Abstract
Biotin synthase was the first example of what is now regarded as a distinctive enzyme class within the radical S-adenosylmethionine superfamily, the members of which use Fe/S clusters as the sulphur source in radical sulphur insertion reactions. The crystal structure showed that this enzyme contains a [2Fe–2S] cluster with a highly unusual arginine ligand, besides three normal cysteine ligands. However, the crystal structure is at such a low resolution that neither the exact coordination mode nor the role of this exceptional ligand has been elucidated yet, although it has been shown that it is not essential for enzyme activity. We have used quantum refinement of the crystal structure and combined quantum mechanical and molecular mechanical calculations to explore possible coordination modes and their influences on cluster properties. The investigations show that the protonation state of the arginine ligand has little influence on cluster geometry, so even a positively charged guanidinium moiety would be in close proximity to the iron atom. Nevertheless, the crystallised enzyme most probably contains a deprotonated (neutral) arginine coordinating via the NH group. Furthermore, the Fe···Fe distance seems to be independent of the coordination mode and is in perfect agreement with distances in other structurally characterised [2Fe–2S] clusters. The exceptionally large Fe···Fe distance found in the crystal structure could not be reproduced.
Keywords: Biotin synthase, Fe/S cluster, Radical S-adenosylmethionine enzyme, Quantum mechanics/molecular mechanics, Quantum refinement
Introduction
Biological Fe/S clusters are versatile cofactors in enzymes, well known mainly for their ability to act as electron-transfer sites. In the past few years, an increasing number of other fascinating functions of Fe/S clusters have been discovered [1]. Among those, their use as the source for sulphur atoms in biological radical reactions has been striking with regards to the complex mechanisms of Fe/S cluster assembly [2]. As the Fe/S cluster is destroyed during this reaction, these enzymes are regarded as suicide enzymes and their respective clusters as substrates rather than cofactors [3]. To date, Fe/S clusters have been identified as the source of sulphur atoms in reactions catalysed by four different enzymes belonging to a distinctive class [4] within the radical S-adenosylmethionine superfamily [5]: biotin synthase (BioB) [6], lipoyl synthase (LipA) [7], a transfer RNA-methylthiotransferase (MiaB) [8] and a ribosomal methylthiotransferase (RimO) [9].
Although the latter three enzymes contain [4Fe–4S] clusters as the assumed sulphur source, BioB contains a [2Fe–2S] cluster which has been shown to be destroyed during catalytic turnover [10]. Of the four enzymes, BioB is the most extensively investigated and is also the only one whose crystal structure has been solved [11]. A mechanism for its reaction was first published in 2001 [10] and has been closely investigated since (Scheme 1) [12].
Scheme 1.
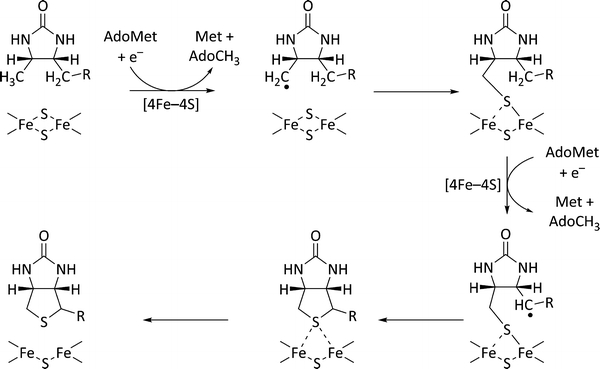
Proposed biotin synthase mechanism [10]. AdoMet S-adenosylmethionine
The proposed mechanism is supported by the close proximity of the [2Fe–2S] cluster to the dethiobiotin molecule found in the crystal structure. The closest bridging sulphide of the cluster is situated 4.6 Å away from C-9 of dethiobiotin, one of the two carbon atoms to which sulphur is attached in the course of the reaction (Fig. 1).
Fig. 1.
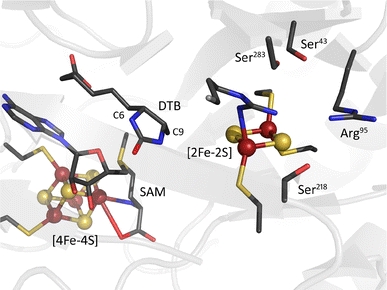
The biotin synthase crystal structure. SAM S-adenosylmethionine, DTB dethiobiotin
Surprisingly, the [2Fe–2S] cluster carries one strictly conserved arginine ligand in addition to three cysteine ligands typical for Fe/S clusters. As arginine is highly exceptional as a ligand coordinating to a metal ion in biological systems [13], its possible importance has been much discussed since publication of the crystal structure. Mutation experiments showed that this arginine ligand is not essential for the catalytic reaction. It has been proposed that it may play electronic, mechanistic or structural roles, possibly related to its bidentate nature or its positive charge in the protonated state [14].
A basic question to be answered is the charge of the arginine guanidine group in the active enzyme. Arginine is usually protonated at physiological pH (the pK a in water solution is approximately 12 [15]), thereby bearing a positive charge. However, a positively charged guanidine group could not be regarded as a true ligand from a coordination chemist’s point of view—for that, it should rather be deprotonated and neutral in BioB. This uncertainty is reflected by suggestions of both NH [13, 16] and NH2 [17] coordination in the literature.
In addition to the protonation issue, the role of the second, non-coordinating NH/NH2 group is to be ascertained. Secondary bonding interactions [18] are conceivable, as well as an involvement of hydrogen bonds with the protein backbone or with the cluster sulphur atoms. Furthermore, the crystal structure shows an Fe···Fe distance of 3.3 Å, which is significantly longer than what is found in any other known [2Fe–2S] clusters (approximately 2.7 Å) [19].
As these issues cannot be solved with the published structural data alone, owing to the low resolution of the crystal structure (3.4 Å), and the importance of the unusual arginine ligand remains elusive, theoretical methods seem to be a promising strategy to answer the above-mentioned questions. This work focuses on the structural properties of the highly unusual [2Fe–2S] cluster of BioB to improve our understanding of its importance for the enzyme mechanism.
Materials and methods
Quantum mechanical/molecular mechanical calculations
The quantum mechanical (QM)/molecular mechanical (MM) calculations were performed with the ComQum software program [20, 21] utilising Turbomole 5.9 [22] for the QM calculations and Amber 9 [23] for the MM calculations. The QM calculations were performed using the BP86 functional [24, 25] and the def2-SV(P) basis sets [26], which have given reasonable results for [2Fe–2S] clusters in previous calculations [18, 27]. For the MM calculations, we used the Amber-99 force field [28, 29]. For the [4Fe–4S] cluster and the dethiobiotin and S-adenosylmethionine ligands, we used force-field parameters previously determined in our group [30–32].
In the QM/MM approach, the protein and solvent are split into three subsystems. The QM region (system 1) contains the most interesting atoms and is relaxed by QM methods. System 2 consists of the residues closest to the QM system and is optimised by MM calculations. The remaining part of the protein and the surrounding solvent molecules (system 3) are kept fixed at the crystallographic coordinates. In the QM calculations, system 1 is represented by a wavefunction, whereas all the other atoms are represented by an array of partial point charges, one for each atom, taken from MM libraries. Thereby, the polarisation of the quantum chemical system by the surroundings is included in a self-consistent manner. When there is a bond between systems 1 and 2 (a junction), the quantum region is truncated by hydrogen atoms, the positions of which are linearly related to the corresponding carbon atoms in the full system (the hydrogen-link-atom approach) [20]. To eliminate the non-physical effect of placing point charges on atoms in the MM region bound to junction atoms (i.e. the closest neighbours of the QM system), those charges are zeroed, and the resulting residual charges are smoothly distributed [20].
The total energy is calculated as
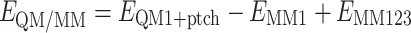 |
1 |
where E QM1+ptch is the QM energy of system 1 truncated by the hydrogen atoms and embedded in the set of point charges (but excluding the self-energy of the point charges). E MM1 is the MM energy of system 1, still truncated by hydrogen atoms, but without any electrostatic interactions. Finally, E MM123 is the classical energy of all atoms with normal atoms at the junctions and with the charges of the quantum system set to zero (to avoid double-counting of the electrostatic interactions). By this approach, which is similar to the one used in the ONIOM method [33], errors caused by the truncation of the quantum system should cancel.
The calculations were based on the crystal structure (Protein Data Bank code 1R30) [11]. As the enzyme was crystallised as a homodimer with little difference in atom positions (less than 0.1 Å differences within the [2Fe–2S] cluster), only the A subunit was used for the investigations and only this subunit is discussed. Hydrogen atoms were added to the crystal structure and the protein was solvated in a sphere of water molecules with a radius of 36 Å using the Leap module in the Amber software suite. The protonation status of all residues was checked by the PROPKA program [34] and it was concluded that no residues have strongly perturbed pK a values (thus, all arginine and lysine residues, except Arg260, see below, were considered in their protonated state, whereas all aspartate and glutamate residues were considered in their deprotonated state). For the histidine residues, the protonation was decided from a detailed study of the solvent exposure and hydrogen-bond pattern. This procedure led to the following assignment: His34 and His107 were protonated on both nitrogen atoms, whereas His31 was protonated on Nε2 only and His152 was protonated on Nδ1 only. The cysteine residues coordinating the Fe/S clusters were assumed to be deprotonated. The [4Fe–4S] cluster, S-adenosylmethionine and the dethiobiotin molecule found in the crystal structure were all included in the calculations. The total charge of the simulated system was −8 (neutral arginine) or −7 (protonated arginine). The positions of the atoms added were optimised by a 90-ps simulated-annealing molecular dynamics simulation, followed by 10,000 steps of conjugate gradient energy minimisation. All bond lengths involving hydrogen atoms were constrained by the SHAKE algorithm [35]. The water solvent was described explicitly using the TIP3P model [36]. The temperature was kept constant at 300 K using the Berendsen weak-coupling algorithm [37] with a time constant of 1 ps. The molecular dynamics time step was 2 fs. The non-bonded cut-off was 15 Å and the pair list was updated every 50 fs. In the QM/MM calculations, an infinite cut-off was used instead.
The entire system was then divided into three subsystems. System 1 contained the [2Fe–2S] cluster and the relevant atoms of the four coordinating amino acids (Cys97, Cys128, Cys188 and Arg260) and was treated with QM methods. The side chains were included as far as Cβ for the cysteine residues (replacing Cα by a hydrogen atom) and as far as Cδ for the arginine residue (replacing Cγ by a hydrogen atom). Thus, it consisted of [(CH3S)3(CH3NHCH(NH)NH2)Fe2S2]− for the calculations with neutral arginine and [(CH3S)3(CH3NHCH(NH2)NH2)Fe2S2] for the calculations with protonated arginine. System 2 included all residues with any atom within 6 Å of any atom in system 1 and was relaxed with MM methods. System 3 included the remaining protein atoms as well as the water molecules and was kept fixed at the crystallographic coordinates.
As both iron atoms of the oxidised [2Fe–2S] cluster are in the FeIII high-spin state (S = 5/2), two spin states are possible (the ferromagnetically coupled, F, state, S = 5, and the antiferromagnetically coupled, AF, state, S = 0). The AF state always had a lower energy than the F state and it is also the one observed experimentally. Therefore, all results presented are AF energies. To ensure that the QM/MM energy differences are stable, the calculations were in general run forth and back between the relevant states until the energies were stable within 4 kJ/mol.
Similar calculations were also performed on one-electron-reduced clusters, i.e. clusters containing one FeII and one FeIII ion (net charge of the QM system −1 or −2, depending on the protonation of the arginine model), on two-electron-reduced clusters (net charge −2 or −3) and on clusters with one of the bridging sulphur atoms removed (the one closest to dethiobiotin; net charge 0 or −1, so this is equivalent to removing an S2− ion and reducing both iron ions to FeII), in all cases in the AF (S = 1/2 or S = 0) state.
For convenience, all geometry optimisations discussed were started from an initial optimisation with structure 2 (Structure 1). To verify that this is acceptable, we tested to what extent the optimised geometry depends on the starting geometry. In addition, the influence of the spin and oxidation states of the [2Fe–2S] cluster on structural properties was investigated. These explorative calculations were performed in a vacuum (i.e. system 1 only), starting from the crystal geometry. Geometry optimisations for structure 2 in different oxidation states (FeII/FeII, FeII/FeIII or FeIII/FeIII) and spin states (AF or F) showed that the final geometries of the intact clusters, especially the Fe···Fe distances and the orientation of the arginine residue, do not depend on the starting geometries. The oxidised AF and F states were also tested for the other protonation states (structures 1, 3 and 4), with similar results. In all calculations, the AF state was energetically favoured (by 18–118 kJ/mol). Short Fe···Fe distances were found in all cases, although they were slightly longer for the F states (AF: 2.57–2.65 Å, F: 2.42–2.89 Å); no additional electronic states with larger Fe···Fe distances were detected. To verify this observation, the Fe···Fe distance was fixed to values between 2.5 and 3.5 Å (structure 2, oxidised, AF state) and the rest of the geometry was optimised. Only one energy minimum was found, at approximately 2.6 Å (61 kJ/mol more stable than the distance in the crystal structure) and no evidence for a second minimum close to the crystal structure distance was found.
Structure 1.
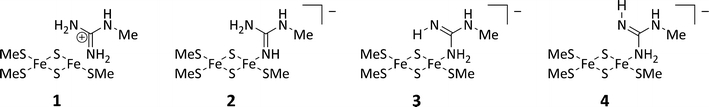
Conceivable coordination modes for arginine in the protonated (1) or neutral (2–4) state as represented in the quantum mechanical system
Similar explorative calculations were performed with the hybrid B3LYP functional [38, 39] (to examine the effect of another functional with exact exchange), giving similar results [E AF − E F = −20 to −55 kJ/mol, d(Fe···Fe) = 2.55–2.72 (AF) and 2.70–2.96 Å (F) and an energy minimum at 2.8 Å, 36 kJ/mol lower than the crystal structure].
Quadrupole splittings were calculated according to
 |
2 |
where Q = 0.16 b (1.6 × 10−29 m2) for 57Fe, η = (V xx − V yy)/V zz, with |V xx| < |V yy| < |V zz|, and 1 mm/s is equivalent to 4.8075 × 10−18 eV.
Quantum-refinement calculations
We also performed a set of quantum-refinement calculations, using the software program ComQum-X [40]. They can be seen as QM/MM calculations in which the structures are restrained towards crystallographic raw data. In ComQum-X, the MM program is replaced by the crystallographic refinement program Crystallography & NMR System (CNS) [41]. In crystallographic refinement, the coordinates, B factors, occupancies, etc. are improved by optimising the fit of the observed and calculated structure-factor amplitudes, typically estimated by the residual disagreement, the R factor. Because of the limited resolution normally obtained with X-ray diffraction of biomolecules, a MM force field is used to supplement the data for the whole protein [42]. This force field ensures that the bond lengths and angles make chemical sense. In ComQum-X, this force field is replaced by more accurate QM calculations for a small, but interesting, part of the protein (system 1), in a manner completely analogous to the use of quantum mechanics in QM/MM calculations. The junctions are handled in the same way as in ComQum.
Thus, the ComQum-X refinement takes the form of a minimisation using an energy function of the form
 |
3 |
Here, E MM1 and E MM123 have the same meaning as in Eq. 1, whereas E QM1 is the energy of the QM system, without any point-charge model of the surroundings. E X-ray is a penalty function, describing how well the model agrees with the experimental X-ray data. We have used the default maximum likelihood refinement target using amplitudes (MLF) in CNS [43]. w A is a weight factor, which is necessary because E X-ray is in arbitrary units whereas the other terms are in energy units. It should be emphasised that the w A factor is nothing special for quantum refinement. On the contrary, it also has to be set in standard crystallographic refinement (which is obtained from Eq. 3 with E QM1 = E MM1 = 0), although it is rarely discussed. The default behaviour of CNS is to determine w A so that the E X-ray and E MM123 forces have the same magnitude during a short molecular dynamics simulation [44], i.e. that the crystallographic raw data and the MM force field have a similar influence on the structure. We tested nine different values for the w A factor between 0 and 30. Unfortunately, we encountered convergence problems if we used the default value of w A (4.87) for some of the structures (because the crystallographically preferred structure of the [2Fe–2S] cluster is so poor at this low resolution that it becomes incompatible with the QM calculations). Therefore, we present results only for the largest value of the w A factor that gave converged structures for all models, viz. w A = 1. The results are qualitatively the same if other values are used, regarding the preferred model and coordination mode of the arginine ligand.
Following crystallographic custom, no hydrogen atoms were included in the MM region of the ComQum-X calculations, because hydrogen atoms are not discernible in the crystal structure. Therefore, polarisation of the quantum system by the surrounding protein is not included in ComQum-X.
Finally, it should be noted that the MM force field used in CNS (protein_rep.param, dna-rna_rep.param, water.param and ion.param) is based on a statistical survey of crystal structures [45], rather than the energy-based force field in Amber and in the QM calculations. Therefore, the CNS energy has to be weighted by a factor of 1/3 to be comparable with the QM and Amber MM energies [40].
The quantum-refinement calculations were based on the same crystal structure as the QM/MM calculations (but both subunits were considered) [11] and the corresponding structure factors were downloaded from the Protein Data Bank. Calculations were performed with the same QM system as with QM/MM [(CH3S)3(CH3NHCH(NH1–2)NH2)Fe2S2], as well as a QM system enlarged with a CH3OH model of Ser43 and a CH3NHCH(NH2)2 model of Arg95 (for both the intact oxidised cluster, as well as the one-electron-reduced cluster without one of the bridging sulphide ions). The QM method and basis sets were the same as in the QM/MM calculations.
Results and discussion
QM/MM calculations
Because of the importance of interactions of the [2Fe–2S] cluster and its four ligands with the surrounding protein and solvent, a QM/MM approach including the protein environment is the theoretical method of choice.
Four different structures were studied depending on the protonation state of Arg260, as is illustrated in Structure 1. In the first (1), Arg260 is protonated, and therefore positively charged. In the other three, one of the terminal NH2 groups of Arg260 is deprotonated. The three structures differ in whether the deprotonated NH group (2) or the protonated NH2 group coordinates to iron. In the latter case, the non-coordinating NH group can have the hydrogen atom pointing either towards the cluster (3) or away from the cluster (4).
Optimised structures obtained from these calculations are shown in Fig. 2. At first glance, the geometry is quite similar in all cases. The Fe–N distances (Table 1) are in a range that agrees with the crystal structure (2.4 Å) considering its low resolution (3.4 Å) in all three structures with neutral arginine (2 2.05 Å, 3 2.25 Å, 4 2.27 Å), as well as in the protonated structure (1 2.45 Å). Judging from these distances, the Fe–N bond is strongest in 2, which could be expected as the NH group has an sp 2-like, nucleophilic lone pair, whereas the p-like lone pair of the NH2 group is involved in π interactions within the guanidine group. The Fe···N distance in 1 is too long to assign an Fe–N bond in the protonated case. Nevertheless, the guanidinium group is still in close proximity to the [2Fe–2S] cluster.
Fig. 2.
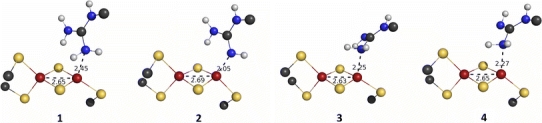
Optimised structures from the quantum mechanical/molecular mechanical calculations
Table 1.
Structural parameters from the crystal structure and from the quantum mechanical/molecular mechanical calculations
| Fe–N (Å) | Fe···Fe (Å) | |
|---|---|---|
| Crystal structure | 2.40, 2.35 | 3.28, 3.24 |
| 1 | 2.45 | 2.65 |
| 2 | 2.05 | 2.69 |
| 3 | 2.25 | 2.63 |
| 4 | 2.27 | 2.65 |
| 1 + e − | 3.26 | 2.59 |
| 2 + e − | 2.11 | 2.66 |
| 3 + e − | 3.26 | 2.57 |
| 4 + e − | 3.42 | 2.58 |
| 1 + 2e − | 3.19 | 2.62 |
| 2 + 2e − | 2.18 | 2.68 |
| 3 + 2e − | 3.24 | 2.62 |
| 4 + 2e − | 3.23 | 2.60 |
| 1 − S | 4.15 | 2.54 |
| 2 − S | 2.01 | 3.08 |
| 3 − S | 2.14 | 2.79 |
| 4 − S | 2.15 | 2.74 |
Recently, Di Costanzo et al. [13] performed a survey of metal–guanidine interactions in the Cambridge structural database. They found 150 such interactions in 45 different structures, but all except four of these involved a diguanidine moiety chelating a single metal, which is quite different from an arginine–metal coordination. They obtained metal–nitrogen distances of 1.84–2.08 Å (average 1.91 ± 0.06 Å), but none of the complexes involved iron. However, it is clear that only structure 2 gives an Fe–N distance that is similar to what is found in small inorganic metal–guanidine complexes. The survey of Di Costanzo et al. also involved the only three protein crystal structures with metal–guanidine coordination, viz. BioB, an H67R carbonic anhydrase I mutant [46] and an arginase l-arginine complex [47]. The latter structure shows a Mn–N distance of 2.5 Å, whereas the Zn–N distance in the carbonic anhydrase mutant is 2.1 Å, i.e., the only protein structure that has a metal–guanidine bond length similar to the bond length of the small inorganic complexes. Di Costanzo et al. assumed that the BioB structure involved a deprotonated (neutral) arginine and did not consider any other possibility.
The Fe···Fe distance is quite independent of the coordination as well as the protonation state of the arginine group (2.63–2.69 Å) and is in perfect agreement with the Fe···Fe distances of other [2Fe–2S] clusters that have been structurally characterised. None of the calculated structures reproduce the long Fe···Fe distance found in the BioB crystal structure (3.24–3.28 Å). As metal sites are often reduced during X-ray experiments, one- or two-electron-reduced clusters containing FeII ions were also optimised, but they do not show any increased Fe···Fe distance. The main geometric change upon reduction is the dissociation of the NH2 group (1, 3, 4). However, when one of the bridging sulphur atoms was removed from the cluster (and the two iron atoms thereby were reduced), the Fe···Fe distance increased in the structures with neutral arginine (2 3.08 Å, 3 2.79 Å, 4 2.74 Å). In this case, only the protonated arginine (1) dissociates from the iron atom. It is possible that one of the sulphur atoms has been removed from the cluster (and inserted into dethiobiotin) in the crystal since the enzyme reaction is started by reduction of the [4Fe–4S] cluster, which could happen owing to radiation damage during the measurement. It is not possible to decide whether or not there is a sulphur atom in the (dethio)biotin molecule in the crystal by examination of the electron density map owing to the low resolution.
The non-coordinating NH/NH2 group acts as a hydrogen-bond donor in the structures with a proton pointing towards the bridging sulphide (1, 2, 3). Additional hydrogen bonds can be found between Arg260 and four surrounding amino acids (Ser43, Ser218, Ser283 and Arg95). Except for Ser283, which accepts a hydrogen bond from the non-terminal NHε group in all cases, these residues seem to be quite flexible. Thus, the positively charged Arg95 can act as a hydrogen-bond donor towards the bridging sulphide of the [2Fe–2S] cluster (1, 2) or the NH group of Arg260 (3, 4). Ser218 acts as an acceptor towards the NH2 group of Arg95 in all structures and can in addition donate a hydrogen bond towards the bridging sulphide (2, 3), whereas Ser43 can accept hydrogen bonds from the non-coordinating NH2 group of Arg260 (1, 2). In all cases, the arginine acts as a monodentate ligand; no evidence for secondary bonding interactions was found.
Of the three structures with a neutral arginine, 2 has the lowest energy, 80 and 88 kJ/mol lower than that of structures 3 and 4, respectively. This shows that coordination by the more nucleophilic NH group is preferred before NH2 coordination. The energy of 1 cannot be compared directly because of the additional proton in the system. Comparing the protonated structure (1) with the best neutral structure (2), one can only find minor differences, besides the difference in the Fe–N distance. The other relevant distances are similar, as are the hydrogen bonds close to the [2Fe–2S] cluster.
Mössbauer parameters of the [2Fe–2S] cluster in BioB have been measured [48–50], showing a single quadrupole doublet with a quadrupole splitting of ΔE Q = 0.51–0.53 mm/s. This is quite unexpected, because for a cluster containing two iron atoms with different coordination environments, two doublets would be expected. Quadrupole splittings (ΔE Q) were calculated from the electric field gradients at the position of the iron atoms for the optimised geometries of structures 1–4 and are presented in Table 2.
Table 2.
| ΔE Q (S4) | ΔE Q (S3N) | |
|---|---|---|
| Experimental | 0.53 | 0.53 |
| 1 | 0.33 | 1.27 |
| 2 | 0.33 | 0.59 |
| 3 | 0.45 | 1.14 |
| 4 | 0.43 | 1.23 |
Although the quadrupole splittings calculated for the sulphur-coordinated iron atom are roughly the same in all four cases (0.33–0.45 mm/s), the other iron atom exhibits very different values depending on the exact coordination mode. Although ΔE Q is quite large with the NH2 coordinating (1 1.27, 3 1.14, 4 1.23 mm/s), it is relatively small in case of NH coordination (2 0.59 mm/s). Taking into account that calculated quadrupole splittings are usually too low in similar cases [18] and that the accuracy of calculated ΔE Q (i.e. the amount by which they are lower than experimental values) seems to depended on the coordination [18, 27], the neutral state with the NH group coordinating (2) fits the experimental data best. As biological samples usually exhibit weak Mössbauer signals owing to their low iron content, especially when another Fe/S cluster is present, it seems reasonable that the experimentally found doublet is the sum of two doublets with similar quadrupole splittings.
Thus, we can conclude that QM/MM calculations predict a neutral arginine with NH coordination (structure 2). Nevertheless, the reason for the experimentally found Fe···Fe distance as well as the significance of the unusual arginine ligand remains elusive.
Quantum-refinement calculations
We also studied the enzyme by quantum refinement, which is standard crystallographic refinement, using the original experimental structure factors, but replacing the MM force field (which is used to supplement the crystallographic raw data and give accurate bond lengths and angles) for the active site by more accurate QM calculations. This will allow us to study what realistic structures of the [2Fe–2S] site actually fit into the electron density. In particular, we will be able to test what protonation state (structures 1–4) fits the crystallographic raw data best. Two sizes of the QM system were tested (with or without models of Ser43 and Arg95), as well as models of both the oxidised state with an intact cluster and the two-electron-reduced state with either an intact cluster or one of the bridging sulphur atoms removed.
The results are summarised in Table 3. It can be seen that all re-refined structures of the intact cluster give Fe···Fe distances (2.58–2.77 Å) that are appreciably shorter than in the crystal structure and therefore similar to those obtained in the QM/MM calculations. In the structures without one of the sulphur atoms, the Fe···Fe distance is longer (2.85–2.99 Å), but not as long as in the crystal structure. However, it should be noted that both the R free and residue (real-space) R factors are slightly lower for the original crystal structure than for any of the re-refined structures. This indicates a misfit between the crystal structure and the QM systems tested, which may indicate that we still have not yet tested the correct QM system or that the crystal structure is a mixture of several different structures, which is expected if the metal site is reduced during data collection.
Table 3.
Results of the quantum-refinement calculations with w A = 0.1
| Fe…Fe | Distance to Fe1 (Å) | Distance to Fe2 (Å) | R free | Residue R | ΔE QM1(kJ/mol) | Δr 1(Å) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Å | N | S1 | S2 | S3 | S2 | S3 | S4 | S5 | |||||
| Small QM system, oxidised state, intact cluster | |||||||||||||
| 1 | 2.58 | 3.29 | 2.24 | 2.21 | 2.19 | 2.25 | 2.22 | 2.25 | 2.25 | 0.3042 | 0.236 | 60.8 | 0.27 |
| 2 | 2.69 | 2.09 | 2.32 | 2.23 | 2.21 | 2.26 | 2.22 | 2.33 | 2.31 | 0.3028 | 0.197 | 41.6 | 0.13 |
| 3 | 2.69 | 2.28 | 2.30 | 2.20 | 2.21 | 2.24 | 2.23 | 2.33 | 2.30 | 0.3032 | 0.212 | 95.3 | 0.29 |
| 4 | 2.69 | 2.37 | 2.31 | 2.18 | 2.21 | 2.22 | 2.24 | 2.33 | 2.30 | 0.3035 | 0.213 | 127.1 | 0.36 |
| Large QM system, oxidised state, intact cluster | |||||||||||||
| 1 | 2.60 | 3.34 | 2.22 | 2.22 | 2.18 | 2.29 | 2.21 | 2.23 | 2.23 | 0.3044 | 0.243 | 178.5 | 0.91 |
| 2 | 2.74 | 2.10 | 2.29 | 2.25 | 2.20 | 2.31 | 2.20 | 2.31 | 2.29 | 0.3028 | 0.199 | 89.8 | 0.20 |
| 3 | 2.72 | 2.33 | 2.26 | 2.20 | 2.21 | 2.27 | 2.23 | 2.31 | 2.28 | 0.3031 | 0.228 | 130.9 | 0.31 |
| 4 | 2.67 | 3.18 | 2.24 | 2.19 | 2.17 | 2.26 | 2.23 | 2.28 | 2.29 | 0.3048 | 0.285 | 133.0 | 0.87 |
| Large QM system, reduced state, intact cluster | |||||||||||||
| 1 | 2.77 | 3.55 | 2.27 | 2.36 | 2.17 | 2.49 | 2.31 | 2.28 | 2.35 | 0.3042 | 0.250 | 125.0 | 0.65 |
| 2 | 2.67 | 2.09 | 2.31 | 2.27 | 2.24 | 2.34 | 2.28 | 2.38 | 2.34 | 0.3029 | 0.206 | 75.2 | 0.11 |
| 3 | 2.77 | 3.08 | 2.29 | 2.23 | 2.21 | 2.32 | 2.30 | 2.33 | 2.36 | 0.3043 | 0.265 | 204.9 | 0.96 |
| 4 | 2.66 | 3.25 | 2.25 | 2.19 | 2.33 | 2.36 | 2.33 | 2.33 | 2.40 | 0.3038 | 0.276 | 191.1 | 1.03 |
| Large QM system, without one S atom | |||||||||||||
| 1 | 2.88 | 2.41 | 2.23 | 2.22 | 2.27 | 2.27 | 2.24 | 0.3034 | 0.212 | 192.4 | 0.26 | ||
| 2 | 2.85 | 2.06 | 2.29 | 2.22 | 2.24 | 2.30 | 2.27 | 0.3040 | 0.196 | 161.6 | 0.20 | ||
| 3 | 2.99 | 2.24 | 2.24 | 2.20 | 2.25 | 2.30 | 2.26 | 0.3030 | 0.193 | 195.3 | 0.33 | ||
| 4 | 2.98 | 2.20 | 2.25 | 2.18 | 2.25 | 2.32 | 2.27 | 0.3030 | 0.196 | 156.4 | 0.14 | ||
| Crystal structure re-refined without quantum mechanics | |||||||||||||
| i | 2.99 | 2.33 | 2.37 | 2.19 | 2.10 | 2.11 | 2.16 | 2.19 | 2.25 | 0.3004 | 0.138 | 1.18 | |
| ii | 2.97 | 2.33 | 2.38 | 2.19 | 2.13 | 2.12 | 2.17 | 2.19 | 2.27 | 0.3004 | 0.140 | 1.11 | |
| Crystal structure | |||||||||||||
| A | 3.24 | 2.35 | 2.32 | 2.23 | 2.22 | 2.22 | 2.23 | 2.28 | 2.30 | 0.3003 | 0.140 | 244.4 | 1.00 |
| B | 3.28 | 2.40 | 2.30 | 2.26 | 2.23 | 2.22 | 2.23 | 2.25 | 2.27 | 1.17 | |||
Distances between the iron ions and the ligands (Å) are given, as well as the R free, residue R factors, ΔE QM1 and Δr QM, which are the differences in the energy and iron–ligand distances of the QM system optimised in the crystal and in a vacuum. Four sets of calculations are presented: with the small QM system or with the QM system enlarged by Ser43 and Arg95, for the oxidised state (FeIII2) or for the two-electron-reduced state (FeII2), and for the intact [2Fe–2S] cluster or for the cluster with one sulphide ion removed. In addition, the data from the crystal structure (both subunits) are presented, as well as a standard crystallographic re-refinement of the [2Fe–2S] cluster with two different treatments of the Fe–S interactions (see the text)
The structure of the cluster also depends on the details of the refinement protocol. Unfortunately, the original publication [11] does not provide such details and we have not been able to obtain them from the authors. Therefore, we tested re-refining the structure of the [2Fe–2S] cluster with standard crystallography (i.e. with E QM1 = E MM1 = 0 in Eq. 3) and with different treatments of the Fe–S interactions in the MM force field (i.e. in the E MM123 term). As can be seen in Table 3, the results are insensitive to whether Fe–S bonds are included with zeroed force constants (the preferred method to allow the site to be determined entirely by the experimental data; protocol i in Table 3) or if no Fe–S bonds are defined (so that there are van der Waals interactions between all iron and sulphur ions; protocol ii in Table 3). This indicates that the default w A factor is so large that the MM force field has only a minor influence on the structure of the [2Fe–2S] site. However, it can also be seen that the structure of the re-refined [2Fe–2S] site is quite different from the original crystal structure, showing that details in the refinement still differ. In particular, the Fe···Fe distance in our re-refined structure (2.97–2.99 Å) is appreciably shorter than in the original crystal structure (3.24 Å). The re-refined results are similar to the quantum-refined results with a cluster without one sulphur atom.
Among the four QM systems tested (models 1–4), it is clear that the one with a deprotonated Arg260 and the NH group coordinated to iron (model 2) fits the crystallographic data best for all structures with an intact cluster: it has the lowest R free and residue (real-space) R factors, and it also gives the lowest strain energy (ΔE QM1, i.e. the energy difference of the QM system when optimised in the crystal or in a vacuum) as well as the lowest difference in geometry when optimised in the crystal or in a vacuum (Δr 1 in Table 3). Thus, all these four criteria point out the same structure as the best one, showing that the results are conclusive. In particular, it is clear that model 2 fits the crystallographic data appreciably better than the structure with a protonated Arg260 (model 1). In fact, Arg260 dissociates from iron in the latter model, giving Fe–N distances of 3.29–3.55 Å, when re-refined with w A < 1. However, when w A ≥ 1, the Fe–N distance is shortened to 2.47–2.26 Å, showing that the crystal structure prefers shorter values. The two deprotonated models coordinating through the NH2 group (models 3 and 4) give intermediate fits to the crystal structure. Figure 3 shows that the quantum-refined structure of model 2 fits the electron density equally well as the original crystal structure, and it also shows the rather poorly defined electron density at this low resolution.
Fig. 3.
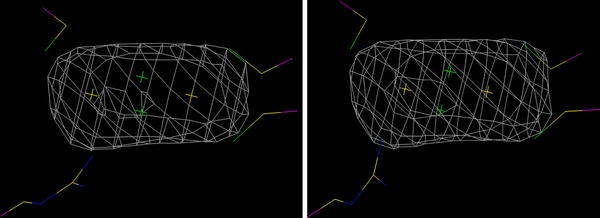
Electron-density omit 2f o – f c maps of the [2Fe–2S] cluster in the original crystal structure (left) and the quantum-refined model 2 (right) at the 2.0σ level
For the structure without one of the sulphur atoms, the results are somewhat different. Then, the various quality criteria give different results: model 3 gives the lowest R free and residue R factors, whereas model 4 gives the smallest strain energy and difference in geometry. This indicates that this is not the correct model of the protein. This is also supported by the higher R free factor (0.3030), compared with the best values obtained for the oxidised models (0.3028). However, the difference is not very large, indicating that the crystal structure might actually be a mixture of oxidised and reduced structures.
Conclusions
We have studied the structure of BioB with both quantum-refinement and QM/MM methods. This gave us the opportunity to interpret the crystal structure as much as its low resolution (3.4 Å) allows us. Several interesting results were obtained. First, it is quite clear that the Arg260 ligand is deprotonated in the crystal structure, because such structures fit the crystallographic raw data best. Likewise, both the QM/MM energies and the quantum refinement strongly indicate that it is more favourable for the deprotonated Arg260 to coordinate to iron via the deprotonated NH group, rather than by the NH2 group, even if hydrogen bonds with the surrounding residues are considered. These conclusions are supported by calculated Mössbauer parameters which also fit the experimental data best for this coordination mode. Finally, it also seems clear that the Fe···Fe distance in the [2Fe–2S] cluster is not as long as the initial report on the crystal structure indicated [11]. Instead, it is most likely similar to what is found in all normal [2Fe–2S] clusters, i.e. approximately 2.7 Å. The reason for the long bond in the crystal structure may either be the low resolution or that the structure is a mixture of different states of the [2Fe–2S] cluster, e.g. caused by a successive reduction of the cluster during data collection. Clearly, more accurate structural data are needed, as well as further theoretical investigations of the reaction intermediates, which should help to understand this fascinating enzyme.
Acknowledgements
Financial support by the Fonds der Chemischen Industrie (Kekulé fellowship for M.G.G.F.), the Deutsche Forschungsgemeinschaft and the Swedish Research Council (International Research Training Group GRK 1422 “Metal Sites in Biomolecules: Structures, Regulation and Mechanisms”; see http://www.biometals.eu) is gratefully acknowledged. The investigation was also supported by the computer resources of Lunarc at Lund University.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Franc Meyer, Email: franc.meyer@chemie.uni-goettingen.de.
Ulf Ryde, Email: ulf.ryde@teokem.lu.se.
References
- 1.Beinert H, Holm RH, Münck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DC, Dean DR, Smith AD, Johnson MK. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 3.Gibson KJ, Pelletier DA, Turner IM., Sr Biochem Biophys Res Commun. 1999;254:632–635. doi: 10.1006/bbrc.1998.9991. [DOI] [PubMed] [Google Scholar]
- 4.Booker SJ, Cicchillo RM, Grove TL. Curr Opin Chem Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Layer G, Heinz DW, Jahn D, Schubert W-D. Curr Opin Chem Biol. 2004;8:468–476. doi: 10.1016/j.cbpa.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Tse Sum Bui B, Florentin D, Fournier F, Ploux O, Méjean A, Marquet A. FEBS Lett. 1998;440:226–230. doi: 10.1016/S0014-5793(98)01464-1. [DOI] [PubMed] [Google Scholar]
- 7.Cicchillo RM, Booker SJ. J Am Chem Soc. 2005;127:2860–2861. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
- 8.Hernández HL, Pierrel F, Elleingand E, García-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- 9.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. Proc Natl Acad Sci USA. 2008;105:1826–1831. doi: 10.1073/pnas.0708608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ugulava NB, Sacanell CJ, Jarrett JT. Biochemistry. 2001;40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrar CE, Jarrett JT. Biochemistry. 2009;48:2448–2458. doi: 10.1021/bi8022569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Costanzo L, Flores LV, Jr, Christianson DW. Proteins. 2006;65:637–642. doi: 10.1002/prot.21127. [DOI] [PubMed] [Google Scholar]
- 14.Broach RB, Jarrett JT. Biochemistry. 2006;45:14166–14174. doi: 10.1021/bi061576p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullmann GM, Knapp E-W. Eur Biophys J. 1999;28:533–551. doi: 10.1007/s002490050236. [DOI] [PubMed] [Google Scholar]
- 16.Taylor AM, Farrar CE, Jarrett JT. Biochemistry. 2008;47:9309–9317. doi: 10.1021/bi801035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarrett JT. Arch Biochem Biophys. 2005;433:312–321. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Ballmann J, Dechert S, Bill E, Ryde U, Meyer F. Inorg Chem. 2008;47:1586–1596. doi: 10.1021/ic702095a. [DOI] [PubMed] [Google Scholar]
- 19.Venkateswara Rao P, Holm RH. Chem Rev. 2004;104:527–559. doi: 10.1021/cr020615+. [DOI] [PubMed] [Google Scholar]
- 20.Ryde U. J Comput Aided Mol Des. 1996;10:153–164. doi: 10.1007/BF00402823. [DOI] [PubMed] [Google Scholar]
- 21.Ryde U, Olsson MHM. Int J Quantum Chem. 2001;81:335–347. doi: 10.1002/1097-461X(2001)81:5<335::AID-QUA1003>3.0.CO;2-Q. [DOI] [Google Scholar]
- 22.Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C. Chem Phys Lett. 1989;162:165–169. doi: 10.1016/0009-2614(89)85118-8. [DOI] [Google Scholar]
- 23.Case DA, Darden TA, Cheatham TE, III, Simmerling TL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
- 24.Becke AD. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/PhysRevA.38.3098. [DOI] [PubMed] [Google Scholar]
- 25.Perdew JP. Phys Rev B. 1986;33:8822–8824. doi: 10.1103/PhysRevB.33.8822. [DOI] [PubMed] [Google Scholar]
- 26.Weigend F, Ahlrichs R. Phys Chem Chem Phys. 2005;7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 27.Ballmann J, Albers A, Demeshko S, Dechert S, Bill E, Bothe E, Ryde U, Meyer F. Angew Chem. 2008;120:9680–9684. doi: 10.1002/ange.200803418. [DOI] [Google Scholar]
- 28.Cornell WD, Cieplak PI, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. doi: 10.1021/ja00124a002. [DOI] [Google Scholar]
- 29.Wang J, Cieplak P, Kollman PA. J Comput Chem. 2000;21:1049–1074. doi: 10.1002/1096-987X(200009)21:12<1049::AID-JCC3>3.0.CO;2-F. [DOI] [Google Scholar]
- 30.Rod TH, Ryde U. J Chem Theory Comput. 2005;1:1240–1251. doi: 10.1021/ct0501102. [DOI] [PubMed] [Google Scholar]
- 31.Söderhjelm P, Ryde U. J Mol Struct Theochem. 2006;770:199–219. doi: 10.1016/j.theochem.2006.06.008. [DOI] [Google Scholar]
- 32.Weis A, Katebzadeh K, Söderhjelm P, Nilsson I, Ryde U. J Med Chem. 2006;49:6596–6606. doi: 10.1021/jm0608210. [DOI] [PubMed] [Google Scholar]
- 33.Svensson M, Humbel S, Froese RDJ, Matsubara T, Sieber S, Morokuma K. J Phys Chem. 1996;100:19357–19363. doi: 10.1021/jp962071j. [DOI] [Google Scholar]
- 34.Li H, Robertson AD, Jensen JH. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 35.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura J, Klein ML. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 37.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 38.Becke AD. J Chem Phys. 1993;98:5648–5652. doi: 10.1063/1.464913. [DOI] [Google Scholar]
- 39.Stephens PJ, Devlin FJ, Frisch MJ, Chabalowski CF. J Phys Chem. 1994;98:11623–11627. doi: 10.1021/j100096a001. [DOI] [Google Scholar]
- 40.Ryde U, Olsen L, Nilsson K. J Comput Chem. 2002;23:1058–1070. doi: 10.1002/jcc.10093. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewsk JI, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (2000) Crystallography & NMR System CNS, version 1.0. Yale University, New Haven [DOI] [PubMed]
- 42.Kleywegt GJ, Jones TA. Methods Enzymol. 1997;277:208–230. doi: 10.1016/S0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- 43.Pannu NS, Read RJ. Acta Crystallogr A. 1996;52:659–668. doi: 10.1107/S0108767396004370. [DOI] [Google Scholar]
- 44.Brünger AT, Rice LM. Methods Enzymol. 1997;277:243–269. doi: 10.1016/S0076-6879(97)77015-0. [DOI] [PubMed] [Google Scholar]
- 45.Engh RA, Huber R. Acta Crystallogr A. 1991;47:392–400. doi: 10.1107/S0108767391001071. [DOI] [Google Scholar]
- 46.Ferraroni M, Tilli S, Briganti F, Chegwidden WR, Supuran CT, Wiebauer KE, Tashian RE, Scozzofava A. Biochemistry. 2002;41:6237–6244. doi: 10.1021/bi0120446. [DOI] [PubMed] [Google Scholar]
- 47.Bewley MC, Jeffrey PD, Patchett ML, Kanyo ZF, Baker EN. Structure. 1999;7:435–448. doi: 10.1016/S0969-2126(99)80056-2. [DOI] [PubMed] [Google Scholar]
- 48.Cosper MM, Jameson GNL, Hernández HL, Krebs C, Huynh BH, Johnson MK. Biochemistry. 2004;43:2007–2021. doi: 10.1021/bi0356653. [DOI] [PubMed] [Google Scholar]
- 49.Tse Sum Bui B, Benda R, Schünemann V, Florentin D, Trautwein AX, Marquet A. Biochemistry. 2003;42:8791–8798. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- 50.Tse Sum Bui B, Marquet A, Benda R, Trautwein AX. FEBS Lett. 1999;459:411–414. doi: 10.1016/S0014-5793(99)01300-9. [DOI] [PubMed] [Google Scholar]


