Google Scholar]
- Wind RD, Uitdehaag JC, Buitelaar RM, Dijkstra BW, Dijkhuizen L. Engineering of cyclodextrin product specificity and pH optima of the thermostable cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1. J Biol Chem. 1998;273:5771–5779. doi: 10.1074/jbc.273.10.5771. [DOI] [PubMed] [Google Scholar]
- Yampayont P, Iizuka M, Ito K. Isolation of cyclodextrin producing thermotolerant Paenibacillus sp. from waste of starch factory and some properties of the cyclodextrin glycosyltransferase. J Incl Phenom Macrocycl Chem. 2006;56:203–207. [Google Scholar]
- Yoon SH, Robyt JF. Addition of maltodextrins to the nonreducing-end of acarbose by reaction of acarbose with cyclomaltohexaose and cyclomaltodextrin glucanyltransferase. Carbohydr Res. 2002;337:509–516. doi: 10.1016/s0008-6215(02)00018-6. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Robyt JF. Bacillus macerans cyclomaltodextrin glucanotransferase transglycosylation reactions with different molar ratios of D-glucose and cyclomaltohexaose. Carbohydr Res. 2002;337:2245–2254. doi: 10.1016/s0008-6215(02)00224-0. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Robyt JF. Optimized synthesis of specific sizes of maltodextrin glycosides by the coupling reactions of Bacillus macerans cyclomaltodextrin glucanyltransferase. Carbohydr Res. 2006;341:210–217. doi: 10.1016/j.carres.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Bruce FD, Robyt JF. Enzymatic synthesis of two salicin analogues by reaction of salicyl alcohol with Bacillus macerans cyclomaltodextrin glucanyltransferase and Leuconostoc mesenteroides B-742CB dextransucrase. Carbohydr Res. 2004;339:1517–1529. doi: 10.1016/j.carres.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K, Abe J, Tanimoto T, Koizumi K, Hizukuri S. Preparation and reactivity of a novel disaccharide, glucosyl 1, 5-anhydro-D-fructose (1, 5-anhydro-3-O-alpha-glucopyranosyl-D-fructose) Carbohydr Res. 2003;338:2221–2225. doi: 10.1016/s0008-6215(03)00341-0. [DOI] [PubMed] [Google Scholar]
- Zhekova B, Dobrev G, Stanchev V, Pishtiyski I. Approaches for yield increase of β-cyclodextrin formed by cyclodextrin glucanotransferase from Bacillus megaterium. World J Microbiol Biotechnol. 2009;25:1043–1049. [Google Scholar]
- Zheng M, Endo T, Zimmermann W. Synthesis of large-ring cyclodextrins by cyclodextrin glucanotransferases from bacterial isolates. J Incl Phenom Macrocycl Chem. 2002;44:387–390. [Google Scholar]