Summary
Degradation by the ubiquitin-proteasome system requires assembly of a polyubiquitin chain upon substrate. However, the structural and mechanistic features that enable template-independent processive chain synthesis are unknown. We show that chain assembly by ubiquitin ligase SCF and ubiquitin-conjugating enzyme Cdc34 is facilitated by the unusual nature of Cdc34-SCF transactions: Cdc34 binds SCF with nanomolar affinity, nevertheless the complex is extremely dynamic. These properties are enabled by rapid association driven by electrostatic interactions between the acidic tail of Cdc34 and a basic ‘canyon’ in the Cul1 subunit of SCF. Ab initio docking between Cdc34 and Cul1 predicts intimate contact between the tail and the basic canyon, an arrangement confirmed by cross-linking and kinetic analysis of mutants. Basic canyon residues are conserved in both Cul1 paralogs and orthologs, suggesting that the same mechanism underlies processivity for all cullin-RING ubiquitin ligases. We discuss different strategies by which processive ubiquitin chain synthesis may be achieved.
Introduction
Regulation of protein stability by the UPS is now appreciated to have at least some impact on nearly all aspects of eukaryotic cell biology. The ubiquitylation cascade first involves the activation of ubiquitin by E1 enzyme and its subsequent transfer to a ubiquitin-conjugating enzyme (E2). E2 charged with ubiquitin (E2~Ub) then coordinates with a ubiquitin ligase (E3) to ubiquitylate the protein substrate. Subsequent interactions between E2~Ub and E3 can result in transfer of additional ubiquitins to the substrate-linked Ub to form a ubiquitin chain. A chain of four ubiquitins linked together via their Lysine 48 residue constitutes a minimal signal for substrate recognition by the 26S proteasome, which then degrades the appended protein (Chau et al., 1989; Thrower et al., 2000).
Some 650 ubiquitin ligases are encoded in the human genome (Deshaies and Joazeiro, 2009). Thus, regulation by ubiquitylation may be comparable to phosphorylation in terms of its impact on post-translational control of protein function. Approximately half of the ~650 human ubiquitin ligases are predicted to be Cullin–Ring Ligases (CRLs). The collective impact of CRLs on protein turnover is substantial: ~20 percent of all proteasome-dependent degradation is CRL-dependent (Soucy et al., 2009).
CRLs are modular multi-subunit complexes that recruit E2 via their RING domain subunit and recruit substrate via a variable subunit linked to the cullin by an adaptor protein. For instance, the human CRL SCFβ-TrCP is composed of four subunits: the RING protein Rbx1/Roc1/Hrt1, the cullin protein Cul1, the adaptor protein Skp1, and the substrate recruitment factor, β-TrCP. CRL activity is stimulated by covalent modification of a conserved lysine on the cullin subunit with the ubiquitin-like protein Nedd8 (Pan et al., 2004). Nedd8 conjugation causes a massive conformational change in the Cul1–Rbx1 complex that modestly improves the affinity of the E2 enzyme Cdc34 for SCF and brings the active site of E2 into close proximity to a substrate bound to β-TrCP (Duda et al., 2008; Saha and Deshaies, 2008; Yamoah et al., 2008).
Cul1-based SCF complexes, which are the best-characterized CRLs, can work with different E2s in vitro, but their only genetically-validated E2 partner is Cdc34 (Schwob et al., 1994). Members of the Cdc34 family are distinguished by an extensive C-terminal tail in which at least 50 % of the residues are acidic. This tail is essential for Cdc34 function in vivo (Mathias et al., 1998), binds to SCF, and when grafted onto the E2 Rad6, can confer Cdc34 function (Kolman et al., 1992; Silver et al., 1992), indicating that the tail is sufficient to redirect the activity of Rad6 to the SCF pathway.
Ubiquitylation by Cdc34-SCF is processive and ~75% of modified substrates acquire a degradation-competent chain with ≥4 ubiquitins in a single encounter with SCF (Saha and Deshaies, 2008). Although processive ubiquitylation of substrates by Cdc34-SCF is likely to be important for their efficient recognition by the proteasome and subsequent proteolysis, the mechanism by which SCF or any other CRL achieves processivity remains unsolved. Indeed, careful inspection of the biochemical parameters of substrate ubiquitylation by Cdc34-SCF reveals a perplexing conundrum. The interface that E2s employ to bind RING domain E3s also mediates interaction with E1 (Eletr et al., 2005). Thus, a discharged E2 must dissociate from the RING domain to be recharged with ubiquitin. For substrate to acquire a long polyubiquitin chain while bound to SCF, it must stay put through multiple cycles of E2~Ub-RING association, ubiquitin transfer, and E2 dissociation. However, this seems incompatible with existing biochemical data. For example, a β-Catenin peptide substrate acquires upwards of 10 ubiquitins in the 2–3 seconds (Saha and Deshaies, 2008) it remains bound to SCF even though Cdc34 binds SCF very tightly with an equilibrium dissociation constant (Kd) of ~20 nM. Protein-protein interactions that are diffusion-limited typically exhibit on-rates ≤ 106 M−1 sec−1 (Alsallaq and Zhou, 2008; Schreiber et al., 2009). If this is the case with Cdc34 binding to Nedd8-conjugated SCF (Saha and Deshaies, 2008) the off-rate would equal 0.02 sec−1, which would be far too slow to sustain assembly of a ubiquitin chain prior to substrate dissociation. Thus, the following question emerges: how does the Cdc34–SCF complex achieve rapid dynamics of assembly and disassembly while maintaining high affinity? Here, we set out to address how the biophysical properties of the E2-E3 complex contribute to processive ubiquitylation. We uncover an exceptionally dynamic, electrostatically-driven interaction between Cdc34 and SCF that we suggest is fundamental to the operation of all CRLs.
Results
Cdc34 and SCF bind and dissociate very rapidly
The known role of the Cdc34 tail in binding SCF first suggested to us that Cdc34 might transiently dissociate from the RING interface while remaining tethered by tail-SCF interaction, such that multiple cycles of ubiquitin discharge and recharging could occur without Cdc34 ever dissociating fully from SCF. To test this hypothesis, we set out to measure the dynamics of Cdc34-SCF interaction using the Cul1–Rbx1 subcomplex that is responsible for recruiting E2. Using a FRET-based read-out (Saha and Deshaies, 2008), we measured a dissociation rate constant (koff) of 35 ± 1 sec−1 and a Kd of 7.4 ± 0.7×10−8 M for the Cdc34-RCCFP (Rbx1 plus Cul1-CFP) complex at 50 mM NaCl (Figure 1a). The association rate constant, kon, was estimated from Kd and koff to be 4.7 ± 0.6×108 M−1sec−1 (we were unable to measure directly the rate at which Cdc34 binds RCCFP due to the limit of sensitivity of the FRET assay coupled with the exceptionally fast kon). Kinetic parameters for complex formation between Cdc34 and Nedd8-conjugated RCCFP varied only ~2-fold regardless of whether or not Cdc34 was charged with ubiquitin (Figure S2a,b). Moreover, koff varied less than 2-fold regardless of whether Cdc34 binding was measured with RCCFP heterodimer or SCF holoenzyme (Figure 1a vs. S2c). As noted in the Introduction, conjugation of Nedd8 to Cul1 enhances affinity SCF for Cdc34 (Saha and Deshaies, 2008), and consistent with this we did see a ~5 fold decrease in Cdc34 off-rate upon Nedd8 conjugation (Figure 1a vs. Figure S2a), but the rate of dissociation from neddylated RCCFP was still faster than the maximal rate of ubiquitin transfer (Saha and Deshaies, 2008). Thus, the rapid dynamics we observed were not peculiar to a particular assembly or modification state of Cdc34 and SCF.
Figure 1.
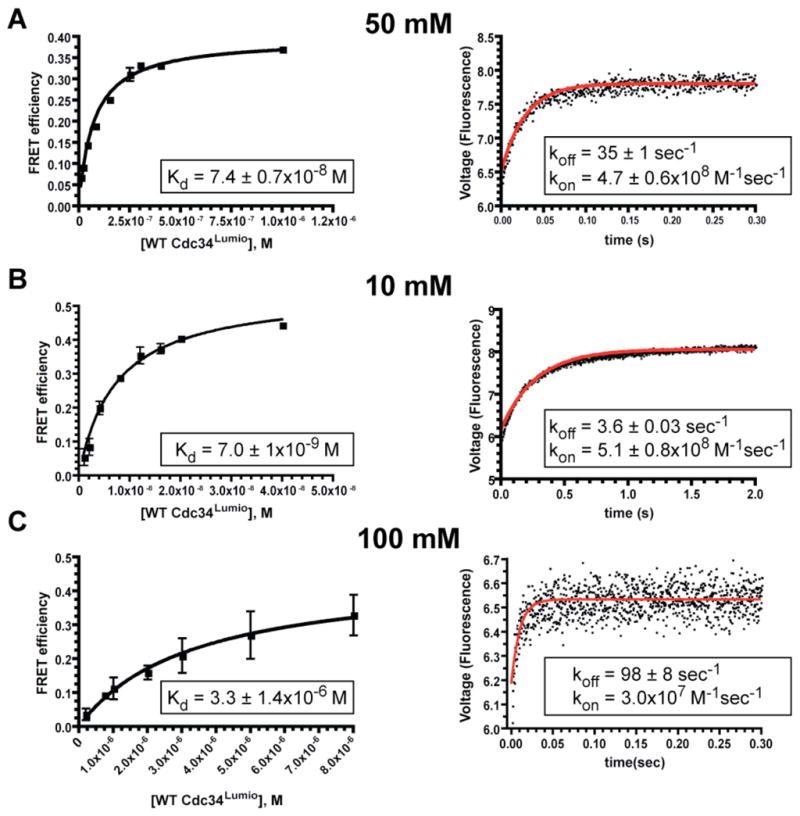
The kinetics of complex formation for human Cdc34 and RCCFP were dependent on the ionic strength of the reaction buffer. The Kd (left panels) and koff (right panels) were measured at equivalent ionic strength. kon was then estimated from kon = koff/Kd. Error bars represent the standard deviation of at least duplicate measurements of all data points. (A) 50 mM NaCl. (B) 10 mM NaCl. (C) 100 mM NaCl.
Our observation that the rate of Cdc34 dissociation (35 ± 1 sec−1 for unmodified RCCFP and 6 sec−1 for neddylated RCCFP) exceeded the maximum rate of ubiquitin-ubiquitin bond formation by Cdc34-SCF (~2–4 sec−1) (Petroski and Deshaies, 2005b; Saha and Deshaies, 2008) by ~2-fold was critical in that it invalidated the ‘tail tether’ hypothesis and suggested instead that multiple ubiquitin transfers can occur during the interval of time that substrate is bound to SCF because E2 cycles on and off the complex extremely rapidly. Given that these rapid dynamics enable ubiquitin chain synthesis to occur on the requisite time-scale, we sought to determine their molecular basis.
Electrostatic binding of Cdc34 to SCF is mediated by its acidic tail
The association kinetics for Cdc34 and RCCFP were faster than is typically seen for proteins that interact via random collision, but such rates have been observed for interacting proteins that pre-align each other through electrostatic interactions, such that a higher than expected fraction of collisions result in productive binding (Sheinerman et al., 2000). Thus, we reasoned that the acidic tail of Cdc34 might mediate the ultra-fast dynamics of Cdc34-RCCFP transactions.
Protein-protein interfaces sustained by electrostatic interactions are extremely sensitive to salt, which shields the attractive force between opposite charges (Schreiber and Fersht, 1993, 1996). Consistently, salt had profound effects on both kon and koff for Cdc34-RCCFP. Increasing the ionic strength from 10 mM (Fig. 1b) to 100 mM (Fig. 1c) resulted in a 470-fold change in Kd, which was driven by a 27-fold increase in koff and estimated 17-fold decrease in kon. Strikingly, even a modest increase in NaCl from 50 mM to 100 mM slowed kon by 16-fold (Fig. 1b,c). Similar results were observed regardless of whether the fluorescent reporter on Cdc34 was appended to the N-terminus or C-terminus (Figure S3).
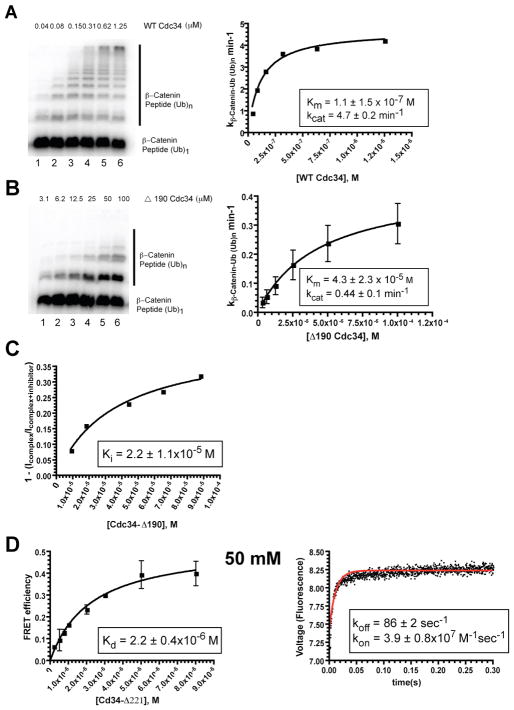
We next sought to test the contribution of the acidic tail to the function and dynamics of Cdc34-SCF by constructing a mutant, Cdc34-Δ190, that lacked the entire acidic region of Cdc34’s tail (see Fig. S1 for an alignment of Cdc34 sequences). Both WT and Cdc34-Δ190 were titrated into multiturnover ubiquitylation reactions containing a mono-ubiquitylated β-Catenin peptide substrate and SCFβ-TrCP complex with the goal of measuring both Km and kcat for these E2s. Note that kcat is defined here in terms of substrate conversion. Thus, a kcat of ~4 per minute means that each enzyme molecule turned over four substrates per minute. Meanwhile, each of those substrates might receive multiple ubiquitins. Thus kcat should not be confused with the rate of ubiquitin transfer. Deletion of the tail resulted in a ~400-fold increase in Km (Figure 2a and 2b), suggesting a severe defect in SCF binding. While direct binding assays were not possible with Cdc34-Δ190, we were able to measure Ki for the competitive inhibition of WT Cdc34-RCCFP complex formation (Figure 2c). The Ki for Cdc34-Δ190 was similar to the Km, which supports the notion that Cdc34’s tail shifts the equilibrium of Cdc34-SCF binding by approximately 2 orders of magnitude. In addition to the binding defect, deletion of the Cdc34 tail resulted in a 10-fold reduction in kcat, possibly by influencing the orientation of the bound Cdc34~Ub.
Figure 2.
The acidic tail promotes rapid assembly of functional Cdc34-SCF complexes. Ubiquitylation of a mono-ubiquitylated β-Catenin peptide by SCFβ-TrCP plus either WT (A) or Cdc34-Δ190 (B). The plot on the right shows substrate turnover (min−1) versus Cdc34 concentration. Error bars in (B) represent the standard deviation for duplicate values of data points. The experiments in panels A and C were performed in triplicate using a slightly different titration series each time. Therefore error bars are not given. The error of the measurement is the standard deviation of the three measurements. Representative data are shown. Note that nearly 100 percent of Cdc34-Δ190 protein became thioesterified with ubiquitin under these assay conditions (Table S2), even when assayed at 100 μM, the highest concentration of Cdc34-Δ190 in the titration series (Figure S7) (C) Competition binding experiment between a RCCFP complex that was saturated with Lumio Green labeled WT Cdc34 and titrated with increasing concentrations of unlabeled Cdc34-Δ190. I is the fluorescence intensity. (D) The Kd for Cdc34 Δ221 (left panel) and koff (right panel) were measured at equivalent NaCl (50 mM) and kon was then estimated as in Figure 1.
To evaluate whether the acidic tail of Cdc34 contributed to the unusual dynamics of Cdc34-RC interaction, we generated a partial truncation that was deleted for amino acids 222–238, Δ221, and measured the kinetics of binding to RCCFP. It was essential to use a partial truncation, because complete deletion of the tail destabilized the Cdc34-SCF complex so profoundly that it was not possible to measure the binding parameters directly. At 50 mM NaCl, koff for Cdc34-Δ221 (86 ± 2 sec−1) was modestly (~2.5-fold) faster than WT Cdc34 and estimated kon was ~10-fold slower (3.9 ± 0.8×107 M−1sec−1) (Figure 2d). Taken together, these results provide evidence that electrostatic interactions were important for complex formation and that the acidic tail of Cdc34 promoted rapid association with RCCFP.
Cdc34’s acidic tail binds a basic surface on Cul1 that is highly conserved
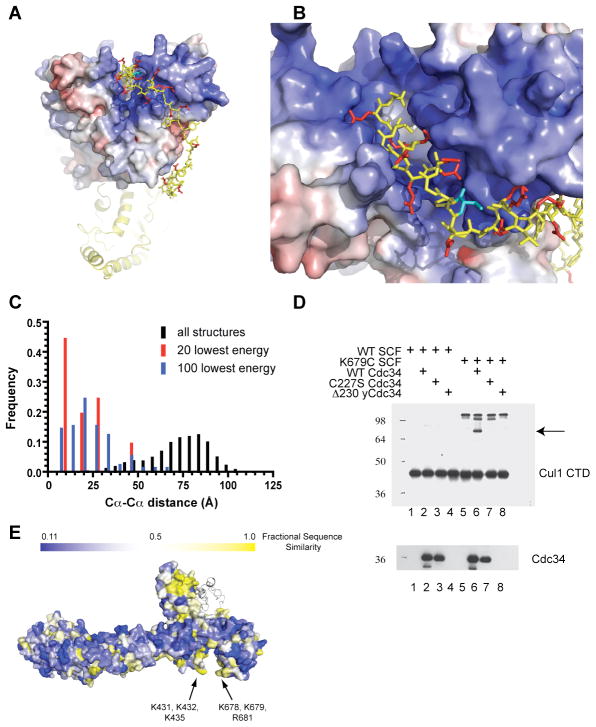
Given that Cdc34’s acidic tail potently enhanced Cd34-SCF interaction, we reasoned that a positively-charged binding site may exist on SCF. To locate candidate docking sites, the electrostatic surface potential of Cul1 was calculated and plotted onto the molecular surface. A broad, positively-charged surface was identified (Figure 3b,c), located on a convex feature resembling a canyon (the ‘basic canyon’) underneath Rbx1 in the C-terminal domain (CTD) of Cul1.
Figure 3.
The underside of cullin-RING complexes is dominated by a basic canyon. (A) Molecular surface representation of a modeled SCF complex with the Cdc34 catalytic domain docked on the RING subunit. Arrows identify the location of the KR claw (see Figures 4e and Figure 5). The model was generated by homology modeling the Cdc34 and Rbx1 subunits to the UbcH7-Cbl x-ray structure and by structural alignment of the cyclin E-Fbw7-Skp1 x-ray structure to the Skp1-Cul1-Rbx1 structure (Hao et al., 2007; Zheng et al., 2002; Zheng et al., 2000) (B,C) Electrostatic surface plot of Cul1 CTD-Rbx1, where blue is positively-charged and red is negatively-charged. For reference, the orientation of the CTD in panel B is the same as the structure in the right panel of (A). (D) Electrostatic surface plot of Cul5 CTD-Rbx1. The orientation of Cul5 is similar to Cul1 in panel B. (E) Electrostatic surface plot of neddylated Cul5 CTD-Rbx1. The electrostatic potential was calculated with APBS (Baker et al., 2001) and figures were made in PYMOL. The range of the electrostatic potential (kT/e) is given at the bottom.
The activity of human CRLs is significantly increased when the ubiquitin-like protein Nedd8 is conjugated to a conserved lysine in the CTD of the cullin subunit (Pan et al., 2004). The atomic structure of the human Cul5 CTD bound to Rbx1 has been solved in both the unmodified and Nedd8-conjugated state (Duda et al., 2008). Whereas neddylation results in significant structural rearrangement of both the CTD as well as the relative orientation of Rbx1 with CTD, the positively-charged basic canyon of Cul1 is retained in both of these structures (Figure 3d,e).
To determine if Cul1’s basic canyon can accommodate Cdc34’s acidic tail, ab initio modeling of the tail was attempted using a custom protocol written with Rosetta (Das and Baker, 2008) (see Supplemental Materials for details). A docked model for the complex was created from the Cdc34 catalytic domain positioned on Rbx1-Cul1 CTD (modeled from the UbcH7-Cbl crystal structure (Zheng et al., 2000)). In silico analysis of the Cdc34 tail revealed little preference for secondary structure (Cole et al., 2008), which is consistent with the lack of alpha helical structure detectable by circular dichroism (Ptak et al., 1994). Thus, we allowed the tail to find its lowest energy configuration without imposing constraints on secondary structure. Some 16 of the 20 models with the lowest binding interaction energy between Cdc34 and CTD-Rbx1 had the acidic tail of Cdc34 nestled into the basic canyon of Cul1 (Figure 4a,b). This result is highly significant given that no additional information was used during the simulations to guide the tail to the canyon. These results indicate that the tail can reach the basic canyon and does so in low-energy conformations. Simulations with the Cdc34 tail and neddylated Cul5 CTD also resulted in a tremendous enrichment for models in which the tail fits snugly into the basic canyon (Figure S4a,b). Whereas no two top-scoring models were identical, the molecular interfaces consistently buried the same residues in proximity to each other. For instance, Cα of Cys 227 of Cdc34 was within 18 Å of Cα of Lys 679 of Cul1 in 9 of the top 20 models, an enrichment of 90-fold compared to all 28,574 models generated (Figure 4c).
Figure 4.
Cdc34’s acidic tail occupies Cul1’s basic canyon. (A) The CTD of Cul1 and Rbx1 are represented as molecular surfaces with the electrostatic potential shown. The catalytic domain of Cdc34, bound to Rbx1, is shown as yellow ribbons. The acidic tail residues are yellow balls-and-sticks. Acidic residues are red. Cys 227 of Cdc34 is cyan. All other residues in the tail are yellow. (B) Magnified view of (A), showing the tight molecular interface between the tail residues and the Cul1 basic canyon. (C) Histogram showing the binned frequencies for the distance between Cα atoms of K679 of Cul1 and Cys 227 of Cdc34 for either the 20 models with the lowest binding interaction energy, the 100 lowest, or all 28,574 models generated. (D) In vitro cross-linking between WT or K679C SCF and either WT or C227S Cdc34. Reactions were resolved by SDS-PAGE and immunoblotted with antibodies against the indicated proteins. The arrow indicates the cross-linked product between Cul1 CTD and WT Cdc34. (E) Molecular surface representation displaying the phylogenetic conservation of residues in Cul1. Residue positions are colored according to the degree of sequence similarity found in a multiple sequence alignment of five Cul1 orthologs (Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Caenorhabditis elegans, human) and human Cul2-Cul5. Bright yellow residues are conserved in all nine cullins whereas deep blue residues are unique to human Cul1. The Rbx1 subunit is shown as a gray ribbon diagram. Residues forming the KR claw are located with black arrows.
To probe the predictive value of our model, we performed in vitro disulfide cross-linking experiments. Since modeling suggested that Cys 227 of Cdc34 was close to Lys 679 of Cul1, Lys 679 was mutated to cysteine and the mutant protein was mixed with WT Cdc34 in the presence of an oxidizing reagent. A cross-linked species between Cdc34 and Cul1-K679C CTD (note that Cul1 was expressed in E. coli as two separate fragments, the N-terminal domain (NTD) and CTD, as previously described (Li et al., 2005)) formed that migrated at the predicted MW and reacted with Cul1 and Cdc34 antibodies (Figure 4d). The cross-linked product was dependent on both the K679C substitution and Cys 227 of Cdc34, as no product formed when WT Cul1 or Cdc34-C227S were used, despite the fact that two cysteine residues including the solvent-exposed active site one remained in the Cdc34-C227S protein. Thus, as predicted by Rosetta, Cdc34’s acidic tail can occupy Cul1’s basic canyon.
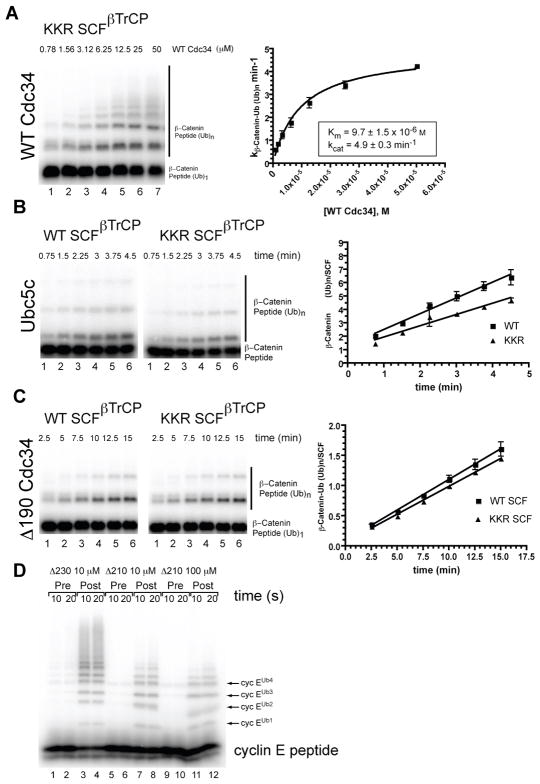
To probe further the functional significance of this interaction, we sought to generate a mutant Cul1 that cannot accommodate the acidic tail. Framing the basic canyon are two clusters of closely apposed basic residues that we refer to as the ‘KR claw’ (Figure 3a, arrows). The conserved (see below) lysine residues 431, 432 and 435 form one side of the pincer and Lys 678, Lys 679 and Arg 681 comprise the other. To evaluate the functional role of the KR claw, double and triple mutants lacking various combinations of its residues were constructed. One mutant (K678E, K679E, and R681E, referred to as ‘KKR’) assembled properly but was profoundly defective in multiturnover ubiquitylation reactions. Strikingly, the Km of WT Cdc34 for KKR SCF was nearly 10 μM (Figure 5a), which is consistent with the idea that the KKR mutation disrupts recruitment of Cdc34 to SCF.
Figure 5.
Cul1’s KR claw mediates the function of Cdc34’s acidic tail. (A) Ubiquitylation of a mono-ubiquitylated β-Catenin peptide by KKR SCF. Titration of WT Cdc34 on KKR SCF is shown. The plot on the right shows substrate turnover (min−1) versus Cdc34 concentration. (B) Time course of β-Catenin peptide ubiquitylation by 0.75 μM UbcH5c in the presence of either WT or KKR SCF. UbcH5c was used below its Km to sensitize detection of subtle perturbations to UbcH5c-SCF complex formation. (C) Same as (B) except 9 μM Cdc34-Δ190 (also below its Km) and mono-ubiquitylated β-Catenin peptide substrate were used. Error bars represent the standard deviation of at least duplicate measurements of all data points. (D) Single encounter ubiquitylation reaction with yeast SCFCdc4 comparing Δ230 and Δ210 mutants of yeast Cdc34. Pre: labeled cyclin E peptide was pre-incubated with SCFCdc4 and unlabeled competitor prior to initiating the reaction by adding a mixture of E1/E2/Ub/ATP. Post: same as Pre, except that the unlabeled competitor peptide was introduced via the E1/E2/Ub/ATP mix.
If the KKR mutation in SCF is truly specific to the acidic tail of Cdc34, KKR SCF should be comparable in activity to WT SCF when assayed with E2s that do not contain acidic tails. UbcH5c, an E2 lacking an acidic tail yet commonly assayed with mammalian SCF, yielded similar kcat/Km values when assayed with WT or KKR-SCF (Figure 5b). Similarly, Cdc34-Δ190 exhibited identical kcat/Km values with WT and KKR-SCF (Figure 5c). As a final control, KKR SCF did not abolish the stimulatory effect of Nedd8 conjugation, further demonstrating the specificity of this mutant (Figure S5).
The major contribution of basic residues in the KR claw to Cdc34 recruitment inspired us to explore more deeply the conservation of this region. A multiple sequence alignment (Figure S6) reveals that the basic residues comprising the KR claw are conserved in Cul1 orthologs from yeast to humans, as well as in human Cul2-Cul5. More generally, the molecular surface of the KR claw is highly conserved in Cul1 orthologs and homologs (yellow regions in Figure 4e), pointing to a general role for this landmark in CRL function. Interestingly, the KR claw is conserved in Schizosaccharomyces pombe, which does not have a detectable Cdc34 ortholog. Thus, multiple SCF cofactors may converge on this striking surface feature.
The acidic tail enables processive ubiquitin chain assembly on substrate
The exceptional dynamics of Cdc34-SCF interaction that are afforded by the electrostatic and structural complementarity of Cdc34’s acidic tail and Cul1’s basic canyon may enable rapid cycles of Cdc34 recruitment during polyubiquitin chain synthesis. To address this hypothesis, we performed ‘single encounter’ reactions at physiological salt concentration with the yeast CRL SCFCdc4 and a phospho-peptide substrate derived from the human cyclin E1 protein (see methods) (Nash et al., 2001; Orlicky et al., 2003). Two yeast Cdc34 derivatives that either retain (Δ230) or lack (Δ210) the acidic region of the tail were tested for processive ubiquitylation of the peptide substrate. A single encounter experiment measures the ubiquitin transfer that occurs before labeled substrate dissociates and is replaced by unlabeled competitor. Whereas Cdc34-Δ230 sustained conversion of a substantial amount of substrate into highly ubiquitylated product, Cdc34-Δ210 activity was significantly perturbed in that little substrate was converted to products containing 4 or more ubiquitins (Figure 5d). Chain synthesis was not rescued by increasing Cdc34-Δ210 to 100 μM, indicating that in addition to promoting rapid binding, the acidic tail influenced the rate of ubiquitin transfer within the substrate-SCF-Cdc34~Ub complex.
Discussion
Fast, electrostatically-driven E2-E3 dynamics underlie processive ubiquitin chain synthesis upon substrate
Ubiquitin ligases that target substrates for degradation by the proteasome have a challenging job. In the specific cases of β-TrCP binding to β-Catenin substrate peptide and Cdc4 binding to cyclin E substrate peptide, the substrates bind with Kd values of 0.1–1.0 μM and dissociate with off-rates between ~0.1 – 1 sec−1 (Saha and Deshaies, 2008) (N. Pierce, unpublished data). These off-rates must strike a balance between being sufficiently slow to allow for ubiquitin chain assembly, but sufficiently fast that the ligase does not become product-inhibited. In the limited time-frame that a substrate is engaged, it must be conjugated with at least four ubiquitins to serve as a competent ligand for the proteasome (Thrower et al., 2000). If a substrate were to dissociate before receiving four ubiquitins, it would run the risk of being ‘edited’ by isopeptidases (Lam et al., 1997) or interacting, prematurely, with the multitude of ubiquitin-binding proteins in the cell that govern non-proteolytic functions of ubiquitin (Kirkin and Dikic, 2007). Given that E2s employ overlapping surfaces to bind RING domains and E1 (Eletr et al., 2005), the E2-RING interface must form four times and dissociate three times in the course of assembly of a minimal tetraubiquitin signal on a bound substrate. Our data indicate that an electrostatically-driven interaction between the acidic tail of Cdc34 and a basic surface on Cul1 makes it possible for Cdc34-SCF complexes to form with high affinity yet cycle with exceptionally rapid kinetics. It is possible that a small fraction of successive ubiquitin transfer events are mediated by a single molecule of Cdc34 that is recharged while being held to SCF by its acidic tail. However, given that the off-rate reported here (≥ 6 sec−1) and the fastest ubiquitin transfer rate that we have been able to measure (~2–4 sec−1; (Petroski and Deshaies, 2005b; Saha and Deshaies, 2008)), at best only a small fraction of transfers could occur in this manner and the synthesis of a ubiquitin chain must require multiple rounds of E2 recruitment.
At the reaction rates we observe, the rate-determining step for ubiquitin chain assembly is the chemical step of ubiquitin transfer (~2–4 sec−1) (Petroski and Deshaies, 2005b). However, depending upon the concentration of Cdc34 in different cells and compartments, it is possible that in some instances Cdc34 binding and dissociation are rate-limiting. Regardless, the fast on-rate and off-rate of Cdc34–SCF interaction establishes conditions conducive to rapid, processive assembly of a substrate-linked polyubiquitin chain. The presence of acidic tail sequences on Cdc34 orthologs and basic surfaces on all cullins indicate that this particular solution to the problem of how to achieve high affinity yet dynamic binding of E2 is broadly conserved throughout eukaryotic evolution. However, patterns of conservation also suggest that at least one other conserved CRL regulator, possibly including Dcn1 (Kurz et al., 2005), Nedd8 E2 (Huang et al., 2009), UbxD7 (Alexandru et al., 2008), or CSN (Lyapina et al., 2001) engages the same notable landmark on cullins.
The role of electrostatic interactions in Cdc34-SCF function
Work on other macromolecular interactions has established the general principle that electrostatic interactions enable association rates that exceed by several orders of magnitude the diffusion-limited on-rate of 105–106 M−1 sec−1 (Fersht, 1999; Schreiber et al., 2009; Sheinerman et al., 2000). To understand how this works requires a brief digression. Protein-protein interaction proceeds via formation of a ‘transient complex’ that is similar in general orientation to the final ‘native complex’, but is held together by long-range interactions (Alsallaq and Zhou, 2008; Schreiber et al., 2009). High affinity binding occurs when the proteins in the transient complex realign and pack against each other to form the numerous short-range interactions that stabilize the native complex (Harel et al., 2009). The partners in a transient complex have a much higher propensity to interact stably with each other compared to molecules that are free in solution, due to the great reduction in the translational and rotational degrees of freedom. Importantly, formation of transient complexes is driven exclusively by attractive electrostatic forces between the interacting species, and it is by this mechanism that electrostatic interactions drive rapid on-rates (Alsallaq and Zhou, 2008; Schreiber et al., 2009).
How does electrostatically-driven interaction benefit Cdc34–SCF? If Cdc34 bound SCF exclusively through short-range interactions (e.g. van der Waals), the kon would fall in the diffusion-limited range. To achieve rates of complex formation of ~10 sec−1 would require an intracellular concentration of 10–100 μM Cdc34. Moreover, for the affinity to be in the 20–100 nM regime reported here, koff would then be 0.002–0.1 sec−1, which is far too slow to complete ubiquitin chain synthesis prior to substrate dissociation ((Saha and Deshaies, 2008), Pierce personal communication). Electrostatically-driven binding presents an optimal solution, in that it occurs rapidly even at low concentrations of Cdc34 and is of high affinity, yet allows for a fast koff. Accordingly, electrostatically assisted macromolecular association is seen in other settings where speed and accuracy are in simultaneous demand, including charging of tRNA by amino-acyl tRNA synthetases (Creighton, 1993; Fersht, 1999).
Ubiquitin chain synthesis: one problem, multiple solutions
In addition to CRLs, other E3s may employ basic surfaces to mediate dynamic, electrostatically-driven interaction with E2. Two of the nine other E2s encoded in the budding yeast genome (Ubc8 and Ubc2/Rad6) contain acidic stretches at their C-termini. Moreover, yeast Cdc34 is also known to function with the E3 San1, raising the possibility of a basic docking site on this protein.
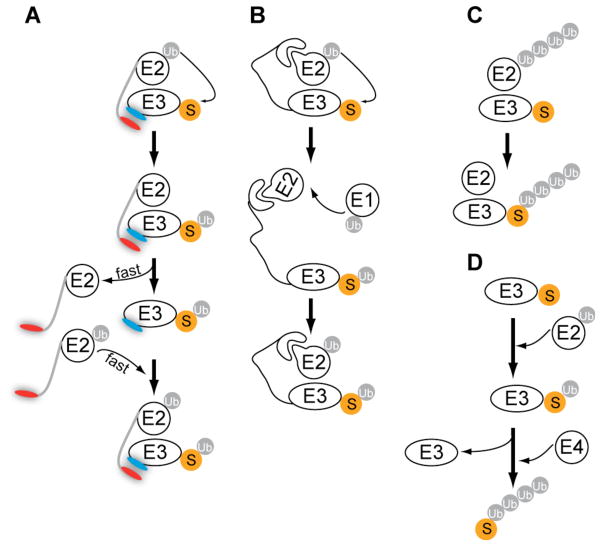
Although the mechanism of rapid E2 cycling described here appears to apply broadly to the CRL family of E3s (Figure 6a), we suggest it is not the only mechanistic innovation that arose during evolution to enable processive ubiquitylation on short timescales. Recharging of spent E2 requires that residues within the H1 α-helix that are normally buried at the E2-E3 interface (Huang et al., 2005; Zheng et al., 2000) must become exposed to engage E1. Whereas complete dissociation of spent E2 is one means to achieve this (Figure 6a), it is also conceivable that the E2-E3 pair be held together indefinitely by a non-dissociable tether that allows E2 to disengage from the RING interface without completely dissociating from E3 (Figure 6b). An extreme version of this strategy is presented by the apoptosis regulator BRUCE, which contains E2 and E3 modalities fused in a single polypeptide (Pohl and Jentsch, 2008). This may also be the case for both Ube2g2 (Chen et al., 2006) and Ubc2/Rad6 (Xie and Varshavsky, 1999), which bind tightly to gp78 and Ubr1, respectively, via sequences other than the RING domain. Likewise, Ubc1 could potentially use its UBA domain to remain bound to ubiquitylated substrate while the RING-E2 interface dissociates to enable recharging by E1 (Rodrigo-Brenni and Morgan, 2007). Another possible solution is to build the ubiquitin chain on E2 and transfer it en bloc to substrate (Li et al., 2007) (Figure 6c). Finally, there may be cases where the E3 recruits multiple E2s (which may be of a distinct nature), which then take turns in transferring ubiquitin to substrate (Tang et al., 2007). We suggest that this diversity of mechanisms came to be because evolution acted to maximize the rate of polyubiquitin assembly and not the specific means for achieving this end. This has resulted in multiple solutions to a singular problem. The task of how to build a ubiquitin chain before substrate dissociates from its cognate ligase is likely to be a pervasive one in the ubiqutin system. This challenge may be experienced most acutely by quality-control E3s, which by their very nature are likely to recognize relatively degenerate, low-affinity motifs in mis-folded substrates. This could explain the existence of secondary E2 tethers in Ubr1 and gp78, both of which are implicated in quality control (Chen et al., 2006; Eisele and Wolf, 2008), and ‘downstream’ E3s such as Ufd2, Ufd4, and Hul5 that can recognize a monoubiqutin conjugate as a degron (Crosas et al., 2006; Johnson et al., 1992) (Figure 6d). Because the second ligase recognizes a defined degron (ubiquitin), the dynamics can readily be tuned so that all substrates remain bound long enough to acquire a degradation-competent chain, regardless of their initial affinity for E3.
Figure 6.
Schematic of four distinct mechanistic solutions that may enable polyubiquitylation of protein substrates in a single encounter with enzyme. See text for details. (A) Rapid dynamics of E2 and E3 association and dissociation, an example of which is the CRLs and Cdc34. The protein substrate is orange and labeled with the letter S. In this example, rapid dynamics are facilitated through the negatively charged acidic tail on E2 (red) and the positively charged region on the E3 (blue). (B) E2 never fully dissociates and is recharged while bound to ubiquitin ligase. (C) en bloc transfer of pre-assembled chain. (D) Substrate is first mono-ubiquitylated by a conventional ubiquitin ligase, after which substrate can bind a chain-elongating ubiquitin ligase to complete polyubiquitin chain formation.
Materials and Methods
Expression and Purification of Recombinant Proteins
All proteins were recombinantly expressed in either E. coli or Hi5 insect cells and purified using standard procedures (Table S1). The final buffer condition for all proteins except those to be labeled with Lumio Green was 30 mM Tris-Cl (pH 7.5), 100 mM NaCl, 1 mM DTT and 10 percent glycerol. The final buffer condition for proteins to be labeled was 20 mM Tris-Cl (pH 7.5), 100 mM NaCl, 2 mM TCEP (Thermo Scientific), 1 mM EDTA, and 5 percent glycerol.
The Rbx1-Cul1 (RC) and Rbx1-Cul1-CFP (RCCFP) complexes were expressed using a previously described ‘Split-n-Coexpress’ protocol (Li et al., 2005) where the Cul1 protein is expressed as two fragments, referred to as the N-terminal domain (NTD) and the C-terminal domain (CTD). This system allows higher expression of these complexes in E. coli. With the exception of Figure 5d, all experiments in the paper were performed with human proteins. The yeast SCFCdc4 complex used in the experiment to produce Figure 5d was generated and purified from baculovirus infected insect cells as previously described (Petroski and Deshaies, 2005a). Mammalian ubiquitin was used for all relevant assays (Boston Biochem).
FRET measurement and quantification
Cdc34-labeling reactions were performed using Lumio Green (Invitrogen) as described previously (Saha and Deshaies, 2008). Lumio Green reacts irreversibly with the amino acid sequence CCPGCC. Briefly, purified Cdc34 (WT Cdc34 with the amino acid sequence CCPGCCHHHHHH appended to the C-terminus, WT Cdc34 with MGCCPGCCGSG appended to the N-terminus and HHHHHH at the C-terminus, or Cdc34-Δ221 with CCPGCCHHHHHH at the C-terminus) were incubated at 30 μM with 40 μM Lumio Green in a buffer containing 20 mM Tris-Cl (pH 7.5), 100 mM NaCl, 2 mM TCEP (Thermo Scientific),1 mM EDTA and 5 percent glycerol at room temperature (20–22°C) for 2 hours. The efficiency of coupling was determined by measurement of the intact mass of reacted proteins. C-terminally labeled WT and Δ221 Cdc34 reacted to completion with Lumio Green. WT human Cdc34 with the Lumio Green at the N-terminus reacted to 80 percent completion.
Equilibrium fluorescence measurements were carried out on a FluoroLog-3 or FluoroLog-4 spectrofluorimeter (Jobin Yvon). Binding reactions containing RCCFP (at a concentration below Kd) and various concentrations of Cdc34 proteins were incubated for 20 minutes at room temperature (20–22°C) in a buffer containing 20 mM Tris-Cl (pH 7.5), various concentrations of NaCl to regulate the ionic strength of the solution, 0.5 mM DTT and 5 percent glycerol. Samples were excited at 430 nm, and the emission spectra were acquired from 450 to 570 nm. FRET efficiency was calculated as described (Saha and Deshaies, 2008). Kd was estimated by fitting titration curves to a hyperbolic equation assuming a one-site binding model (Prism). All measurements were done in at least duplicate.
Competitive inhibition experiments of Lumio Green labeled WT Cdc34 (1 μM) and RCCFP (50 nM) complex formation were carried out on the same instrument as above. Various concentrations of Cdc34-Δ190 were introduced to individual reactions and incubated at room temperature (20–22°C) for 20 minutes. Identical buffer conditions as above with 50 mM NaCl were used. Data were processed using custom PERL software and Ki was measured using Prism.
Measurement of koff was performed using a Kintek stopped-flow instrument. Complex formation between Lumio Green labeled Cdc34 and RCCFP (1 μM for WT Cdc34 and 0.15 μM RCCFP at 10 and 50 mM NaCl, 6 μM for WT Cdc34 and 0.3 μM RCCFP at 100 mM NaCl, and 6 μM for Cdc34-Δ221 and 0.3 μM RCCFP at 50 mM NaCl) was accomplished by incubating the proteins in the same reaction buffer used for equilibrium binding experiments. Dissociation was initiated by mixing a 10-fold excess of unlabeled Cdc34 protein with the labeled Cdc34-RCCFP complex. At least 8 independent measurements were taken and averaged before fitting the data to a single exponential equation (Prism). We were unable to measure directly the rate at which Cdc34 binds RCCFP due to the limit of sensitivity of the FRET assay coupled with the exceptionally fast kon.
In vitro cross-linking
Cdc34 (10 μM of WT or C227S) was mixed with 0.8 μM RC in PBS. Cross-linking was initiated by adding 10 μM Sodium tetrathionate (Sigma) for 1 minute at 22°C. Reactions were quenched by adding 625 μM NEM and evaluated by non-reducing SDS-PAGE followed by Western Blot analysis. Because the Cul1 subunit is expressed as two fragments in E. coli, Cdc34 forms cross-links to the CTD fragment of Cul1. WT Cdc34 and Cul1-CTD migrate upon SDS-PAGE at apparent MWs of ~ 35 kDa and ~45 kDa, respectively. Therefore the expected migration of a Cdc34-CTD cross-linked species is approximately 80 kDa.
Multiple Sequence Alignments and Calculation of amino acid sequence similarity
All multiple sequence alignments (MSAs) were performed using the MUSCLE algorithm and online server (Edgar, 2004). For the Cul1 MSA, custom PERL software was written to analyze the sequence similarity for each position of the alignment. Fractional sequence similarity was calculated by counting each amino acid at a given position in the MSA that was identical or similar to the amino acid occupying that position for Cul1. This sum was then divided by 9, the total number of sequences in the MSA. Amino acid pairs were considered similar in the following cases: Arg and Lys, Glu and Asp, Asn and Gln, Ser and Thr, and Leu and Ile.
Ubiquitylation Assays
Ubiquitylation assays were performed using either the β-Catenin peptide or a β-Catenin peptide with one conjugated ubiquitin as previously described (Saha and Deshaies, 2008), or an N-terminally acetylated (Ac) cyclin E peptide containing a Protein Kinase A phosphorylation site and a six histidine tag at the C-terminus (N-term-AcKAMLSEQNRASPLPSGLL(phospho Thr)PPQ(phospho Ser)GRRASY HHHHHH-C-term). Note that a cyclin E peptide with nearly identical amino acid sequence binds with high affinity to yeast SCFCdc4 and that the cyclin E degron in this peptide can replace the natural phosphodegron in Sic1 and support SCFCdc4-dependent degradation of Sic1 in vivo (Nash et al., 2001; Orlicky et al., 2003). Moreover, human cyclin E is degraded in a Cdc4-dependent manner when expressed in yeast (Strohmaier et al., 2001). Briefly, 50 μM β-Catenin peptide, 30 μM Ub-β-Catenin peptide, or 0.6 μM cyclin E peptide were labeled with 5 kU of cAMP-dependent protein kinase (NEB) in the presence of [γ32P]ATP for one hour at 30°C.
All ubiquitylation experiments were performed at room temperature (20–22°C) in the following buffer: 30 mM Tris-Cl (pH 7.5), 20 mM NaCl, 5 mM MgCl2, 2 mM DTT and 2 mM ATP (See Table S2 for the concentration of proteins). Reactions were initiated by adding labeled substrate and quenched by the addition of 2X reducing SDS-PAGE buffer. Note that reactions in Figure 2a and 2b were assayed with purified neddylated-Cul1-Rbx1 (Saha and Deshaies, 2008). Reactions with WT Cdc34 (Figure 2a) and Cdc34-Δ190 (Figure 2b) were quenched after 30 seconds and 30 minutes, respectively. Reactions with WT Cdc34 and KKR SCF (Figure 5a) were quenched after 5 minutes. All samples were resolved by SDS-PAGE, phosphor-imaged, and quantified using Image Quant (G.E. HealthCare). Estimation of Km and kcat were done by fitting at least duplicate measurements of data points to the Michaelis-Menten equation (Prism software).
For the ubiquitylation reactions with neddylated WT and KKR SCF (Figure S5), 20 nM APPBP1-UBA3 (Nedd8 E1), 2.5 μM Ubc12, 1.5 μM Nedd8, and 0.6 μM Cul1-Rbx1 were incubated in the presence of ATP for 20 minutes. These reactions were then used to assemble a final reaction with the following components: 5 μM Ub-β-Catenin peptide, 1.3 μM human E1, 0.5 μM WT Cdc34, 60 μM ubiquitin, and 0.15 μM βTrCP-Skp1. Reactions were initiated and processed in an identical manner to the previously described ubiquitylation reactions.
For the single encounter experiment, reactions were performed with yeast proteins in the following buffer: 30 mM Tris-Cl (pH 7.5), 150 mM NaCl, 2 mM DTT, 5 mM MgCl2, and 2 mM ATP. E1, Ub, Cdc34 and ATP were pre-incubated to form Cdc34-ubiquitin thioesters (tube 1). In a separate tube, yeast SCF and labeled substrate were incubated (tube 2). The reactions were initiated when tubes 1 and 2 were mixed.
The addition of the cold chase peptide to either tube 1 or tube 2 determined product formation. When chase peptide was added to tube 2, it competed with the labeled peptide for binding to SCF. Very little product was formed under these conditions since the chase peptide was in great excess over the labeled one (Table S2). When chase peptide was added to tube 1, it competes with labeled peptide for SCF binding only after the labeled peptide dissociated from SCF.
Supplementary Material
Acknowledgments
We would like to thank Sonja Hess and members of the Caltech PEL for mass spectrometry analysis. We also thank Jost Vielmetter and members of the Caltech PEC for protein expression. We thank Shu-ou Shan for expect advice on the kinetic analysis of the data, and members of the Deshaies lab for comments on the manuscript. G.K. is a recipient of a National Institutes of Health Ruth Kirschstein Postdoctoral Fellowship (F32 GM074471-01). R.J.D. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- CTD
Cul1 C-terminal domain
- NTD
Cul1 N-terminal domain
- RCCFP
Rbx1-Cul1/Cyan Fluorescent Protein
- UPS
Ubiquitin Proteasome System
- CRL
Cullin-RING-ligase
- SCF
Skp1 Cullin Fbox ligase
- MSA
Multiple Sequence Alignment
- E2
Ubiquitin Conjugating Enzyme
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsallaq R, Zhou HX. Electrostatic rate enhancement and transient complex of protein-protein association. Proteins. 2008;71:320–335. doi: 10.1002/prot.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton TE. Proteins: structures and molecular properties. 2. New York: W.H. Freeman; 1993. [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Fersht A. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. New York: W.H. Freeman; 1999. [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Harel M, Spaar A, Schreiber G. Fruitful and futile encounters along the association reaction between proteins. Biophys J. 2009;96:4237–4248. doi: 10.1016/j.bpj.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol. 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Kolman CJ, Toth J, Gonda DK. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11:3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Li T, Pavletich NP, Schulman BA, Zheng N. High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 2005;398:125–142. doi: 10.1016/S0076-6879(05)98012-9. [DOI] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Mathias N, Steussy CN, Goebl MG. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J Biol Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. In vitro reconstitution of SCF substrate ubiquitination with purified proteins. Methods Enzymol. 2005a;398:143–158. doi: 10.1016/S0076-6879(05)98013-0. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005b;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Regulation of apoptosis and cytokinesis by the anti-apoptotic E2/E3 ubiquitin-ligase BRUCE. Ernst Schering Found Symp Proc; 2008. pp. 115–126. [DOI] [PubMed] [Google Scholar]
- Ptak C, Prendergast JA, Hodgins R, Kay CM, Chau V, Ellison MJ. Functional and physical characterization of the cell cycle ubiquitin-conjugating enzyme CDC34 (UBC3). Identification of a functional determinant within the tail that facilitates CDC34 self-association. J Biol Chem. 1994;269:26539–26545. [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Fersht AR. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- Schreiber G, Haran G, Zhou HX. Fundamental Aspects of Protein-Protein Association Kinetics. Chem Rev. 2009 doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Silver ET, Gwozd TJ, Ptak C, Goebl M, Ellison MJ. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial Orientation of the SCF(Cdc4) Dimer Accommodates Multiple Geometries for Substrate Ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.