Abstract
Each Sertoli cell can support a finite number of developing germ cells. During development of the testis, the cessation of Sertoli cell proliferation and the onset of differentiation determine the final number of Sertoli cells and, hence, the number of sperm that can be produced. We hypothesize that the transition from proliferation to differentiation is facilitated by E-box transcription factors that induce the expression of differentiation-promoting genes. The relative activities of E-box proteins were studied in primary Sertoli cells isolated from 5-, 11-, and 20-day-old rats, representing proliferating, differentiating, and differentiated cells, respectively. E-box DNA-binding activity is almost undetectable 5 days after birth but peaks with initiation of differentiation 11 days after birth and remains elevated. Upstream stimulatory factors 1 and 2 (USF1 and USF2) were found to be the predominant E-box proteins present within DNA-protein complexes formed after incubating E-box-containing probes with nuclear extracts from developing Sertoli cells. The known potentiator of Sertoli cell differentiation, thyroxine, increases USF DNA-binding activity in Sertoli cells before differentiation (5-day-old Sertoli cells) but not after differentiation is initiated (11- and 20-day-old Sertoli cells). The developmental-specific increase in USF1 and USF2 DNA-binding activity may facilitate the switch from proliferation to differentiation and, thus, determine the ultimate number of Sertoli cells present within the testes and the upper limit of fertility.
Keywords: bHLH proteins, E-box factors, follicle-stimulating hormone, follicle-stimulating hormone receptor, gene regulation, ID proteins, spermatogenesis, testis
USF1 and USF2 transcription factor DNA-binding activity increases during the differentiation of rat Sertoli cells.
INTRODUCTION
Sertoli cells are required to support male germ cell development (spermatogenesis) in the mammalian testis. Sertoli cells are capable of supporting only a finite number of germ cells [1]. Therefore, the final number of Sertoli cells sets the upper limit for testicular sperm production and determines the level of male fertility [2, 3]. Sertoli cells do not divide after puberty, and apoptosis of these cells is rare; therefore, the number of Sertoli cells is primarily determined by the expansion of Sertoli cells before the completion of puberty [4, 5]. Sertoli cell proliferation and the decision to differentiate are regulated by a number of hormonal cues, including thyroid hormone, retinoic acid, activin, FSH, and testosterone [6]. The regulation of Sertoli cell development is also dependent on cell-cell interactions, signals from the extracellular matrix, and factors from developing germ cells [6]. Although numerous factors regulating Sertoli cell development have been identified, little is known about the molecular mechanisms in Sertoli cells that halt cell division and trigger differentiation.
During the period in which proliferation decreases, the Sertoli cell undergoes a developmental process that results in a fully differentiated, nonproliferating cell that is capable of supporting spermatogenesis. The decrease in Sertoli cell proliferation during the differentiation period has been well documented in rats [4, 7, 8] and mice [9]. The percentage of thymidine-labeled Sertoli cells is 33% at birth and 32% at 5 days after birth but decreases to 18% and 7% at 10 and 11 days, respectively. By 16 days after birth, labeled Sertoli cells can no longer be detected [10].
The Sertoli cell differentiation process includes morphological changes, the induction of secreted proteins that are required by germ cells, and the formation of specialized tight junctions between Sertoli cells that establish the blood-testis barrier [6]. Potential mediators of Sertoli cell differentiation signals include members of the basic helix-loop-helix (bHLH) family of transcription factors. The bHLH transcription factors previously shown to be expressed in Sertoli cells include scleraxis (Scx), upstream stimulatory factors 1 and 2 (Usf1 and Usf2), REBα isoform (the rat homologue of the human HeLa E-box-binding protein, HEB [Tcf12]), and E47 but not E12 proteins that are derived from the Tcfe2a gene (formerly known as E2a) [11–13]. The bHLH transcription factors bind to specific DNA sequence motifs known as E-boxes (CANNTG), closely related N-boxes (CACNAG), and Ets-binding sites (GGAA/T) [14–16]. In general, two categories of proteins bind to E-boxes based on the sequence of the binding motif. E-boxes having the sequence CAGCTG bind E47, E12, and REBα, whereas E-boxes having the sequence CACGTG bind USF proteins and MYC [17]. E-box motifs are known to regulate the promoters of a number of Sertoli cell-specific genes that are markers for differentiated status, including transferrin (Trf), steroidogenic factor 1 (Nr5a1), and the follicle-stimulating hormone receptor (Fshr) [18–22].
E-box protein actions can be inhibited by repressor proteins called inhibitors of cell differentiation or inhibitors of DNA binding (ID). Four ID proteins have been characterized in mammalian cells, ID1, ID2, ID3, and ID4 [23–26]. All four ID proteins contain a conserved helix-loop-helix (HLH) dimerization motif that mediates interactions with other HLH proteins [14, 16]. However, these proteins lack the basic DNA-binding domain and function as dominant negative repressors of bHLH transcription factors by forming non-DNA-binding, inactive heterodimers. A major consequence of ID protein expression is the inhibition of bHLH protein-mediated activation of genes required for the differentiation of tissues during development [27]. ID proteins have been shown to repress the activity of at least two E-box protein-activated genes in Sertoli cells, namely, Trf and Fshr [18, 20].
We tested the hypothesis that E-box protein activity and E-box-mediated transcription are regulated during the transition from proliferation to differentiation within Sertoli cells. We examined E-box DNA-binding activity and the major E-box proteins responsible for DNA-protein interactions within Sertoli cell nuclei from 5-, 11-, and 20-day-old Sertoli cells, corresponding to proliferating, differentiating, and differentiated Sertoli cells, respectively. The levels of E-box protein expression and E-box-mediated transcription were assessed. Finally, the effects of thyroid hormone, a promoter of differentiation, on E-box protein DNA-binding ability and on ID and E-box mRNA expression were evaluated.
MATERIALS AND METHODS
Animal Care and Use
Male Sprague-Dawley rat pups were obtained from Charles River Laboratories (Boston, MA). Animals used in these studies were maintained and euthanized according to the principles and procedures described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. These studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Isolation of Sertoli Cells for Direct Assay
Rats were euthanized 5 and 11 days after birth, and Sertoli cells were isolated as described by Anway et al. [28], with slight modifications. Briefly, decapsulated testes were digested with collagenase (0.5 mg/ml, 34°C, 10–15 min, 80 oscillations/min) in enriched Krebs-Ringer bicarbonate medium (EKRB [118.5 mM NaCl, 4.7 mM KCl, 2.1 mM CaCl2-H2O, 1.0 mM KH2Po4, 1.2 mM MgSO4-7H2O, 25 mM NaHCO3, 10 mM Hepes, and 0.1% BSA]), followed by three settlings in EKRB to isolate seminiferous tubules. Tubules were dispersed through digestion with trypsin (0.5 mg/ml) in the presence of DNase (1 μg/ml) for 5–10 min at 37°C without shaking. Following the digestion, tubule fragments were washed with soybean trypsin inhibitor (0.3 mg/ml), followed by two washes with EKRB. Sertoli cells were separated from germ cells by incubation with 0.1% collagenase, 0.2% hyuronidase, 0.04% DNase I, and 0.03% trypsin inhibitor (40 min, 34°C, 80 oscillations/min). Following digestion, cells were pelleted and washed three times with EKRB. To remove contaminating germ cells, the suspension of single cells and clusters of 5–10 cells was subjected to hypotonic shock by resuspending the pellet in EKRB diluted with water (final concentration, 0.2× EKRB), gently inverting three times to disperse cells, followed by immediate centrifugation at 700 rpm (63 × g) for 10 min. Cells were then washed three times with serum-free media containing 50% Dulbecco modified Eagle medium (DMEM), 50% Ham F-12, 5 mg/ml of insulin, 5 mg/ml of transferrin, 10 ng/ml of epidermal growth factor, 3 μg/ml of cytosine β-D-arabinofuranosidase, 1 mM sodium pyruvate, 200 U/ml of penicillin, and 200 μg/ml of streptomycin. The resuspended cells were counted and directly aliquoted for preparation of protein extracts or plated for analysis of purity.
Isolation of Sertoli Cells for Cell Culture
Rats were euthanized 5, 11, and 20 days after birth as described previously [29]. Decapsulated testes were digested with collagenase (0.5 mg/ml, 33°C, 12 min) in EKRB, followed by three settlings in EKRB to isolate seminiferous tubules. The tubules were digested with trypsin (0.5 mg/ml, 32°C, 12 min) in the presence of DNase (2 μg/ml), and cell aggregates were passed repeatedly through a Pasteur pipette. An equal volume of DMEM containing 10% fetal bovine serum was added to the Sertoli cells, which were then pelleted (500 × g, 5 min) and resuspended in supplemented serum-free media. Sertoli cells were cultured on dishes coated with Matrigel (Collaborative Research, Bedford, MA) (33°C, 5% CO2). Sertoli cells were routinely 95% pure as determined by phase microscopy and alkaline phosphatase staining [30]. Cells were cultured 3 days before treatment. Sertoli cells were stimulated with vehicle (ethyl alcohol) or thyroxine (T4 [10−6 M]) for 24 h.
Determining Purity of Sertoli Cells
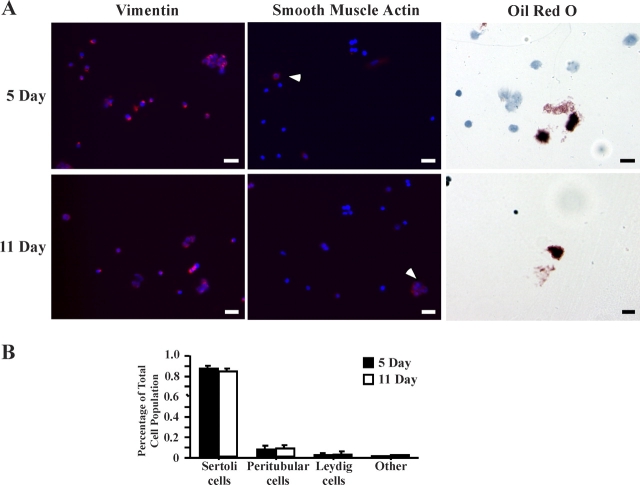
In a 24-well plate, 12-mm2 glass coverslips were coated with poly-l-lysine and allowed to dry. Coverslips were washed with PBS, and 2.5 × 104 cells were plated per well. Cells were allowed to attach to coverslips for approximately 2 h, after which Sertoli cells and peritubular cells were identified by immunofluorescence. Briefly, coverslips were washed with PBS and fixed with 4% formalin for 15 min at room temperature and stored in 70% ethanol overnight. The coverslips were stained with antisera against Sertoli cell-specific vimentin (V6630; Sigma-Aldrich, St. Louis, MO) or with antisera against peritubular cell-specific alpha smooth muscle actin (A2547; Sigma-Aldrich). Primary antisera were detected by secondary antibodies tagged with Alexa 488 or Cy5. Nuclei were visualized using Hoechst 33258 (861405; Sigma-Aldrich). Leydig cells were detected by staining with the lipid-specific stain Oil Red O (O0625; Sigma-Aldrich). The cells were fixed in 2% paraformaldehyde for 5 min, washed three times with distilled water, incubated in propylene glycol for 5 min, and stained with 0.5% Oil Red O in propylene glycol for 8 min at 60°C. Cells were rinsed in 85% propylene glycol for 5 min and washed with distilled water before staining with hematoxylin. Cells were washed repeatedly with water during 3 min, followed by two changes of distilled water before mounting. All images were taken on an Olympus Provis AX70 Microscope (Olympus, Tokyo, Japan), and quantification was performed using Northern Eclipse v. 7.0 Software (MVIA Inc., Monaca, PA). Ten random fields from multiple coverslips were analyzed to determine the total number of nuclei and the number of cells expressing each marker. The mean (±SEM) percentage of cells expressing the cell-specific marker was calculated. More than 500 cells were counted for each detection method.
Nuclear Protein Extracts
Freshly isolated Sertoli cells (1.5×106 cells/sample) or three 100-mm2 plates of cultured Sertoli cells were collected in a total of 1 ml of 1× PBS and used for the preparation of nuclear extracts. Nuclear and cytoplasmic extracts were prepared by detergent lysis [31]. Briefly, after pelleting, the cells were incubated in buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol (DTT), 0.5 mM PMSF, and a protease inhibitor cocktail) for 15 min on ice, followed by the addition of 0.06% Nonidet P40 (Sigma-Aldrich). Cells were vortexed for 10 sec, and nuclei were collected by centrifugation (12 000 × g, 30 sec). The nuclei pellet was then washed once with buffer A and resuspended in buffer C (20 mM Hepes, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 20% glycerol, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail). Nuclei were incubated for 15 min at 4°C with shaking. The cell debris was pelleted (12 000 × g, 5 min), and the supernatant containing nuclear proteins was stored at −80°C. Protein concentrations were determined by the Bradford method (Bio-Rad protein assay; Bio-Rad Laboratories, Inc., Hercules, CA).
DNA-Protein Binding Studies
Radiolabeled DNA probes were generated by annealing nucleotide templates containing protein-binding motifs plus flanking promoter sequences to 10-nucleotide primers that are complementary to the 5′ end of the templates (Table 1). The overhangs were filled in with Klenow enzyme (Promega, Madison, WI) in the presence of [α-32P] dATP and 5 mM each of dCTP, dGTP, dTTP and 5-bromo-2′-deoxyuridine-5′-triphosphate (BrdU). Electrophoretic mobility shift assays (EMSAs) were performed as described [32]. Briefly, 32P-labeled probes were incubated with nuclear extracts (0.2–1.0 μg). Binding reactions were incubated for 15 min at room temperature in the presence of 1 μg of poly deoxyinosinic-deoxycytidylic acid (dI-dC), 0.25 mg/ml of BSA, 5 mM DTT, 50–100 mM KCl, 20 mM Hepes, pH 7.9, 20% glycerol, and 0.2 mM EDTA. For supershift assays, antiserum was added to the mixture for 15 min at room temperature before the addition of the probe. All of the following antisera were obtained from Santa-Cruz Biotechnologies (Santa Cruz, CA): α-USF1 (sc-229), α-USF2 (sc-862), α-E47 (sc-763), α-E2A.E12 (sc-349), and α-MYC (sc-40). Protein-DNA complexes were resolved via 5% PAGE under nondenaturing conditions in a Tris borate/EDTA buffer. The gels were dried and exposed to Classic X-Ray Film, Blue Sensitive (Laboratory Products Sales, Rochester, NY) at −80°C for 1 h to 1 wk. DNA-protein complex formation was quantified from digitized images of the x-ray films using ImageJ v. 1.37 software [33, 34]. Results from at least three independent experiments were analyzed by ANOVA with Fisher probable least-squares difference at a 5% significance level using Statview 4.5 (Abacus Concepts, Inc., Berkeley, CA).
TABLE 1.
Electrophoretic mobility shift assay probes.
RNA Isolation and cDNA Preparation
RNA was obtained from Sertoli cells directly isolated from testis tissue using RNA STAT60 (Tel-Test, Friendswood, TX) and from cultured Sertoli cells using RNeasy Mini Kit (Quiagen, Inc., Valencia, CA). After digestion with RNase-free DNase, the RNA was subjected to reverse transcription using random hexamers [35]. For reverse transcription, 250 ng of RNA was incubated with 100 μl of reaction mix containing 7.5 mM MgCl2, 400 μM deoxynucleotide triphosphates (Promega, Madison, WI), 10× PCR II Buffer (Applied Biosystems, Foster City, CA), 40 U of RNasin RNase inhibitor (Promega), 2.25 μM random hexamers (Integrated DNA Technologies, Coralville, IA), 250 U of Superscript RT II (Invitrogen, San Diego, CA), and nuclease-free water (Ambion, Austin, TX). Parallel reactions were performed without RT to control for the presence of contaminant DNA. The samples were incubated at 25°C for 10 min, at 48°C for 30 min, and at 95°C for 5 min, followed by 4°C for 5 min.
Quantitative PCR Analysis of Gene Expression
Real-time PCR (quantitative PCR [qPCR]) amplifications were performed in a 96-well plate in the ABI Prism 7900HT Sequence Detection System v. 2.3 (Applied Biosystems) in a total volume of 25 μl, which included 2 μl of cDNA, 12.5 μl of ABsolute SYBR Green ROX Mix (ThermoFisher Scientific, ABgene House, Surrey, England), and 600 μM of each primer. Primers used for each gene are listed in Table 2. Previously undescribed primers were designed using Primer Express Software v. 3.0 (Applied Biosystems). All primers were independently validated for use in the ΔΔCt method of gene expression analysis [40] through the use of a standard curve derived from serial dilutions of the cDNA obtained from the reverse transcription reactions. The resulting cycle threshold (Ct) values for each sample were plotted vs. the log of the mRNA concentration present in each cDNA dilution. Ct is the cycle number at which the fluorescent signal of the amplification product exceeds background levels. The slope of the line was used to calculate the efficiency of amplification (Efficiency = 10[1/−Slope] − 1). Primers with a mean (±SEM) efficiency of 1 ± 0.1 were considered acceptable. Ppia (peptidylprolyl isomerase A, commonly known as cyclophillin) was used as an endogenous control. The qPCR analysis was initiated with melting of cDNA at 95°C for 15 min, followed by 40 amplification cycles (15 sec at 95°C and 1 min at 60°C). A dissociation curve was performed immediately after amplification to ensure that there was only one (gene specific) amplification peak.
TABLE 2.
Primers for qPCR.
Ct values were recorded and analyzed via the ΔΔCt method and via the efficiency-corrected ΔCt method [40]. The ΔΔCt method was used to characterize relative fold changes in mRNA expression between treatment groups. The following terms were defined before calculation: GOI indicates gene of interest; reference, Ct value for GOI of 5-day-old untreated sample; unknown, Ct value for GOI of samples from any time point or treatment; and control, Ct value of Ppia for a given treatment. The ΔCt was calculated for each sample according to the following formula: ΔCt = Unknown − Control. The ΔΔCt was calculated by comparing each sample with the reference according to the following equation: ΔΔCt = ΔCtUnknown − ΔCtReference. The fold change was then calculated relative to the reference according to the following formula: Fold Change = 2(−ΔΔCt). The means (±SEMs) of 3 individual experiments were determined for each GOI in each treatment group. The relative quantity of mRNA for each GOI was determined by the efficiency-corrected ΔCt method. The relative quantity is derived from the following equation: Quantity = (Efficiency + 1)−Ct. For each sample, the calculated quantity is then normalized to the quantity found for cyclophillin. The means (±SEMs) of 3 individual experiments were determined for each treatment group for each gene of interest. Results were analyzed by ANOVA with Fisher probable least-squares difference at a 5% significance level using Statview 4.5.
RESULTS
Isolation of Highly Enriched Fractions of Proliferating and Differentiating Sertoli Cells for Immediate Biochemical Analysis
To test the hypothesis that E-box transcription factor activities are up-regulated during the development of the Sertoli cell, the DNA-binding activity of E-box proteins was assayed in Sertoli cells isolated from the testes of 5- and 11-day-old rats, representative of proliferating and differentiating Sertoli cells, respectively. To examine Sertoli cell biochemical properties as close to in vivo conditions as possible, enriched fractions of Sertoli cells were assayed immediately after isolation from rat testes using an adaptation of the protocol by Anway and colleagues [28]. Using this protocol, we obtained approximately 1.7 × 107 cells from ten 5-day-old rat pups and 9.0 × 107 cells from ten 11-day-old rat pups. Imaging analysis confirmed that the isolation procedure resulted in highly enriched Sertoli cell fractions (Fig. 1A). Approximately 85% of the cells stained with the Sertoli cell-specific marker vimentin, whereas approximately 9% stained with the peritubular cell marker alpha smooth muscle actin, and 3% to 9% stained positive for the Leydig cell marker Oil Red O (Fig. 1B). One percent of cells were not stained and are most likely immature germ cells.
FIG. 1.
Enriched fractions of Sertoli cells are obtained immediately after enzymatic digestion of testis tissue. Cell suspensions obtained immediately after enzymatic digestion of rat testes were cultured on coverslips for 2 h and then fixed. A) Sertoli cells were detected using antisera against vimentin (red, left column), peritubular cells were detected by antisera against alpha smooth muscle actin (red, white arrows, middle column), and nuclei were stained with Hoechst dye (blue). Leydig cells were stained with the lipid stain Oil Red O (red stain, right column). Bar = 20 μm. B) Quantitation of the mean (±SEM) percentage of cell-specific markers of 10 random fields from multiple coverslips is provided.
DNA-Binding Activities of USF Proteins Increase During Sertoli Cell Differentiation
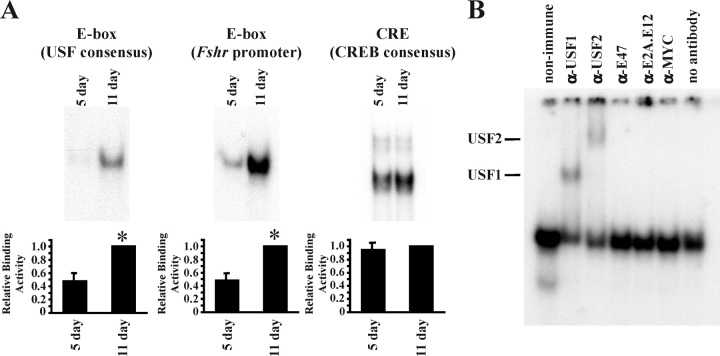
EMSAs were performed with nuclear extracts from the freshly isolated Sertoli cells. The extracts were incubated with radiolabeled, double-stranded oligonucleotide probes containing E-box motifs that are selective for the binding of specific E-box proteins. For probes containing a consensus USF-binding site or a region of the Fshr promoter containing an E-box known to bind USF [13, 41], protein binding was low or undetectable in extracts from 5-day-old Sertoli cells, but binding activity increased at least 2-fold at 11 days (Fig. 2A). As expected, CREB1 binding to a consensus CREB-binding motif, which is generally stable under most conditions, was unchanged at 5 and 11 days [42].
FIG. 2.
DNA-binding activity increases during Sertoli cell differentiation. Nuclear extracts isolated from Sertoli cells immediately after enzymatic digestion were assayed for DNA-protein interactions. A) In EMSAs, radiolabeled probes containing 1) a consensus E-box that is selective for USF binding, 2) a region of the Fshr promoter containing an E-box known to bind USF proteins, or 3) a CREB-binding site (CRE) were incubated with Sertoli cell nuclear extracts isolated from 5- and 11-day-old rats. Representative images of the resulting DNA-protein complexes are shown, and quantitation of the mean (±SEM) of five independent experiments is provided. The relative binding activity for each probe is normalized to that of 11-day-old Sertoli cells (=1). Statistically significant differences (P < 0.05) are indicated by an asterisk (*). B) Nuclear extracts of enriched Sertoli cells from 11-day-old rats were incubated with nonimmune sera or antisera against USF1, USF2, E47, E2A, or MYC. Supershifted DNA-protein complexes containing USF1 or USF2 are indicated. Exposure times for all complexes are less than 48 h.
To identify E-box-binding proteins that are responsible for forming the DNA-protein complexes, supershift studies were performed. Sertoli cell nuclear extracts prepared from 11-day-old testes were preincubated with nonimmune sera or antisera against USF1, USF2, E47, E2A, or MYC before incubation with the E-box region of the Fshr promoter (Fig. 2B). The supershift assay demonstrated that both USF antisera upshifted or disrupted DNA-protein complexes, whereas the E47, E2A, and MYC antisera had little effect. These data suggest that USF1 and USF2 proteins are the major E-box factors that account for increased binding to E-box motifs during Sertoli cell differentiation.
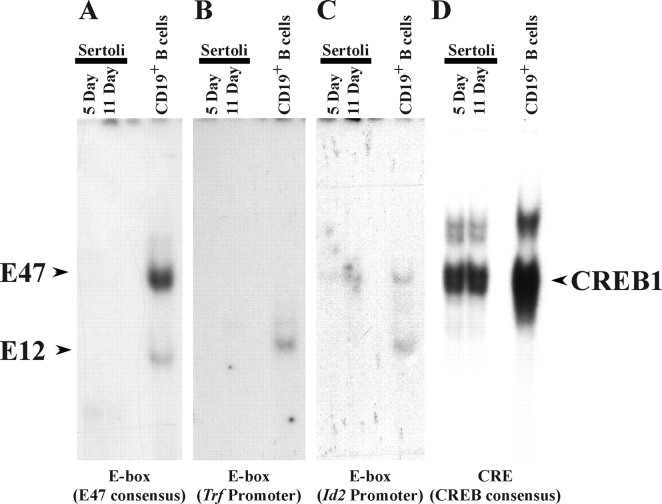
DNA-Binding Activity of the E47 E-box Protein Is Low in 5- and 11-Day-Old Sertoli Cells
The E47 E-box protein has been implicated in numerous activities in Sertoli cells, including FSH-induced regulation of Trf [22] and the influencing of hormone-induced activation of the Fos promoter through the serum response element [43]. To investigate further the binding activity of E47 and the related E12 E-box factors, nuclear extracts of Sertoli cells isolated from 5- and 11-day-old rats were incubated with probes that were previously shown to bind E47, including a consensus binding site for E47, the Trf promoter E-box, or an E-box from the Id2 promoter region [22, 43, 44]. Incubation of the E47-selective probes with Sertoli cell nuclear extracts did not result in detection of any DNA-protein complexes (Fig. 3, A–C). In contrast, all probes formed complexes with nuclear extracts derived from splenic origin CD19+ B cells activated with lipopolysaccharide, which are known to contain high levels of E47- and E12-binding activity [45–47]. Using a probe containing a binding site for the CREB1 transcription factor, the protein-binding activity of the Sertoli nuclear extracts was confirmed by the finding that DNA-protein complexes were formed (Fig. 3D). Together, these studies indicate that E47-binding activity is below detectable limits in the nuclei of 5- and 11-day-old Sertoli cells.
FIG. 3.
E47 DNA-binding activity is not detected in developing Sertoli cells. EMSAs using probes containing a consensus E-box selective for E47 binding (A), the Trf promoter E-box (B), the Id2 promoter E-box (C), or a CREB-binding site (D) were incubated with nuclear extracts from enriched Sertoli cells isolated 5 and 11 days after birth or with CD19+ B cells isolated from mouse spleen. The DNA-protein complexes formed by E47, E12, and CREB1 are indicated. The complexes were identified from the results of separate supershift assays using E47- and E12-selective antisera (data not shown). Assays for E47-selective-binding probes were exposed for 7 days, while CREB1 binding complexes were detected after 12 h.
USF DNA-Binding Activity Is Similar in Freshly Isolated and Cultured Sertoli Cell Models
To facilitate studies of factors that may regulate Sertoli cell differentiation and E-box-binding activity, Sertoli cells from 5- and 11-day-old rats were studied after being placed in culture. In addition, fully differentiated Sertoli cells from 20-day-old rats were cultured. By using a gentler isolation procedure and maintaining the Sertoli cells in culture for 4 days, cell survival was increased compared with the protocol by Anway et al. [28]. Previous investigations have shown that Sertoli cell populations are more than 95% pure 4 days after initiating the cultures [29].
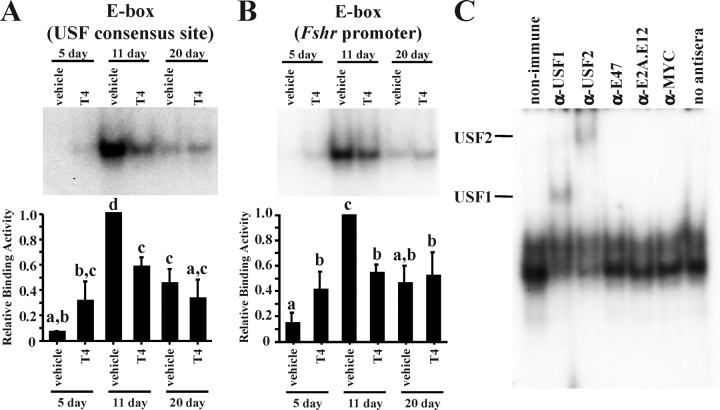
EMSA analysis of nuclear extracts from Sertoli cells cultured from 5- and 11-day-old rats was similar to the analysis using freshly isolated cells in that E-box protein binding to the USF consensus and Fshr promoter probe was low at 5 days but increased 5- to 10-fold at 11 days (Fig. 4, A and B). E-box-binding activity decreased at 20 days but was still 4- to 5-fold greater than E-box-binding activity present in 5-day-old Sertoli cells. As shown for freshly isolated Sertoli cells, supershift analysis of the cultured Sertoli cell nuclear extracts indicated that USF1 and USF2 proteins were the predominant E-box factors interacting with the Fshr promoter E-box (Fig. 4C). In agreement with the results obtained from freshly isolated Sertoli cells, E47 DNA-binding activity assayed using three E47-selective probes was at or below the level of detection in extracts from cultured 5-, 11-, or 20-day-old Sertoli cells (data not shown). Together, these data confirm that the developmental profile of E-box protein-binding activities and the prevalence of USF-binding activity are similar in freshly isolated and cultured Sertoli cells.
FIG. 4.
E-box DNA-binding activity increases during Sertoli cell differentiation. In EMSAs, radiolabeled probes containing a consensus E-box that is selective for USF binding (A) or a region of the Fshr promoter containing an E-box known to bind USF proteins (B) were incubated with nuclear extracts isolated from cultured 5-, 11-, and 20-day-old Sertoli cells that were treated with vehicle or T4 (10−6 M). Representative images of the resulting DNA-protein complexes are shown, and quantitation of the mean (±SEM) of three independent experiments is provided. The relative binding activity for each condition is normalized to that of vehicle-treated 11-day-old Sertoli cells (=1). Values with different lowercase letters differ significantly (P < 0.05). C) USF1 and USF2 are responsible for the increase in DNA-binding activity during Sertoli cell differentiation. DNA-protein complexes from nuclear extracts isolated from cultured 11-day-old Sertoli cells were incubated with nonimmune sera or antisera for USF1, USF2, E47, E2A, or MYC. Supershifted DNA-protein complexes containing USF1 or USF2 are indicated. Exposure times for all complexes are less than 48 h.
Thyroid Hormone Induces USF DNA-Binding Activity in Proliferating Sertoli Cells
To determine whether thyroid hormone, a known Sertoli cell-differentiating agent, could increase the DNA-binding activity of USF proteins, cultured Sertoli cells were incubated with vehicle or T4 (10−6 M) for 24 h. Thyroxine was used because T4 levels are 100-fold greater than triiodothyronine (T3) levels in the testis during the period of Sertoli cell differentiation [48]. Furthermore, Sertoli cells have been shown to express iodothyronine deiodinase type 2, which converts T4 to T3, the more potent form of thyroid hormone [49]. The dose of T4 that was used is 12.5-fold greater than the peak T4 serum level (8 × 10−8 M) reached in rats during differentiation [48]. A 24-h stimulation of cultured Sertoli cells from 5-day-old rats with T4 resulted in induced protein binding to the USF consensus and Fshr promoter E-box probes (Fig. 4, A and B). In contrast, T4 stimulation was unable to further induce E-box protein binding in Sertoli cells from 11- and 20-day-old rats. Furthermore, at 11 days, T4 caused a significant reduction in protein binding to the USF consensus and Fshr promoter E-box probes. Results of these studies suggest that T4 is able to induce USF DNA-binding activity in Sertoli cells before the commitment to differentiation.
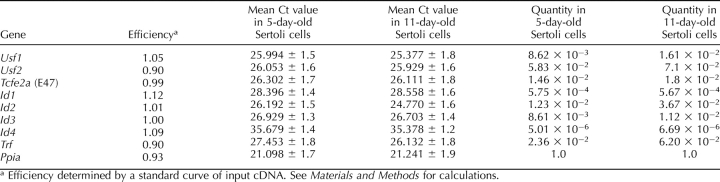
Usf1 mRNA Expression Increases During Sertoli Cell Differentiation
To determine whether increased USF DNA-binding activity during Sertoli cell differentiation corresponded with increased Usf expression, the expression of mRNAs encoding E-box proteins was assayed during Sertoli development using qPCR. Assays of mRNA preparations from Sertoli cells immediately after isolation from 5- and 11-day-old rat testes revealed a 62% increase in Usf1 mRNA levels 11 days after birth. Usf2 and Tcfe2a (E47) mRNA levels also trended higher (24% for both) but did not reach a level of statistical significance (Fig. 5, A–C). Trf mRNA levels increased from 5 to 11 days, in agreement with previous studies [50, 51] showing that Trf mRNA levels increase during differentiation (Fig. 5D). The mRNA levels of the housekeeping gene Ppia did not change during the course of differentiation (data not shown). The use of the efficiency-corrected ΔCt method allowed us to determine that Usf2 mRNA levels in Sertoli cells are 4- to 6-fold greater than Usf1 mRNA levels (Table 3).
FIG. 5.
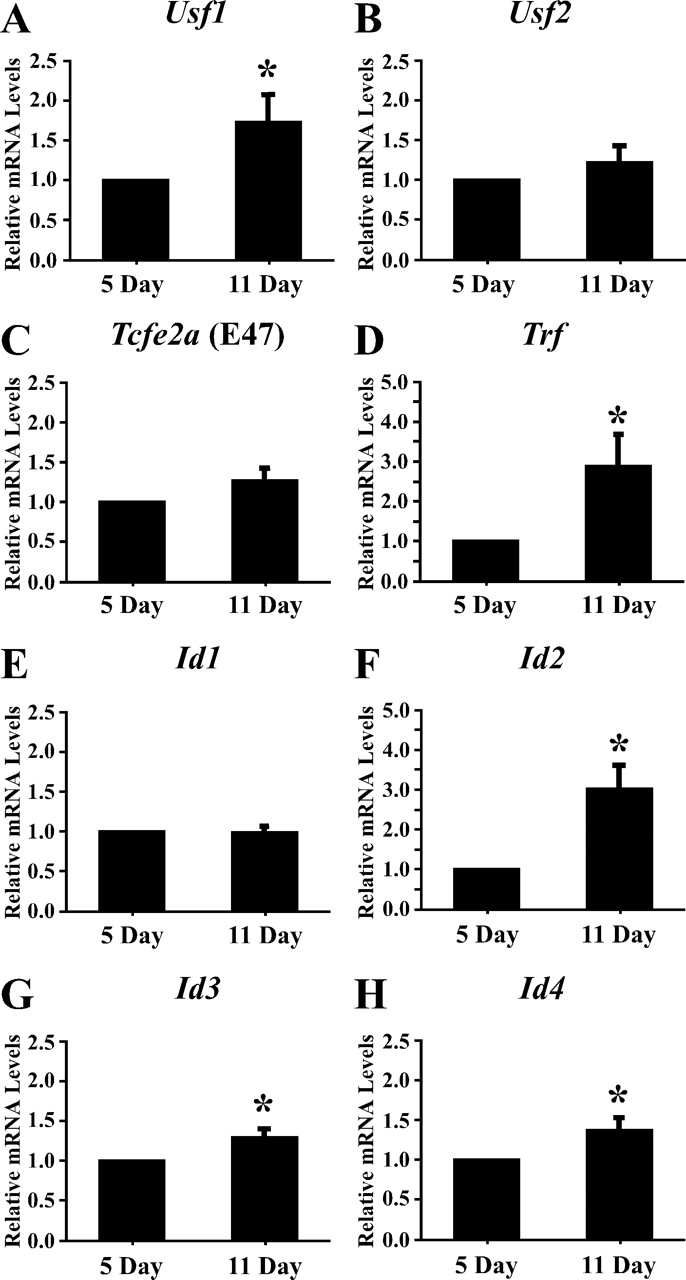
Expression of E-box and ID protein mRNAs during Sertoli cell development in freshly isolated cells. The mRNAs isolated from 5- and 11-day-old Sertoli cells were analyzed by qRT-PCR using primers for Usf1 (A), Usf2 (B), Tcfe2a (E47) (C), Trf (D), and Id1, Id2, Id3, and Id4 (E–H). Also analyzed were PCR reactions performed on RT reactions lacking RT (−RT) and PCR reactions performed in the absence of cDNA (no DNA) (data not shown). Data were analyzed using the ΔΔCt method, and quantitation of the mean (±SEM) of three individual experiments for each condition is provided for each primer set. The relative mRNA levels were normalized to Ppia levels and were made relative to 5-day-old Sertoli cells (=1). Statistically significant differences (P < 0.05) are indicated by an asterisk (*).
TABLE 3.
qPCR primer validation and relative quantity of mRNAs in freshly isolated rat Sertoli cells.
Id2, Id3, and Id4 mRNA Levels Increase During Sertoli Cell Differentiation
Increased USF protein binding to E-box motifs during Sertoli cell differentiation could result from decreased expression of ID proteins that inhibit E-box proteins from dimerizing and binding to DNA [14]. Examination of Id1 mRNA levels from freshly isolated Sertoli cells revealed no significant differences in the levels of mRNA expression between 5 and 11 days after birth (Fig. 5E), whereas Id2, Id3, and Id4 mRNA levels increased 149%, 23%, and 30%, respectively, from 5 to 11 days after birth (Fig. 5, F–H). Together, these data suggest that changes in Id expression do not account for the increase in DNA-binding activity of USF1 and USF2 during Sertoli cell development, as USF1 and USF2 binding to DNA increases in spite of an increase in mRNA expression of the ID inhibitors. Further analysis of the qPCR results using the efficiency-corrected ΔCt method revealed that the relative Id2 and Id3 mRNA levels in Sertoli cells are similar to those of Usf1 and Usf2, but the levels of Id1 and Id4 mRNAs are 100- and 1000-fold lower, respectively (Table 3).
Thyroid Hormone Does Not Greatly Alter the Expression of mRNAs Encoding E-box or ID Proteins
The ability of thyroid hormone to regulate the gene expression of E-box and ID proteins was tested using cultured Sertoli cells. Using qPCR, an initial analysis of basal Usf1, Usf2, and Tcfe2a (E47) mRNA levels in cultured 5- and 11-day-old Sertoli cells indicated that the expression patterns of these mRNAs in cultured Sertoli cells were similar to those found in freshly isolated Sertoli cells (Fig. 6, A–C). In cultured cells, Usf1, Usf2, and Tcfe2a (E47) mRNA levels all increased from 5 to 11 days after birth (41%, 43%, and 59%, respectively) and then decreased to or below the level of 5-day mRNA levels in cells isolated 20 days after birth. Trf mRNA levels increased from 5 to 11 days and again from 11 to 20 days after birth, in agreement with the results obtained from freshly isolated cells (Fig. 6D). Thyroxine stimulation (10−6 M) did not significantly alter the expression of Usf2, Tcfe2a (E47), or Trf but increased Usf1 mRNA levels by 28% at 11 days. The mRNA levels of the housekeeping gene Ppia did not change during the course of differentiation (data not shown).
FIG. 6.
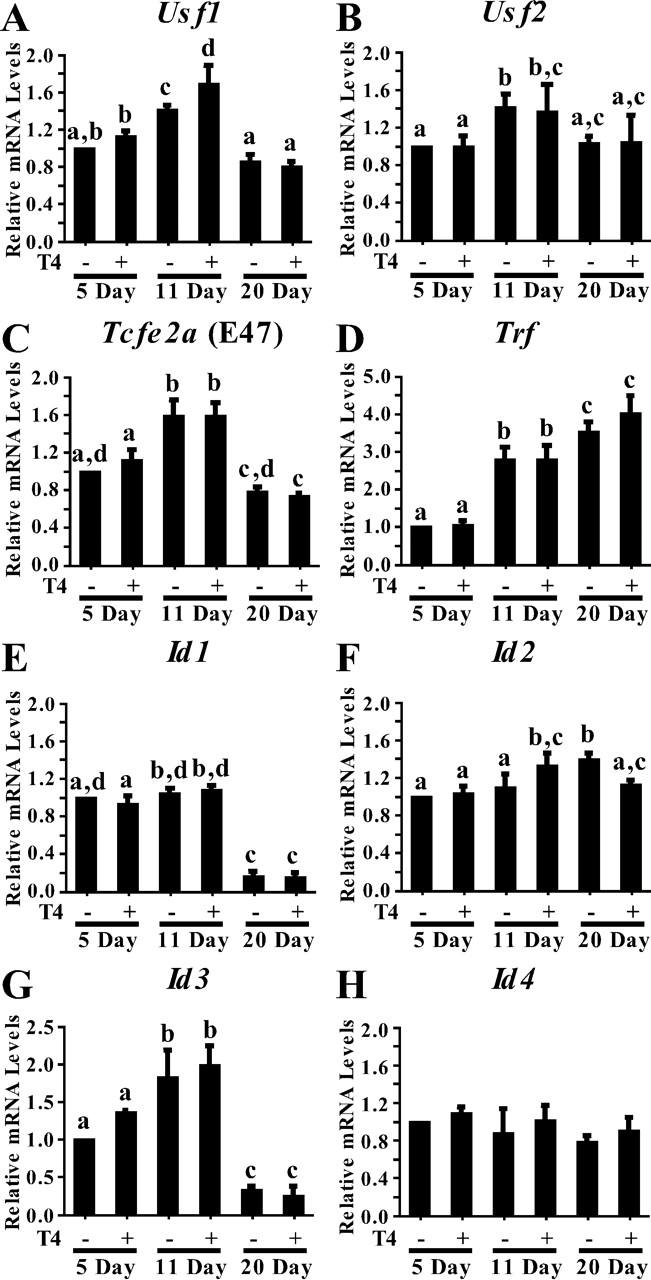
Expression of E-box and ID protein mRNAs during Sertoli cell development. The mRNAs isolated from cultured 5-, 11-, and 20-day-old Sertoli cells treated with vehicle (−) or T4 (10−6 M) for 24 h were analyzed by qRT-PCR using primers for Usf1 (A), Usf2 (B), Tcfe2a (E47) (C), Trf (D), and Id1, Id2, Id3, and Id4 (E–H). Also analyzed were PCR reactions performed on RT reactions lacking RT (−RT) and PCR reactions performed in the absence of cDNA (no DNA) (data not shown). Data were analyzed using the ΔΔCt method, and quantitation of the mean (±SEM) of three individual experiments for each condition is provided for each primer set. The relative mRNA levels were normalized to Ppia levels and were made relative to untreated 5-day-old Sertoli cells (=1). Values with different lowercase letters differ significantly (P < 0.05).
Basal Id1 mRNA levels were not altered between 5 and 11 days after birth in cultured Sertoli cells, as was found for Sertoli cells immediately after isolation (Fig. 6E), but between 11 and 20 days after birth, Id1 mRNA levels decreased significantly (84%). In contrast to the increasing mRNA levels detected in freshly isolated cells, Id2 mRNA levels remained constant throughout the differentiation period in cultured cells (Fig. 6F). In agreement with the results from the freshly isolated cells, Id3 mRNA levels increased in cultured Sertoli cells between 5 and 11 days after birth. However, Id3 mRNA levels decreased between 11 and 20 days after birth, similar to the Id1 pattern of expression (Fig. 6G). Id4 mRNA levels in cultured Sertoli cells did not change during the differentiation period, despite showing a small increase between 5 and 11 days in freshly assayed Sertoli cells (Fig. 6H). Stimulation of the cultured Sertoli cells with T4 resulted in a slight further stimulation of Id2 mRNA levels (24%) at 11 days but not at 5 or 20 days after birth (Fig. 6A). Thyroxine stimulation did not result in any significant changes to Id1, Id3, or Id4 mRNA expression at any time point.
Together, the analysis of mRNA levels in freshly isolated and cultured Sertoli cells indicates that most measurements of mRNA expression patterns did not differ greatly for the two model systems. Basal levels of Usf1, Usf2, Tcfe2a (E47), and Id3 mRNA levels increase between 5 and 11 days after birth, whereas Id1 and Id3 mRNA levels decrease between 11 and 20 days during the later stages of Sertoli cell differentiation. Id2 and Id4 mRNA levels increase from 5 to 11 days after birth in freshly isolated Sertoli cells, but the levels of these mRNAs do not change significantly in cultured cells. Finally, thyroid hormone has a small positive effect only on Usf1 and Id2 mRNA expression and only 11 days after birth.
DISCUSSION
Many differentiation-associated genes are regulated by E-box proteins in Sertoli cells [12, 13, 18, 22, 41, 43, 52, 53]. We have found that during the differentiation of Sertoli cells, E-box protein binding to DNA probes containing a consensus USF-binding site or an Fshr promoter E-box increases from 5 to 11 days after birth. The E-box proteins binding to the Fshr E-box during differentiation were identified as USF1 and USF2. These findings were similar for Sertoli cells immediately after isolation and for cultured Sertoli cells. These results are consistent with findings by Heckert et al. [13, 41, 53] that USF proteins bound to the Fshr promoter E-box in fully differentiated Sertoli cells (27 or 50 days old). We also determined that the DNA-binding activity of the ubiquitous E47 protein was below detection limits in freshly isolated and cultured Sertoli cell nuclear extracts from 5-, 11-, or 20-day-old rats. The inability to detect E47 binding to any of three E47-selective E-box probes is in agreement with findings by Chaudhary and Skinner [22], who noted that E47 binding to the Trf promoter E-box was not observed except after stimulation for 48 h by FSH in 20-day-old Sertoli cells. Together, these data indicate that USF1 and USF2, but not E47 or E12 E-box proteins, are potential regulators of Sertoli cell differentiation. The findings that USF proteins are the predominant E-box transcription factors that are able to bind E-box motifs during Sertoli cell differentiation, as well as that USF proteins are the major E-box regulators of Fshr, are consistent with previous studies [54, 55] showing that fertility is compromised in the absence of FSHR and USF2. Our data suggest that signals transmitted to the Sertoli cell between 5 and 11 days after birth increase the DNA-binding activity of USF proteins. It is assumed that the increase in USF DNA-binding activity will promote transcription of genes regulated by USF1 and USF2. Therefore, regulatory signals likely act through USF proteins to ensure that the correct differentiation-associated genes are expressed. USF-induced transcription was not investigated in this study but will be the focus of future work.
It is possible that the increase in USF-binding activity observed during differentiation could be due to an increase in the amount of USF protein present within the cell. Because of the low levels of USF expression in isolated Sertoli cells, Western blot data of whole-cell or nuclear extracts could not be obtained to determine how the levels of these proteins change during Sertoli cell differentiation. However, qPCR was used to evaluate basal mRNA levels for E-box proteins. Usf1 mRNA levels increased from 5 to 11 days after birth in Sertoli cells assayed immediately after isolation (62%) and in cultured Sertoli cells (41%). Usf2 and Tcfe2a (E47) remained unchanged in freshly isolated cells but increased between 5 and 11 days after birth in cultured Sertoli cells (43% and 59%, respectively). It is possible that the increases in Usf mRNA levels and any subsequent increased USF protein expression could account for the 2-fold and 7- to 10-fold elevations of USF DNA-binding activity observed during differentiation in freshly isolated and cultured Sertoli cells, respectively. However, it cannot yet be ruled out that another mechanism such as posttranslational modification of USF proteins or activation of accessory proteins is responsible for the increase in USF-binding activity that is observed with the initiation of differentiation.
ID proteins were investigated as potential negative regulators of E-box protein-DNA interactions that would be active before the initiation of Sertoli cell differentiation. However, instead of observing decreased Id expression during differentiation, the levels of all Id mRNAs increased or remained constant between 5 and 11 days after birth. Because ID proteins block E-box protein function, an increase in expression of the ID proteins would not be expected to contribute to the activation of USF DNA-binding activity found in 11-day-old Sertoli cells. Although ID mRNA expression patterns do not suggest a role for Id proteins in regulating USF-mediated Sertoli cell differentiation 5 to 11 days after birth, the possibility remains that ID proteins regulate other USF-mediated processes in Sertoli cells at other stages of development. Previously, overexpression of ID1 was found to inhibit expression of the USF-regulated Fshr promoter [20]. Id1 and Id3 mRNA levels decreased dramatically (85% and 70%, respectively) during the later stages of differentiation between 11 and 20 days after birth, but the significance of the decrease in Id1 and Id3 expression, as well as any relationship to the activity of the Fshr promoter, is not clear at this time. Additional studies are required to determine whether ID proteins directly or indirectly inhibit USF protein and E-box interactions, as well as the regulation of USF-mediated transcription.
Our studies using cultured Sertoli cells indicated that T4 increased the DNA-binding activity of USF proteins in undifferentiated Sertoli cells from 5-day-old rats. The increase in USF protein-DNA interactions in 5-day-old Sertoli cells in response to T4 is further evidence that cells at this early stage of differentiation are receptive to thyroid hormone signals. The timing of the T4-mediated increases in USF DNA-binding activity is consistent with previous data showing that thyroid hormone inhibits Sertoli cell proliferation but promotes differentiation between 4 and 8 days after birth [48, 56, 57]. Thyroid hormone stimulation of cultured Sertoli cells isolated from 11-day-old rats decreased USF binding to a consensus E-box probe and to the Fshr promoter E-box probe. This decrease in binding activity occurred in spite of T4-mediated increases in Usf1 mRNA levels 11 days after birth. It is possible that the small increase in Usf1 mRNA levels observed after T4 stimulation may be negated by other factors that inhibit USF1 DNA-binding activity by a posttranslational mechanism [58–61]. The exact mechanism of how thyroid hormone contributes to changes in USF activity during the transition from proliferation to differentiation within Sertoli cells remains to be determined.
Identification of the mechanisms that regulate Sertoli cell differentiation is crucial to understanding the establishment of male fertility. To assay the biochemical and gene expression changes that occur during Sertoli cell differentiation, we used two model systems. For the first time (to our knowledge), enriched fractions of proliferating and differentiating Sertoli cells assayed directly after isolation from testis tissue were used to complement studies of cultured Sertoli cells. Comparison of the two models confirmed that Sertoli cells retain their differentiation status in culture because the protein activities and mRNA expression patterns in cultured Sertoli cells closely mimicked those in freshly isolated Sertoli cells. Our results suggest one possible mechanism to initiate the onset of differentiation in Sertoli cells. We propose that signals, including thyroid hormone, initiate a cascade of molecular events that allows transcription factors, including USF1 and USF2 E-box-binding proteins, to activate the transcription of differentiation-promoting genes. The differentiation-promoting genes would then enable the Sertoli cell to support germ cell development and may facilitate exiting the cell cycle. Further characterization of USF1 and USF2 regulation during Sertoli cell development will identify how these bHLH proteins function in the regulation of a molecular switch from proliferation to differentiation in Sertoli cells.
Acknowledgments
We thank Dr. Anthony Zeleznik for valuable advice and Ppia primers. We thank Dr. Lisa Borghesi and Anthony St. Leger (Department of Immunology) for nuclear extracts from CD19+ B cells of splenic origin. We also thank Kathrin Gassei, Steven Reisenweber, John Shupe, and the Center for Biologic Imaging at the University of Pittsburgh for technical assistance.
Footnotes
1Supported by cooperative U54 ( HD008610) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
REFERENCES
- Orth JM, Gunsalus GL, Lamperti AA.Evidence from Sertoli cell-depleted rats indicates that spermatid number in adults depends on numbers of Sertoli cells produced during perinatal development. Endocrinology 1988; 122: 787–794. [DOI] [PubMed] [Google Scholar]
- Sharpe RM.Regulation of spermatogenesis. Knobil E, Neil JD.The Physiology of Reproduction New York, NY:Raven Press;1994: 1363–1434. [Google Scholar]
- Sharpe RM, McKinnell C, Kivlin C, Fisher JS.Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003; 125: 769–784. [DOI] [PubMed] [Google Scholar]
- Orth JM.Proliferation of Sertoli cells in fetal and postnatal rats: a quantitative autoradiographic study. Anat Rec 1982; 203: 485–492. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Wreford NG, De Kretser DM.Determination of Sertoli cell numbers in the developing rat testis by stereological methods. Int J Androl 1989; 12: 58–64. [DOI] [PubMed] [Google Scholar]
- Gondos B, Berndston WE.Postnatal and pubertal development. Russell LD, Griswold MD.The Sertoli Cell Clearwater, FL:Cache River Press;1993: 115–154. [Google Scholar]
- Steinberger A, Steinberger E.Replication pattern of Sertoli cells in maturing rat testis in vivo and in organ culture. Biol Reprod 1971; 4: 84–87. [DOI] [PubMed] [Google Scholar]
- Bortolussi M, Zanchetta R, Belvedere P, Colombo L.Sertoli and Leydig cell numbers and gonadotropin receptors in rat testis from birth to puberty. Cell Tissue Res 1990; 260: 185–191. [DOI] [PubMed] [Google Scholar]
- Baker PJ, O'Shaughnessy PJ.Role of gonadotrophins in regulating numbers of Leydig and Sertoli cells during fetal and postnatal development in mice. Reproduction 2001; 122: 227–234. [DOI] [PubMed] [Google Scholar]
- Clermont Y, Perey B.Quantitative study of the cell population of the seminiferous tubules in immature rats. Am J Anat 1957; 100: 241–267. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Kim G, Skinner MK.Expression of the basic helix-loop-helix protein REBalpha in rat testicular Sertoli cells. Biol Reprod 1999; 60: 1244–1250. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Skinner M.The basic helix-loop-helix E2A gene product E47, not E12, is present in differentiating Sertoli cells. Mol Reprod Dev 1999; 52: 1–8. [Google Scholar]
- Heckert LL, Daggett MA, Chen J.Multiple promoter elements contribute to activity of the follicle-stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. Mol Endocrinol 1998; 12: 1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM.Id proteins in cell growth and tumorigenesis. Cancer Cell 2003; 3: 525–530. [DOI] [PubMed] [Google Scholar]
- Zebedee Z, Hara E.Id proteins in cell cycle control and cellular senescence. Oncogene 2001; 20: 8317–8325. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R.Id proteins in development, cell cycle and cancer. Trends Cell Biol 2003; 13: 410–418. [DOI] [PubMed] [Google Scholar]
- Corre S, Galibert MD.Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res 2005; 18: 337–348. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Cupp AS, Skinner MK.Role of basic-helix-loop-helix transcription factors in Sertoli cell differentiation: identification of an E-box response element in the transferrin promoter. Endocrinology 1997; 138: 667–675. [DOI] [PubMed] [Google Scholar]
- Daggett MA, Rice DA, Heckert LL.Expression of steroidogenic factor 1 in the testis requires an E-box and CCAAT box in its promoter proximal region. Biol Reprod 2000; 62: 670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz TL, Lloyd TL, Griswold MD.Role of E box and initiator region in the expression of the rat follicle-stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. J Biol Chem 1996; 271: 33317–33324. [DOI] [PubMed] [Google Scholar]
- Nomura M, Bartsch S, Nawata H, Omura T, Morohashi K.An E box element is required for the expression of the ad4bp gene, a mammalian homologue of ftz-f1 gene, which is essential for adrenal and gonadal development. J Biol Chem 1995; 270: 7453–7461. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK.E-box and cyclic adenosine monophosphate response elements are both required for follicle-stimulating hormone-induced transferrin promoter activation in Sertoli cells. Endocrinology 1999; 140: 1262–1271. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H.The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 1990; 61: 49–59. [DOI] [PubMed] [Google Scholar]
- Sun XH, Copeland NA, Jenkins NA, Baltimore D.Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 1991; 11: 5603–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D.An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci U S A 1991; 88: 1815–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, van Cruchten I, Sablitzky F.The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res 1994; 22: 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M, Murre C.Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol Cell Biol 2000; 20: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Folmer J, Wright WW, Zirkin BR.Isolation of Sertoli cells from adult rat testes: an approach to ex vivo studies of Sertoli cell function. Biol Reprod 2003; 68: 996–1002. [DOI] [PubMed] [Google Scholar]
- Walker WH, Fucci L, Habener JF.Expression of the gene encoding transcription factor cyclic adenosine 3′,5′-monophosphate (cAMP) response element-binding protein (CREB): regulation by follicle-stimulating hormone-induced cAMP signaling in primary rat Sertoli cells. Endocrinology 1995; 136: 3534–3545. [DOI] [PubMed] [Google Scholar]
- Chapin RE, Phelps JL, Miller BE, Gray TJ.Alkaline phosphatase histochemistry discriminates peritubular cells in primary rat testicular cell culture. J Androl 1987; 8: 155–161. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W.Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res 1989; 17: e6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard DW, Walker WH, Doerre S, Sista P, Molitor JA, Dixon EP, Peffer NJ, Hannink M, Greene WC.The v-rel oncogene encodes a kappa B enhancer binding protein that inhibits NF-kappa B function. Cell 1990; 63: 803–814. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ.Image processing with ImageJ. Biophotonics Int 2004; 11: 36–42. [Google Scholar]
- Rasband WS. ImageJ. Bethesda, MD:: US National Institutes of Health;; pp. 1997–2007. [Google Scholar]
- Innis MA. PCR Protocols: A Guide to Methods and Applications. San Diego, CA:: Academic Press;; 1990. [Google Scholar]
- Zhu Y, Casado M, Vaulont S, Sharma K.Role of upstream stimulatory factors in regulation of renal transforming growth factor-beta1. Diabetes 2005; 54: 1976–1984. [DOI] [PubMed] [Google Scholar]
- Ciarrocchi A, Jankovic V, Shaked Y, Nolan DJ, Mittal V, Kerbel RS, Nimer SD, Benezra R.Id1 restrains p21 expression to control endothelial progenitor cell formation. PLoS ONE 2007; 2: e1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M, Hamzeh M, Robaire B.Expression, localization, and regulation of inhibitor of DNA binding (Id) proteins in the rat epididymis. J Androl 2006; 27: 212–224. [DOI] [PubMed] [Google Scholar]
- Shan L, Yu M, Qiu C, Snyderwine EG.Id4 regulates mammary epithelial cell growth and differentiation and is overexpressed in rat mammary gland carcinomas. Am J Pathol 2003; 163: 2495–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF.High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol 2006; Chapter 15: Unit 15.8. [DOI] [PubMed] [Google Scholar]
- Heckert LL, Sawadogo M, Daggett MA, Chen JK.The USF proteins regulate transcription of the follicle-stimulating hormone receptor but are insufficient for cell-specific expression. Mol Endocrinol 2000; 14: 1836–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, Tsien R, Montminy MR.Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol 1993; 13: 4852–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary J, Skinner M.Basic helix-loop-helix proteins can act at the E-box within the serum response element of the c-fos promoter to influence hormone induced activation in Sertoli cells. Mol Endocrinol 1999; 13: 774–786. [DOI] [PubMed] [Google Scholar]
- Neuman K, Nornes HO, Neuman T.Helix-loop-helix transcription factors regulate Id2 gene promoter activity. FEBS Lett 1995; 374: 279–283. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D.A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989; 56: 777–783. [DOI] [PubMed] [Google Scholar]
- Nelson C, Shen LP, Meister A, Fodor E, Rutter WJ.Pan: a transcriptional regulator that binds chymotrypsin, insulin, and AP-4 enhancer motifs. Genes Dev 1990; 4: 1035–1043. [DOI] [PubMed] [Google Scholar]
- Henthorn P, Kiledjian M, Kadesch T.Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science 1990; 247: 467–470. [DOI] [PubMed] [Google Scholar]
- Dussault JH, Labrie F.Development of the hypothalamic-pituitary-thyroid axis in the neonatal rat. Endocrinology 1975; 97: 1321–1324. [DOI] [PubMed] [Google Scholar]
- Wajner SM, dos Santos Wagner M, Melo RC, Parreira GG, Chiarini-Garcia H, Bianco AC, Fekete C, Sanchez E, Lechan RM, Maia AL.Type 2 iodothyronine deiodinase is highly expressed in germ cells of adult rat testis. J Endocrinol 2007; 194: 47–54. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Griswold MD.Sertoli cells synthesize and secrete transferrin-like protein. J Biol Chem 1980; 255: 9523–9525. [PubMed] [Google Scholar]
- Perez-Infante V, Bardin CW, Gunsalus GL, Musto NA, Rich KA, Mather JP.Differential regulation of testicular transferrin and androgen-binding protein secretion in primary cultures of rat Sertoli cells. Endocrinology 1986; 118: 383–392. [DOI] [PubMed] [Google Scholar]
- Muir T, Sadler-Riggleman I, Stevens JD, Skinner MK.Role of the basic helix-loop-helix protein ITF2 in the hormonal regulation of Sertoli cell differentiation. Mol Reprod Dev 2006; 73: 491–500. [DOI] [PubMed] [Google Scholar]
- Hermann BP, Hornbaker K, Rice DA, Sawadogo M, Heckert LL.In vivo regulation of follicle-stimulating hormone receptor by the transcription factors upstream stimulatory factor 1 and upstream stimulatory factor 2 is cell specific. Endocrinology 2008; 149: 5297–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR.What have we learned about gonadotropin function from gonadotropin subunit and receptor knockout mice? Reproduction 2005; 130: 293–302. [DOI] [PubMed] [Google Scholar]
- Sirito M, Lin Q, Deng JM, Behringer RR, Sawadogo M.Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci U S A 1998; 95: 3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaster LH, de Jong FH, Docter R, de Rooij DG.High neonatal triiodothyronine levels reduce the period of Sertoli cell proliferation and accelerate tubular lumen formation in the rat testis, and increase serum inhibin levels. Endocrinology 1993; 133: 755–760. [DOI] [PubMed] [Google Scholar]
- Cooke PS, Porcelli J, Hess RA.Induction of increased testis growth and sperm production in adult rats by neonatal administration of the goitrogen propylthiouracil (PTU): the critical period. Biol Reprod 1992; 46: 146–154. [DOI] [PubMed] [Google Scholar]
- Qyang Y, Luo X, Lu T, Ismail PM, Krylov D, Vinson C, Sawadogo M.Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol Cell Biol 1999; 19: 1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert MD, Carreira S, Goding CR.The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced Tyrosinase expression. EMBO J 2001; 20: 5022–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Cultaro CM, Segal S, Spiegel S.The potent lipid mitogen sphingosylphosphocholine activates the DNA binding activity of upstream stimulating factor (USF), a basic helix-loop-helix-zipper protein. Biochim Biophys Acta 1998; 1390: 225–236. [DOI] [PubMed] [Google Scholar]
- Cheung E, Mayr P, Coda-Zabetta F, Woodman PG, Boam DS.DNA-binding activity of the transcription factor upstream stimulatory factor 1 (USF-1) is regulated by cyclin-dependent phosphorylation. Biochem J 1999; 344(pt 1):145–152. [PMC free article] [PubMed] [Google Scholar]