Abstract
We previously reported on the purification and characterization of a functional multi-protein DNA replication complex (the DNA synthesome) from human cells and tissues. The synthesome is fully competent to carry-out all phases of the DNA replication process in vitro. In this study, DNA primase, a component of the synthesome, is examined to determine its activity and processivity in the in vitro synthesis and extension of RNA primers. Our results show that primase activity in the P4 fraction of the synthesome is 30-fold higher than that of crude cell extracts. The synthesome synthesizes RNA primers that are 7–10 ribonucleotides long and DNA primers that are 20–40 deoxyribonucleotides long using a poly(dT) template of exogenous single-stranded DNA. The synthesome-catalyzed RNA primers can be elongated by E. coli DNA polymerase I to form the complementary DNA strands on the poly(dT) template. In addition, the synthesome also supports the synthesis of native RNA primers in vitro using an endogenous supercoiled double-stranded DNA template. Gel analysis demonstrates that native RNA primers are oligoribonucleotides of 10–20 nt in length and the primers are covalently link to DNA to form RNA-primed nascent DNA of 100–200 nt. Our study reveals that the synthesome model is capable of priming and continuing DNA replication. The ability of the synthesome to synthesize and extend RNA primers in vitro elucidates the organizational and functional properties of the synthesome as a potentially useful replication apparatus to study the function of primase and the interaction of primase with other replication proteins.
Keywords: PRIMASE, RNA PRIMER, DNA REPLICATION, DNA SYNTHESOME, IN VITRO
DNA replication in eukaryotic cells is a multi-step and tightly coordinated process that requires a specific set of active proteins to participate and interact with one another in each reaction [Malkas, 1998; Frouin et al., 2003]. The initiation of DNA replication involves the synthesis of a short RNA primer of about 10 ribonucleotides in length that is catalyzed by DNA primase [Waga and Stillman, 1998]. RNA primers play a central role in the initiation of DNA replication on both the leading and lagging strands at the replication fork [Wang, 1991]. After the synthesis of RNA primers, DNA polymerase α(pol α) elongates the primers by polymerizing deoxyribonucleotide triphosphates (dNTPs) to generate DNA primers of about 20–40 nucleotides [Baker and Bell, 1998; Arezi and Kuchta, 2000]. Pol α and primase are thought to be dissociated from the DNA template after the synthesis of DNA primers. DNA pol δ and probably also DNA pol ε extend DNA primers for DNA replication to produce one continuous DNA fragment on the leading strand, or many discontinuous DNA fragments (Okazaki fragments) on the lagging strand [Lee et al., 1989; Bullock et al., 1991]. An Okazaki fragment on the lagging strand is a relatively short DNA fragment of 100–200 nucleotides in length in eukaryotes containing an RNA primer at its 5′ terminus [Ogawa and Okazaki, 1980].
The eukaryotic DNA primase is comprised of two subunits of about 49 and 58 kDa (p49 and p58). The p49 subunit has a high and unstable primase activity [Santocanale et al., 1993; Schneider et al., 1998]. The p58 subunit is necessary for the stability and activity of the p49 subunit [Zerbe and Kuchta, 2002]. DNA primase is not capable of processive DNA synthesis, but it can switch the activity of DNA polymerase for DNA replication [Kuchta et al., 1990]. DNA primase with unique features usually exists in a tightly bound complex of DNA pol α and primase (pol-prim, p180-p70-p58-p49) or as two tightly bound primase subunits (prim2, p58-p49) [Weisshart et al., 2000]. DNA polymerase subunits p180 and p70 produce a short DNA primer of about 40 nucleotides long [Collins et al., 1993; Stadlbauer et al., 1996]. The p70 subunit does not possess a known enzymatic activity but it plays an important role in the assembly of the pol-prim complex [Ott et al., 2002]. The pol-prim complex can catalyze the synthesis of oligoribonucleotide primers on the poly(dT) or poly(dC) templates of synthetic single-stranded DNA [Conaway and Lehman, 1982]. The synthesis of RNA primers mediated by pol-prim complex is strongly ATP-dependent and DNA template-dependent [Kuchta et al., 1990].
Various models of eukaryotic DNA replication have been used to investigate the mechanism of RNA primer synthesis and extension catalyzed by DNA primase. The highly purified primase system utilizes exogenous single-stranded DNA template to synthesize RNA primer [Santocanale et al., 1993; Bakkenist and Cotterill, 1994]. The nuclear matrix replication system of whole cell lysates has been used to examine the synthesis and distribution of native RNA primers and RNA-primed nascent DNA in cell nuclei using endogenous nuclear matrix-bound double-stranded DNA templates [Tseng and Goulian, 1977; Paff and Fernandes, 1990]. However, in many previous studies, the purified primase model does not drive the synthesis of native RNA primer and RNA-primed nascent DNA due to the absence of many DNA replication proteins and native double-stranded DNA template. The nuclear matrix replication model of whole cell lysates does not support the in vitro synthesis and extension of RNA primers on exogenous single-stranded DNA template because of a low primase activity in this system. The synthesis and extension of RNA primers in vitro not only requires highly active DNA primase, but also involves the activity of other DNA replication proteins. Therefore, we propose that an interesting and plausible cell-free DNA replication model that is competent to carry out all phases of the DNA synthetic process should be more physiologically relevant and closer to the cellular environment than a highly purified primase model system, and far less complex than a whole cell lysate system, in the study of DNA primase function and primase-mediated RNA primer synthesis. The essential homogeneous DNA synthetic apparatus has been isolated and purified from human cells and tissues [Malkas et al., 1990; Applegren et al., 1995]. This functional multiprotein complex was termed the DNA synthesome [Coll et al., 1996; Tom et al., 1996], and is fully competent to carry-out all phases of the DNA synthesis process in vitro; including the initiation, elongation and termination phases of the double-stranded DNA replication process [Wu et al., 1994; Jiang et al., 2002]. In this study, we characterize the synthesome-associated primase activity and processivity during the synthesis and extension of RNA primers in vitro using exogenous single-stranded DNA or supercoiled double-stranded DNA as the replicated template. The features of different primase products catalyzed by the DNA synthesome associated DNA primase are analyzed and discussed.
MATERIALS AND METHODS
MATERIALS
The nucleoside triphosphates (NTPs) and deoxyribonucleoside triphosphates (dNTPs), actinomycin D and α-amanitin were purchased from the Sigma Chemical Company. The poly(dT)290, 8–32 base pair (bp) nucleotide marker ladder and 25 bp DNA markers were obtained from Amersham. (α-32P)NTP, (α-32P)dNTP, (3,000 Ci/mmol), and (3H)dATP (17.5 Ci/mmol) were purchased from PerkinElmer. DNA primase antibodies (Ab-1(p49), Ab-2(p58)) were purchased from LabVision.
ISOLATION OF THE DNA SYNTHESOME
The DNA synthesome was isolated and purified from the human breast cancer cell line MCF-7, according to the procedure previously described [Malkas et al., 1990; Lin et al., 1997]. All fractions containing the DNA synthesome were stored at −80°C and the P4 fraction was used for all primase experiments.
IMMUNODETECTION OF DNA PRIMASE IN THE DNA SYNTHESOME PREPARATIONS
Fifty micrograms of total protein from different fractions of the DNA synthesome purification were electrophoresed through a 10% SDS–polyacrylamide gel as previously described [Jiang et al., 2002]. The proteins were transferred to nitrocellulose membranes. Polyclonal antibodies for DNA primase subunits p49 and p58 were used at a 1:500 dilution and the secondary antibody, conjugated to horse-radish peroxide, was used at a dilution of 1:3,000 (Amersham), prior to the addition of ECL reagent.
RNA PRIMER SYNTHESIS ASSAY
RNA primers were synthesized with the incorporation of radiolabeled rATP on the poly(dT) template with some modifications of the procedure described by Kuchta et al. [1990]. The reaction was performed at 37°C for 60 min in 25 μl mixtures containing 60 nM poly(dT), 50 mM Tris–HCl (pH.8.0), 10 mM magnesium acetate, 50 mM sodium acetate, 0.5 mg/ml bovine serum albumin (BSA), 1 mM DTT, 100 μM (α-32P)ATP, 15% glycerol and 50 μg P4 fraction.
DNA PRIMER SYNTHESIS ASSAY
DNA primers were synthesized with incorporation of non-radioactive ATP and radioactive dATP on the poly(dT) template according to the procedure described [Kuchta et al., 1990; Copeland and Wang, 1993]. The reaction was performed at 37°C for 60 min in 25 μl mixtures containing 60 nM poly(dT), 50 mM Tris–HCl (pH 8.0), 10 mM magnesium acetate, 50 mM sodium acetate, 0.5 mg/ml BSA, 1 mM DTT, 15% glycerol, 0.5 mM ATP, 100 μM (α-32P)dATP or 100 μM (3H)dATP and the 50 μg P4 fraction.
RNA PRIMER EXTENSION ASSAY
RNA primer synthesis, catalyzed by the synthesome, was extended by E. coli DNA polymerase I with the incorporation of radiolabeled dATP to the RNA primer on the poly(dT) template in order to form a radiolabeled polynucleotide strand using the procedure described [Copeland and Wang, 1993]. The reaction was performed at 37°C for 60 min in 25 μl mixtures containing 60 nM poly(dT), 50 mM Tris–HCl (pH 8.0), 10 mM magnesium acetate, 50 mM sodium acetate, 0.5 mg/ml BSA, 1 mM DTT, 15% glycerol, 0.5 mM ATP, 100 μM (α-32P)dATP or 100 μM (3H)dATP, 10 μg of the P4 fraction, and 0.5 U Klenow fragment E. coli DNA polymerase I (USB).
THE SYNTHESIS OF RNA-PRIMED DNA AND THE IN VITRO SV40 DNA REPLICATION ASSAY
RNA-primed DNA and nascent DNA were synthesized using a two radionucleotide labeling method according to the procedure described with modification [Malkas et al., 1990; Abdel-Aziz et al., 2000]. The reaction was performed at 37°C for 3 h in 25 μl reaction mixtures containing 30 mM HEPES, 7.5 mM MgCl2, 0.5 mM DTT, 100 μM (α-32P)-ATP (3,000 Ci/mmol) and 100 μM (α32P)-dTTP, 200 μM of GTP, CTP, UTP, 100 μM of dATP, dGTP, dCTP, 40 mM phosphocreatine, 1 μg creatine phosphokinase, 0.5 μg SV40 large T-antigen, 50 ng pSVO+ plasmid DNA containing a 200 bp insert of the SV40 replication origin DNA sequence [Stillman and Gluzmen, 1985] and 50 μg of the P4 fraction. The reaction products were extracted by phenol/chloroform, and then precipitated by 2 M ammonium acetate and ethanol. Newly replicated DNA and RNA products were analyzed in 8% denaturing polyacrylamide gels after being heated to 90°C for 5 min. In order to further characterize native RNA primer and RNA-primed nascent DNA, the reaction of RNA synthesis catalyzed by the synthesome was performed in the presence of rNTPs and dNTPs at 25°C for 10 min using a single radionucleotide (α32P)ATP labeling method. The newly replicated RNA products were extracted by phenol/chloroform and resuspended in 10 μl TE buffer (pH 7.8) containing 1 U/μl RNase inhibitor. RNA products were analyzed in 22% denaturing polyacrylamide gels after treatment with 10 U of DNase I, 10 U of RNase A or 0.3 N KOH at 37°C for 1 h, respectively. All RNA experiments were carried out under RNase-free conditions.
SCINTILLATION COUNTING AND GEL ELECTROPHORESIS ANALYSIS
Primase reaction products were spotted onto DE81 filters (Whatman) and quantified by liquid scintillation counting. Primase products were analyzed by electrophoresis through denaturing gels of 8–22% polyacrylamide-7 M urea in TBE buffer. The gels were dried and used to expose Kodak X-OMAT AR-5 film at −70°C for 1–7 days. The markers of DNA and oligonucleotide were labeled using radioactive (γ-32P)-ATP and T4 polynucleotide kinase.
ENZYME KINETIC ANALYSIS OF THE DNA SYNTHESOME-ASSOCIATED PRIMASE ACTIVITY
The amount of radiolabeled nucleotide incorporated during the primase enzyme assay was measured as the number of disintegrations per minute (DPM) (DPM = CPM/0.68), and then converted to microcuries (μCi) (1 μCi = 2.22 × 106 DPM). Enzyme kinetic parameters, Vmax (maximum velocity) and Km (substrate concentration in 1/2 Vmax), were obtained by Lineweaver–Burke analysis using a plot of the reciprocal of the initial velocity versus the reciprocal of the substrate concentration. The Y and X intercepts of a straight line in the graph represented the reciprocal of Vmax and Km, respectively. (Y intercept = 1/Vmax; X intercept = −1/Km.) All calculations and graphs were performed using Microsoft Excel software to determine the kinetics of the enzymatic reaction.
RESULTS
DISTRIBUTION AND ACTIVITY OF DNA PRIMASE IN DIFFERENT FRACTIONS OF THE DNA SYNTHESOME
Some different fractions of the synthesome from the crude cell extract (homogenate, H) of human breast cancer cell line (MCF-7) were isolated according to the fractionation scheme described in the previously published procedure [Applegren et al., 1995]. The different fractions obtained during the synthesome preparation included cytosolic supernatants 1–3 (S1–3), nuclear extract (NE), and the upper 70% (S4) and the low 30% (P4) of the soluble protein fraction obtained from an overnight ultracentrifugation of the combined NE/S3 at 100,000g [Lin et al., 1997]. The expression of DNA primase subunits p49 and p58 in the different fractions were examined using western blot analysis of p49 and p58 antibodies (Fig. 1A). The Ab-p49 and Ab-p58 were polyclonal antibodies that did not cross react with each other or with DNA polymerase gamma. The P4 fraction of the synthesome contained a high concentration of the primase subunits p49 and p58 as compared to other fractions of the synthesome preparation. DNA primase activities in these fractions were measured using the RNA primer extension assay (Fig. 1B). The results showed that the DNA primase specific activity in the P4 fraction was about 30-fold higher than that of the crude cell extract.
Fig. 1.
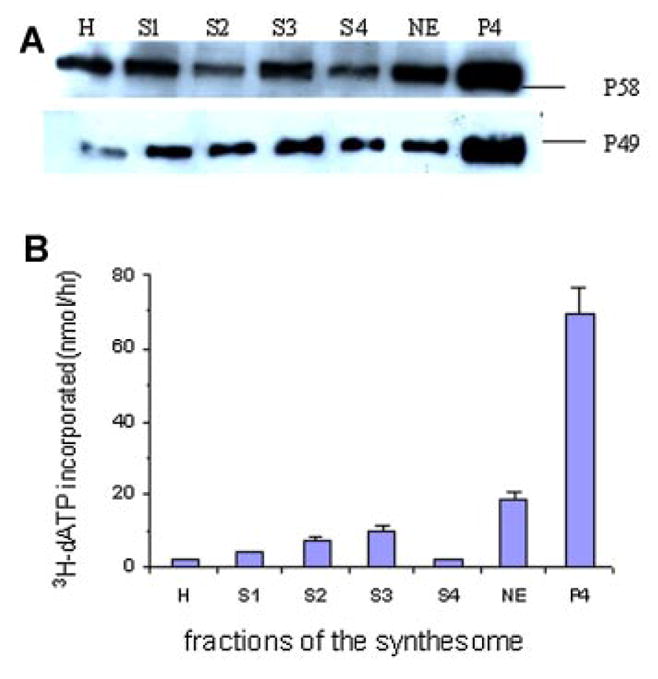
A: Immunodetection of DNA primase subunits p49 and p58 by Western blot analysis in different fractions during the purification of the DNA synthesome. The fractions include homogenate (H), low to high speed cytosolic supernatants 1–3 (S1–3), nuclear extract (NE), and the upper 70% (S4) and the lower 30% (P4) of the soluble protein derived from a 16 h ultracentrifugation of the combined NE/S3 [Lin et al., 1997]. B: DNA primase activity in different fractions of the synthesome was examined using a primer extension assay (Materials and Methods Section). The reaction was performed at 37°C for 1 h in 25 μl mixtures contained 60 nmol poly(dT), 10 μg of each different fraction, 500 μM ATP, 100 μM (3H)-dATP and 0.5 U of the Klenow fragment of E. coli DNA polymerase I. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
SYNTHESOME-CATALYZED RNA PRIMER SYNTHESIS
The DNA synthesome synthesized RNA primers by incorporating (α-32P)ATP onto the single-stranded DNA poly(dT) template. The abundance of the reaction products was quantified by liquid scintillation counting (Fig. 2A,B) and analyzed by electrophoresis through 20% denaturing polyacrylamide gels (Fig. 2C). Newly synthesized RNA primers consisted of unit-length primers of 7–10 nucleotides (nt) and short primers of 4–6 nt. The results show the synthesis of RNA primer as a function of the reaction time at 37°C and the synthesome concentration. The synthesis of RNA primer was enhanced significantly following an increase of the synthesome concentration (Fig. 2A). Gel analysis results further demonstrated that the abundance of unit-length primers of 7–10 nt increased, while the abundance of short primers of 4–6 nt decreased (Fig. 2C). The synthesis of RNA primers reached steady-state levels rapidly (15 min), and the relative amount of RNA primers remained constant from 15 to 120 min (Fig. 2B,C).
Fig. 2.
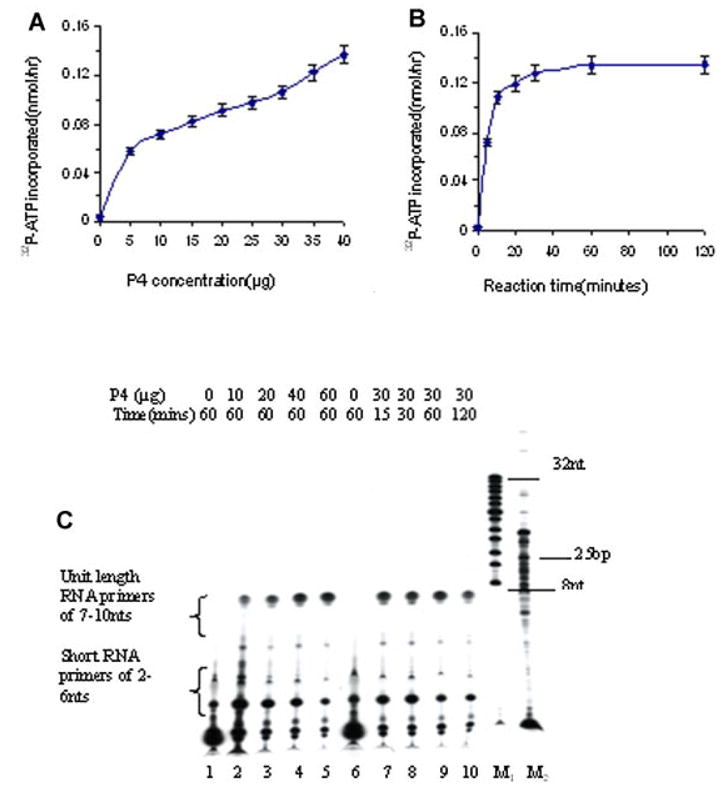
A,B: The amount of RNA primer synthesized by the P4 fraction was determined as a function of the concentration of the P4 fraction (A), for different lengths of reaction time (B). C: Twenty percent denaturing gel electrophoretic analysis of RNA primer products formed by various concentrations of the P4 fraction (lanes2–5: 10–60 μg) or formed as a function of reaction time (lanes 7–10: 15–120 min). Lanes M1 and M2 contained a 8–32 bp marker and a 25–2,600 bp DNA marker, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
SYNTHESOME-CATALYZED DNA PRIMER SYNTHESIS
The synthesome extended RNA primers by incorporating deoxyribonucleotides, to the 3′ end of the RNA primer, to form a DNA-primer nucleotide strand. The effects of synthesome concentration and reaction time on the extent of synthesis of the DNA-primers nucleotide strands were examined (Fig. 3A,B). DNA primers catalyzed by the synthesome were analyzed by electrophoresis through 20% denaturing polyacrylamide gels (Fig. 3C). The results showed that the DNA-primer strand was an oligonucleotide of 20–40 nt in length, and that it was synthesized rapidly (15 min). Synthesis of the DNA-primer band was influenced by the synthesome concentration and the length of reaction time. Increasing the synthesome concentration and the reaction time, enhanced the abundance of the DNA-primer strands but did not change the length of the DNA-primer strand synthesized.
Fig. 3.
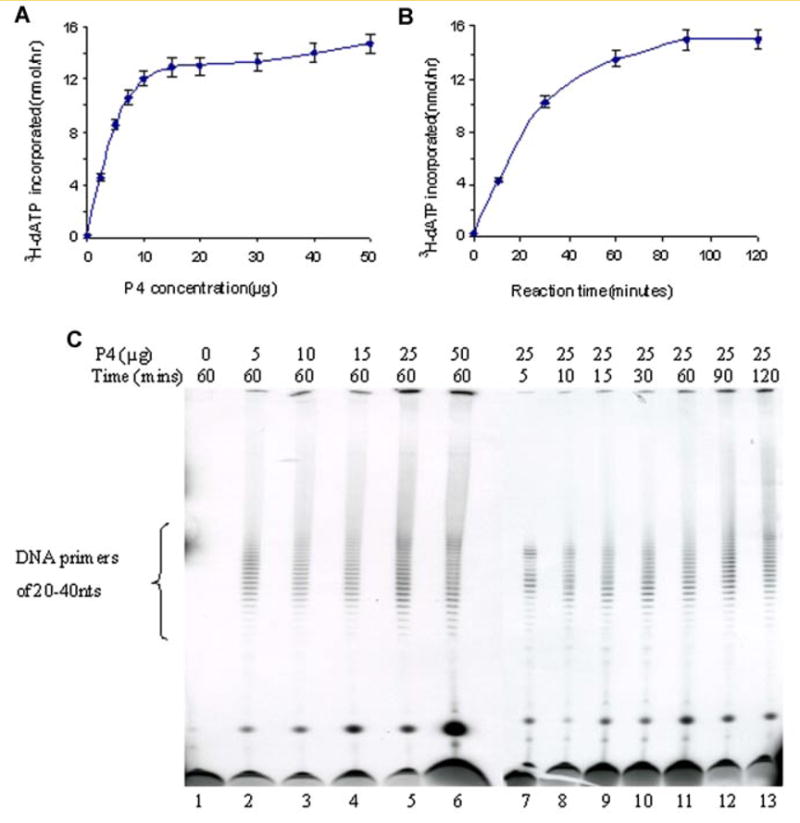
A,B: The abundance of DNA primers synthesized by the P4 fraction was measured using liquid scintillation counting as a function of either different concentrations of the P4 fraction (A) and or for different lengths of reaction time (B). C: Twenty percent denaturing gel electrophoretic analysis of DNA-primer products synthesized by various concentrations of the P4 fraction (lanes 2–6: 5–50 μg) or formed during reactions performed for various lengths of reaction time (lanes 7–13: 5–120 min). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
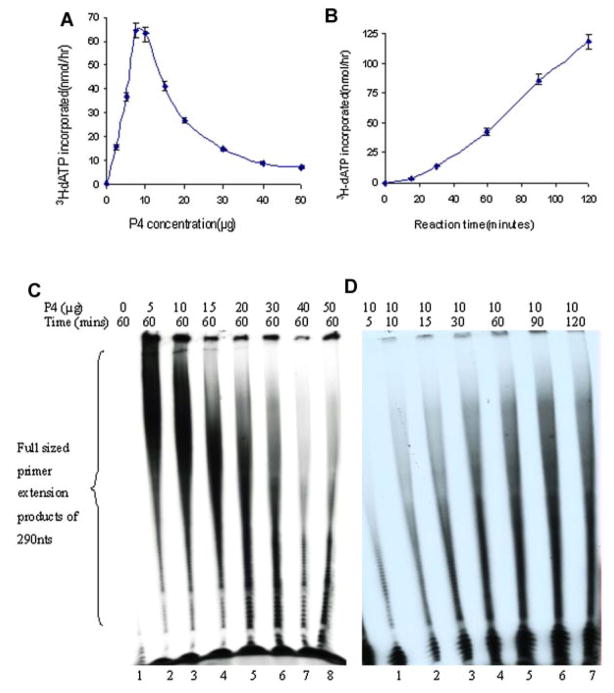
SYNTHESOME-MEDIATED RNA PRIMER EXTENSION
The synthesome-catalyzed RNA primers were extended by E. coli DNA polymerase I by incorporation of deoxyribonucleotides to form complementary poly(dA) strands on the poly(dT) templates. The effects of the synthesome concentration and reaction time on RNA primer extension were examined and quantified by liquid scintillation counting (Fig. 4A,B) and denaturing gel electrophoresis (Fig. 4C,D). The extended primer products were full-size polynucleotide strands of 290 nt. The extension of RNA primers was synthesome-dependent because E. coli DNA polymerase I lacks DNA primase activity and cannot synthesize RNA primers. The omission of the synthesome abolished the formation of the reaction products (Fig. 4C). The extension of the RNA primers was affected significantly by reaction time and the synthesome concentration. Increasing the reaction time significantly enhanced the amount and size of the reaction products (Fig. 4B,D). In contrast to the synthesis of RNA primers and DNA-primer strands, RNA primer extension required low concentrations of the DNA synthesome. The extension of RNA primer was inhibited dramatically when the concentration of the synthesome was above 10 μg in the reaction mixture (Fig. 4A,C).
Fig. 4.
A,B: The extent of RNA primed template extension catalyzed by different concentrations of the P4 fraction (A) and for different lengths of reaction time (B) was determined by liquid scintillation counting. Ten percent denaturing gel electrophoretic analysis of RNA primed-template extension as a function of several concentrations of the P4 fraction (lanes 2–8: 5–50 μg) (C) and for varying lengths of reaction time (lanes 1–7: 5–120 min) (D). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
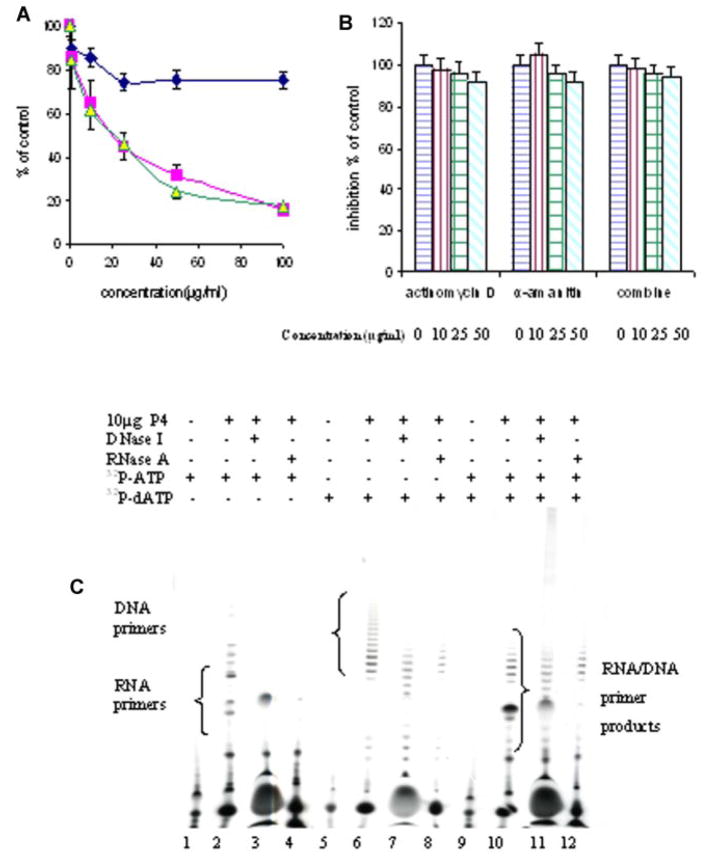
CHARACTERIZATION OF THE SYNTHESOME-ASSOCIATED PRIMASE AND PRIMASE PRODUCTS
The primer extension assay mediated by E. coli DNA polymerase I has been widely used to measure DNA primase activity [Paff and Fernandes, 1990; Copeland and Wang, 1993]. The effect of the DNA primase antibodies on the activity of the p49 and p58 subunits within the synthesome were examined. The primase activity was inhibited by these antibodies, but was not inhibited by BSA, which served as a control (Fig. 5A). The result showed that these antibodies bound the synthesome-associated primase and blocked its activity. The effect of an RNA polymerase inhibitor on synthesome-associated primase activity was also investigated. The result confirmed that primase activity was not inhibited by high concentrations of the RNA polymerase inhibitors, actimomycin D and α-amanitin or by both combinations (50–100 μg/ml; Fig. 5B). In order to characterize the synthesome-catalyzed primers, RNA primer and DNA-primer strands were synthesized using the double radio-nucleotide labeling method (Fig. 5C), and analyzed by gel electrophoresis. The results showed that DNA-primer strands consisted of two parts, one containing ribonucleotides of 7–10 nt in length and the other deoxyribonucleotides of 10–40 nt in length. The effect of reaction temperature on the synthesis of RNA primer and DNA primer was also measured (Fig. 5C). The results showed that a lower reaction temperature (25°C) increased RNA primer length (from 10 to 20 nt), but did not affect DNA primer length. The effects of DNA/RNA hydrolytic enzymes, deoxyribonuclease (DNase) and ribonuclease (RNase) on the length of the RNA primer and DNA primer were investigated (Fig. 5C). The unit-length RNA primers of 7–10 nt were fully hydrolyzed by RNase to form short RNA fragments of 2–4 nt, but they were not affected by DNase. In contrast, DNA primers were shortened by both RNase and DNase from 20–40 nt to 10–20 nt. These results further confirmed: (1) The unit-length RNA primers synthesized by the synthesome were ribonucleotide primers of 7–10 nt, because RNA primers were fully hydrolyzed by RNase, but were not shortened by DNase; (2) DNA-primer strands contained two parts consisting of ribonucleotides and deoxyribonucleotides. They were partially hydrolyzed by both RNase and DNase to form shorter primers than untreated DNA-primer strands; (3) Decreasing the reaction temperature increased DNA primase efficiency to increase RNA primer length, but it did not affect DNA polymerase function and DNA-primer strand length.
Fig. 5.
A: The effect of DNA primase antibodies, Ab-1(p49) (■) and Ab-2(p58) (▲), on DNA primase activity of P4 fraction. (◆) BSA served as a negative control. B: The effect of different concentrations (0–50 μg/ml) of actimomycin D, α-amanitin and both on the DNA primase activity of the P4 fraction. DNA primase activities were determined the using primer extension assay described in Materials and Methods Section. C: The effect of DNase I and RNase A on RNA primer and DNA primer products length. The reaction was performed at 25°C for 1 h in 25 μl reaction mixtures containing 0.5 μg poly(dT), 10 μg P4 fraction, 100 μM (32P)-ATP (lanes 1–4, 9–12), 100 μM (α-32P)-dATP (lanes 5–12) with 0.5 mM ATP. The primer products were analyzed in 22% denaturing gels after being treated at 37°C for 1 h with 10 U of DNase I (lanes 3, 7, 11) or 10 U of RNase A (lanes 4, 8, 12). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
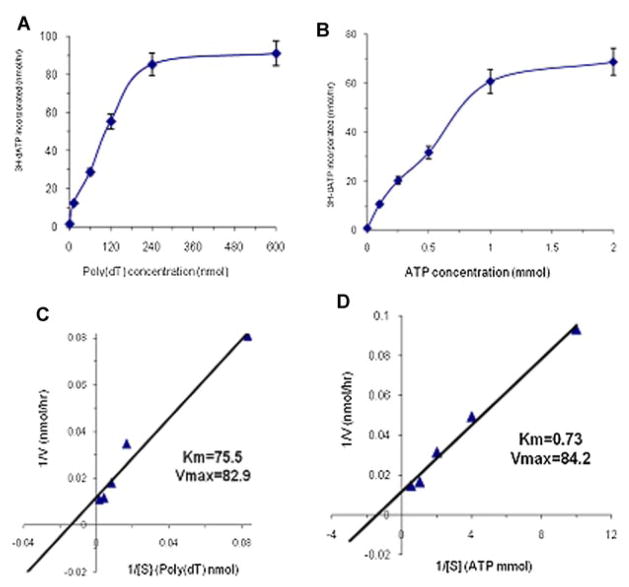
To further characterize the DNA synthesome-associated primase activity, an enzyme kinetic analysis was performed using the RNA primer extension assay carried-out by the DNA synthesome. The graphs in Figure 6A,B showed the initial reaction velocity as a function of varying substrate concentration [i.e., poly(dT) template or nucleotide ATP]. Enzyme kinetic constants of the synthesome-associated primase, Vmax and Km, were determined using a linear Lineweaver–Burke analysis (Fig. 6C,D). The values of Km for the substrates, poly(dT) and ATP, were 75.5 nM and 730 μM, respectively. The values of Vmax for the substrates, poly(dT) and ATP, were 82.9 and 84.2 nmol/h, respectively.
Fig. 6.
The kinetics of synthesome-associated DNA primase activity was determined by Michaelis–Menton Kinetic Analysis using the RNA primer extension assay described in Materials and Methods Section. The reactions were performed at 37°C for 60 min in 25 μl reaction mixtures containing 10 μg of the P4 fraction, 100 μM (3H)-dATP, 0.5 U of the Klenow fragment of E. coli DNA polymerase I and various concentrations of substrates, [poly(dT) template or nucleotide ATP]. The reaction velocity (V: the amount of 3H-dATP incorporation per hour) was determined as a function of substrate concentration ([S]) for poly(dT) (A) or ATP (B). The kinetic constants Vmax and Km were determined from the Lineweaver–Burke analysis by plotting the reciprocal of the initial reaction velocity (1/V) versus the reciprocal of the substrate concentration ([1/S]) of poly(dT) (C) or ATP(D). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
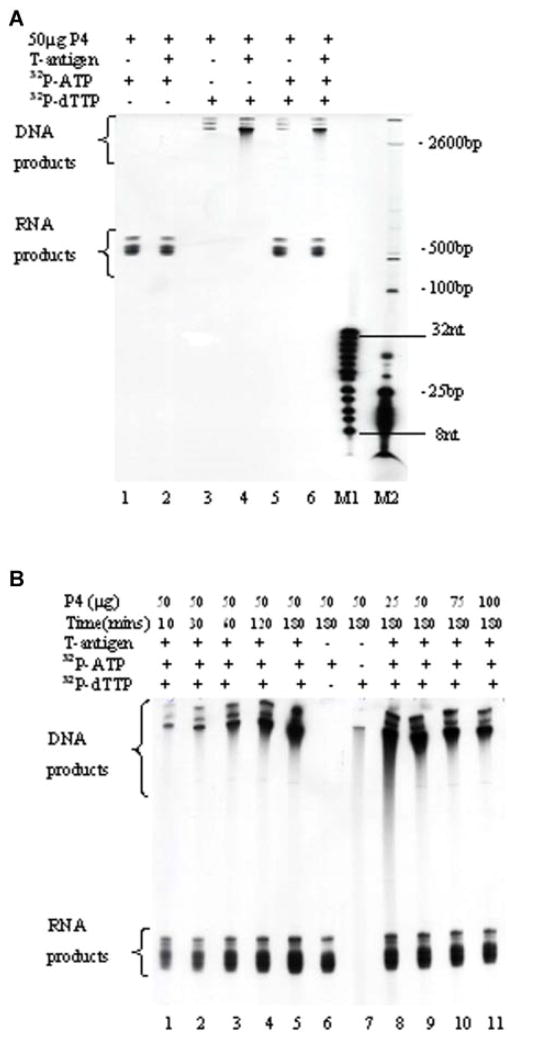
SYNTHESIS OF RNA-PRIMED NASCENT DNA AND IN VITRO SV40 ORIGIN DEPENDENT DNA REPLICATION MEDIATED BY THE SYNTHESOME
The synthesome catalyzed the synthesis of RNA-primed nascent DNA and the replication of SV40 origin containing double-stranded plasmid DNA in the presence of SV40 large, T-antigen, ribonucleoside/triphosphates, and dNTPs using the double radionucleotide labeling method. Newly replicated DNA and RNA products were analyzed using electrophoresis through 8% polyacrylamide-urea gels (Fig. 7A). The length of RNA-primed nascent DNA and DNA products was determined by comparison to the oligonucleotide and DNA markers described in the Materials and Methods Section. The results indicated that the maximal sizes of the DNA and RNA primed DNA were 2,800 and 600 bp long, respectively. The effect of the synthesome concentration and reaction time on the synthesis of DNA and RNA products was determined (Fig. 7B). The DNA and RNA products formed by the synthesome exhibited different features. The synthesis of DNA and RNA was synthesome-dependent, but in vitro DNA replication of plasmid DNA also required the presence of large T-antigen and an SV40 replication origin containing exogenous double-stranded DNA template. Increasing the reaction time from 10 to 180 min significantly increased the extent of DNA replication, but it did not affect RNA synthesis (Fig. 7B). High concentrations of the synthesome (75–100 μg) reduced overall in vitro SV40 origin dependent DNA replication under these reaction conditions, but it did not change RNA synthesis.
Fig. 7.
A: DNA and RNA products synthesized in the in vitro DNA replication reaction by the P4 fraction were analyzed by denaturing gel electrophoresis through an 8% polyacrylamide gel. The reaction was carried out at 37°C for 3 h in a 25 μl reaction mixture containing 50 ng double stranded DNA template containing an intact SV40 origin of replication, 0.5 μg large T-antigen (lanes 2, 4, 6), rNTP, dNTP, 100 μM (α-32P)-ATP (lanes 1, 2, 5, 6), (α-32P)-dTTP (lanes 3, 4, 5, 6), and 50 μg of the P4 fraction. Lanes M1 and M2 contain the 8–32 bp marker and the 25–2,600 bp DNA marker, respectively. B: Twelve percent denaturing gel electrophoretic analysis of newly replicated DNA and RNA products as a function of the reaction time (lanes 1–5: 10–180 min) and at various concentrations of the P4 fraction (lanes 8–11: 25–100 μg). Lane 6 contains RNA products without (α-32P)-dTTP added to the reaction mixture. Lane 7 contains DNA products without T-antigen and (α-32P)-ATP added to the reaction mixture. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
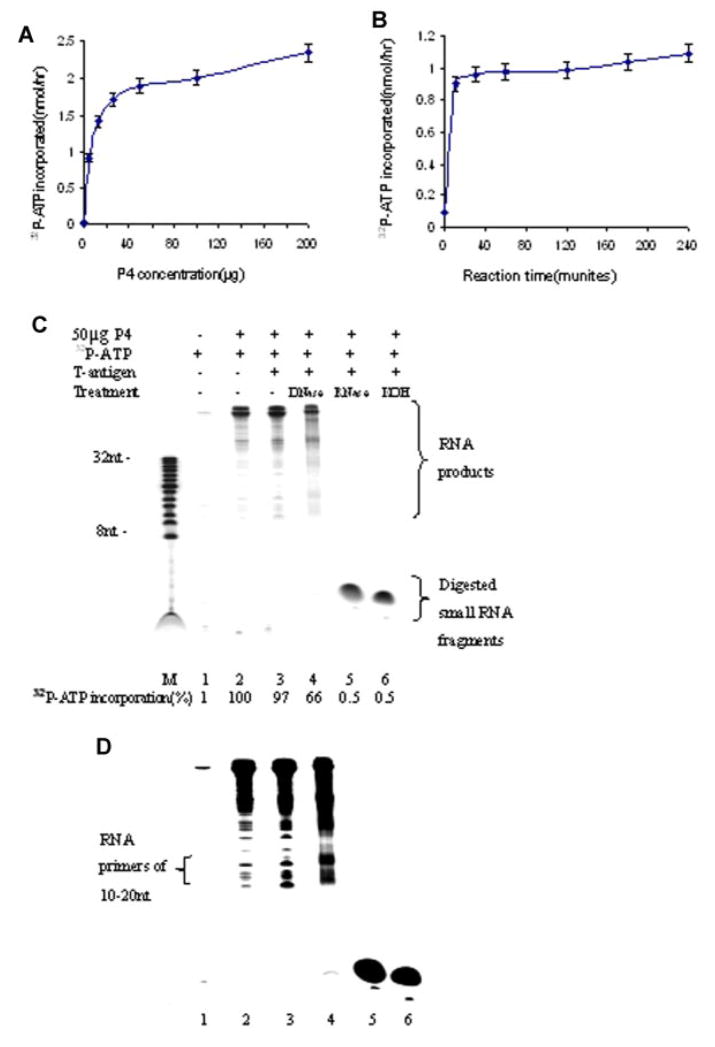
To further analyze the composition of newly replicated RNA products, the synthesis of RNA primer and RNA-primed nascent DNA, catalyzed by the synthesome, was performed using the single radionucleotide labeling procedure described in the Materials and Methods Section. The effects of the synthesome concentration and reaction time on the synthesis of RNA primers was quantified by liquid scintillation counting (Fig. 8A,B). The synthesis of RNA primers reached steady-state levels quickly (15 min), and increasing the reaction time and the synthesome concentration did not enhance the synthesis of RNA primers. The synthesome-synthesized RNA products were analyzed in 22% denaturing polyacrylamide gels (Fig. 8C). Newly replicated RNA containing products ranged from 10 to ~600 bp in length. RNA products included a major long product of 600 bp, and a minor (lower abundance) short product ranging from 10 to 200 bp upon prolonged auto-radiographic exposure time for 7 days (Fig. 8D). The short RNA products of 10–200 bp contained some RNA primers of 10–20 nt and RNA-primed nascent DNA of 100–200 bp. In order to remove any attached DNA from total RNA products, newly replicated RNA products were treated with DNase (RNase-free) prior to electrophoresis. After DNase treatment, total RNA containing products decreased significantly (66%) while RNA primers of 10–20 nt clearly increased in abundance (Fig. 8D). This result suggested that some native RNA primers were covalently link to DNA to form RNA-primed nascent DNA. RNA-primed nascent DNA was partially hydrolyzed by DNase resulting in a decrease of total RNA products and an increase in the abundance of RNA primers of 10–20 nt. Newly replicated RNA products were fully hydrolyzed by RNase and strong alkali (0.3 N KOH) into smaller ribonucleotides of 2–4 nt in length.
Fig. 8.
Quantification of the extent of RNA primer synthesis catalyzed by the human DNA synthesome as a function of the concentration of the P4 fraction added to the reaction mixture (A), and as a function of the length of the reaction time (B). Autoradiographic analysis of newly replicated RNA products following gel electrophoretic analysis through 22% polyacrylamide gels. C: Autoradiographic exposure time was 2 days. Lane 1 contains reaction products formed in the absence of the synthesome. Lanes 2 and 3 were RNA products formed in the absence or presence of SV40 large T-antigen, respectively. Lanes 4–6 contain different RNA products treated at 37°C for 1 h with either 10 U DNase I, or 10 U RNase A, or 0.3 N KOH, respectively. Lane M contains an 8–32 bp nucleotide marker. Newly replicated RNA products were quantified by liquid scintillation counting using 2 μl samples from each 25 μl reaction mixture. The percentage of (α-32P)-ATP incorporation into RNA was presented at the bottom of subpart (C). D: The same RNA products are shown as in subpart (C), with the exception that radiographic exposure time was increased to 7 days. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
The DNA synthesome model system contains all of the proteins needed to support each phase of the DNA replication process, and these proteins range in size from 20 to 240 kDa under current assay conditions, and can carry out all the phases of the DNA replication process in vitro [Wu et al., 1994; Tom et al., 1996]. In this study, synthesome-associated DNA primase activity was characterized by examining the synthesis and extension of RNA primers catalyzed in vitro by the DNA synthesome. The results of our functional analyses show that synthesome associated DNA primase activity in the P4 fraction derived from breast cancer cells is 30-fold higher than that of crude breast cancer cell extracts. The P4 fraction, containing the DNA synthesome, was derived from the combined nuclear and cytoplasmic soluble protein fractions prepared from MCF7 cells and was used to perform the SV40 in vitro DNA replication assay [Wu et al., 1994; Abdel-Aziz et al., 2000]. The activities of DNA polymerase α and δ and DNA primase in the P4 fraction are 20–40 times higher than that of crude cell extracts [Applegren et al., 1995; Coll et al., 1996; Jiang et al., 2002]; making it a useful tool for these studies because it is fully competent to support the synthesis and extension of RNA primers in an in vitro model system that has been shown to support all phases of the DNA replication process [Malkas et al., 1990].
DNA primase exhibits a unique processivity in the synthesis and extension of RNA primers. This processivity is defined by a competition between the addition of the next correct nucleotide and dissociation of the primer-template complex [Kuchta et al., 1990]. DNA primase is the only enzyme capable of synthesizing RNA primers during the initiation of DNA synthesis [Arezi and Kuchta, 2000]. Neither of the single primase subunits is capable of synthesizing or extending an RNA primer on their own [Copeland and Wang, 1993]. The synthesome contains both primase subunits, (i.e. p49 and p58) and can catalyze the synthesis of RNA primers in vitro using an exogenous poly(dT) template. The synthesome-synthesizes a functional RNA primer of unit-length (7–10 nt). These RNA primers are completely hydrolyzed by RNase into short fragments of 1–4 bp, but they are not hydrolyzed by DNase. The hallmark feature of the synthesome associated primase, which distinguishes it from an isolated DNA primase, is its specific ability to extend RNA primer to form a DNA-primer strand. The synthesis of a DNA-primer strand also requires DNA polymerase activity in addition to DNA primase activity. DNA polymerase activity is normally switched to start the elongation of an RNA primer after the synthesis of the primer. Only RNA primers ≥7 nucleotides long are utilized by the polymerase; regardless of the dNTP concentration [Kuchta et al., 1990]. Thus, the synthesis and extension of unit-length RNA primer not only requires primase but also involves the DNA polymerase α subunit p180 [Copeland and Wang, 1993], and the synthesome contains highly active DNA polymerases, which catalyze the incorporation of deoxyribonucleotides to extend the RNA primer. RNA primers are extended by the synthesome associated polymerases from a DNA-RNA oligoprimer of 20–40 nt in length (DNA primer). DNA-primer strands contain two components: (1) a ribonucleotide of 10 nt and, (2) a deoxyribonucleotide of 20–40 nt. RNase and DNase separately hydrolyze the appropriate components of DNA-primer strands resulting in shortening but incomplete hydrolysis of the DNA-primer strand.
Another feature of the synthesome is the rapid synthesis and distinct sizes of the RNA primer and DNA-primer strand. The synthesome catalyzes the synthesis of both an RNA primer and a DNA-primer strand rapidly, and steady-state levels appear to accumulate within (5–15 min). The sizes of the unit-length RNA primer (7–10 nt) and DNA-primer strand (20–40 nt) catalyzed by the DNA synthesome always remains constant, regardless of the synthesome concentration or reaction time. This study on the mechanism of DNA primase activity implies that the synthesis of RNA primers, catalyzed by the synthesome associated primase, occurs via a slow initiation, rapid polymerization and “intelligent” termination of the primer [Sheaff and Kuchta, 1993]. DNA primase binds the DNA template and slowly initiates the synthesis of a dinucleotide. After the synthesis of the dinucleotide, additional NTPs are rapidly added to the dinucleotide polymerized by DNA primase. DNA primase activity is characterized by synthesis of each RNA primer in a single “burst” of activity, rather than via distributive synthesis [Kuchta et al., 1990]. Once unit-length primers of 7–10 nt have been generated, the primase-template acts as a termination signal and inhibits further RNA synthesis. This inhibition is due to the generation of a stable primer-template, which likely remains associated with the pol α-primase complex. Interestingly, DNA-primer strands synthesized by the DNA synthesome in vitro are similar to the synthesome-catalyzed RNA primers synthesized in vitro. The synthesis of a DNA-primer strand reaches steady state levels rapidly and the size of the DNA-primer strand remains relatively constant (between 20 and 40 nt), regardless of the reaction time or the concentration of the synthesome used during the synthesis reaction. DNA-primer strand synthesis requires DNA polymerase α activity besides DNA primase [Frick and Richardson, 2001]. Our results suggest that the synthesome-associated polymerase α activity is also characterized by a rapid polymerization and “intelligent” termination in the synthesis of the DNA-primer strand. The synthesome, with the feature of rapid polymerization and intelligent DNA-primer strand termination, differs from synthesis medicated by the Klenow fragment of E. coli DNA polymerase I during the extension of the RNA primer. In contrast to the synthesome, RNA primer extension catalyzed by polymerase I depends strongly on the length of the reaction time. Increasing the reaction time significantly enhances the amount and size of the RNA primers extended.
Two potential mechanisms have been proposed to account for the switch from primer synthesis to primer extension during the DNA replication process [Copeland and Wang, 1993]. The first proposes that the primase-polymerase complex dissociates from the primer-template and re-associates with the polymerase active site. The second proposes that the switch occurs in an intrastrand-translocation of pol α-primase complex from the primase active site to the active site of polymerase α. This switch does not involve dissociation of the pol α-primase complex from the DNA template. Our experiments demonstrate that polymerase I-catalyzed RNA primer extension is inhibited by high concentrations of the DNA synthesome. The ability of the Klenow fragment to extend an RNA primer depends on the synthesis of RNA primers catalyzed by the synthesome, which also needs an available binding site at the RNA primer terminus. Therefore, the Klenow fragment of E. coli polymerase I requires the dissociation of primase-polymerase complex from the primer-template. However, high concentrations of the synthesome compete with polymerase I for the binding site on the primer-template so that the extension of the RNA primer is inhibited. The synthesome forms a dead-end complex at the RNA primer terminus; resulting in the inability of the Klenow fragment to participate in the RNA primer extension process. This result suggests that the polymerase-primase complex of the DNA synthesome must dissociate from the primer-template after the synthesis of the RNA primer. Our result shows that the synthesome increases the size of the unit-length RNA primer when the reaction temperature is reduced. Decreasing the reaction temperature increases the efficiency of switching the polymerase-primase complex and decreases primer-template denaturation [Kuchta et al., 1990]. In addition, the DNA primase p49 subunit contains the RNA polymerase catalytic activity, needed to increase the size of the unit-length RNA primer [Copeland and Wang, 1993]. Therefore, our results suggest that the DNA synthesome has the appropriate RNA polymerase activity needed to form RNA-primed DNA fragments.
The kinetic analysis of RNA primer initiation and elongation demonstrates that synthesome associated DNA primase is a slow and relatively weak enzyme with respect to template-binding [Swart and Griep, 1995]. DNA primase is also an error-prone nucleic acid polymerase because this enzyme has a very poor nucleotide discrimination ability; resulting in misincorporation of nucleotides [Sheaff and Kuchta, 1994]. Therefore, our analysis of the enzymatic kinetics of synthesome associated DNA primase activity could be used to determine the relation of synthetic rates and error frequencies during the synthesis of RNA primers. The discontinuous synthesis of DNA (Okazaki) fragments on the lagging strand of the replication fork is an important biochemical process during DNA replication. In order to ensure effective coordination between the continuous synthesis of DNA on the leading strand and the discontinuous synthesis of DNA on the lagging strand, DNA primase requires a precisely timed series of enzymatic steps that control the synthesis of the RNA primer. A kinetic study of a multiprotein replication complex from bacteriophage T7 shows that the primase acts as a molecular brake and transiently halts progression of the replication fork [Lee et al., 2006]. Because of the physical interaction of DNA polymerase δ with pol α [Coll et al., 1996] the result presented by Lee et al. [2006] suggests that DNA primase may be able to prevent leading strand synthesis from outpacing lagging strand synthesis during the slow enzymatic steps on the lagging strand.
Various eukaryotic DNA replication models have been used in the study of DNA primase function. The nuclear matrix replication system of whole cell lysate has been used to examine the synthesis and distribution of native RNA primer and RNA-primed nascent DNA in eukaryotic cell nuclei using endogenous nuclear matrix-bound double-stranded DNA template [Tseng and Goulian, 1977; Paff and Fernandes, 1990]. RNA primers are tightly associated with the nuclear matrix of rapidly proliferating mammalian cells [Kitani et al., 1985]. RNA primers are covalently attached to newly replicated DNA to form RNA-primed nascent DNA. RNA-primed DNA is degraded into some oligoribonucleotides of ~10 nt in length after DNase digestion [Tseng and Goulian, 1977], and at least 94% of native RNA primers and RNA-primed nascent DNA are located within the insoluble matrix fraction of the nucleus. RNA primers of 8–10 nt in length that are found associated with the nuclear matrix, make up <1% of the total RNA in the cell [Paff and Fernandes, 1990]; making it very difficult to examine RNA primer and RNA-primed nascent DNA in the cell nuclear matrix fraction due to a very low concentration of RNA primers and the relative large amount of other RNAs. Moreover, the isolation of RNA primers and RNA-primed nascent DNA from radio-nucleotide labeling whole cell lysates is a complicated and non-trivial purification procedure when applied to the analysis of nuclear matrix replication model-mediated RNA primer synthesis. In contrast, the DNA synthesome purified from the combined nuclear and cytoplasmic fractions contains highly active DNA primase and DNA polymerases, and fully supports the synthesis of native RNA primers in vitro using a double-stranded SV40 origin containing DNA template. The DNA synthesome-synthesized RNA primers are of normal length (10–20 nucleotides) and appear to function properly to support primer extension by DNA polymerase. These RNA primers can be readily extended to form RNA-primed nascent DNA of 100–200 nt. The discontinuous synthesis of DNA (Okazaki) fragments on the lagging strand of the replication fork is an important biochemical process involved in DNA replication, and full-length RNA-primed DNA (Okazaki) fragments synthesized by the DNA synthesome are also about 100–200 nucleotides in length [Ogawa and Okazaki, 1980]. We observed that the DNA synthesome could synthesize different RNA primed products ranging in size from 10 to 200 nt in length. This may be considered reasonable, as native RNA primers and RNA-primed nascent DNA are also of this approximate length. The above observation, that the synthesis of RNA primer and RNA-primed nascent DNA, catalyzed by the DNA synthesome, is essentially equivalent to that of the study published using the nuclear matrix replication model of whole cell lysate [Paff and Fernandes, 1990], suggests that the DNA synthesome is an excellent model system capable of carrying out a physiologically meaningful DNA replication process in vitro.
The DNA synthesome model is therefore a unique and valuable tool in the study of DNA primase function. Primase mediated synthesis and extension of RNA primer synthesis and elongation carried-out by the DNA synthesome closely resembles that carried-out by other cell-free replication models. The synthesome model supports the synthesis and extension of RNA primers in vitro using exogenous single-stranded DNA templates as well as supercoiled double-stranded DNA containing an intact SV40 replication origin. Therefore, the DNA synthesome is fully capable of mediating the synthesis and extension of RNA primers in vitro, and may facilitate the further investigation of the mechanisms initiating DNA synthesis in eukaryotic cells. Together our results suggest that the synthesome model provides a unique tool for studying the interaction of DNA primase and other replication proteins during the DNA replication process.
Acknowledgments
National Institute of Health; Grant numbers: CA57350, CA74904.
This work is supported in part by research award CA57350 from the National Institute of Health/National Cancer Institute (to L.H.M.) and research award CA 74904 from the National Institute of Health/National Cancer Institute (to R.J.H.). The authors wish to thank the Vera Bradley Foundation for their initial support of this research work. The authors also wish to thank Dr. Carita Lanner for her assistance in reviewing and discussing the content of the manuscript.
References
- Abdel-Aziz W, Jiang HY, Hickey RJ, Malkas LH. Ara-C affects formation of cancer cell DNA synthesome replication intermediates. Cancer Chemother Parmacol. 2000;45:312–319. doi: 10.1007/s002800050046. [DOI] [PubMed] [Google Scholar]
- Applegren N, Hickey RJ, Kleinschmidt AM, Zhou Q, Coll J, Wills P, Swaby R, Wei Y, Quan JY, Lee MY, Malkas L. Further characterization of the human cell multiprotein DNA replication complex. J Cell Biochem. 1995;59:91–107. doi: 10.1002/jcb.240590111. [DOI] [PubMed] [Google Scholar]
- Arezi B, Kuchta RD. Eukaryotic DNA primase. Trends Biochem Sci. 2000;25:572–576. doi: 10.1016/s0968-0004(00)01680-7. [DOI] [PubMed] [Google Scholar]
- Baker TA, Bell SP. Polymerases and the replisome: Machines within machines. Cell. 1998;93:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Cotterill S. The 50-kDa primase subunit of Drosophila melanogaster DNA polymerase alpha. Molecular characterization of the gene and functional analysis of the overexpressed protein. J Biol Chem. 1994;269:26759–26766. [PubMed] [Google Scholar]
- Bullock PA, Seo YS, Hurwitz J. Initiation of simian virus 40 DNA synthesis in vitro. Mol Cell Biol. 1991;11:2350–2361. doi: 10.1128/mcb.11.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll JM, Sekowski JW, Hickey RJ, Schnaper L, Yue W, Brodie AM, Uitto L, Syvaoja JE, Malkas LH. The human breast cell DNA synthesome: Its purification from tumor tissue and cell culture. Oncol Res. 1996;8:435–447. [PubMed] [Google Scholar]
- Collins KL, Russo AA, Tseng BY, Kelly TJ. The role of the 70 kDa subunit of human DNA polymerase alpha in DNA replication. EMBO. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Lehman IR. Synthesis by the DNA primase of Drosophila melanogaster of a primer with a unique chain length. Proc Natl Acad Sci USA. 1982;79:4585–4588. doi: 10.1073/pnas.79.15.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC, Wang TS. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem. 1993;268:26179–26189. [PubMed] [Google Scholar]
- Frick DN, Richardson CC. DNA primase. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- Frouin I, Montecucco A, Spadari S, Maga G. DNA replication: A complex matter. EMBO. 2003;4:666–670. doi: 10.1038/sj.embor.embor886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Hickey RJ, Abdel-Aziz W, Tom TD, Wills PW, Liu J, Malkas LH. Human cell DNA replication is mediated by a discrete multiprotein complex. J Cell Biochem. 2002;85:762–774. doi: 10.1002/jcb.10182. [DOI] [PubMed] [Google Scholar]
- Kitani T, Yoda K, Ogawa T, Okazaki T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J Mol Biol. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- Kuchta RD, Reid B, Chang LM. DNA primase. Processivity and the primase to polymerase alpha activity switch. J Biol Chem. 1990;265:16158–16165. [PubMed] [Google Scholar]
- Lee SH, Eki T, Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases alpha and delta. Proc Natl Acad Sci USA. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- Lin S, Hickey RJ, Malkas LH. The isolation of a DNA synthesome from human leukemia cells. Leuk Res. 1997;21:501–512. doi: 10.1016/s0145-2126(96)00103-8. [DOI] [PubMed] [Google Scholar]
- Malkas LH. DNA replication machinery of the mammalian cell. J Cell Biochem Suppl. 1998:30–31. 18–29. [PubMed] [Google Scholar]
- Malkas LH, Hickey RJ, Li CJ, Pedersen N, Baril EF. A 21S enzyme complex from HeLa cells functions in simian virus 40 DNA replication in vitro. Biochemistry. 1990;29:6362–6374. doi: 10.1021/bi00479a004. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Okazaki T. Discontinuous DNA replication. Annu Rev Biochem. 1980;49:421–457. doi: 10.1146/annurev.bi.49.070180.002225. [DOI] [PubMed] [Google Scholar]
- Ott RD, Rehfuess C, Podust VN, Clark JE, Fanning E. Role of the p68 subunit of human DNA polymerase alpha-primase in simian virus 40 DNA replication. Mol Cell Biol. 2002;22:5669–5678. doi: 10.1128/MCB.22.16.5669-5678.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paff MT, Fernandes DJ. Synthesis and distribution of primer RNA in nuclei of CCRF-CEM leukemia cells. Biochemistry. 1990;29:3442–3450. doi: 10.1021/bi00466a004. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Foiani M, Lucchini G, Plevani P. The isolated 48,000-dalton subunit of yeast DNA primase is sufficient for RNA primer synthesis. J Biol Chem. 1993;268:1343–1348. [PubMed] [Google Scholar]
- Schneider A, Smith RW, Kautz AR, Weisshart K, Grosse F, Nasheuer HP. Primase activity of human DNA polymerase alpha-primase. Divalent cations stabilize the enzyme activity of the p48 subunit. J Biol Chem. 1998;273:21608–21615. doi: 10.1074/jbc.273.34.21608. [DOI] [PubMed] [Google Scholar]
- Sheaff RJ, Kuchta RD. Mechanism of calf thymus DNA primase: Slow initiation, rapid polymerization and intelligent termination. Biochemistry. 1993;32:3027–3037. doi: 10.1021/bi00063a014. [DOI] [PubMed] [Google Scholar]
- Sheaff RJ, Kuchta RD. Misincorporation of nucleotides by calf thymus DNA primase and elongation of primers containing multiple noncognate nucleotides by DNA polymerase alpha. J Biol Chem. 1994;269:19225–19231. [PubMed] [Google Scholar]
- Stadlbauer F, Voitenleitner C, Bruckner A, Fanning E, Nasheuer HP. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase alpha-primase. Mol Cell Biol. 1996;16:94–104. doi: 10.1128/mcb.16.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman BW, Gluzmen Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol Cell Biol. 1985;5:2051–2060. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart JR, Griep MA. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry. 1995;34:16097–16106. doi: 10.1021/bi00049a025. [DOI] [PubMed] [Google Scholar]
- Tom TD, Malkas LH, Hickey RJ. Identification of multiprotein complexes containing DNA replication factors by native immunoblotting of HeLa cell protein preparations with T-antigen-dependent SV40 DNA replication activity. J Cell Biochem. 1996;63:259–267. doi: 10.1002/(sici)1097-4644(19961201)63:3<259::aid-jcb1>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Tseng BY, Goulian M. Initiator RNA of discontinuous DNA synthesis in human lymphocytes. Cell. 1977;12:483–489. doi: 10.1016/0092-8674(77)90124-6. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Wang TSF. Eukaryotic DNA polymerases. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- Weisshart K, Forster H, Kremmer E, Schlott B, Grosse F, Nasheuer HP. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J Biol Chem. 2000;275:17328–17337. doi: 10.1074/jbc.M000717200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Hickey R, Lawlor K, Wills P, Yu F, Ozer H, Starr R, Quan JY, Lee M, Malkas L. A 17S multiprotein form of murine cell DNA polymerase mediates polyomavirus DNA replication in vitro. J Cell Biochem. 1994;54:32–46. doi: 10.1002/jcb.240540105. [DOI] [PubMed] [Google Scholar]
- Zerbe LK, Kuchta RD. The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry. 2002;41:4891–4900. doi: 10.1021/bi016030b. [DOI] [PubMed] [Google Scholar]