Abstract
Conventional group analysis of functional MRI (fMRI) data usually involves spatial alignment of anatomy across participants by registering every brain image to an anatomical reference image. Due to the high degree of inter-subject anatomical variability, a low-resolution average anatomical model is typically used as the target template, and/or smoothing kernels are applied to the fMRI data to increase the overlap among subjects’ image data. However, such smoothing can make it difficult to resolve small regions such as subregions of auditory cortex when anatomical morphology varies among subjects. Here, we use data from an auditory fMRI study to show that using a high-dimensional registration technique (HAMMER) results in an enhanced functional signal-to-noise ratio (fSNR) for functional data analysis within auditory regions, with more localized activation patterns. The technique is validated against DARTEL, a high-dimensional diffeomorphic registration, as well as against commonly used low-dimensional normalization techniques such as the techniques provided with SPM2 (cosine basis functions) and SPM5 (unified segmentation) software packages. We also systematically examine how spatial resolution of the template image and spatial smoothing of the functional data affect the results. Only the high-dimensional technique (HAMMER) appears to be able to capitalize on the excellent anatomical resolution of a single-subject reference template, and, as expected, smoothing increased fSNR, but at the cost of spatial resolution. In general, results demonstrate significant improvement in fSNR using HAMMER compared to analysis after normalization using DARTEL, or conventional normalization such as cosine basis function and unified segmentation in SPM, with more precisely localized activation foci, at least for activation in the region of auditory cortex.
Keywords: functional group analysis, functional MRI, inter-subject variability, spatial normalization, HAMMER, DARTEL, SPM2, SPM5, template effect, spatial smoothing, human auditory cortex, Heschl’s gyrus
1. Introduction
Inter-subject variability in the spatial location of activation foci in functional neuroimaging studies can result from variability in anatomical structure, variability in functional organization, or both. To the extent that variability in functional localization reflects variability in anatomical structure, it may be decreased by improving anatomical registration across subjects. The conventional approach for functional localization is to find the correspondence among brain volumes of all participants within the study by registering every brain image to a given template brain. Such inter-subject registration in group studies is referred to as “spatial normalization”. Minimizing the contribution of variable anatomical structure to variability in the spatial location of activation foci has several advantages: First, it increases experimental power, so that small, focal functional activations can be more easily detected. Second, it reduces the need for smoothing in group studies, and improves spatial resolution, permitting activation foci to be localized to specific anatomical locations with greater precision.
Several brain normalization techniques have been proposed to register anatomy across subjects. Registration techniques vary from linear transformations of rigid-body registration that have few parameters and match size and shape [14, 9], to high degrees-of-freedom deformable registration methods that match residual details on the cortical surface and internal brain structures [24, 10, 13, 35]. An overview of different normalization techniques proposed for brain functional data analysis can be found in [18].
The accuracy of inter-subject registration using normalization methods has been assessed using both landmark-based [1, 42] and intensity-based [19] measures. Both types of studies confirm the effectiveness of spatial normalization for reducing inter-subject anatomical variability. In addition, the effect of inter-subject registration on the accuracy of functional group analysis has received some attention. Gee et al. [17] evaluated three different registration techniques (Bayesian volumetric warping proposed by him, SPM96 [5] and a 9-parameter affine registration) using t-statistics from a functional group analysis. Ardekani et al. [3] presented a quantitative comparison between three registration techniques (SPM’99, AFNI [9] and ART [2]) and examined the effect of registration method on the reproducibility of the fMRI activation maps. Both Gee and Ardekani concluded that increased accuracy in inter-subject registration results in a significant increase in the sensitivity of activation detection. Recently, Wu et al. [41] compared the performance of AIR [39], SPM95 [16], and their custom-developed demons-based registration in a region-of-interest (ROI)-based functional analysis. Similarly, they concluded that improving the normalization step in fMRI data analysis improves the reliability of the colocalized fMRI results, but at a cost of increased complexity of registration and computation time.
However, these published studies suffer from a number of limitations including: 1) the selected registration techniques are relatively low-dimensional and the impact of using a high-dimensional registration method in functional analysis has not been evaluated thoroughly; 2) the use of low-resolution anatomical templates and spatial filtering (smoothing) in current techniques may, in any case, compromise the effectiveness of using a high-dimensional inter-subject registration in group analysis; and 3) the cognitive tasks investigated in previous studies appear to activate large, distributed brain networks. To assess improvements in spatial resolution, it would be better to choose a task that is known to activate an anatomically circumscribed region, so that improvements in structural anatomical registration and in functional signal-to-noise ratio (fSNR) can be assessed concurrently. Here, we assess activity in auditory and speech regions of the temporal cortex in response to auditory and speech stimuli. The fSNR is defined as the ratio between the intensity of a signal associated with changes in brain function and the variability in the data due to all sources of noise. fSNR is conceptually very similar to t-statistics as calculated by SPM (Statistical Parametric Mapping: Wellcome Department of Cognitive Neurology, London, UK) software, which we shall use as an index of fSNR.
In this study, we evaluate and compare the effectiveness of several registration techniques. We compare a high-dimensional technique known as HAMMER (Hierarchical Attribute Matching Mechanism for Elastic Registration) [33] to DARTEL [4], a high-dimensional inverse-consistent diffeomorphic image registration method and also to commonly used low-dimensional normalizations, such as the normalization methods provided with SPM software (version 2 [6]: deformable modeling using discrete cosine transform basis functions, and version 5 [7]: unified segmentation). We evaluate: (a) the effects of the normalization technique; (b) the effects of the normalization template; and (c) the effects of conventional isotropic spatial smoothing of functional data, on fSNR. We assess the accuracy of the registration in reducing macroanatomical differences among subjects both qualitatively (i.e., visually inspecting the average of the registered volumes resulting from the application of each of the registration techniques) and quantitatively (i.e., comparing the average of the normalized cross-correlation (NCC) values calculated between the normalization template image and the warped image data for all registration methods). Cross-correlation is a simple but effective way to assess similarity between a registered volume and a reference template. This metric is intended to be used in images of the same modality where the relationship between the intensities of the two images is given by a linear equation. For applications in which the brightness of the image and the template can vary due to lighting and exposure conditions, normalized cross-correlation is used. Moreover, we compare peak activation values (t-statistics) resulting from statistical analysis on the group (treating the subject’s variable as a random effect) in both smoothed and unsmoothed fMRI data that has been normalized using different registration techniques.
We also evaluate the effect of the normalization template. Standard normalization techniques use a low-resolution average anatomical model as the target template. In this work, we have selected four well-known templates: (1) ICBM152 [27], a population-based template; (2) Colin27 [20], a high-resolution anatomical reference; (3) ICBM452 Tissue Probabilistic Atlas [21]; and finally, (4) A custom-built group template, generated using the DARTEL tool of the SPM5 package.
It is standard in conventional whole-brain fMRI analysis to apply isotropic three-dimensional filtering kernels of 6 – 10 mm typically to the functional data [28]. The spatial smoothing is done for many reasons one of which is to reduce the effect of inter-subject variability in group analysis. Although often helpful and necessary, smoothing has the undesirable effect of reducing the spatial resolution, blurring and/or shifting activations and merging adjacent peaks of activation. In this work, we examine whether using a high-dimensional normalization will reduce or eliminate the need for spatial smoothing to increase anatomical overlap among subjects, while maintaining a similar fSNR to that obtained with spatial smoothing.
This paper is further evidence that high-dimensional non-rigid registration methods are needed for group analysis of functional activation in the auditory cortex, as previously demonstrated by Desai et al. [11], and Kang et al. [22], which dealt with flattened 2D cortical surfaces and furthermore, by Viceic et al. [36], which dealt with 3D volumes. Moreover, this paper explores the effects of the normalization template and spatial smoothing on subsequent group analysis of functional data from an auditory imaging experiment. This experiment examined whether functional networks supporting production of speech and perception of speech overlap (Zheng et al. submitted). We wished specifically to determine whether a subset of regions in the superior temporal region are particularly sensitive to a mismatch between the actual auditory consequences of speaking and the predicted consequences based on the motor speech command. A more complete description of the experiment can be found in Zheng et al., submitted. Only aspects of the experimental design relevant to the methodological question are described here.
2. Materials and Methods
2.1. Image Acquisition
Seventeen normal healthy volunteer subjects (13 female, 4 male, ages 23 ± 3 years (mean±std), right-handed, native English speakers) participated in this study. All subjects gave written informed consent for their participation. The experimental protocol was cleared by the Queen’s University Health Sciences Research Ethics Board.
MR imaging was performed on the 3.0 Tesla Siemens Trio MRI system in the Queen’s University Centre for Neuroscience Studies, MRI Facility, Kingston, Ontario. -weighted GE-EPI sequences were acquired with a typical field of view of 211 × 211 mm2, in plane resolution of 3.3 × 3.3 mm2, slice thickness of 4.0 mm, TA = 1600 msec per acquired volume, TE = 30 msec, and TR = 3000 msec. In order to present sounds and record the verbal responses without any acoustic interference, a visual cue instructing the subject to listen or speak was presented at the beginning of the 1400 msec silent period between successive scans: trials were always complete by the end of this period. In addition to the functional data, a whole-brain 3D MPRAGE T1-weighted anatomical image was acquired for each participant (voxel resolution of 1.0 ×1.0 ×1.0 mm3, flip angle α = 9°, TR = 1760 msec, and TE = 2.6 msec).
2.2. fMRI Experimental Paradigm
The experiment was designed as a 2 × 2 factorial:
Whispering “TED”, with concomitant clear auditory feedback,
Whispering “TED”, while hearing masking Gaussian white noise,
Listening to the stimuli of the first condition without speaking,
Listening to the stimuli of the second condition without speaking,
A resting baseline condition was also scanned.
On each trial, the subject was told to produce “TED” if a green cross was presented at the start of the trial, or to remain silent (listen/rest trials) if a red cross was presented.
In every set of five trials, each of the five conditions was presented once in pseudo-random order. Thirty-six such sets of trials were presented in each of three nine-minute runs. The auditory stimuli in listening trials (conditions 3 and 4) were yoked to those in the corresponding production trials of conditions 1 and 2 (i.e., listening trials consisted of utterances generated on previous production trials, or their white-noise masks).
2.3. Stimulus Generation Hardware
Two PC workstations (one for stimulus delivery and one for realtime speech signal processing) communicated via a fast Ethernet switch. Auditory stimuli were delivered via high-fidelity magnet compatible headphones (Nordic Neuro Lab, Norway). Verbal responses were recorded via a dual-channel microphone system. Participants spoke into the optical microphone and their voices were digitized, transmitted, and saved on the second workstation. Real-time analysis of the signal was achieved using an embedded controller (National Instruments Co., Texas) and LabView software (National Instruments, Inc.). Processed signals were converted back to analogue using a multifunction I/O data acquisition board (National Instruments Co., Texas) and played back over the headphone in realtime, either intact or as a modulated noise burst. The processing delays were negligible (imperceptible to the listeners).
Here, we concentrate on two contrasts; (1) Four sound conditions vs. rest, which should reveal activity in auditory regions; and (2) listening to speech (condition 3) compared to rest (condition 5), which should reveal auditory activity as well as activity in speech-sensitive regions of the superior temporal gyrus and sulcus.
2.4. Data Preprocessing
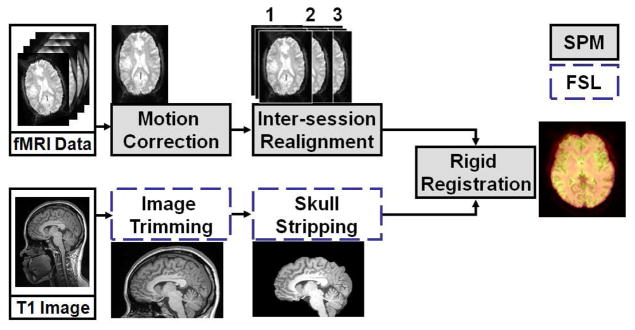
Structural and functional image data were passed through a series of preprocessing steps before normalization; see Figure 1. Data were motion-corrected with respect to the first volume of the first session using the realignment tool of SPM (i.e., using a least squares approach and a 6 parameter rigid-body spatial transformation with 4th degree B-Spline interpolation). Structural MR data were edited to remove skull and scalp using the Brain Extraction Tool (BET) of the FSL software package (Oxford Centre for Functional MRI, Oxford University, UK). Next, the structural images were rigidly registered to the functional time series using the Mutual Information coregistration tool of SPM5.
Figure 1.
Preprocessing: The SPM package was used for fMRI data realignment (motion correction), and structural-to-functional coregistration. Skull-stripping and volume trimming were performed using FSL’s BET.
2.5. Normalization Template
In this study, we consider four common templates for the normalization: (1) ICBM152, which was generated by registering 152 normal brain images to the MNI305 template [12] using a nine-parameter affine transformation. This template has been incorporated into several software packages such as SPM. ICBM152 is an average volume and therefore lacks anatomical details. The original ICBM152 template provided in SPM package has the resolution of 2.0×2.0×2.0 mm3; however, we resampled this template to 1.0×1.0×1.0 mm3 for comparison with the other templates. (2) Colin27 or CJH27, in which structural details are preserved. Colin27 was created by registering 27 high-resolution scans (1.0 × 1.0 × 1.0 mm3) of a single subject to the more spatially blurred ICBM152 average brain. (3) ICBM Tissue Probabilistic Atlas, which is used in SPM5 segmentation-based registration. 452 subject T1-weighted MR images were aligned with the MNI305 template, and classified into gray matter, white matter, and cerebrospinal fluid. The 452 tissue maps were separated into their separate components and each component was averaged across the subjects to create a probability map for each tissue type. These maps give the prior probability of any voxel in a registered image being of any of the tissue classes - irrespective of its intensity. (4) Finally, a custom-built group template, generated using DARTEL tool of SPM5 package. Structural T1-weighted scans of 17 subjects within the study were segmented into different tissue types using segmentation tool of SPM5. Intensity averages of the grey and white matter images were generated to serve as an initial template for DARTEL registration. The initial smooth template is then iteratively sharpened after each phase of registration.
2.6. Spatial Smoothing
Normalized fMRI data may also undergo spatial smoothing. Spatial smoothing is an averaging process in which the intensity at a given voxel is replaced by a weighted average of the values of voxels in the spatial neighborhood of that voxel. Smoothing is implemented as a convolution of the imaging data with a Gaussian kernel described by a parameter of full width at half maximum (FWHM). In fMRI studies, spatial smoothing is performed to render the data more normally distributed and therefore, appropriate for parametric tests. Also, smoothing reduces the effect of residual inter-subject anatomical variability remaining after spatial normalization. Besides suppressing the influence of anatomical variability across individual subjects, spatial smoothing of fMRI data prior to the statistical analysis can enhance the signal-to-noise ratio and increase sensitivity to signals of specific shapes and sizes depending on filter design [26, 32]. Smoothing also results in fewer statistically independent tests within a given volume in functional group studies, and thus a smaller correction for multiple comparisons is required. Moreover, smoothing is useful in reducing resampling-related artifacts after image registration [25]. Finally, determination of thresholds for statistical inference in SPMs (if the family-wise-error correction is used) depends on the theory of Gaussian random fields and the assumption that the image data are good lattice representations of a smooth Gaussian field [15, 40]. This only holds when the voxel size is appreciably smaller than the smoothness, and it has been suggested that smoothing be applied such that the effective FWHM is at least twice the size of the voxel [40].
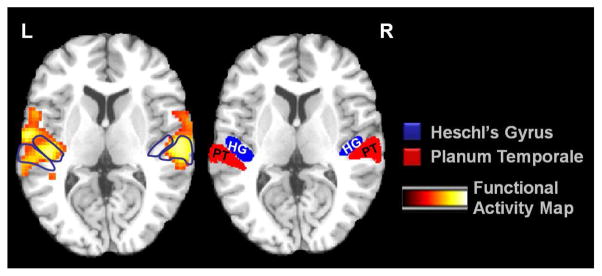
Spatial smoothing of fMRI data has some drawbacks as well. Undesirable effects of smoothing include decreased effective spatial resolution, blurring and/or shifting of activations and merging of adjacent peaks of activation. Figure 2 demonstrates an example of the undesirable effect of spatial smoothing in merging functional activation foci from two anatomically adjacent regions of Heschl’s gyrus (HG) and planum temporale (PT) in human auditory cortex. The labeled regions in blue and red are extracted from HG [30] and PT [38] probabilistic maps, respectively, which are thresholded at 40%. The most recent work of Weibull et al. [37] investigates how the choice of spatial resolution and smoothing kernel width affects the temporal signal-to-noise ratio in single-subject fMRI data analysis. Their conclusion is that in studies requiring detailed localization and quantification of small activation clusters, high resolution and limited smoothing kernel is most appropriate. However, when expecting large volumes of activation or when the precise localization of a activation focus is less important than simply observing it, lower resolution and smoothing may be used more generously. Mikl et al. [28], and Reimold et al. [31], in separate works, also investigate the related artifacts of spatial smoothing on t-maps and conclude that spatial smoothing may lead to critical, sometimes counterintuitive artifacts in t-maps, especially in subcortical brain regions. In this work, we evaluate the effect of such smoothing on t-statistics and functional signal-to-noise.
Figure 2.
Overlapping functional activation map shown for the anatomically neighboring regions of Heschl’s gyrus (HG) and planum temporale (PT). Right: The labeled regions in blue and red are extracted from HG and PT probabilistic maps, respectively, which are thresholded at 40%. Left: The colormap depicts the activation map resulting from statistical group analysis of an auditory functional task using spatially smoothed fMRI data.
2.7. Inter-subject Registration
Four different registration techniques were considered for normalization of structural images and the corresponding functional image data. All 17 subjects’ structural data were aligned using these four registration techniques:
HAMMER is an elastic registration technique that utilizes an attribute vector for every voxel of the image. The attribute vector reflects the geometric features of the underlying anatomy at different scales. Our application of the HAMMER algorithm proceeded in two steps: First, the brain data were segmented into gray matter, white matter and cerebrospinal fluid using FMRIB’s Automated Segmentation Tool (FAST) of the FSL software package. Second, HAMMER registration is applied to warp the brain images to the 1 mm3 Colin27 template. HAMMER uses every voxel’s information in its hierarchical multi-resolution approach, and therefore, the number of parameters for the deformation field is equal to the number of voxels within the volume. Consequently, HAMMER registration is not appropriate for use with an anatomically smooth template like ICBM152 as the anatomical details that are used as the reference to guide the registration, does not exist in the smooth template. HAMMER’s voxel-based approach for registration preserves the local amount of signal concentration unlike modulated voxel-based morphology analyses in which the normalization is expected to compensate for structural differences while preserving the total amount of signal. A copy of the HAMMER-based normalized fMRI dataset was smoothed with an isotropic Gaussian kernel of FWHM 8 mm.
Normalization in SPM2 [6] includes global linear (affine transform) and local nonlinear (3D discrete cosine transform basis functions) transformations. The deformation field for each subject is described by 1176 parameters. We consider two templates for the normalization of the structural image from each fMRI subject: (i) the default ICBM152 template with 2 mm3 spatial resolution, which was resampled to 1 mm3 and (ii) the high-resolution single-subject 1 mm3 Colin27 template. In both cases, a copy of the registered fMRI data was smoothed using an isotropic Gaussian kernel (FWHM 8 mm) to compensate for the inter-subject variability remaining after the normalization procedure, created four sets of normalized data. Condition (i) with smoothing is a standard implementation of SPM2 image processing procedures. One should note that SPM-based normalization includes spatial smoothing of the images prior to registration. This smoothing is due to the fact that the templates supplied with SPM have been smoothed by 8 mm, and that smoothness combines by Pythagoras’ rule (refer to SPM manual [34]).
SPM5’s normalization [7], referred to as “unified segmentation”, includes a probabilistic framework, which integrates image registration, tissue classification, and bias correction within the same generative model. The number of deformation parameters is in the order of 103; however, the exact number depends on the image field of view (FOV) (John Ashburner, communication on SPM forum, March 24, 2009). The unified segmentation technique requires tissue probability maps as the priors and for the current evaluation, ICBM452 tissue probability maps were used. Because we could not define tissue probability maps for the single-subject template, we could not use the Colin27 template as the target for normalization with SPM5. A copy of the registered fMRI data was smoothed using an isotropic Gaussian kernel with FWHM 8 mm.
DARTEL (Diffeomorphic Anatomical Registration Through Exponential Lie Algebra) has been proposed by Ashburner [4] as an alternative method of normalization in the SPM package. DARTEL is an algorithm for diffeomorphic image registration, which utilizes large deformations in an inverse consistent framework. DARTEL’s deformations are parameterized by a time-invariant velocity field. Similar to the unified segmentation method, DARTEL also requires tissue classification of the brain images. Intensity averages of the grey and white matter images were generated to serve as an initial template for DARTEL registration. The template is iteratively updated after each step of the registration. DARTEL encodes the spatial transforms using roughly around 6 × 103 parameters per subject (John Ashburner, communication on SPM forum, March 24, 2009). DARTEL was primarily designed for voxel-based morphometry (VBM) studies, which is well-suited to its diffeomorphic nature. Similar to other conditions, a copy of the registered fMRI data was smoothed using an isotropic Gaussian kernel with FWHM 8 mm.
Overall, we will be evaluating 10 different conditions, which are presented in Table 1.
Table 1.
10 different conditions considered for evaluating the effects of the normalization method, the template resolution and the spatial smoothing on the functional group analysis.
| Condition | Normalization | Template | Smoothing |
|---|---|---|---|
| HMR w. s. | HAMMER | Colin27 | ✓ |
| HMR w/o s. | HAMMER | Colin27 | × |
| SPM2c w. s. | cosine basis functions | Colin27 | ✓ |
| SPM2c w/o s. | cosine basis functions | Colin27 | × |
| SPM2i w. s. | cosine basis functions | ICBM152 | ✓ |
| SPM2i w/o s. | cosine basis functions | ICBM152 | × |
| SPM5 w. s. | unified segmentation | ICBM452 tissue maps | ✓ |
| SPM5 w/o s. | unified segmentation | ICBM452 tissue maps | × |
| DARTEL w. s. | diffeomorphic | custom-built | ✓ |
| DARTEL w/o s. | diffeomorphic | custom-built | × |
2.8. Assessment of Registration Accuracy
The accuracy of the inter-subject registration techniques and their impact on the localization of activation patterns, the influence of the normalization template, and the effect of smoothing in functional group analysis were assessed for all four selected registration techniques. Average (n = 17) brain images were generated from the warped structural data for all four registration conditions. Normalized cross-correlation was used as the measure of similarity between the template volume and the warped volume data. Moreover, statistical analysis of fMRI was conducted using Analysis of Variance (ANOVA).
2.8.1. Average Volume
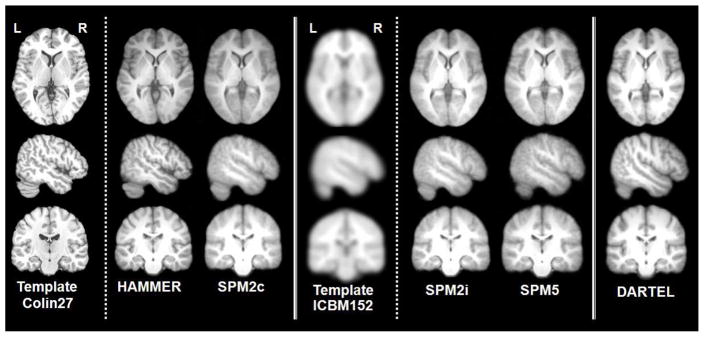
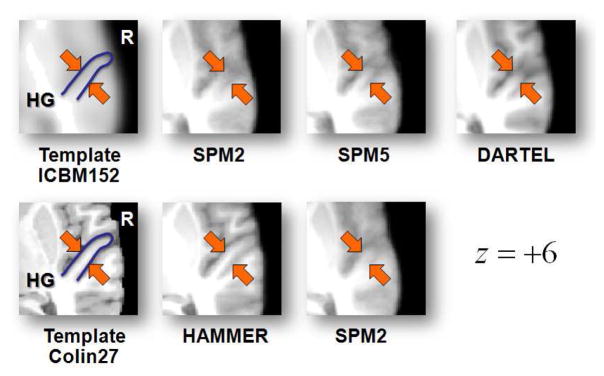
Mean volumes were generated by averaging all 17 registered T1-weighted structural volumes for four different registration conditions. Figure 3 shows the cross-sections derived from the mean volumes obtained with each registration condition. Figure 4 provides a closer look at a region of interest around Heschl’s gyrus, the approximate location of primary auditory cortex, for comparison. It can be observed that HAMMER improves the delineation of sulci and gyri and consequently, the spatial homogeneity between individual subject brains and the reference template (Colin27). This was confirmed using normalized cross-correlation measure as shown in Section 3.
Figure 3.
From top to bottom: axial (z = +4), sagittal (x = +50), and coronal (y = −16) sections of (from left to right) Colin27 template, inter-subject average volumes computed for HAMMER-based registration, and SPM2-based normalization using Colin27 as template (SPM2c), SPM2-based normalization using ICBM152 template (SPM2i), unified segmentation-based normalization using ICBM452 tissue probability maps, and DARTEL normalization using the custom-built template. Coordinates are in ICBM152 frame.
Figure 4.
Zoomed window over Heschl’s gyrus region on average brain volumes generated using five different registration conditions; HAMMER, SPM2 using two templates: ICBM152 (upper row) and Colin27 (lower row), SPM5 unified segmentation, and DARTEL.
2.8.2. Similarity Measure: Normalized Cross-Correlation
NCC is computed by first normalizing each image to have zero mean and unit variance, and then multiplying each voxel of one volume by the corresponding voxel in the other volume, and summing the products (Eq. 1).
| (1) |
where S and T refer to the intensity values in the subject and template volumes, respectively.
NCC values were computed for the entire brain volume of every subject considering different registration techniques. In addition to the entire volume, we created rectangular cuboidal regions of interest around auditory cortex, extending into the superior temporal sulcus, in both hemispheres. These ROIs were defined using the following coordinate ranges: (Left: x = −66: −20, y = −50: +15, z = −15: +20, Right: x = +20: +66, y = −50: +15, z = −15: +20 with respect to the ICBM152 coordinate frame).
2.8.3. Statistical analysis of fMRI
Statistical analysis of fMRI data was accomplished with SPM5. Deformation parameters resulting from registration of each structural image to the desired template were used to warp the corresponding fMRI data. The warped fMRI data were resampled to 3.0×3.0×3.0 mm3 after normalization for all methods of registration prior to conducting single-subject and second-level group analysis. Next, fMRI data were entered into a fixed-effects general linear model for each subject using an event-related analysis procedure. The hemodynamic response function was selected as the basis function. Contrast images were created for each subject and these were entered into a second-level group analysis, treating the subject’s factor as a random effect [15]. One-sample t-tests were calculated for contrasts of ‘listening to speech vs. rest’ and ‘four sound conditions vs. rest’ for each of the 10 different processing methods mentioned in Section 2.7, enabling us to examine the effects of three factors:
Registration method: HAMMER, nonlinear warping using cosine basis functions (SPM2), unified segmentation (SPM5), diffeomorphic registration (DARTEL).
Template: Colin27, ICBM152.
Spatial smoothing: with or without 8 mm isotropic Gaussian spatial smoothing.
2.8.4. Euclidean Distance Error
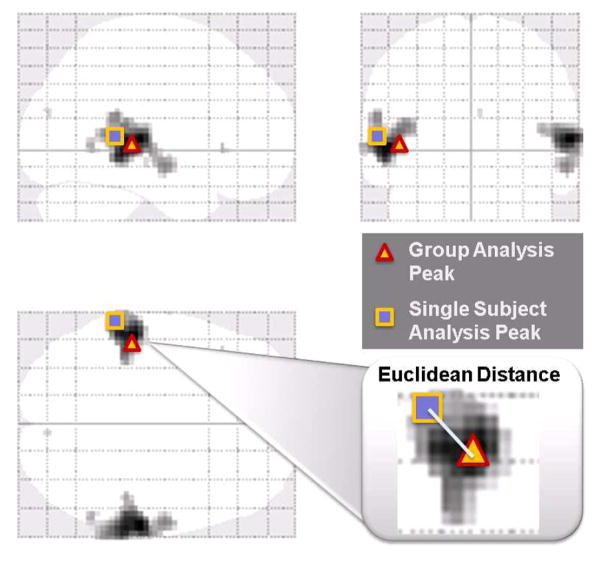
The Euclidean distances between the highest activation peak obtained from the group analysis and the ‘listening vs. rest’ contrast and the closest activation peak in each individual from single-subject analysis of the same contrast were calculated for all 10 different processing methods [29, 23, 8] as depicted in Figure 5. A smaller discrepancy between the group and individual peaks indicates better inter-subject alignment among all subjects after registration.
Figure 5.
Euclidean distances between the highest activation peak obtained from the group analysis and the ‘listening vs. rest’ contrast and the closest activation peak in each individual from single-subject analysis for the same contrast were calculated for all 10 different processing conditions.
2.8.5. Analysis of Variance: Normalization vs. Smoothing
A 3×2 repeated-measures ANOVA was conducted on the Euclidean distance error data. The factors were (1) registration method (four levels: HAMMER, cosine basis functions (SPM2), unified segmentation (SPM5), DARTEL), and (2) smoothing (two levels: with and without smoothing).
3. Results and Discussion
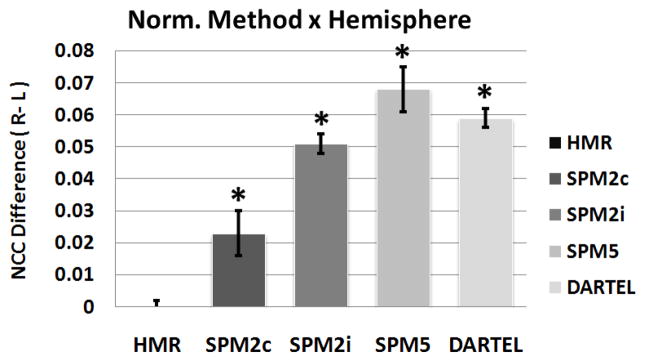
NCC scores were computed for the entire brain volume as well as the specified ROIs as defined in Section 2.8.2 for each of 17 subjects considering five different registration conditions. NCC results are shown in the form of mean±std in Table 2. One-way ANOVA analysis on whole-brain NCC scores (five levels: HAMMER, SPM2c, SPM2i, SPM5, and DARTEL), p < 0.05, was performed using SPSS software (Statistical Package for the Social Sciences). Results showed a significant main effect of the normalization method; HAMMER and DAR-TEL slightly outperformed SPM2 and unified segmentation-based normalizations (i.e., less than 3% improvement). There was no significant difference in performance between SPM5-based normalization and SPM2-based normalization using the Colin27 template (SPM2c). In fact, SPM2c performed worse than other methods. Pairwise comparisons using Sidak-correction between SPM2c and SPM2i revealed no significant difference in performance. Therefore, it can be concluded that using a high-resolution template such as Colin27 does not improve registration accuracy for SPM2-based registration. A second ANOVA was performed on NCC scores for the left and right ROIs with two factors: normalization method (five levels) and hemisphere (Left/Right), p < 0.05. There was a significant main effect of hemisphere (i.e., a greater correlation (higher NCC score) for the right hemisphere compared to left hemisphere). There was also a significant main effect of the normalization method. Pairwise comparisons using Sidak-correction showed that HAMMER significantly outperformed other techniques (i.e., over 11% improvement) and SPM2 with the Colin27 template yielded the lowest NCC scores among all methods. DARTEL slightly outperformed the other SPM-based normalization methods; however, unlike the full volume case, DARTEL did not match up HAMMER’s performance at ROI level. Such difference in performance is due to the diffeomorphic nature of DARTEL registration, in which the method is given enough freedom to estimate quite large deformations. Such freedom of deformation may result in unrealistic shrinkage/expansion of some structures in the brain image volume. Consequently, diffeomorphic normalization can not capture small deformations required for matching areas with small residual anatomical details such as the selected ROI in this study. There was no significant difference in performance between SPM2 using the ICBM152 template and SPM5 using the probabilistic tissue maps. Comparing SPM2c and SPM2i results, it was reconfirmed that SPM2-based normalization cannot take advantage of the high-resolution template. There was also a significant interaction between the two factors; NCC scores were higher for the right hemisphere than the left hemisphere for SPM2c, SPM2i, SPM5, and DARTEL but not for HAMMER, which yielded no difference between the two hemispheres (Figure 6).
Table 2.
Comparing mean and std. (%) of normalized cross-correlation values (17 subjects) among different registration techniques; HAMMER, cosine basis function of SPM2 using two templates: SPM2c (Colin27), SPM2i (ICBM152), unified segmentation (SPM5), and diffeomorphic registration (DARTEL) considering the entire brain and a region-of-interest around the primary auditory cortex.
| Volume | Registration Method |
||||
|---|---|---|---|---|---|
| HMR mean ± std | SPM2c mean ± std | SPM2i mean ± std | SPM5 mean ± std | DARTEL mean ± std | |
| Entire | 98.2 ± 0.0 | 94.1 ± 0.1 | 95.7 ± 0.1 | 94.1 ± 0.2 | 97.6 ± 0.4 |
| Left ROI | 95.3 ± 0.1 | 68.8 ± 0.4 | 77.8 ± 0.3 | 77.8 ± 0.7 | 81.8 ± 1.0 |
| Right ROI | 95.3 ± 0.2 | 71.1 ± 0.4 | 82.9 ± 0.4 | 84.6 ± 0.4 | 87.7 ± 0.7 |
Figure 6.
Significance test of NCC score differences between left and right hemispheres (R - L) among five types of normalization. (*) indicates a significant difference in NCC score between left and right hemispheres.
Figure 7 illustrates the region of activation produced in the contrast of ‘four sound conditions vs. rest’, across five registration conditions. Activation resulting from the HAMMER-based normalization is more intense (lighter color) and more tightly localized over auditory cortex, compared to other techniques. The increase in t-values comparing smoothed vs. unsmoothed data for HAMMER-based, SPM2c-based (using Colin27), SPM2i-based (using ICBM152), unified segmentation-based, and finally, DARTEL-based normalized data are 1.5%, 22.6%, 13.5%, 85.7% and 20.3%, respectively. This implies that smoothing unnecessarily expands the region of activation for the condition using HAMMER since there is no significant increase in t-values for smoothed vs. unsmoothed HAMMER-normalized data; however, for analyses conducted on SPM2, DAR-TEL, and specifically unified segmentation normalized data, smoothing substantially improved fSNR.
Figure 7.
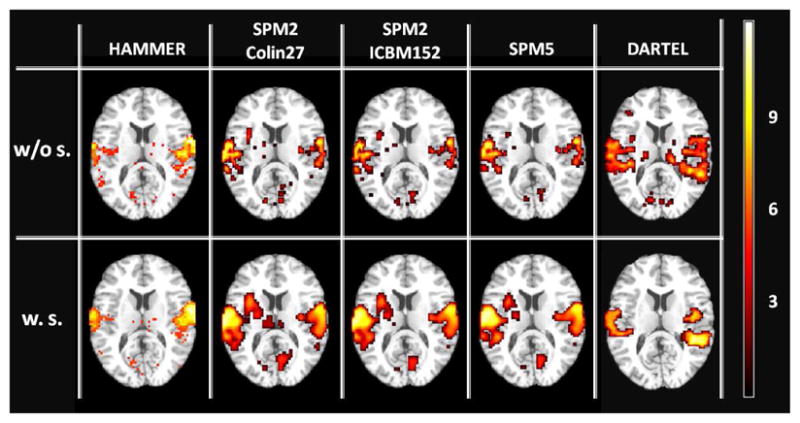
Comparing activation maps (axial view) corresponding to an auditory-related fMRI task for conditions given in Table 1. The colormap depicts the activation maps resulting from group analysis of 17 subjects for the contrast of ‘four sound conditions vs. rest’.
The statistical analysis of fMRI data was performed in MATLAB® using SPM functions. Analysis of the ‘four sound conditions vs. rest’ contrast revealed significant activation in the superior temporal region bilaterally using False Discovery Rate (FDR) correction for multiple comparisons, p < 0.05. Highest activation peaks (i.e., t-value + 3D coordinates in MNI space) for two contrasts of ‘four sound conditions vs. rest’ and ‘listening vs. rest’ observed in each hemisphere in the group analyses (in which the subject was treated as a random effect) are listed in Table 3.
Table 3.
Coordinates of peak activation (in MNI space) for 10 different processed fMRI datasets and for two different contrasts; (1) listening vs. rest, and (2) four sound conditions vs. rest.
| Condition | Registration | Template | Listening vs. Rest | Four Sound Conditions vs. Rest | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Left | Right | Left | Right | |||||||
| t value | (x, y, z) mm | t value | (x, y, z) mm | t value | (x, y, z) mm | t value | (x, y, z) mm | |||
| 1 | HMR. w/o s. | Colin27 | 11.57 | −63, −24, 9 | 11.28 | 57, −9, −6 | 10.89 | −45, −33, 18 | 10.02 | 57, −9, 12 |
| 2 | HMR. w. s. | Colin27 | 12.57 | −66, −36, 3 | 10.81 | 57, −12, −6 | 11.05 | −57, −6, 27 | 10.96 | 57, −12, 15 |
| 3 | SPM2c w/o s. | Colin27 | 7.33 | −51, −33, 6 | 6.66 | 63, −24, 3 | 8.67 | −48, −12, 27 | 8.77 | 54, −9, 21 |
| 4 | SPM2c w. s. | Colin27 | 8.24 | −51, −36, 9 | 7.59 | 66, −24, 3 | 10.63 | −45, −12, 24 | 9.55 | 54, −6, 18 |
| 5 | SPM2i w/o s. | ICBM152 | 5.44 | −51, −33, 6 | 6.09 | 63, −33, 6 | 6.63 | −42, −33, 9 | 3.94 | 54, −27, 0 |
| 6 | SPM2i w. s. | ICBM152 | 8.83 | −51, −36, 6 | 7.64 | 66, −24, 3 | 10.81 | −60, −30, 9 | 7.53 | 63, −33, 12 |
| 7 | SPM5 w/o s. | ICBM152 | 5.28 | −51, −33, 3 | 4.60 | 63, −27, 0 | 4.06 | −51, −24, 0 | 5.10 | 60, −27, 0 |
| 8 | SPM5 w. s. | ICBM152 | 6.28 | −54, −33, 3 | 6.09 | 60, −36, 3 | 7.54 | −48, −36, 9 | 6.61 | 60, −33, −3 |
| 9 | DARTEL w/o s. | custom-built | 6.79 | −51, −39, 6 | 6.75 | 66, −21, 3 | 8.65 | −54, −42, 12 | 8.85 | 54, −3, 15 |
| 10 | DARTEL w. s. | custom-built | 7.40 | −54, −30, 0 | 5.79 | 63, −33, 6 | 10.41 | −45, −39, 12 | 8.75 | 66, −21, 3 |
The following can be observed from the data presented in Table 3: (1) Group analysis of HAMMER-normalized data, with or without smoothing, yields higher t-values in both hemispheres compared to normalized data using other techniques. Considering NCC comparison results (i.e., higher NCC scores for higher-d registration), one can conclude that increased fSNR is due to increased overlap across subjects; (2) Analysis conducted using SPM2 and Colin27 as the template, without smoothing, yields higher t-values compared to the ICBM152 template without smoothing; however, the opposite is true if data are smoothed; (3) Smoothed fMRI data yields higher t-values compared to unsmoothed data (except for one case; HAMMER, listening vs. rest, Right Hemisphere); however, the smoothing-related increase in t-values is more substantial for SPM2-, SPM5-, and DARTEL-based normalizations. One should note that one application of spatial smoothing kernels in functional group analysis, as mentioned in Section 2.6, is to render the data more normally distributed and therefore, suitable for parametric tests. To check the validity of the normality condition for the non-smoothed normalized data, we checked the effective smoothing of our data using SPM software tools - even without any smoothing applied, our data has an average effective smoothness of 6 mm in all directions, which is approximately twice the voxel size (3 mm3). We judge this to be sufficient to render the data appropriate for parametric tests.
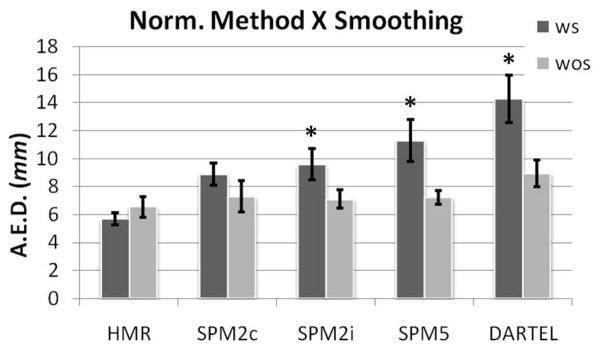
Average Euclidean Distances (A.E.D.) between the highest activation peak obtained from group analysis and the ‘listening vs. rest’ contrast and the closest activation peak observed in individual analyses for the same contrast are shown in Table 4. ANOVA on Euclidean distances with the two factors: normalization method (four levels) and smoothing (two levels) revealed a significant main effect of smoothing (F(1, 16) = 17.52, p < 0.05). Smoothing yielded significantly higher distance values than no smoothing. We also observed a significant main effect of normalization method (F(4, 64) = 7.16, p < 0.05), which we followed up using Sidak-corrected pairwise comparisons (see Figure 8); HAMMER registration yielded significantly smaller distances between group location estimates and peaks in individuals compared to SPM2i, SPM2c, unified segmentation, and DARTEL. There was no significant difference between SPM2c, SPM2i and SPM5; however, DARTEL yielded significantly higher distances compared to the rest. Further, there was no significant difference between smoothed and un-smoothed data when normalized using HAMMER or SPM2c registration. On the other hand, smoothing yielded higher distance values when the normalization method is either SPM2i, SPM5’s unified segmentation, or DARTEL. One may conclude that using a high-resolution template for normalization (such as in HAMMER and SPM2c-based normalization) results in more accurate alignment of the functional activation foci and therefore suppresses the impact of spatial smoothing which is applied for increasing overlap among subject data; however, validation of such conclusion requires further investigation by adding an extra factor for template type within the statistical analysis. Finally, there was no significant interaction between normalization method and smoothing.
Table 4.
Average Euclidean Distances (A.E.D.) between the highest activation peak obtained from the group analysis and the closest activation peak in each individual from the ‘listening vs. rest’ contrast.
| Condition | Highest Second-Level Peak Coord. mm | Ave. Closest First-Level Peak Coord. (mean ± std) |
A.E.D. (mean ± std) mm | ||
|---|---|---|---|---|---|
| x mm | y mm | z mm | |||
| HAMMER w/o s. | (−63, −24, 9) | −63 ± 4 | −25 ± 5 | 7 ± 3 | 6.56 ± 0.74 |
| HAMMER w. s. | (−66, −36, 3) | −67 ± 2 | −34 ± 3 | 5 ± 4 | 5.70 ± 0.43 |
| SPM2c w/o s. | (−60, −39, 15) | −60 ± 4 | −38 ± 4 | 12 ± 5 | 7.09 ± 0.65 |
| SPM2c w. s. | (−54, −36, 9) | −55 ± 5 | −34 ± 5 | 8 ± 5 | 9.60 ± 1.11 |
| SPM2i w/o s. | (−51, −33, 6) | −51 ± 4 | −32 ± 5 | 6 ± 3 | 6.49 ± 3.05 |
| SPM2i w. s. | (−51, −36, 6) | −53 ± 5 | −35 ± 6 | 7 ± 6 | 8.75 ± 4.81 |
| SPM5 w/o s. | (−51, −33, 3) | −53 ± 3 | −34 ± 5 | 4 ± 5 | 7.21 ± 0.51 |
| SPM5 w. s. | (−54, −33, 3) | −53 ± 8 | −31 ± 7 | 5 ± 8 | 11.29 ± 1.50 |
| DARTEL w/o s. | (−51, −39, 6) | −59 ± 4 | −29 ± 6 | 5 ± 3 | 8.92 ± 3.94 |
| DARTEL w. s. | (−54, −30, 0) | −56 ± 11 | −33 ± 9 | 5 ± 4 | 14.26 ± 6.89 |
Figure 8.
Average Euclidean distance MANOVA results from the ‘listening vs. rest’ contrast for five normalization methods with and without smoothing. (*) indicates the significant difference between smoothed and unsmoothed data.
Based on NCC scores, t-values, and analysis of Euclidean distances, it can be concluded that higher t-test values resulting from HAMMER registration are due to an increased activation overlap among all subjects. The use of the high-resolution template with the low-dimensional SPM normalization procedure neither increased the activation peak value nor improved the localization of activation foci. Clearly, the SPM2 normalization technique cannot take advantage of the spatial detail in a high-resolution template to allow matching of morphologically variable regions. Spatial smoothing of fMRI data prior to group analysis does increase the magnitude of peak activation in most cases. However, such smoothing degrades spatial resolution, so that activation foci cannot be localized as precisely.
4. General Discussion
In this work, we compared the effect of a high-dimensional elastic registration technique (i.e., HAMMER) to other normalization methods on group-level statistics in an auditory fMRI experiment. The accuracy of the registration techniques was assessed using normalized cross-correlation. Moreover, the functional contrast-to-noise estimates, and measures of distance between estimates of the location of peak activation for the group and estimates for each individual, were also assessed and compared between different methods. Importantly, the functional peak assessment is derived from data (i.e., fMRI data) that is independently acquired from the structural data involved in the registration. Therefore, besides NCC-based comparison, the functional peak assessment provides an independent converging evidence for the overall pattern of results. We took higher normalization coefficients, higher t-values, and smaller group-individual differences to be consistent with a greater anatomical homogeneity (overlap) among experimental participants.
HAMMER outperformed SPM2, unified segmentation in SPM5, and DAR-TEL normalization techniques according to all three aforementioned measures. A better match across subjects in brain morphology resulted in better functional signal-to-noise (higher t-statistics) and more focal regions of activation that were also more precisely located with respect to Heschl’s gyrus. DAR-TEL’s deformation-based registration makes it a suitable choice for voxel-based morphometry analysis; however, in applications such as normalization of fMRI data for group analysis where the focus of the study is more localized to a specific region-of-interest, DARTEL-based registration does not have the freedom of small deformation to match macroanatomy in small subregions of the brain. This may reduce the sensitivity to signal change in that particular region. The effect of using a high-resolution template (Colin27) for normalization was also examined. The use of the high-resolution template with the low-dimensional SPM2 normalization procedure neither increased the t-statistics nor improved the registration of activation foci across subjects; we conclude that the SPM2 normalization technique cannot take advantage of the spatial detail in a high-resolution template to improve alignment of morphological details across individuals. Spatial smoothing was effective at increasing t-statistics and functional signal-to-noise. However, (1) such smoothing decreases spatial resolution so that activation foci cannot be localized as precisely, and (2) the effect of smoothing in terms of yielding higher t-values is more significant for SPM-based analyses.
Inter-individual variability in the location of activation foci has at least three components - variability in sulcal/gyral morphology; variability in the extent and topography of microanatomically defined regions with respect to gross morphology, and functional variability. To the extent that brains in a common reference space differ in sulcal and gyral morphology, activation foci can be expected to be at different spatial coordinates. It is this component of functional variability that we can overcome, in part, with high-dimensional anatomical registration. However, brains also differ in microanatomical structure, which is correlated with gross morphology although not entirely. Thus, patches of tissue which may be microanatomically and functionally homologous across individuals may have somewhat different relationship with sulcal and gyral morphology. This anatomical variability cannot be eliminated with high-dimensional normalization, nor can variability in location due to the recruitment of different perceptual/cognitive processes (and thus, anatomically and functionally different cortical regions) across individuals. However, high-dimensional registration techniques like HAMMER do provide a tool for assessing whether variability in the location and extent of activation foci among individuals is due primarily to sulcal and gyral morphological variation among individuals, since it can be used to eliminate this component. Residual inter-subject variability can then be attributed either to anatomical variability at a microscopic scale (i.e., variability in the location and extent of cytoarchitectonically or chemoarchitectonically or hodologically defined areas) or to variability in function due to a different processing network being recruited. Using high-dimensional registration techniques will permit the analysis and removal of much of the anatomical component from current estimates of functional variability thereby providing a more precise identification of regions related to elemental cognitive functions.
Acknowledgments
The authors would like to thank Drs. J. Ashburner, D. Shen, X. Wu, and G. Ridgeway for their help. This work is supported by the Canadian Institutes of Health Research (CIHR) through an operating grant to I.S.J., the Natural Sciences and Engineering Research Council of Canada (NSERC) discovery grant to I.S.J. and P.A., Ontario Graduate Scholarship (OGS) to A.M.T. and an early research award to I.S.J.
References
- 1.Ardekani B, Guckemus S, Bachman A, Hoptman M, Wojtaszek M, Nierenberg J. Quantitative comparison of algorithms for inter-subject registration of 3D volumetric brain MRI scans. Journal of Neuroscience Methods. 2005;142:67–76. doi: 10.1016/j.jneumeth.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Ardekani BA. An improved method for intersubject registration in 3D volumetric brain MRI. Sydney, Australia. World Congress on Medical Physics and Biomedical Engineering; 2003. Abstract 452. [Google Scholar]
- 3.Ardekani BA, Bachman AH, Strother S, Fujibayashi Y, Yonekura Y. Impact of inter-subject image registration on group analysis of fMRI data. International Congress Series, Elsevier. 2004;1265:49–59. [Google Scholar]
- 4.Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner J, Friston K. Fully three-dimensional nonlinear spatial normalization: A new approach. NeuroImage. 1996;3:S111. [Google Scholar]
- 6.Ashburner J, Friston K. Nonlinear spatial normalization using basis functions. Human Brain Mapping. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner J, Friston K. Unified segmentation. NeuroImage. 2005;26 (3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Bakker A, Kirwan C, Miller M, Stark C. Pattern separation in the human hippocampal CA3 and dentate gyrus. Science. 2008;319 (5870):1640–1642. doi: 10.1126/science.1152882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox R. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research. 1996;29 (3):162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 10.Davatzikos C. Spatial normalization of 3D brain images using deformable models. Journal of Computed Assisted Tomography. 1996;20:656–665. doi: 10.1097/00004728-199607000-00031. [DOI] [PubMed] [Google Scholar]
- 11.Desai R, Liebenthal E, Possing E, Waldron E, Binder J. Volumetric vs. surface-based alignment for localization of auditory cortex. NeuroImage. 2005;26 (4):1019–1029. doi: 10.1016/j.neuroimage.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Evans AC, Collins DL. A 305-member MRI-based stereotactic atlas for CBF activation studies. Proceedings of the 40th Annual Meeting of the Society for Nuclear Medicine.1993. [Google Scholar]
- 13.Fischl B, Sereno M, Tootell R, Dale A. High-resolution inter-subject averaging and a coordinate system for the cortical surface. Human Brain Mapping. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox P, Perlmutter S, Raichle M. A stereotactic method of anatomical localization for positron emission tomography. Journal of Computed Assisted Tomography. 1985;9:141–153. doi: 10.1097/00004728-198501000-00025. [DOI] [PubMed] [Google Scholar]
- 15.Friston K, Holmes A, Poline JB, Grasby P, Williams S, Frackowiak R, Turner R. Analysis of fMRI time-series revisited. NeuroImage. 1995;2 (1):45–53. doi: 10.1006/nimg.1995.1007. [DOI] [PubMed] [Google Scholar]
- 16.Friston K, Holmes A, Worsley K, Poline J, Frith C, Frackowiak R. Statistical Parametric Maps in functional imaging: A general linear approach. Human Brain Mapping. 1995;2:189–210. [Google Scholar]
- 17.Gee J, Alsop D, Aguirre G. Effect of spatial normalization on analysis of functional data. SPIE Medical Imaging 1997: Image Processing. 1997:550–560. [Google Scholar]
- 18.Gholipour A, Kehtarnavaz N, Briggs R, Devous M, Gopinath K. Brain functional localization: A survey of image registration techniques. IEEE Transaction on Medical Imaging. 2007;26 (4):427–451. doi: 10.1109/TMI.2007.892508. [DOI] [PubMed] [Google Scholar]
- 19.Hellier P, Barillot C, Corouge I, Gibaud B, Le Goualher G, Collins D, Evans A, Malandain G, Ayache N, Christensen G, Johnson H. Retrospective evaluation of intersubject brain registration. IEEE Transaction on Medical Imaging. 2003;22 (9):1120–1130. doi: 10.1109/TMI.2003.816961. [DOI] [PubMed] [Google Scholar]
- 20.Holmes C, Hoge R, Collins L, Woods R, Toga A, Evans A. Enhancement of MR images using registration for signal averaging. Journal of Computed Assisted Tomography. 1998;22 (2):324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- 21.ICBM452. 2009 Available from http://www.loni.ucla.edu/ICBM/ICBM_Probabilistic.html.
- 22.Kang X, Bertrand O, Alho K, Yund E, Herron T, Woods D. Local landmark-based mapping of human auditory cortex. NeuroImage. 2004;22 (4):1657–1670. doi: 10.1016/j.neuroimage.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Kirwan C, Jones C, Miller M, Stark C. High-resolution fMRI investigation of the medial temporal lobe. Human Brain Mapping. 2006;28 (10):959–966. doi: 10.1002/hbm.20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang M-C, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage. 2009 doi: 10.1016/j.neuroimage.2008.12.037. In Press. Corrected Proof. URL http://www.sciencedirect.com/science/article/B6WNP-4VC7DX1-1/2/8d9dffe89b9981c86fcebf256c21cd04. [DOI] [PMC free article] [PubMed]
- 25.Maas L, Renshaw P. Post-registration spatial filtering to reduce noise in functional MRI data sets. Magnetic Resonance Imaging. 1999;17 (9):13711382. doi: 10.1016/s0730-725x(99)00059-4. [DOI] [PubMed] [Google Scholar]
- 26.Maisog J, Chmielowska J. An efficient method for correcting the edge artifact due to smoothing. Human Brain Mapping. 1998;6 (3):128–136. doi: 10.1002/(SICI)1097-0193(1998)6:3<128::AID-HBM2>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Pike GSB, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Goualher NKGL, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philosophical Transactions of the Royal Society B: Biological Sciences. 2001;356 (1412):1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikl M, Marecek R, Hluštik P, Pavlicováe M, Drastich A, Chlebus P, Brázdil M, Krupa P. Effects of spatial smoothing on fMRI group inferences. Magnetic Resonance Imaging. 2008;26 (4):490–503. doi: 10.1016/j.mri.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Miller M, Beg M, Ceritoglu C, Stark C. Increasing the power of functional maps of the medial temporal lobe by using large deformation diffeomorphic metric mapping. Proceedings of National Academy of Sciences. 2005;102 (27):9685–9690. doi: 10.1073/pnas.0503892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penhune V, Zatorre R, MacDonald J, Evans A. Interhemispheric anatomical differences in human primary auditory cortex: probabilistic mapping and volume measurement from magnetic resonance scans. Cerebral Cortex. 1996;6 (5):661–672. doi: 10.1093/cercor/6.5.661. [DOI] [PubMed] [Google Scholar]
- 31.Reimold M, Slifstein M, Heinz A, Mueller-Schauenburg W, Bares R. Effect of spatial smoothing on t-maps: arguments for going back from t-maps to masked contrast images. Journal of Cerebral Blood Flow and Metabolism. 2006;26:751–759. doi: 10.1038/sj.jcbfm.9600231. [DOI] [PubMed] [Google Scholar]
- 32.Scouten A, Papademetris X, Constable R. Spatial resolution, signal-to-noise ratio, and smoothing in multi-subject functional MRI studies. NeuroImage. 2006;30 (3):787–793. doi: 10.1016/j.neuroimage.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 33.Shen D, Davatzikos C. HAMMER: Hierarchical Attribute Matching Mechanism for Elastic Registration. IEEE Transaction on Medical Imaging. 2002;22 (11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 34.SPM. 2009 manual available from http://www.fil.ion.ucl.ac.uk/spm/doc/manual.pdf.
- 35.Thompson P, Woods R, Mega M, Toga A. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Human Brain Mapping. 2000;9:81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viceic D, Campos R, Fornari E, Spierer L, Meuli R, Clarke S, Thiran J. Local landmark-based registration for fMRI group studies of nonprimary auditory cortex. NeuroImage. 2008;44 (1):145–153. doi: 10.1016/j.neuroimage.2008.07.051. [DOI] [PubMed] [Google Scholar]
- 37.Weibull A, Gustavsson H, Mattsson S, Svensson J. Investigation of spatial resolution, partial volume effects and smoothing in functional MRI using artificial 3D time series. NeuroImage. 2008;41 (2):346–353. doi: 10.1016/j.neuroimage.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Westbury C, Zatorre R, Evans A. Quantifying variability in the planum temporale: a probability map. Cerebral Cortex. 1999;9 (4):392–405. doi: 10.1093/cercor/9.4.392. [DOI] [PubMed] [Google Scholar]
- 39.Woods R, Grafton S, Watson J, Sicotte N, Mazziotta J. Automated image registration: II. intersubject validation of linear and nonlinear models. Journal of Computed Assisted Tomography. 1997;22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- 40.Worsley K, Friston K. Analysis of fMRI time-series revisited-again. NeuroImage. 1995;2 (3):173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- 41.Wu M, Carmichael O, Lopez-Garcia P, Carter C, Aizenstein H. Quantitative comparison of AIR, SPM, and the fully deformable model for atlas-based segmentation of functional and structural MR images. Human Brain Mapping. 2006;27 (9):747–754. doi: 10.1002/hbm.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yassa M, Stark C. A quantitative evaluation of cross-participant registration techniques for MRI studies of the medial temporal lobe. NeuroImage. 2009;44:319–327. doi: 10.1016/j.neuroimage.2008.09.016. [DOI] [PubMed] [Google Scholar]