Abstract
Hexavalent chromium Cr(VI) is known to be a carcinogenic metal ion, with a complicated mechanism of action. It can be found within our environment in soil and water contaminated by manufacturing processes. Cr(VI) ion is readily taken up by cells, and is recognized to be both genotoxic and cytotoxic; following its reduction to the stable trivalent form of the ion, chromium(Cr(III)), within cells. This form of the ion is known to impede the activity of cellular DNA polymerase and polymerase-mediated DNA replication. Here, we report the effects of chromium on the activity and fidelity of the DNA replication process mediated by the human cell DNA synthesome. The DNA synthesome is a functional multiprotein complex that is fully competent to carry-out each phase of the DNA replication process. The IC50 of Cr(III) toward the activity of DNA synthesome-associated DNA polymerases α, δ and ε is 15, 45 and 125 μM, respectively. Cr(III) inhibits synthesome-mediated DNA synthesis (IC50 =88 μM), and significantly reduces the fidelity of synthesome-mediated DNA replication. The mutation frequency induced by the different concentrations of Cr(III) ion used in our assays ranges from 2–13 fold higher than that which occurs spontaneously, and the types of mutations include single nucleotide substitutions, insertions, and deletions. Single nucleotide substitutions are the predominant type of mutation, and they occur primarily at GC base-pairs. Cr(III) ion produces a lower number of transition and a higher number of transversion mutations than occur spontaneously. Unlike Cr (III), Cr(VI) ion has little effect on the in vitro DNA synthetic activity and fidelity of the DNA synthesome, but does significantly inhibit DNA synthesis in intact cells. Cell growth and proliferation is also arrested by increasing concentrations of Cr(VI) ion. Our studies provide evidence indicating that the chromium ion induced decrease in the fidelity and activity of synthesome mediated DNA replication correlates with the genotoxic and cytotoxic effects of this metal ion; and promotes cell killing via inhibition of the DNA polymerase activity mediating the DNA replication and repair processes utilized by human cells.
Keywords: Chromium ion, Human cell DNA synthesome, DNA replication fidelity, Orgin dependent in vitro DNA replication
Introduction
Both epidemiological and laboratory studies support the perception that chromium ion is a genotoxic and cytotoxic metal ion in humans (Gibb et al., 2000). Chromium is found in the environment in two predominant forms, hexavalent chromium (Cr(VI)) and trivalent chromium(Cr(III)). Cr(VI) is transported into cells through the sulfate transporter (Arslan et al., 1987), and subsequently exerts a number of toxic effects on the cell. Exposure to chromium ion increases the risk for developing respiratory tract cancer (IARC, WHO, 1990), and is known to inhibit cell proliferation and arrest the cell division cycle (Xu et al., 1996). It also induces processes leading to cell transformation (Biedermann and Landolph, 1990) and apoptosis (D’Agostini et al., 2002), alters signal transduction pathways (Kim and Yurkow, 1996) and induces chromosomal abnormalities and gene mutations (Sr. John et al., 2002; Liu et al., 1999). Cr(VI) also induces growth arrest in cultured cells and is accompanied by the generation of reactive oxygen species that presumably trigger oxidative DNA damage (Zhang et al., 2001). Oxidative damage is associated with the generation of free radicals in cells exposed to Cr(VI) ion, and the propensity of cells to generate mutations in response to free radical damage has been reported (Singh et al., (1998); Ding and Shi (2002)). However, compounds containing Cr(VI) have in themselves little or no affect on DNA integrity and nuclear structure in vitro (Hneihen et al., 1993); suggesting that the genotoxic and cytotoxic effects of Cr(VI) are associated with the reduction of Cr(VI) to stable Cr(III) via the reactive intermediates Cr(IV) and Cr(V); following transport of the ion into the cell. Reduction of Cr(VI) requires its interaction with intracellular reducers, such as ascorbate, cysteine and glutathione (Tsou et al., 1999; Kortenkamp et al.,1990). Unlike Cr(VI) however, Cr(III) does not readily cross cell membranes and is relatively inactive in vivo. However, once inside a cell, Cr(III) binds to purified DNA to form adducts with DNA. Cr(III) is known to promote DNA cross-linking and DNA polymerase arrest (O’Brien et al., 2001). Cr(III)-induced DNA lesions decrease the fidelity of DNA replication in vitro (Snow, 1991, 1994; Singh and Snow, 1998), and have been shown to arrest in vitro DNA replication via inhibition of the activity of DNA polymerases involved in the replication process, (Bridgewater et al., 1994a, 1998). Following its transfection into human cells, Cr(III)-treated plasmid DNA also generates specific types of mutations, while undergoing replication (Tsou et al., 1997). Thus the carcinogenic properties of Cr (VI) appear to result from the induction of DNA damage following the reduction of Cr(VI) to Cr(III). Reduction of Cr(VI) ion leads to the generation of an assortment of lesions including Cr-DNA binary (mono) adducts, Cr-DNA ternary adducts, DNA protein crosslinks (DPC’s), bi-functional (DNA interstrand crosslinks (ICLs)) adducts, single-strand breaks (SSB’s) and oxidized bases (O’Brien et al., 2003). Chromium ion-DNA binding studies indicates a preference towards the formation of ternary complexes, such as guanine-Cr(III)-DNA adducts (Arakawa et al 2000), and cysteine-Cr(III)-DNA adducts (Zhitkovich et al., 2001). The large ternary ascorbate-Cr(III)-DNA cross-links are much more mutagenic and more potent replication-blocking DNA lesions than the smaller binary Cr(III)-DNA adducts, and is reported to account for more than 90% of Cr(VI) mutagenicity (Quievryn et al., 2003).
Although many different experimental approaches have been utilized to determine the mechanisms mediating the genotoxic and cytotoxic effects of chromium compounds; how chromium ion specifically induces its toxic effects remains unclear. Studies performed previously, focusing on how chromium ion affects the fidelity and activity of cellular DNA polymerases have primarily utilized highly purified individual DNA polymerase enzymes to drive a single-strand form of DNA synthesis. However, in human cells, replication of the DNA duplex results in the simultaneous synthesis of two identical daughter stands that are complementary to each of the parental DNA “template” strands. The coordinated synthesis of these nascent DNA strands requires the concerted action of two different DNA polymerases (polymerase α on the lagging DNA strand, and polymerase δ on the leading DNA strand), and a series of other proteins, which are needed to coordinate the synthesis of both the leading and lagging strands at the replication fork. This is because the replication process leading to the semi-conservative replication of double stranded DNA requires the processing and maturation of Okazaki fragments formed on the lagging DNA strand, unwinding of the parental double helix so that the replication fork can move along the DNA strand being replicated, resolution of torsional strain in the processing replication fork, as the fork moves along the DNA helix, sequestration of the individual semi-conservatively replicated DNA strands within the cell prior to cell division, and ligation of nascent DNA strands once replication of two adjacent sections of DNA (i.e., replicons) is completed. Hence, in order to more accurately explore the molecular mechanisms involved in mediating the carcinogenic effects of chromium ion, a cell-free model system that more closely resembles the intact cellular DNA synthetic machinery of cells needs to be employed. The advantages of such a model system are: 1) the tight association of all of the proteins required to perform semi-conservative DNA replication; 2) the ability of the replication products to point toward specific proteins within the model system that are specifically affected by the chromium ion, and 3) the ability to monitor the extent of damage induced by the simultaneous exposure of both the leading and lagging strand polymerases to the metal ion.
Our in vitro DNA replication model system (employing the DNA synthesome) exhibits these advantages, and has been extensively characterize and is fully competent to carry-out all phases of the DNA replication process mediated by human cells (Malkas et al., 1990; Applegren et al., 1995). The DNA synthesome contains DNA polymerases α, δ and ε, DNA primase, topoisomerases I and II, proliferation cell nuclear antigen (PCNA), replication factor C (RFC), replication protein A (RPA), 3′>5′ and 5′>3′ DNA helicases, DNA methyltransferase, poly(ADP)ribose polymerase and DNA ligase (Coll et al., 1996; Jiang et al., 2002). We have also shown it to be capable of supporting, in vitro, the semi-conservative replication of SV40 viral origin containing DNA, when assayed in the presence of SV40 large T-antigen (Tom et al., 1996). In addition, a fully competent DNA synthesome has been isolated from murine cells, and was subsequently shown to be fully competent to replicate polyoma virus origin containing DNA, (Wu et al., 1994) via a semi-conservative mechanism that was origin dependent. We therefore selected this unique and powerful model system for our investigation into the effects of Chromium ion on the DNA replication process because it is more physiologically relevant, and more closely mimics the intracellular environment, than other cell-free models used for this purpose. Because the DNA synthesome supports all phases of the DNA replication process, we believe that it can more accurately define, the effects of chromium ion on the DNA synthetic process than model systems employing highly purified DNA polymerases.
In this study, we therefore utilized the DNA synthesome model to determine the effects of chromium on the in vitro activity of synthesome-associated DNA polymerases α, δ, ε during the origin dependent replication of DNA. The effects of chromium ion on DNA replication fidelity were also determined by transformation of E. coli cells with DNA that had been newly replicated by the human cell DNA synthesome in the absence or presence of chromium ion. The frequency and spectrum of mutations induced by Cr(III) ion were examined, and compared to the effects of Cr(III) and Cr(VI) ion on human cell proliferation, cell growth cycle and intact cell DNA synthesis. Our studies provided evidence that the genotoxicity and cytotoxicity of chromium ion on human cells resulted, in part, from a reduction of the fidelity and inhibition of the activity of the DNA polymerases mediating DNA synthesis in intact cells.
Methods
Cell culture and reagents
HeLa cells were grown in DMEM medium with 10% newborn bovine serum. The rNTP, dNTP, activated calf thymus DNA, Methylthiazoleterzolium (MTT), CrCl3, Na2Cr2O7, NaCl, ZnCl2 and NaNO3 were purchased from Sigma Chemical Company (St. Louis, MO). CrCl3 (Cr(III)) and Na2Cr2O7 (Cr(VI)) were freshly prepared in deionized water prior to each experiment. Poly(dA-dT).poly(dA-dT), poly(dA). oligo(dT)12–18 DNA were bought from Amersham (GE Healthcare) (Piscataway, NJ). The radioactive nucleotides (α-32P)-dCTP, (3H)-dTTP and (3H)-thymidine were purchased from Perkin Elmer Life Science (Boston, MA).
Preparation of the DNA synthesome, SV40 large T-antigen and pBK-CMV plasmid DNA
The DNA synthesome was isolated from HeLa cells as previously described (Malkas et al., 1990; Lin et al., 1997; Jiang et al., 2000). The Q-Sepharose fraction containing the purified synthesome was isolated from the P4 fraction, using the BioLogic Duo-Flo Chromatography System (BIO-RAD, Hercules, CA). Proteins bound to the chromatography column were eluted using a salt gradient buffer (50–500 mM KCl). The single preparation of the synthesome was stored as aliquots at −80 °C and later thawed as needed and used for all of the experiments described in this manuscript. The SV40 T-antigen protein was purchased from Molecular Biology Resources, (Milwaukee, WI). pBK-CMV plasmid DNA was extracted from the XLM Blue MRF’ strain of E. coli, carrying the pBK-CMV vector (Stratagene, La Jolla. CA), and purified using CsCl-Ethidium Bromide equilibrium sedimentation (Sekowski et al., 1997).
Cell proliferation assay
HeLa cells were seeded in 96 well plates at a density of 5×104 cells/well, and incubated in 5% CO2 at 37 °C for 1 day. A range of concentrations of CrCl3, Na2Cr2O7 or NaCl were incubated with cells in each well for either 24 or 48 h. Cells were then washed in PBS and incubated in 200 μl of medium containing MTT (0.5 mg/ml) for 4 h, and the media was replaced with 200 μl DMSO to release the colored product formed by the reduction of MTT in viable cells. Cell survival correlated directly with the extent of absorbance at 570 nm.
Intact cell DNA synthesis assay
Monolayer HeLa cells (5×105) were seeded in 60 mm dishes, and grown in 5% CO2 at 37 °C for 1 day, prior to the addition of either CrCl3, Na2Cr2O7, NaCl, ZnCl2 or NaNO3 over a range of concentrations, and then incubated with these metal ions for 1, 2.5 or 8 h. The cells were washed with PBS and incubated for 1 h with (3H)-thymidine. Following incubation with radiolabeled thymidine, cells were lysed, and the DNA precipitated by the addition of 1% TCA prior to being filtered onto GF/A glass fiber filters. The amount of 3H-thymidine incorporated into DNA was quantified in a scintillation counter.
Measurement of cell cycle
5×105 HeLa cells were grown in 60 mm dishes overnight, and a range of different concentrations of Cr(III) or Cr(VI) salts were added to the culture media for an additional 24 h. Cells were then collected by centrifugation, and fixed for 1 h in 70% cold ethanol, before being suspended in 200 μl of PBS containing 0.1% Triton X-100, 100 μg/ml of RNase A, and 100 mg/ml propidium iodide (PI) for 30 min. Cell division cycle analysis was performed by flow cytometry on 10,000 cells for each sample. The percentages of cells in the G0/G1, S and G2/M phases were measured using ModFit LT software.
The synthesome-associated DNA polymerase α, δ and ε activity assays
The DNA polymerase α, δ and ε activities were measured under distinct buffer conditions using the appropriate DNA templates, (3H)-dTTP, and the Q-Sepharose fraction derived from cultured human cervical cancer cells (HeLa). The assays were performed either in the absence or presence of various concentrations of Cr(III) or Cr(VI) ion. Polymerase activity was measured as previously described with minor modifications (Malkas et al., 1990; Syvaoja et al., 1990). The assay for DNA polymerase α was performed at 37 °C for 1 h and contained 1 μg of calf thymus DNA, 0.11 μCi (3H)-dTTP, 20 μg of purified DNA synthesome, 20 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 1 mM DTT, 0.1 mM dATP, dGTP, dCTP. The assay for polymerase δ was performed at 37 °C for 30 min and contained 0.5 μg poly(dA-dT).poly(dA-dT) DNA, 0.5 μCi (3H)-dTTP, 200 μM dATP, 20 μg synthesome, 40 mM HEPES (pH6.5),1 mM MgCl2,10 mM KCl, 2 mM DTT, and 2% Glycerol. The assay for DNA polymerase ε was performed at 37 °C for 15 min and contained 0.5 μg poly(dA).Oligo(dT), 0.5 μCi (3H)-dTTP,10 μM dTTP,10 μg purified DNA synthesome, 5% glycerol, 2 mg/ml bovine serum albumin, 25 mM HEPES (pH 7.6), and 10 mM MgCl2. The radiolabeled reaction products were spotted onto DE81 filters, and the amount of (3H)-dTTP retained on the filters was quantified by liquid scintillation counting and used to measure the amount of radiolabeled nucleotide incorporated into the newly synthesized DNA.
In vitro DNA replication mediated by the synthesome
The SV40 origin dependent in vitro DNA replication assay was preformed as described previously (Abdel-Aziz et al., 2000). 25 μl reaction mixtures containing 30 mM HEPES, 7.5 mM MgCl2, 0.5 mM DTT, 1 μg of affinity purified SV40 large T-antigen, 50 μg synthesome derived from the P4 fraction of HeLa cells, 50 ng pBK-CMV plasmid DNA containing the intact SV40 replication origin, 1 μCi (α-32P)-dCTP (3000Ci/mmol), 100 μM each of dATP, dGTP, dTTP, 10 μM dCTP, 200 μM of rNTP, 40 mM phosphocreatine, and 1 μg creatine phosphokinase were incubated at 37 °C for 4 h. Nascent DNA was phenol extracted and ethanol precipitated from the reaction mixtures, and analyzed by electrophoresis through a 1% agarose gel, prepared in TBE buffer. The extent of replication was measured by spotting an aliquot of the replication reaction mixture onto DE81 filters, just prior to phenol extraction of the remainder of the reaction mixture. The filters were washed with 0.1 M sodium pyrophosphate, and sodium formate pH 7.4, and then dried, prior to quantify the amount of newly synthesized radioactive DNA retained on the filter, using liquid scintillation counting.
Mutagenesis assay
The fidelity of the in vitro DNA replication assay mediated by the human cell DNA synthesome was determined as previously described, (Sekowski et al., 1997). The in vitro DNA replication assay was performed as described, except that the radioactive nucleoside triphosphate [(α-32P)-dCTP] was replaced with 100 μM dCTP. After extracting and precipitating the nascent DNA synthesized in the reaction, the newly replicated DNA was digested with 0.5 units of Dpn I for 1 h at 37 °C, to remove any unreplicated parental template DNA. The Dpn I-resistant DNA was transfected into the XL-1 Blue MRF’ strain of E. coli cells, at a setting of 1.4 KV, 25 μF and 200 μ, using a Bio-Rad electroporation instrument. Bacterial cells were placed into 20 ml of LB agar containing 1.25 mg kanamycin, 4 μl IPTG (200 mg/ml) and 40 μl X-gal (20 mg/ml) and incubated overnight at 37 °C. The mutation frequencies were determined as the percentage of (white)/(blue+white) colonies developed on the agar plates. To identify the types of mutation being expressed by bacterial cells, individual mutant colonies (white) were picked and grown in LB medium containing kanamycin. Mutant plasmid DNA was purified using a Qiaprep spin miniprep kit (Qiagen; Valencia, CA), and DNA sequencing of the LacZα gene contained in the prepared plasmids was performed commercially by Retrogen Inc., (San Diego).
Results
Chromium ion inhibits cell growth
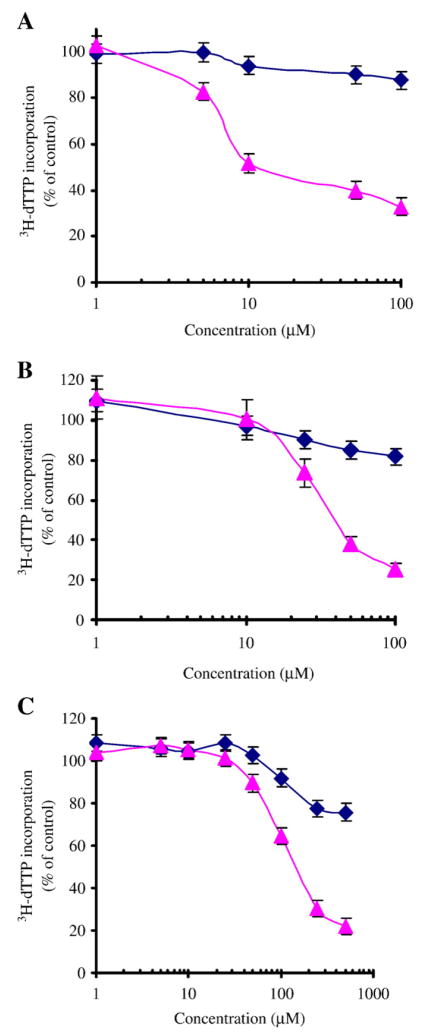
Our first step toward examining the mechanism by which chromium ion, induces DNA damage and inhibits cell proliferation was to expose HeLa cell cultures to a range of concentrations of both the hexavalent and trivalent forms of chromium ion, and to quantify the viability of these cells using the MTT assay. The assay is based upon the ability of only viable cells to reduce MTT to a colored product, which can then be released from cells using DMSO, and quantified by determining the absorbance of the reduced MTT assay product spectrophotometrically. Our results, which are shown in Fig. 1A and Table 1, indicate that cell proliferation was inhibited by Cr(VI), but not by Cr(III) or NaCl when cells were incubated in the presence of these salts for 8, 24, or 48 h. Cell proliferation was inhibited significantly by exposure to as little as 10 μM sodium dichromate for 24 h, and inhibition was even more pronounced when incubation was continued for an additional 24 h. The IC50 for inhibiting HeLa cell proliferation was found to be 7.5 μM of sodium dichromate after a 24 h exposure, and 3.5 μM of sodium dichromate after a 48 h exposure to the metal ion. In contrast, HeLa cell proliferation was not affected by culturing cells with either CrCl3 or NaCl at the same concentrations and over the same time periods.
Fig. 1.
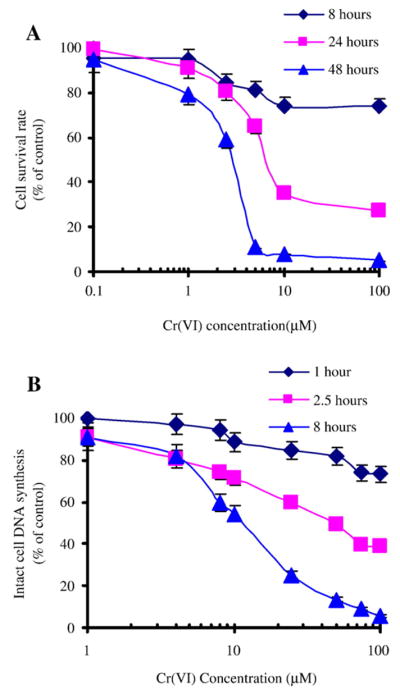
Effect of Cr(VI) on HeLa cell proliferation. Cell proliferation and intact cell DNA synthesis reactions were performed as described in the Methods. (A) The effects of sodium dichromate on HeLa cell proliferation were measured in cultured cells using the MTT assay following 8, 24 and 48 h of incubation with a range of concentrations of the metal ion. (B) The effects of sodium dichromate on intact cell DNA synthesis were measured by monitoring the incorporation of (3H)-thymidine into cells following 1, 2.5 or 8 h of incubation with a range of concentrations of the metal ion. The values reported were the average of three separate experiments. The degree to which Cr(VI) ion inhibits cell proliferation and intact cell DNA synthesis was calculated relative to an untreated control group and was reported as a percentage of the control.
Table 1.
Effect of CrCl3, Na2Cr2O7 and NaCl on HeLa cell proliferation at 8, 24, and 48 h
| Time Conc (μM) | 8 h |
24 h |
48 h |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 100 | 1 | 10 | 100 | 1 | 10 | 100 | |
| Na2Cr2O7 | 94.8±1.8 | 74.2±2.1 | 73.8±3.8 | 90.6±2.6 | 35.3±5.2 | 27.2±4.1 | 79.2±5.6 | 11.2±7.4 | 8.1±3.2 |
| CrCl3 | 99.6±2.7 | 111±3.4 | 121±5.2 | 106.7±3.8 | 114.2±2.9 | 115±4.2 | 92.9±3.1 | 98.8±4.8 | 81.4±2.3 |
| NaCl | 109±5.1 | 106±2.9 | 107±4.4 | 101±4.6 | 108±3.1 | 121±5.2 | 98.4±3.6 | 94.2±4.1 | 87.4±3.7 |
HeLa cell proliferation was determined by using the MTT assay as described in the Methods. The values reported in the table represent the average of three independent experiments and are given as a percent of control, (mean value±SE). Control cultures were grown in media as described in the Methods, but without the addition of any of these salts.
Chromium ion inhibits intact cell DNA synthesis
Intact cell DNA synthesis was examined by measuring the extent to which 3H-thymidine could be incorporated into newly synthesized DNA after exponentially growing HeLa cells were incubated in the presence of different concentrations of CrCl3, Na2Cr2O7, NaCl, ZnCl2 and NaNO3 for 1, 2.5 or 8 h. Our results, shown in Fig. 1B and Table 2, indicate that DNA synthesis was decreased significantly by exposure to sodium dichromate. The inhibitory effects of dichromate on intact cell DNA synthesis could be measured in cell cultures within 1 h of exposing these cultures to high concentrations (100 μM) of this metal ion. We observed that the inhibition of cellular DNA synthesis was proportional to the length of time the cells were exposed to the Cr(VI) ion; regardless of the concentration of the ion in the culture media. We also observed that the inhibition of intact cell DNA synthesis increased greatly, as a function of time, at low concentrations of the sodium dichromate (10 μM). Furthermore, following 8 h of exposure to the metal ion, cellular DNA synthesis decreased to 91.2%, 54.5%, and 5.6% when cells were cultured in the presence of 1, 10 and 100 μM sodium dichromate, respectively. The IC50 for inhibition of intact cell DNA synthesis by sodium dichromate was 65 μM and 15 μM, when cells were cultured with this salt for 2.5 and 8 h, respectively. In contrast to these effects, CrCl3, NaCl, ZnCl2 and NaNO3 did not inhibit intact cell DNA synthesis; regardless of the concentration of these salts or the duration of the exposure to these metal ions. These results demonstrated that intact HeLa cell DNA synthesis was differentially affected by Cr(III) and Cr(VI) ion, and that the inhibition of cell proliferation by Cr(VI) was mediated at least in part by inhibition of intact cell DNA synthesis.
Table 2.
Effect of CrCl3, Na2Cr2O7, NaCl, ZnCl2 and NaNO3 on intact HeLa cell DNA synthesis
| Time Conc. (μM) | 1 h |
2.5 h |
8 h |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 100 | 1 | 10 | 100 | 1 | 10 | 100 | |
| Na2Cr2O7 | 100±4.9 | 88.7±3.9 | 73.3±2.2 | 91.3±3.1 | 74±4.6 | 9.1±3.9 | 91.2±6.0 | 54.5±2.8 | 5.6±2.5 |
| CrCl3 | 100±3.1 | 96.2±5.4 | 90.8±3.1 | 111±1.6 | 111.7±1.4 | 98.0±2.6 | 106±7.0 | 107±2.4 | 105±2.6 |
| NaCl | 105±4.1 | 93.8±2.2 | 99.2±4.7 | 93.8±6.0 | 95.8±4.0 | 95.3±4.3 | 96.1±6.4 | 102±5.1 | 93.7±6.9 |
| ZnCl2 | 98.6±5.2 | 99.6±6.2 | 91.0±2.9 | 99.6±6.2 | 90.3±3.2 | 93.4±2.1 | 101±5.2 | 91.0±5.6 | 98.0±4.3 |
| NaNO3 | 103±4.9 | 99.2±3.5 | 92.6±5.5 | 91.4±3.9 | 89.1±2.9 | 91.2±3.6 | 97.8±4.5 | 98.1±3.4 | 97.7±3.9 |
HeLa cells were grown in culture and incubated with a range of concentrations of these salts, as described in the Methods. Intact cell DNA synthesis was determined by measuring the incorporation of 3H-thymidine into acid insoluble and retention on GF/A filters as described in the Methods. The mean values±S.E are reported in the table, and represent the average of 3 independent experiments, and are expressed as a percentage of radiolabel incorporated into acid insoluble material from cultures grown in the absence of these metal salts. (% of control, Mean).
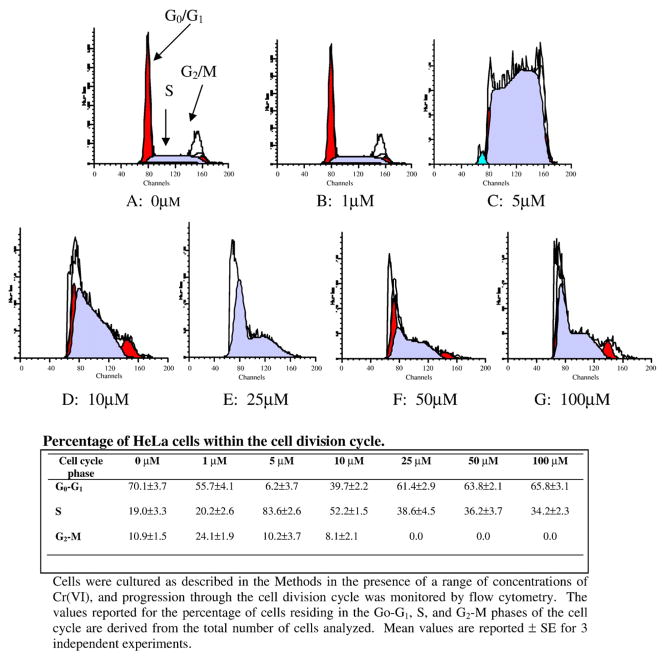
Chromium ion inhibits progression through the cell division cycle
Inhibition of both intact cell DNA synthesis and consequently cell proliferation by sodium dichromate suggested that cells exposed to the hexavalent form of chromium ion become arrested either within, or just prior to, the S-phase of the cell cycle. We therefore performed a cell cycle analysis of our HeLa cell cultures, following a 24 h exposure of the cultures to various concentrations of sodium dichromate and chromium chloride. Our results are shown in Fig. 2, and indicate that progression through the cell division cycle was significantly affected by increasing concentrations of sodium dichromate but not chromium chloride. In the absence of chromium ion, the cell populations were 70.1%, 19% and 10.9% in the G0/G1, S, and G2/M phases, respectively,. With exposure to as little as 1 μM sodium dichromate, the population of cells in the G2/M-phase of the cell cycle increased ~15% from 10.9% to 24.1%; while the percent of cells in S-phase was essentially unchanged, and the percentage of Go/G1 phase cells dropped ~15%. Increasing the concentration of sodium dichromate to 5–10 μM, further increased the number of S-phase cells from 19% to 84% at 5 μM and to only 52.2% at 10 μM. The percentage of cells in G0/G1 phase decreased from 70% to 6.2% at 5 μM and to ~40% at 10 μM. When the sodium dichromate concentration was increased to 25–100 μM, the percentage of cells accumulating in the G2/M phase of the cell cycle decreased significantly from 10.9% to 0%; indicating that while the cells were cleared from the G2/M phase they appeared to become locked into either the Go/G1- or S-phases of the cell cycle. Our results suggest that at very low concentrations of Cr(VI) ion, (i.e., 2 μM), Cr(VI) may slightly stimulate cell proliferation and promote cell mitosis; while at higher concentrations (i.e., 5–10 μM) sodium dichromate appears to arrest cell growth during the DNA synthetic phase (S-phase)of the cell cycle. At still higher concentrations of the dichromate ion (i.e., 25–100 μM), cells in either Go/G1 or S-phase appeared to stop their progression through the cell cycle at the specific point within the cycle where they were exposed to the metal ion; while cells already in the G2/M phase of the cell cycle undergo cell division and then stop proliferating. Cell proliferation appeared to be rapidly and completely inhibited by exposure to high concentrations of the metal ion, as these cells appeared to neither complete DNA replication nor carry-out cell division even though the S-phase cells have clearly passed the commitment point that is known to reside within the late G1 phase of the cell cycle. During our cell cycle analysis, we observed a rapid decrease in G1 phase cells and a corresponding rise in S-phase cells when cells were exposed to 5 μM sodium dichromate. However, even at higher concentrations of this metal ion, our flow-cytometric data did not appear to identify the presence of apoptotic cells.
Fig. 2.
Effect of Cr(VI) ion on cell cycle progression. Cultured HeLa cells were incubated at 37 °C for 24 h with a range of concentrations of sodium dichromate. Cells were then collected, stained with propidium iodide, and DNA content was measured by flow cytometry as described in the Methods.
Chromium ion selectively inhibits the synthesome-associated DNA polymerase α, δ and ε activities
We determined the effect of chromium ion on the activity of the synthesome-associated α, δ and ε DNA polymerases using assay specific DNA templates and (3H)-dTTP. The amount of radioactive nucleotide (3H)-dTTP incorporated into nascent DNA formed on these templates was quantified by liquid scintillation counting, as described in the Methods. The polymerase α activity was examined using an activated calf thymus DNA template. Cr(III) significantly inhibited the DNA synthesome-associated polymerase α activity (Fig. 3A). The IC50 for chromium chloride inhibition of DNA polymerase α activity was 15 μM; while the same concentration of sodium dichromate did not inhibit the activity of the DNA polymerase α enzyme. DNA synthesome-associated polymerase δ activity was detected using a poly(dA-dT).poly(dA-dT) template. The DNA synthesome-associated polymerase δ activity was also inhibited by Cr(III) ion, but only slightly inhibited by Cr(VI) ion, (Fig. 3B). The IC50 for inhibition of DNA polymerase δ activity by chromium chloride was 45 μM. The DNA synthesome-associated polymerase ε activity was examined using a poly(dA).oligo(dT) template, under slightly different buffer conditions from that used to monitor DNA polymerase δ activity. The synthesome-associated DNA polymerase ε activity was also inhibited by chromium chloride (IC50 of 125 μM); while the same concentration of sodium dichromate did not significantly affect the activity of DNA polymerase ε, (Fig. 3C). These results indicated that each of these three synthesome-associated DNA polymerases was uniquely sensitive to inhibition by Cr(III) ion, and that the DNA polymerase, thought to carry-out synthesis of the lagging DNA strand (i.e., DNA polymerase α), was the most sensitive to inhibition by Cr(III) ion.
Fig. 3.
Effect of Cr(III) and Cr(VI) on the in vitro activity of synthesome-associated DNA polymerase α, δ and ε. Polymerase assays were performed in the absence or presence of a range of concentrations of chromium chloride (▲) or sodium dichromate (◆) ion, under the reaction conditions described in the Methods. (A) Polymerase α activity was reported as the percentage of 3H-dTTP incorporated into an activated calf thymus DNA template. (B) Polymerase δ activity was reported as the percentage of 3H-dTTP incorporated into a poly(dA-dT).poly(dA-dT) DNA template. (C) Polymerase ε activity was reported as the percentage of 3H-dTTP incorporated into a poly(dA).oligo(dT) DNA template. All values reported in the figure are the average of three independent experiments, and are reported as a percentage of the radiolabeled nucleotide incorporated into the nascent DNA during the polymerase reaction, relative to a control reaction which was performed in the absence of chromium ion.
Chromium ion inhibits the origin dependent in vitro DNA synthetic activity of the human cell DNA synthesome
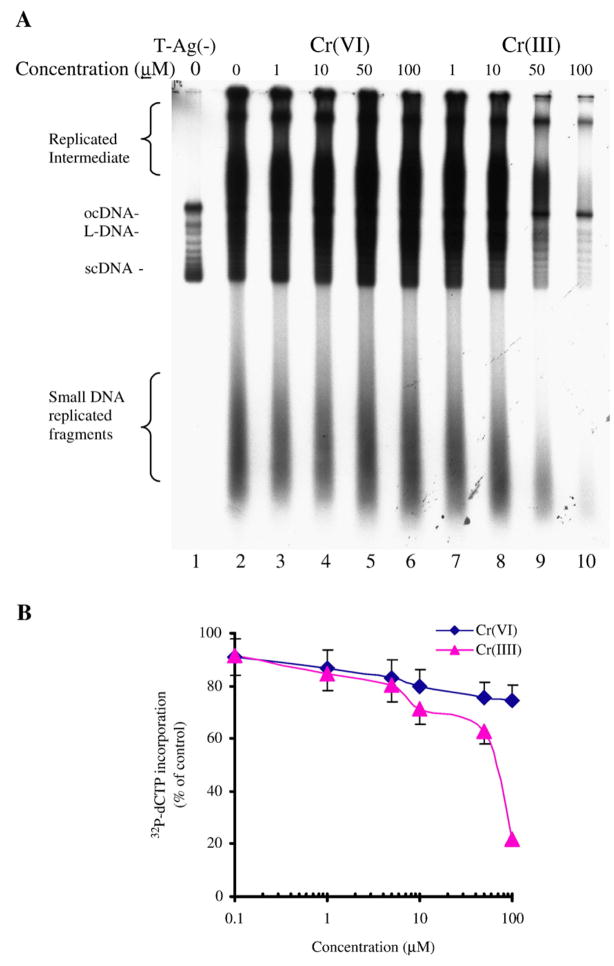
We performed assays to monitor the origin dependent replication of double stranded DNA by the human cell DNA synthesome in the absence and presence of a range of concentrations of Cr(III) and Cr(VI) ion, as previously described (Malkas et al., 1990). The effect of chromium (III) ion on in vitro DNA synthesis were determined by measuring the incorporation of (α32P)-dCTP into the newly replicated DNA products using liquid scintillation counting (Fig. 4B), and the products of the replication assay were analyzed using neutral agarose gel electrophoresis, (Fig. 4A). We observed that incorporation of radiolabeled dCTP into newly synthesized DNA, formed in the synthesome mediated in vitro DNA replication assay, was significantly inhibited by chromium chloride, but not by sodium dichromate over the same range of concentrations. The IC50 for inhibition of origin dependent synthesome mediated DNA replication in vitro by chromium chloride, was found to be 88 μM. Cr(III) ion decreased all of the forms of the nascent DNA produced in the in vitro DNA replication reaction, with synthesis of Okazaki fragments (i.e., small DNA replication fragments) being inhibited above 10uM, and a corresponding decrease in replication intermediate production, as well as linear DNA replication products, nicked open circular DNA formation, and the production of fully replicated supercoiled DNA.
Fig. 4.
Effect of Cr(III) and Cr(VI) on synthesome-mediated and SV40 origin dependent in vitro DNA replication. In vitro DNA replication reactions were performed in the absence or presence of a range of concentrations of chromium chloride and sodium dichromate ion using a plasmid DNA template, (pBK-CMV), and affinity purified T-antigen, as described in the Methods. (A) Newly replicated DNA products were analyzed by electrophoresis through 1% neutral agarose gels following extraction and precipitation of the nascent DNA. Lane 1 shows the control reaction products formed in the absence of T-antigen. Replication reaction products include open circle DNA (oc-DNA), linear DNA (L-DNA) and supercoiled DNA (sc-DNA), and are shown in lanes 2–10. Reactions carried-out in the presence of sodium dichromate are shown in lanes 3–6; while reactions carried-out in the presence of chromium chloride are shown in lanes 7–10. The position of supercoiled DNA (scDNA); linear DNA (L-DNA); and open circle DNA (ocDNA) are noted to the left of the gel, as are the positions of higher order replication intermediates and the smaller Okazaki fragments formed during the reaction. (B) As described in the Methods, the amount of (32P)-dCTP incorporated into in vitro DNA replication reaction products was quantified by scintillation counting. The values reported are the average of three independent experiments.
Chromium reduces the fidelity of synthesome-mediated DNA replication
The effect of chromium ion on the fidelity of DNA replication mediated in vitro by the HeLa cell DNA synthesome was examined using the mutagenesis assay, previously described by us (Sekowski et al., 1997). The assay employs a highly purified supercoiled plasmid DNA template (i.e., pBK-CMV), that when transfected into bacteria and grown on agar in the presence of X-gal and IPTG, results in the formation of blue bacterial colonies. Mutations sustained in the LacZα gene, during in vitro replication of the plasmid, result in the inability of the bacteria harboring this plasmid to convert X-gal into its colored reaction product; thus forming a white bacterial colony on the agar plate. The frequency of mutation is scored by counting only those colonies that are pure white, and expressing their abundance, relative to the total number of bacterial colonies growing on the plate, as a percentage of the total number of bacterial colonies on the culture plate. Mutations that partially inactivate the LacZα gene product, lead to the formation of bacterial colonies that are light blue-green, and thus are scored by us as wild-type rather than mutant colonies. This methodological approach provides a conservative estimate of the actual number of mutations sustained by the LacZα gene during synthesome mediated in vitro DNA synthesis.
pBK-CMV contains an intact SV40 origin of replication, in addition to the lacZα reporter gene and the lacZ promoter; which together makes it suitable for use as a template in this fidelity assay. Because of these features of the plasmid, we have also used pBK-CMV to determine the fidelity of the DNA synthesome mediated in vitro DNA replication reaction, (Sekowski et al., 1997 and 1998) employing the DNA synthesome isolated from malignant and from non-malignant breast cells, or synthesome isolated from HeLa cells exposed to low levels of mercuric ion. In addition, pBK-CMV has also been used by other laboratories to monitor the fidelity of in vitro SV40 DNA replication, (Servant et al., 2002) Newly replicated DNA was obtained from the reaction products by first digesting any non-replicated template DNA with the restriction endonuclease DpnI, (under optimal conditions, as described in the Methods). DpnI cuts the double stranded parental DNA template into multiple fragments because the plasmid used in the reaction is fully methylated on both DNA strands. DpnI however, spares the newly replicated DNA because only the template strand remains methylated, making the newly replicated DNA duplex hemi-methylated. This is a result of the semi-conservative nature of the DNA replication process carried-out by the DNA synthesome, during the in vitro DNA replication assay. The restriction enzyme reaction mixture is then extracted using phenol/chloroform, and the extracted DNA is concentrated by ethanol precipitation at −20 °C.
We then tested the mutagenic properties of chromium ion by performing the DNA synthesome-mediated in vitro DNA replication assay either in the absence or presence of a range of concentrations of Cr(III) or Cr(VI) ion. The newly replicated DNA products were digested by 0.5 units of Dpn I, and transfected into electro-competent E. coli cells, in order to determine the frequency of mutation sustained by the nascent DNA, during semi-conservative replication of the pBK-CMV template. We found that chromium chloride significantly increased the frequency of mutation sustained by the lacZα gene, and decreased the colony formation rates in direct proportion to the concentration of Cr(III) ion in the reaction mixture (Table 3). In contrast to these results with Cr(III), the frequency of mutation sustained by the newly replicated plasmid DNA and the number of colonies formed when the in vitro DNA replication assay was performed in the presence of Cr(VI) ion was significantly higher than that previously observed for the reaction run in the presence of Cr(III) ion. Both the frequency of mutation and the number of bacterial colonies formed in the presence of sodium dichromate did not significantly differ from that observed when an equivalent amount of unreplicated pBK-CMV plasmid DNA was transfected into E. coli. Thus, the spontaneous-mutation frequency exhibited by the transformed E. coli cells used in this assay was essentially equivalent to that observed when the in vitro DNA replication assay was performed in the presence of sodium dichromate. The mutation frequencies induced by Cr(III) over the range of concentrations examined were approximately 2–13 fold higher than that observed for the spontaneous mutation frequency. Our results are consistent with our observations that Cr(III) significantly reduces the activity and fidelity of DNA replication mediated by the human cell DNA synthesome in vitro; resulting in a concentration dependent lowering of the colony formation rate and concentration dependent increase in the occurrence of mutation of the lacZα reporter gene during the DNA replication process.
Table 3.
The effect of Cr(III) and Cr(VI) ion on the generation of mutations by the DNA synthesome during SV40 origin dependent in vitro DNA replication
| Chromium Concentration (μM) | Colonies scored |
Mutation frequency (× 10−4) | Error ratea (× 10−5) | Mutationb fold increase | ||
|---|---|---|---|---|---|---|
| Total | Mutant | |||||
| CrCl3 | 0 | 53,058 | 58 | 10.9 | 0.44 | 1 |
| 1 | 50,208 | 84 | 16.7 | 0.68 | 1.53 | |
| 5 | 48,308 | 122 | 25.3 | 1.02 | 2.30 | |
| 10 | 47,126 | 138 | 29.3 | 1.19 | 2.68 | |
| 50 | 42,105 | 229 | 54.4 | 2.20 | 4.97 | |
| 100 | 12,458 | 185 | 148.5 | 6.01 | 13.57 | |
| Na2Cr2O7 | 0 | 43,240 | 64 | 14.8 | 0.60 | 1 |
| 1 | 42,840 | 58 | 13.5 | 0.55 | 0.92 | |
| 5 | 45,728 | 72 | 15.7 | 0.63 | 1.31 | |
| 10 | 43,289 | 84 | 19.4 | 0.79 | 1.28 | |
| 50 | 39,456 | 79 | 20.0 | 0.81 | 1.35 | |
| 100 | 38,870 | 82 | 21.1 | 0.85 | 1.42 | |
SV40 origin dependent DNA replication assays, mediated by the DNA synthesome, were performed in the absence or presence of a range of concentrations of chromium chloride or sodium dichromate as described in the Methods. Newly replicated pBK-CMV plasmid DNA, containing the LacZα and lac promoter elements was transformed into E. coli cells, and the frequency of mutation was scored by counting the number of white colonies growing on the agar plates and expressing this number relative to the total number of colonies bacterial colonies growing on the plate, (Sekowski et al., 1997, 1998.). The target sequence leading to the creation of white colonies in the pBK-CMV plasmid includes the lac Zα and lac promoter elements (494 bp). The error rate,
represents the relative number of errors created in the target sequence of the nascent DNA during the in vitro DNA replication reaction. The values were calculated by subtracting the background mutant frequency (5×10−8) from the average mutation frequency, dividing this number by 0.5, and the number of nucleotides (494) within the DNA being sequenced.
Fold increase in mutation frequency was determined by comparing the mutation frequency observed for reactions performed in the absence of chromium ion with those performed in the presence of a range of chromium ion concentrations. The values reported represent the mean of 3 independent experiments.
Identification of the types of mutation induced by chromium ion during origin dependent in vitro DNA replication
To determine whether chromium ion induces specific types of mutations during the DNA replication process, we performed a DNA sequence analysis of the lacZα and lac promoter regions of the plasmid DNA isolated from 75 white bacterial colonies, which grew from bacteria that had been previously transfected with the newly synthesized DNA, that had been recovered from the in vitro DNA replication assay reaction mixtures. The mutant replicated DNA (white phenotype) was isolated and purified as previously described (Sekowski et al., 1997) and sequenced by a commercial DNA sequencing vendor (Retrogen, Inc.; San Diego, CA). We identified three types of mutations that were produced by the DNA synthesome during the origin dependent DNA replication reaction. These three types of mutations included nucleotide insertions, deletions, and substitutions, and these types of mutations occurred irrespective of whether the DNA replication reaction was performed in the absence or presence of either Cr(III) or Cr(VI) ion. Our results, shown in Table 4, indicate that the abundance of each type of mutation was as follows: nucleotide insertions (27.8% (Cr(III)); 27.3% (Cr(VI)); 24% (no Cr ion)), nucleotide deletions (8.3% (Cr(III)); 9.1% (Cr(VI)); 5.9% (no Cr ion)) and nucleotide substitutions (63.4% (Cr(III)); 63.6% (Cr(VI); 70.6% (no Cr ion), (Table 4). Base substitutions occurred primarily in GC base-pairs, and nucleotide substitutions occurred more frequently during replication reactions performed in the presence of Cr(III), than in the presence of Cr(VI) ion or in the absence of chromium ion. It should be noted that Cr(VI) also leads to the formation of the same types of lesions observed when reactions are performed in the presence of Cr(III) ion; including nucleotide substitutions, but the frequency of these lesions formed in the presence of the Cr(VI) ion is approximately half that observed in the presence of Cr(III). Cr(III) ion also produced a higher frequency of nucleotide transversions (44.4%) than transitions (19.4%) and a lower frequency of nucleotide transitions (19.4%) than would be expected to arise spontaneously (41.2%), [X2=81.72, p<0.005]. It should also be noted that the relative percentage of the types of mutations arising in newly replicated DNA is not significantly different (p>0.05) when comparing the types of mutations being generated by the synthesome in reactions containing Cr(III), Cr(VI), or no chromium ion. What differs is the increased number of nucleotide substitutions and insertions formed when reactions are performed in the presence of Cr(III) ion. Our results suggest that when the DNA synthesome within cells replicates DNA in the presence of Cr(III) ion, their DNA synthesomes likely produce a lower frequency of nucleotide transitions and a higher frequency of nucleotide transversions during the replication process, relative to the frequency of these mutations arising either spontaneously or following exposure to Cr(VI) ion.
Table 4.
Summary of the frequency and type of mutations produced by the DNA synthesome during the in vitro DNA replication reactions performed in the presence of chromium ion
| Mutation type | Cr(III)-treated number (%) | Cr(VI)-treated number (%) | Spontaneous number (%) | All |
|---|---|---|---|---|
| Substitutionsb | 23(63.4%) | 14(63.6%) | 12(70.6%) | 49(65.3%) |
| Transitiona | 7(19.4) | 6(27.3) | 7(41.2) | 20 |
| GcàAT | 4(11.1) | 4(18.2) | 7(41.2) | 15 |
| AtàGC | 3(8.3) | 2(9.1) | 0 | 5 |
| Transversiona | 16(44.4) | 8(36.4) | 5(29.4) | 29 |
| GcàTA | 7(19.4) | 2(9.1) | 2(11.8) | 11 |
| GCàCG | 5(13.8) | 3(13.6) | 2(11.8) | 10 |
| AtàCG | 3(8.3) | 2(9.1) | 1(5.9) | 6 |
| AtàTA | 1(2.8) | 1(4.5) | 0 | 2 |
| Deletionsb | 3(8.3%) | 2(9.1%) | 1(5.9%) | 6(8.0%) |
| Insertionsb | 10(27.8%) | 6(27.3%) | 4(23.5%) | 20(26.7%) |
| All Mutations | 36(100%) | 22(100%) | 17(100%) | 75(100%) |
The types and frequency of mutations induced by Cr(III) and Cr(VI) ion, during the in vitro DNA replication reaction was determined as described in the Methods. The data reported were obtained from three independent experiments.
indicates that there is a significant difference between the frequency of a specific type of mutation arising from synthesome mediated DNA replication reactions containing either chromium chloride, sodium dichromate, or lacking chromium ion; (p<0.005)
indicates that there is no significant difference between the frequency of specific types of mutations arising from synthesome mediated DNA replication reactions either containing or lacking these metal ions; (p>0.05).
Discussion
Cr(VI) ion is an environmental toxicant that is known to be cytotoxic to cells, and to exhibit genotoxic effects. These effects of Cr (VI) ion inhibit cell proliferation, block the cell division cycle during S-phase, and induce chromosomal abnormalities and gene mutations (Xu et al.,1996; Sr. John et al., 2002; Liu et al.,1999). The mechanism by which chromium ion mediates its cytotoxic effects on human cells is therefore both complicated and multi-faceted. At one level, Cr(VI) ion is thought to induce cell growth arrest by stimulating the production of hydrogen peroxide within cells. H2O2 is known to give rise to hydroxyl radicals, which can in-turn alkylate DNA, (Zhang et al., 2001). Cr(VI) is also known to undergo reductive activation to convert Cr(VI) into a stable end-product Cr(III); which is known to participate in a variety of reactions that can cause DNA damage (Arslan et al., 1987). Cr(III) has been shown to produce DNA-DNA strand cross-links (O’Brien et al., 2001), and the ion can arrest the activity of individually isolated DNA polymerases in vitro, (Bridgewater et al., 1998). Studies examining the ability of chromium ion to induce mutations during DNA synthesis have been performed using individually purified DNA polymerases (Snow, 1991, 1994; Jatinder and Snow, 1998), and these suggest that Cr (III) can alter the fidelity with which individually purified DNA polymerases synthesize DNA on primed artificial DNA templates. Unfortunately, none of these individually purified DNA polymerases can support true bi-directional semi-conservative DNA replication, nor can they mimic, as closely, the intact cellular DNA synthetic process exhibited by the DNA synthesome. These studies have therefore provided us with a critical first approximation of how chromium ion could induce DNA damage during the DNA synthetic process, but, by their very nature, these studies were unable to accurately compare either the sensitivity of individual DNA polymerases to chromium ion, or the effect chromium ion has on the activity and fidelity of these polymerases; while they are linked to the other components of the cellular DNA synthetic apparatus. So to better understand the relationship between these studies which examined the effect of chromium ion on the DNA synthetic process using individually purified DNA polymerase and the effect of chromium ion on intact cellular DNA synthesis, we performed the studies described using the replication competent DNA synthesome. The DNA synthesome used in these studies was derived from human cervical cancer cells, (HeLa), and is a fully-functional multiprotein form of DNA polymerase, that has been demonstrated to competently perform, in vitro, each phase of the DNA replication process, (i.e., the initiation, elongation and termination). The synthesome replicates DNA bi-directionally, from a functional and clearly defined mammalian origin of DNA replication (in this case the simian virus 40 origin of replication), and creates semi-conservatively replicated DNA (Malkas et al., 1990; Applegren et al., 1995), which it can wind into supercoiled (form I plasmid) DNA, once replication is completed. Our studies therefore provide a missing element in the analysis of how chromium ion exerts it genotoxic and cytotoxic effects, because unlike the pioneering studies employing purified DNA polymerases, our analysis employs a replication competent multi-protein form of polymerase that essentially mimics the replication process carried-out by intact cells.
The DNA synthesome is a large multiprotein complex consisting of at least 45 polypeptides. It has a defined size of 17–18S in sucrose gradients (Malkas et al., 1990; Wu et al., 1994), and internally consistent molecular dimension of 69 nm×89 nm when examined by transmission electron microscopy, (unpublished data). The DNA synthesome consists of a loosely associated, but organized, small group of proteins involved in the initiation phase of the DNA replication process, and these proteins, which are involved in the initiation of DNA replication, are themselves associated with a tightly bound group of proteins involved in the elongation and termination phases of the DNA replication process. This tightly associated core component of the DNA synthesome contains the leading and lagging strand DNA polymerases (i.e., polymerase delta and alpha, respectively), which are linked to one another by a protein known as Replication Factor C, DNA primase, Proliferating Cell Nuclear Antigen (PCNA), DNA ligase, topoisomerase II, DNA helicase I and IV, and poly (ADP) ribose polymerase (Applegren et al., 1995). DNA polymerase epsilon is also found in the core of the DNA synthesome, but unlike other core components identified thus far, polymerase epsilon does not appear to have a role in SV40 origin dependent DNA synthesis (Pospiech et al., 1999). The initiation component of the DNA synthesome consists of the human cell single stranded DNA binding protein, RPA, and topoisomerase I, which together are involved in removing torsional strain that develops in the DNA duplex; ahead of the moving replication fork. Thus, the DNA synthesome is a highly organized well characterized multiprotein form of DNA polymerase, which is fully competent to support each phase of the DNA replication process, and thus makes the purified human cell DNA synthesome an excellent model system for examining how chromium ion induces mutations during the DNA replication process. The use of the human cell DNA synthesome in our studies bridges the gap between studies utilizing individually isolated DNA polymerases and those utilizing intact cells, and enables us to explore the molecular basis for the toxic effects of chromium ion in a way that was not possible using individually isolated DNA polymerases.
Our initial results utilizing the DNA synthesome, and reported in Tables 1–2 of this manuscript, were designed to be confirmatory in nature; in that we felt that it was important to demonstrate that the DNA synthesome model system should respond appropriately to both hexavalent and trivalent chromium ion before utilizing the synthe-some to examine the effect of chromium ion on the fidelity of the DNA synthetic process carried-out by the DNA synthesome. We therefore expected that the in vitro DNA replication and DNA polymerase activities of the DNA synthesome would be inhibited when incubated with the trivalent form of chromium ion, but not the hexavalent form of the ion; while intact cell DNA synthesis would be inhibited by the hexavalent form of chromium ion, but not by the trivalent form of the ion. Using the purified human cell DNA synthesome, we demonstrated that Cr(III) ion differentially inhibits the elongation activity of all three synthesome-associated DNA polymerases when measured in vitro; while hexavalent chromium ion was found to have essentially no effect on the activity of the synthesome associated DNA polymerases. We found that DNA polymerase α is the most sensitive to inhibition by Cr(III) ion, (IC50 = 15 μM), followed by DNA polymerase δ (IC50 =45 μM). DNA polymerase ε, was found to be the least sensitive to inhibition by trivalent chromium (IC50 =125 μM). This observation suggests that DNA polymerase α is the first polymerase to be inhibited by Cr(III) ion, resulting in the slowing of replication fork movement because of the slowing of lagging strand synthesis. Curiously, this observation is consistent with our observation that intact cellular DNA synthesis can be inhibited after an 8 h exposure to 15 μM sodium dichromate (i.e., 30 uM Cr(VI) ion), and suggests that transport of Cr (VI) into the cell, and its subsequent reduction to the trivalent form of the ion, results in the inhibition of intact cell DNA synthesis at a concentration of chromium ion that is comparable to that added to the growth media and that is consistent with inhibition of lagging strand DNA synthesis in our in vitro assay. In contrast to the effect of Cr(III) on the in vitro activity of the DNA polymerases and the origin dependent replication activity of the synthesome, the trivalent form of chromium ion has no effect on cell proliferation or the DNA replication activity of intact cells; while the hexavalent form of chromium ion strongly inhibits intact cell proliferation, as well as intact cell DNA synthesis. The results of our cell culture studies (i.e., in vivo results) and our enzymatic assays (i.e., in vitro results) are consistent with those described by others, and thus support the use of the DNA synthesome as an appropriate model for directly evaluating how chromium ion exerts its genotoxic effects. The DNA synthesome is therefore a useful model for monitoring the induction of a metal induced “mutator phenotype”, (Bielas et al., 2006), that reduces the fidelity with which the synthesome replicates DNA.
Our results using the DNA synthesome model, are shown in Fig. 3, and support the conclusions of Bridgewater et al., (1994b) regarding the ability of chromium ion to inhibit specific cellular DNA polymerases to different extents. Our finding that DNA polymerase α is the DNA polymerase that is most sensitive to inhibition by Cr(III) ion is intriguing because DNA polymerase α is believed to have a critical role in the initiation of synthesis of both the leading and lagging DNA strands during origin dependent DNA replication. In addition, DNA polymerase α has long been thought to have a primary role in the formation of Okazaki fragments, which are relatively short DNA fragments formed during lagging strand DNA synthesis. DNA primase, which is associated with DNA polymerase α initiates synthesis (Waga and Stillman, 1998) by laying down a short RNA primer on the DNA template before being displaced by DNA polymerase δ. Increasing concentrations of Cr(III), or Cr(VI) in the presence of reducing agents such as ascorbic acid, result in the arrest of DNA synthesis mediated in vitro by DNA polymerase α (Bridgewater et al.,1998). Furthermore, during the initiation of DNA synthesis, it has been proposed that polymerase α is displaced shortly after beginning synthesis of the nascent DNA strand on both the leading and lagging DNA template strands (Waga and Stillman, 1998), and is replaced by polymerase δ. In contrast to DNA polymerases α and δ, a precise role for DNA polymerase ε during the replication process remains unclear. But, studies using the yeast S. cerevissea have shown that the yeast homolog of DNA polymerase ε is none-the-less essential for both cell division and survival (Waga and Stillman, 1998)), and appears to have an important role in DNA repair in intact cells (Zlotkin et al., 1996). The results of our studies demonstrating that DNA polymerase α is the most sensitive of the three polymerases to inhibition by chromium ion, also correlates with the results of work previously reported by this laboratory (Han et al., 2000). In this work, Han et al., (2000) showed that the in vitro activity of the synthesome-associated polymerase α is nearly 10 times more sensitive than that of the synthesome-associated DNA polymerase δ or ε to the DNA synthesis inhibitor araCTP. This observation suggests that there is something fundamentally different about the structure/activity of DNA polymerase α, relative to that of polymerases δ and ε. One explanation could be that the intrinsic 3′-5′ exonuclease activity inherent to the DNA polymerase δ and ε enzymes can efficiently removed a non-complementary nucleotide following its incorporation into the nascent DNA strand. The polymerase associated 3′>5′ exonuclease activities can perform a proofreading function that can remove inappropriately paired nucleotides formed during the DNA replication process, and thus help maintain a high degree of fidelity during DNA replication (Tran et al., 1999). Hence, the effects of Cr(III) on the activity and fidelity of DNA replication could have resulted from the selective inhibition of either the proof-reading function of these polymerases or their ability to correctly match the complementary nucleotide being added to the nascent DNA strand across from its corresponding nucleotide on the DNA template. Differential sensitivity of the synthesome-associated polymerases α, δ and ε to Cr(III) ion suggested that inhibition of synthesome mediated DNA replication occurred mainly during the initiation and elongation phases of the DNA replication process. Therefore, selective inhibition of these DNA polymerases by chromium ion could be the primary cause of the metal’s cytotoxicity and genotoxicity.
Chromium ion is known to influence both the fidelity and activity of the DNA replication process in a concentration-dependent manner. At low concentrations, Cr(III) increases DNA polymerase α and β processivity, and results in a decrease of DNA replication fidelity (Snow and Xu, 1991; Jatinder and Snow, 1998). Low concentrations of Cr(III) ion could also substitute for Mg2+ ion, weakly activating the DNA polymerase. This may account for the slight stimulatory effect of very low Cr(III) ion concentrations on in vitro DNA polymerase activity, and it could be related to the observed enhancement of non-complementary nucleotide incorporation at these concentrations of the metal ion (Snow, 1994). At high concentrations of the metal ion however, the inhibition of in vitro DNA replication, correlates with both the inhibition of DNA polymerase α and β activity and formation of Cr(III)-DNA lesions (Bridgewater et al., 1998); suggesting that Cr (III) ion inhibits the replication process at multiple levels. Chromium (III) forms two major types of adducts with DNA, one involves formation of a binary complex involving interaction with the phosphate backbone of the DNA strand, while the second involves formation of a ternary complex between the DNA strand, chromium ion, and proteins binding to this DNA. Both complexes can be easily imagined to disrupt the ability of the DNA polymerases to inhibit the DNA synthetic process. The first by disrupting utilization of the DNA template; the second by directly interfering with the activity of the proteins bound to the DNA. In our studies, both types of complexes are likely to contribute to the overall inhibition of the DNA polymerase activity of individual DNA polymerases and the in vitro DNA replication activity of the DNA synthesome.
It has been observed that reduction of Cr(VI), (the in vivo carcinogenic form of chromium ion), to Cr(III) by ascorbate leads to a dose-dependent increase of both mutagenic and replication-blocking DNA lesions (Quievryn et al., 2003). Ascorbate was shown to significantly increase the formation of Cr-DNA adducts at 50–100 μM Cr(VI); resulting in the production of mutagenic DNA damage and a large, dose-dependent increase in the frequency of supF mutants; following the replication of Cr(VI)-treated pSP189 plasmid DNA transfected into human fibroblasts. These results parallel those reported by us for the fold increase in the number of mutations sustained by DNA replicated by the DNA synthesome in the presence of chromium ion, (data not shown).
In vivo, chromium(VI) ion increased the number of cells in the G2/M phase of the cell cycle, and slightly stimulated cell proliferation at low concentrations (i.e.,1–5 μM), but significantly inhibited both DNA synthesis and cell proliferation at higher concentrations, (i.e., ≥5 μM) when cells were incubated in the presence of Cr(VI) for at least 48 h. In vitro chromium(III) ion, at 1–10 μM, slightly decreased the fidelity with which the purified DNA synthesome carried-out DNA synthesis, while significantly inhibiting the activity of DNA polymerase α, but not that of the δ or ε polymerase. At higher concentrations (50–100 μM) Cr(III) ion strongly inhibited both the activity and fidelity of the DNA polymerases-mediating in vitro DNA replication. We therefore propose that the mutagenic (or genotoxic) effects of chromium ion, associated with the reduction in DNA synthetic fidelity during the DNA replication process could be the result of inhibition of the intrinsic activity of the DNA polymerase α with only a slowing of the DNA replication process as DNA polymerase α mediates lagging strand DNA synthesis at low concentrations of this metal ion; while inhibition of the activity of the DNA replication process itself at higher metal ion concentration results in the metal ion’s cytotoxic effects as a result of the increased number of mutations sustained by the DNA and the concomitant inability of the DNA repair processes to maintain genomic integrity. Consistent with this suggestion is the observation that at higher Cr(III) ion concentrations, the frequency of mutation increases from 2 to 13 fold, when compared to the spontaneous mutation frequency. The increased mutation frequency in-turn correlates with a decrease in colony formation rates by up to 4 fold at these higher chromium ion concentrations.
This suggestion can in principal be supported by the following observation. Two of the major pathways for repairing damaged DNA, (i.e., base excision repair (BER), which eliminates single damaged-base residues, and nucleotide excision repair (NER), which excises damage within oligomers that are 25–32 nucleotides long) are involved in the repair of Cr-associated genetic lesions (O’Brien et al., 2003). NER efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity (Reynolds et al., 2004). Elimination of DNA damage by NER is a highly coordinated process involving the recognition of the damaged site, excision of a short oligonucleotide containing the DNA lesion, and filling in of the repair-generated gap in the DNA strand by a DNA repair polymerase. Cell-free, as well as cell culture, based studies led to the conclusion that DNA polymerase ð and ε have a direct role in mediating NER associated DNA synthesis (Wood 1996). PCNA and RPA, which are DNA polymerase accessory factors, are also known to participate in the NER process (Kunkel and Erie, 2005). The preparation of the human cell DNA synthesome, a multi-protein complex used in these studies, contains proteins, which are known to participate in both of these repair processes, [polymerase δ, PCNA, etc.]. Thus, it is not surprising that inhibition of the activity of DNA polymerases ð and ε, produced by Cr(III), inhibits the DNA replication and repair processes in cells. The decreased ability of the cell to repair DNA damage sustained during the replication process, could be the result of Cr(III) ion mediated inhibition of synthesome associated DNA polymerase δ and ε activity within the cell. In support of this suggestion, Cr(VI)-induced formation of binary Cr-DNA or DNA-DNA cross-links, and ternary Cr-DNA-protein complexes was shown to correlate with a metal ion induced increase in DNA damage (Mattagajasingh and Misra, 1996). Thus, the decrease in the activity and fidelity with which the DNA synthesome mediates DNA replication in the presence of Cr(III) ion, could be the result of the DNA synthesome forming DNA-protein-chromium complexes at these higher concentrations of Cr(III) ion. These complexes could then inhibit the normal function of specific proteins required to support a high fidelity DNA replication process; such as that typically carried-out by the synthesome in the absence of Cr(III) ion.
Consistent with this suggestion is the observation that chromium induced single nucleotide substitutions, insertions and deletions were the primary types of lesions occurring in the LacZα gene analyzed following synthesome mediated in vitro DNA replication of the pBK-CMV template. Single nucleotide substitutions were the predominant mutation type and occurred mainly in GC base-pairs, Table 4. Although mutation frequencies induced by Cr(III) were higher than that induced by Cr(VI) or than that arising spontaneously in reactions carried out in the absence of metal ions; none-the-less the distribution of each type of mutation formed in the presence of Cr(III) was essentially the same as those formed in its absence. In addition, the types of mutations formed during the in vitro DNA synthetic reaction, in the presence of chromium ion and their distribution into each category, were essentially the same as the types and distribution of mutations formed, in vitro, by the DNA synthesome derived from malignant or nonmalignant breast cells (Sekowski et al., 1998). Our observations with the DNA synthesome mediated reactions are also consistent with those of other laboratories which showed that single base substitutions at the G:C base pairs were the predominant type of Cr(III) induced mutation formed during the replication of pSP189 plasmids in intact human cells (Voitkun and Zhithkovich, 1998). Formation of ternary protein-Cr(III)-DNA adducts containing amino acids such as histidine or cysteine are thought to be of importance in mediating the Cr(VI) to Cr(III)-dependent carcinogenesis pathway (Zhithkovich et al., 1995), and ternary complex formation can be accompanied by non-RedOx associated reduction of Cr(VI) to Cr(III) in the presence of glutathione (Zhithkovich et al., 1995). Our results indicate that chromium ion produced fewer transition mutations and more transversion mutations than arose by spontaneous mutation. The analysis of Cr(VI)-induced mutations following replication of pSP189 plasmids in human cells shows that G/C-targeted point mutations fall into the transition (30%) and transversion (51%) categories. The specificity of the mutation spectrum induced by Cr (III) ion during the synthesome-mediated in vitro DNA replication reaction, appeared to be consistent with the types of mutations observed when intact cells replicated cysteine-treated plasmids in the presence of Cr(VI), (Zhitkovich et al., 2001). Our studies with the DNA synthesome not only confirmed that single-base substitutions at G/C pairs were the predominant type of Cr-induced mutations, but it also showed that the presence of chromium ion led to more nucleotide transversions, and fewer nucleotide transitions when compared to the types of mutations that arise spontaneously.
In summary, our results showed clearly that chromium ion reduced the in vitro activity and fidelity of the DNA replication process mediated by the human cell DNA synthesome, and extended the pioneering studies exploring how chromium ion decreases the activity and fidelity of individually purified DNA polymerases (Snow, 1991 and 1994; Bridgewater et al., 1994b and 1998). In addition, our studies demonstrated that the synthesome-associated in vitro DNA polymerase α, δ and ε were differentially inhibited by Cr(III), which resulted in the preferential inhibition of the activity of DNA polymerase α at low concentrations of the metal ion, and inhibition of both the leading and lagging strand DNA polymerases at higher concentrations of the metal ion. Thus at lower concentrations of the metal ion, the genotoxic effects of chromium ion could be expressed, in part, as an increase in the frequency of mutation produced during the DNA synthetic process; perhaps due to inhibition of the intrinsic activity of DNA polymerase α. While at higher concentrations of the metal ion, both the DNA replication and DNA repair process could be inhibited, via direct inhibition of the polymerizing activity of DNA polymerases α, δ and ε and inhibition of the intrinsic proof-reading activity associated with DNA polymerase δ and ε; resulting in both the genotoxic and the cytotoxic effects of the metal ion.
Acknowledgments
This work was supported in part by research awards from the National Institute of Health RO1 CA74904 to RJH, and RO1 CA57350 to LHM. The authors wish to thank the Vera Bradley Foundation for their initial support of this work, and also wish to express their gratitude to Dr. Carita Lanner for her participation in the initial discussions related to these studies and for helping to edit an initial draft of this manuscript.
References
- Abdel-Aziz W, Jiang H, Hickey R, Malkas L. Ara-C affects formation of cancer cell DNA synthesome replication intermediates. Cancer Chemother Pharmacol. 2000;45:312–319. doi: 10.1007/s002800050046. [DOI] [PubMed] [Google Scholar]
- Applegren N, Hickey RJ, Kleinschmidt AM, Zhou Q, Coll J, Wills P, Swaby R, Wei Y, Quan JY, Lee WYWT, Malkas LH. Further characterization of the human cell multiprotien DNA replication complex. J Cell Biochem. 1995;59:91–107. doi: 10.1002/jcb.240590111. [DOI] [PubMed] [Google Scholar]
- Arakawa H, Ahmad R, Naoui M, Tajmir-Riahi HA. A comparative study of calf thymus DNA binding to Cr(III) and Cr(VI) ions. Evidence for the guanine N-7-chromium-phosphate chelate formation. J Biol Chem. 2000;275:10150–10153. doi: 10.1074/jbc.275.14.10150. [DOI] [PubMed] [Google Scholar]
- Arslan P, Beltrame M, Tomasi A. Intracellular chromium reduction. Biochim Biophys Acta. 1987;931:10–15. doi: 10.1016/0167-4889(87)90044-9. [DOI] [PubMed] [Google Scholar]
- Biedermann KA, Landolph JR. Role of valence state and solubility of chromium compounds on induction of cytotoxicity mutagenesis anchorage independence in diploid human fibroblasts. Cancer Res. 1990;50:7835–7842. [PubMed] [Google Scholar]
- Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgewater LC, Manning FC, Patierno SR. Base-specific arrest of in vitro DNA replication by carcinogenic chromium: relationship to DNA interstrand cross-linking. Carcinogenesis. 1994a;15:2421–2427. doi: 10.1093/carcin/15.11.2421. [DOI] [PubMed] [Google Scholar]
- Bridgewater LC, Manning FC, Woo ES, Patierno SR. DNA polymerase arrest by adducted trivalent chromium. Mol Carcinog. 1994b;9:122–133. doi: 10.1002/mc.2940090304. [DOI] [PubMed] [Google Scholar]
- Bridgewater LC, Manning FCR, Patierno SR. Arrest of replication by mammalian DNA polymerases α and β caused by chromium-DNA lesions. Mol Carcinog. 1998;23:201–206. [PubMed] [Google Scholar]
- Coll JM, Sekowski JW, Hickey RJ, Schnaper L, Yue W, Brodie AM, Uitto L, Syvaoja JE, Malkas LH. The human breast cell DNA synthesome: its purification from tumor tissue and cell culture. Oncol Res. 1996;8:435–447. [PubMed] [Google Scholar]
- D’Agostini F, Izzotti A, Bennicelli C, Camoirano A, Tampa E, De Flora S. Induction of apoptosis in lung but not in the liver of rats receiving intra-tracheal instillation of chromium(VI) Carcinogenesis. 2002;23:587–593. doi: 10.1093/carcin/23.4.587. [DOI] [PubMed] [Google Scholar]
- Ding M, Shi X. Molecular mechanisms of Cr(VI)-induced carcinogenesis. Mol Cell Biochem. 2002;234–235(1–2):293–300. [PubMed] [Google Scholar]
- Gibb HJ, Lees PS, Pinsky PF, Rooney BC. Lung cancer among workers in chromium chemical production. Am J Int Med. 2000;38:115–126. doi: 10.1002/1097-0274(200008)38:2<115::aid-ajim1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Han S, Hickey RJ, Tom TD, Wills PW, Syvaoja JE, Malkas LH. Differential inhibition of the human cell DNA replication complex-associated DNA polymerases by the antimetabolite 1-beta-D-arabinofuranosylcytosine triphosphate (ara-CTP) Biochem Pharmacol. 2000;60:403–411. doi: 10.1016/s0006-2952(00)00336-1. [DOI] [PubMed] [Google Scholar]
- Hneihen PS, Standeven AM, Wetterhahn KE. Differential binding of chromium (VI) and chromium (III) complexes to salmon sperm nuclei and nuclear DNA and isolated calf thymus DNA. Carcinogenesis. 1993;14:1795–1803. doi: 10.1093/carcin/14.9.1795. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. IARC monograph on the evaluation of carcinogenic risks to humans. Vol. 49. World Health Organization; Lyon, France: 1990. Chromium nickel and welding; pp. 49–256. [PMC free article] [PubMed] [Google Scholar]
- Jatinder S, Snow ET. Chromium (III) decreases the fidelity of human DNA polymerase β. Biochemistry. 1998;37:9371–9378. doi: 10.1021/bi9731551. [DOI] [PubMed] [Google Scholar]
- Jiang HY, Hickey RJ, Abdel-Aziz W, Malkas LH. Effects of Gemcitabine and araC on in vitro DNA synthesis mediated by the human breast cell DNA synthesome. Cancer Chemother Pharmacol. 2000;45:320–328. doi: 10.1007/s002800050047. [DOI] [PubMed] [Google Scholar]
- Jiang HY, Hickey RJ, Abdel-Aziz W, Tom TD, Wills PW, Liu J, Malkas LH. Human cell DNA replication is mediated by a discrete multiprotein complex. J Cell Biochem. 2002;85:762–774. doi: 10.1002/jcb.10182. [DOI] [PubMed] [Google Scholar]
- Kim G, Yurkow EJ. Chromium induces a persistent activation of mitogen-activated protein kinases by a redox-sensitive mechanism in H4 rat hepatoma cells. Cancer Res. 1996;56:2045–2051. [PubMed] [Google Scholar]
- Kortenkamp A, Oetken G, Beyersmann D. The DNA cleavage induced by a chromium(V) complex and by chromate and glutathione is mediated by activated oxygen species. Mutat Res. 1990;232:155–161. doi: 10.1016/0027-5107(90)90120-s. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lin S, Hickey RJ, Malkas LH. The isolation of a DNA synthesome from human leukemia cells. Leuk Res. 1997;21:501–512. doi: 10.1016/s0145-2126(96)00103-8. [DOI] [PubMed] [Google Scholar]
- Liu S, Medvedovic M, Dixon K. Mutational specificity in shuttle vector replicating in chromium(VI)-treated mammalian cells. Environ Mol Mutagen. 1999;33:313–319. doi: 10.1002/(sici)1098-2280(1999)33:4<313::aid-em8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Malkas LH, Hickey RJ, Li CJ, Pederson N, Baril EF. A 21 S enzyme complex from HeLa cells functions in Simian Virus 40 DNA replication in vitro. Biochemistry. 1990;29:6362–6374. doi: 10.1021/bi00479a004. [DOI] [PubMed] [Google Scholar]
- Mattagajasingh SN, Misra HP. Mechanisms of the carcinogenic chromium(VI)-induced DNA-protein cross-linking and their characterization in cultured intact human cells. J Biol Chem. 1996;271:33550–33560. doi: 10.1074/jbc.271.52.33550. [DOI] [PubMed] [Google Scholar]
- O’Brien T, Xu J, Patierno SR. Effects of glutathione on chromium-induced DNA crosslinking and DNA polymerase arrest. Mol Cell Biochem. 2001;222:173–182. [PubMed] [Google Scholar]
- O’Brien TJ, Ceryak S, Patierno SR. Complexities of chromium carcinogenesis: role of cellular response, repair and recovery mechanisms. Mutat Res. 2003;533:3–36. doi: 10.1016/j.mrfmmm.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Pospiech H, Kursula I, Abdel-Aziz W, Malkas L, Uitto L, Kastelli M, Vihinen-Ranta M, Eskelinen S, Syväoja JE. A neutralizing antibody against human DNA polymerase epsilon inhibits cellular but not SV40 DNA replication. Nucleic Acids Res. 1999;27:3799–3804. doi: 10.1093/nar/27.19.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quievryn G, Peterson E, Messer J, Zhithkovich A. Genotoxicity and mutagenicity of chromium(VI)/ascorbate-generated DNA adducts in human and bacterial cells. Biochemistry. 2003;42:1062–1070. doi: 10.1021/bi0271547. [DOI] [PubMed] [Google Scholar]
- Reynolds M, Peterson E, Quievryn G, Zhitkovich A. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J Biol Chem. 2004;279:30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- Sekowski JW, Malkas LH, Wei Y, Hickey RJ. Mercuric ion inhibits the activity and fidelity of the human cell DNA synthesome. Toxicol Appl Pharmacol. 1997;145:268–276. doi: 10.1006/taap.1997.8185. [DOI] [PubMed] [Google Scholar]
- Sekowski JW, Malkas LH, Schnaper L, Bechtel PE, Long BJ, Hickey RJ. Human breast cancer cells contain an error-prone DNA replication apparatus. Cancer Res. 1998;58:3259–3263. [PubMed] [Google Scholar]
- Servant L, Bieth A, Hayakawa H, Cazaux C, Hoffmann JS. Involvement of DNA polymerase beta in DNA replication and mutagenic consequences. J Mol Biol. 2002;315:1039–1047. doi: 10.1006/jmbi.2001.5307. [DOI] [PubMed] [Google Scholar]
- Singh J, Snow ET. Chromium III decreases the fidelity of human DNA polymerase beta. Biochemistry. 1998;37 (26):9371–9378. doi: 10.1021/bi9731551. [DOI] [PubMed] [Google Scholar]
- Singh J, Carlisle DL, Pritchard DE, Patierno SR. Chromium-induced genotoxicity and apoptosis: relationship to chromium carcinogenesis. Oncol Rep. 1998;5 (6):1307–1318. doi: 10.3892/or.5.6.1307. [DOI] [PubMed] [Google Scholar]
- Snow ET. A possible role for chromium(III) in genotoxicity. Environ Health Perspect. 1991;92:75–81. doi: 10.1289/ehp.919275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow ET. Effect of chromium on DNA replication in vitro. Environ Health Perspect. 1994;102:41–44. doi: 10.1289/ehp.94102s341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow ET, Xu LS. Chromium (III) bound to DNA templates promotes increased polymerase processivity and decreased fidelity during replication in vitro. Biochemistry. 1991;30:11238–11245. doi: 10.1021/bi00111a007. [DOI] [PubMed] [Google Scholar]
- John PW, Sr, Wise SS, Little JE. The cytotoxicity and genotoxicity of particulate and soluble hexavalent chromium in human lung cells. Mutat Res. 2002;517:221–229. doi: 10.1016/s1383-5718(02)00071-2. [DOI] [PubMed] [Google Scholar]
- Syvaoja J, Suomensaari S, Nishida C, Goldsmith JS, Chui GS, Jain S, Linn S. DNA polymerases alpha, delta, and epsilon: three distinct enzymes from HeLa cells. Proc Natl Acad Sci USA. 1990;87:6664–6668. doi: 10.1073/pnas.87.17.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom TD, Malkas LH, Hickey RJ. Identification of multiprotein complexes containing DNA replication factors by native immunoblotting of HeLa cell protein preparations with T-antigen-dependent SV40 DNA replication activity. J Cell Biochem. 1996;63:259–267. doi: 10.1002/(sici)1097-4644(19961201)63:3<259::aid-jcb1>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Tran HT, Gordenin DA, Resnick MA. The 3′-5′exonucleases of DNA polymerases delta and epsilon and the 5′-3′exonuclease Exo1 have major roles in post-replication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou TC, Lin RJ, Yang JL. Mutational spectrum induced by chromium(III) in shuttle vectors replicated in human cells: relationship to Cr(III)-DNA interactions. Chem Res Toxicol. 1997;10:962–970. doi: 10.1021/tx970040p. [DOI] [PubMed] [Google Scholar]
- Tsou TC, Lai HJ, Yang JL. Effects of mannitol or catalase on the generation of reactive oxygen species leading to DNA damage by chromium(VI) reduction with ascorbate. Chem Res Toxicol. 1999;12:1002–1009. doi: 10.1021/tx9802264. [DOI] [PubMed] [Google Scholar]
- Voitkun V, Zhithkovich A, Costa M. Cr(III)-mediated crosslink’s of glutathione or amino acids to the DNA phosphate backbone are mutagenic in human cells. Nucleic Acids Res. 1998;26:2024–2030. doi: 10.1093/nar/26.8.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- Wu Y, Hickey R, Lawlor K, Wills P, Yu F, Ozer H, Starr R, Quan JY, Lee M, Malkas L. A 17S multiprotein form of murine cell DNA polymerase mediates polyomavirus DNA replication in vitro. J Cell Biochem. 1994;54:32–46. doi: 10.1002/jcb.240540105. [DOI] [PubMed] [Google Scholar]
- Xu J, Bubley GJ, Detrick B, Blankenship LJ, Patierno SR. Chromium(VI) treatment of normal human lung cells results in guanine-specific DNA polymerase arrest, DNA-DNA cross-links and S-phase blockade of cell cycle. Carcinogenesis. 1996;7:1511–1517. doi: 10.1093/carcin/17.7.1511. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Leonard SS, Wang S, Vallyathan V, Castranova V, Shi X. Cr(VI) induces cell growth arrest through hydrogen peroxide-mediated reactions. Mol Cell Biochem. 2001;222:77–83. [PubMed] [Google Scholar]
- Zhithkovich A, Voitkun V, Costa M. Glutathione and free amino acids form stable complexes with DNA following exposure of intact mammalian cells to chromate. Carcinogenesis. 1995;16:907–913. doi: 10.1093/carcin/16.4.907. [DOI] [PubMed] [Google Scholar]
- Zhitkovich A, Song Y, Quievryn G, Voitkun V. Non-oxidative are responsible for the induction of Mutagenesis by reduction of Cr(VI) with cysteine: role of ternary DNA adducts in Cr(III)-dependent mutagenesis. Biochemistry. 2001;40:549–560. doi: 10.1021/bi0015459. [DOI] [PubMed] [Google Scholar]
- Zlotkin T, Kaufmann G, Jiang Y, Lee MY, Uitto L, Syvaoja J, Dornreiter I, Fanning E, Nethanel T. DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J. 1996;15:2298–2305. [PMC free article] [PubMed] [Google Scholar]