Abstract
Mammalian neural stem cells generate transit amplifying progenitors that expand the neuronal population, but these type of progenitors have not been studied in Drosophila. The Drosophila larval brain contains ~100 neural stem cells (neuroblasts) per brain lobe, which are thought to bud off smaller ganglion mother cells (GMCs) that each produce two post-mitotic neurons. Here we use molecular markers and clonal analysis to identify a novel neuroblast cell lineage containing "transit amplifying GMCs" (TA-GMCs). TA-GMCs differ from canonical GMCs in several ways: each TA-GMC has nuclear Deadpan, cytoplasmic Prospero, forms Prospero crescents at mitosis, and generates up to 10 neurons; canonical GMCs lack Deadpan, have nuclear Prospero, lack Prospero crescents at mitosis, and generate two neurons. We conclude that there are at least two types of neuroblast lineages: a type I lineage where GMCs generate two neurons, and a type II lineage where TA-GMCs have longer lineages. Type II lineages allow more neurons to be produced faster than type I lineages, which may be advantageous in a rapidly developing organism like Drosophila.
Keywords: neuroblast, transit amplifying cell, Prospero, GMC, type II lineage
INTRODUCTION
In many mammalian tissues, stem cells generate lineage-restricted "transit amplifying cells" that can proliferate to expand the number of differentiated progeny made from a single precursor (Morrison and Kimble, 2006; Nakagawa et al., 2007). Teasing out the mechanisms that regulate stem cell proliferation and self-renewal from those regulating proliferation of transit amplifying progenitors is an important goal of stem cell biology, and has been complicated by the difficulty in identifying each type of progenitor in vivo or in vitro.
The Drosophila CNS develops from neural precursors called neuroblasts, which have recently become a model for studying neural stem cell self-renewal (Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006a; Lee et al., 2006b; Lee et al., 2006c; Wang et al., 2006) (reviewed in Doe, 2008). Neuroblasts divide asymmetrically in cell size and fate to form a larger neuroblast and a smaller ganglion mother cell (GMC). The neuroblast continues to proliferate, whereas the GMC typically generates just two post-mitotic neurons (Goodman and Doe, 1993; Lee and Luo, 1999; Pearson and Doe, 2003). Many proteins are asymmetrically segregated during neuroblast mitosis: the apical proteins Bazooka, aPKC, Par-6, Partner of Inscuteable (Pins), and Inscuteable (Insc) are segregated into the neuroblast, whereas the basal proteins Numb, Miranda (Mira), Prospero (Pros), and Brain tumor (Brat) are localized into the GMC (reviewed in Caussinus and Hirth, 2007). aPKC promotes neuroblast self-renewal, whereas the basal proteins Numb, Mira, Brat, and Pros all act to inhibit self-renewal and promote neuronal differentiation (Bello et al., 2006; Betschinger et al., 2006; Choksi et al., 2006; Lee et al., 2006a; Lee et al., 2006c; Wang et al., 2006). Neuroblast transcription factors include the basic-helix-loop-helix protein Deadpan (Dpn), which promotes optic lobe proliferation (Wallace et al., 2000), but has not been assayed for a role in neuroblast proliferation. In contrast, the Pros transcriptional repressor is nuclear in GMCs and young neurons (Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995; Li and Vaessin, 2000), where it down-regulates cell cycle gene expression to restrict GMCs to one terminal mitosis (Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995; Li and Vaessin, 2000).
Here we identify a novel "type II" neuroblast lineage that contains transit amplifying GMCs (TA-GMCs) that can each generate up to 10 neurons. These neuroblast lineages provide a model system for studying the similarities and differences between transit amplifying neural progenitors in Drosophila and mammals, and may help explain the phenotypic variation previously observed in wild type and mutant Drosophila larval brains. While this paper was in review, similar reports were published (Bello et al., 2008; Bowman et al., 2008), and our data are consistent with these studies.
MATERIALS AND METHODS
Fly stocks and clonal analysis
To generate mosaic analysis with repressible cell marker (MARCM) clones we crossed hs-flp ; tubP-gal80, FRT40A / CyO ; tubP-gal4, UAS-mcd8∷GFP / TM6 Tb to FRT40A (ovoD) / CyO and assayed clones in progeny of the genotype hs-flp ; tubP-gal80, FRT40A / FRT40A (ovoD) ; tubP-gal4, UAS-mcd8∷GFP / +. We picked first or second instar larvae by morphology and incubated them at 37°C for 25–30 min, aged them for 48h, and then dissected, fixed, and stained the brains (see below). This protocol generates a low frequency of clones per brain lobe; any brain lobe containing clones that could not be individually identified was discarded.
Immunostaining and confocal analysis
Larval brains were dissected in Schneider’s medium (Sigma, St. Louis, MO); fixed in 100 mM Pipes (pH 6.9), 1 mM EGTA, 0.3% Triton X-100, and 1 mM MgSO4 containing 4% formaldehyde for 25 min; washed 30 min in phosphate buffered saline (PBS); washed 30 min in PBS containing 0.3% Triton X-100 (PBS-BT); and incubated with primary antibodies in PBS-BT overnight at 4°C. Primary antibodies were rat Dpn monoclonal (1:1), rabbit phosphohistone H3 (1:1000; Upstate, Billerica, MA), mouse Pros monoclonal (purified MR1A, 1:1000), rabbit GFP (1:1000; Sigma, St. Louis, MO), rabbit Pins (1:1000), guinea pig Mira (1:500), mouse BrdU (1:50; Sigma, St. Louis, MO), mouse Fasciclin II (1:100; Developmental Studies Hybridoma Bank, DSHB), and rat Elav (1:10; DSHB). Secondary antibodies were from Molecular Probes (Eugene, OR). Antibodies without named sources were made in the laboratory; details are available on request. Images were captured with a Biorad Radiance or Leica SP2 confocal microscope and processed in Photoshop 7 (Adobe, San Jose, CA). Three-dimensional brain reconstructions, mushroom body iso-surface representations, and movies were generated using Imaris software (Bitplane, Zurich, Switzerland).
BrdU pulse/chase experiments
Bromodeoxyuridine (BrdU) was purchased from Roche (Basel, Switzerland), dissolved in 1:1 DMSO:acetone, and mixed with food media at a final concentration of 1 mg/mL. Larvae were fed on BrdU-containing food for 4.5 h and immediately fixed for pulse experiments, or allowed to develop on food lacking BrdU for 18h before fixation for chase experiments. Larval brains were dissected, fixed, and antibody stained as described above with the addition of a 2N HCl treatment for 30 min prior to BrdU staining.
Identifying type I and type II neuroblast and GMC lineages
Clonal analysis (Figure 1)
Figure 1. Clonal analysis identifies two types of larval neuroblast lineages.
(A–E) Neuroblast (NB) and GMC clones stained for Deadpan (Dpn, green), Prospero (Pros, red), and the clone marker GFP (green, outlined). Right panel shows summary of markers: green, nuclear Dpn cytoplasmic Pros (Dpn+ Proscyto); red, Dpn-negative nuclear Pros (Dpn− Prosnucl). Type I clones were assayed in the DAL brain region; type II clones were assayed in the DPM brain region.
(A) Type I neuroblast clone containing one large Dpn+ Proscyto NB and many Dpn− Prosnucl GMCs.
(B) Type I GMC clone containing two Prosnucl immature neurons that lack GFP+ axons (data not shown).
(C) Type II neuroblast clone containing one large Dpn+ Proscyto NB and smaller progeny including two Dpn− Proscyto cells closely-associated with the neuroblast (arrows), several Dpn+ Proscyto cells, and several Dpn− Prosnucl cells.
(D) Type II TA-GMC small clone containing one Dpn+ Proscyto cell (arrowhead) and three Dpn− Prosnucl cells.
(E) Type II TA-GMC large clone containing several Dpn− Prosnucl cells, and a pool of Dpn− Pros− mature neurons (based on their GFP+ axon projections).
Scale bar = 6.24 µm.
Type I neuroblast clones were uniquely identified by the presence of one large (>8µm diameter) neuroblast containing nuclear Dpn and cytoplasmic Pros (Dpn+ Proscyto) together with many small (<5µm diameter) progeny that lacked Dpn and had nuclear Pros (Dpn− Prosnucl). Cells furthest from the neuroblast were Dpn− Pros− mature neurons that extended GFP+ axons into the brain. Type I GMC clones were identified by lack of a large neuroblast, and were assayed only in the dorsoanterior lateral (DAL) region, where no type II neuroblasts exist (see Figure 2). Type I GMC clones never had more than two cells.
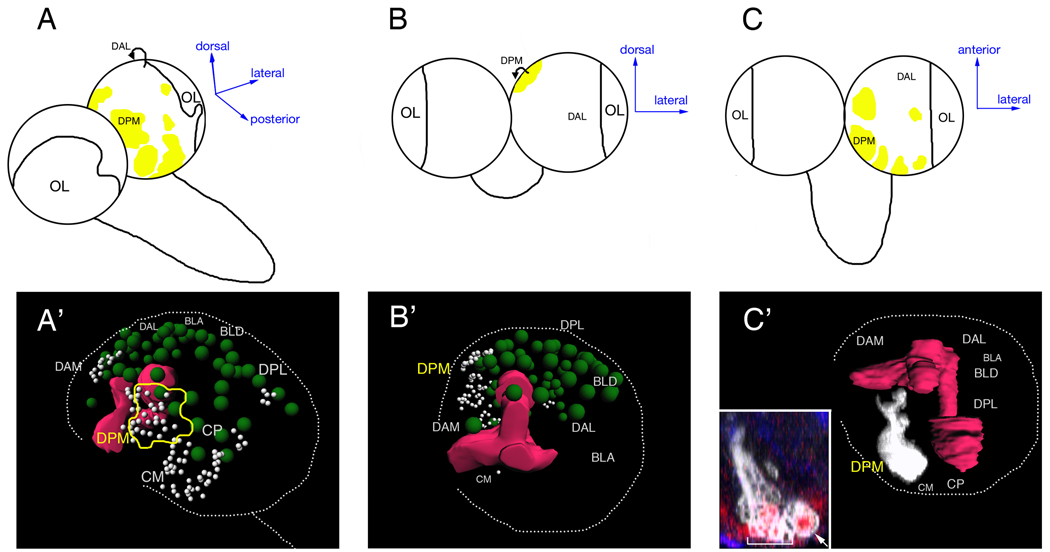
Figure 2. Type I and type II neuroblast lineage locations within the brain.
(A–C) Schematics of the third instar larval brain showing brain regions according to Pereanu and Hartenstein (2006). Type I neuroblast are found in all brain regions, but only type I neuroblasts are located in the dorsoanterior lateral (DAL) region, which is where we performed all type I lineage assays. Type II neuroblast are found in several brain regions (yellow shading); the largest number are in the dorsoposterior medial (DPM) region. The orientation of each brain is indicated by the axial arrows. OL, optic lobe.
(A'–B') Three-dimensional reconstruction of a confocal image stack of a brain lobe double-label for Dpn (green spheres, neuroblasts; silver spheres, TA-GMCs) and Fasciculin II (mushroom body, red). Orientation is the same as the panel above. This brain lobe is shown in Movie 2.
(C') Three-dimensional reconstruction of a confocal image stack of a brain lobe containing a type II neuroblast clone (white) stained for Fasciculin II (mushroom body lobes, red). The clone extends medially across the midline but the fine axon processes do not show up in this image; they can be seen in Movie 1.
(C' inset) Optical section from brain used to generate the image shown panel C'. Clone marker (GFP, white); Deadpan (red); neuroblast in clone, arrow; TA-GMCs in clone, bracket.
Abbreviations: OL, optic lobe; DPM, dorsoposterior medial (yellow outline); DAL, dorsoanterior lateral; DAM, dorsoanterior medial; DPL, dorsoposterior lateral; BLP, basolateral posterior; BLA, basolateral anterior; BLD, basolateral dorsal; BA, basoanterior; CP, centroposterior; CM, centromedial. Regions in smaller fonts are towards the back of the lobe.
Scale bar = 20 µm.
Type II neuroblast clones were identified by the presence of one large Dpn+ Proscyto neuroblast with the unique and defining feature that the clone also contained many small (<5µm diameter) Dpn+ Proscyto cells. Cells furthest from the neuroblast were Dpn− Pros− mature neurons that extended GFP+ axons into the brain. Type II TA-GMCs clones were identified by (i) their lack of a large neuroblast; (ii) their ability to make >2 progeny, which is never observed in type I GMC clones; and (iii) the presence of small (<5µm diameter) Dpn+ Proscyto cells, which are never observed in type I lineages. We observed one and two cell clones in all regions of the brain; we assume they are made by both type I and type II lineages.
Whole brain analysis (antibody stains and BrdU experiments)
Type I neuroblasts can be uniquely identified as a large Dpn+ or Mira+ cell (>8µm diameter) contacting only small (<5µm diameter) Prosnucl cells (the GMCs). Type I neuroblasts are found in the DAL region of the brain, where no type II neuroblasts exist, and thus for consistency we restricted our analysis of type I lineages to this brain region. Type II neuroblasts can be identified as a large Dpn+ cell contacting small Dpn+ Proscyto cells (TA-GMCs) or in BrdU experiments as a Mira+ neuroblast contacting a large group of small Mira+ cells.
Locating type I and type II lineages in the brain
Central brain regions (i.e. the brain excluding the lateral optic lobes) were identified and named as previously described (Pereanu and Hartenstein, 2006). Briefly, we used Fasciculin II as a positional marker and Dpn as a neuroblast marker; double-labeled brains were oriented according to Pereanu and Hartenstein (2006) to determine neuroblast position relative to the Fasciculin II pattern. In this way, we mapped the approximate location of type I and type II neuroblasts; we found that type I neuroblasts were the sole occupants of the DAL brain region, whereas the type II neuroblasts were located in subsets of the following brain regions: dorsoposterior medial (DPM), dorsoposterior lateral (DPL), dorsoanterior medial (DAM), centromedial (CM), and centroposterior (CP) (yellow patches in Figure 2; Supplemental Table 1).We could individually identify only one type II neuroblast (the DPMpm1 neuroblast; Figure 2C') due to natural variation in neuroblast position (Pereanu and Hartenstein, 2006); relatively few axon projections in the clones; and similarity between closely positioned neuroblasts (Pereanu and Hartenstein, 2006). To minimize regional variation in neuroblast lineages, we restricted our analysis of type I neuroblasts to the DAL region, and type II neuroblasts to the DPM region.
RESULTS
Clonal analysis reveals two types of brain neuroblast lineages
During our clonal analysis of a larval neuroblast self-renewal mutant we realized that wild type brains have two distinct types of neuroblast lineages (J.Q.B. and C.Q.D., in preparation). This paper describes these two types of lineages. We used mosaic analysis with repressible cell marker (MARCM; Lee and Luo, 1999) to generate GFP-marked single cell clones in the larval brain. Depending on the cell in which chromosomal recombination occurs, it is possible to label a single neuroblast and all its progeny, a single GMC and all its progeny, or a single neuron derived from a terminal mitosis (Lee and Luo, 1999). We induced a low density of clones randomly throughout the brain at either mid-first or mid-second larval instar and assayed all clones 48 h after induction (Figure 1). We find two distinct neuroblast lineages: a “type I” lineage that matches previously reported neuroblast lineages (Goodman and Spitzer, 1979; Lee and Luo, 1999; Pearson and Doe, 2003), and a novel "type II" lineage that is larger and more complex.
Type I neuroblast lineages
Type I neuroblast clones always contained one large (>8µm diameter) neuroblast near the surface of the brain that had nuclear Dpn and cytoplasmic Pros (Dpn+ Proscyto) (100%; n=26; Figure 1A; Supplemental Table 1). These clones always contained a column of smaller cells that lacked Dpn and had nuclear Pros (Dpn− Prosnucl), with the occasional presence of a single Dpn+ small cell contacting the neuroblast, which is likely to be a newborn GMC (Supplemental Table 1). The cells furthest from the neuroblast were Dpn− Pros− mature neurons that extend GFP+ axons into the central brain (data not shown). Type I neuroblast lineages are the sole occupants of the dorsoanterior lateral (DAL; Pereanu and Hartenstein, 2006) brain region, but can also be found in all other brain regions (Figure 2). To minimize regional variation in neuroblast lineages, we restricted our analysis of type I neuroblasts to the DAL region.
Type I GMC clones were assayed only in the DAL region, where no type II neuroblasts were observed. All clones lacking a large Dpn+ neuroblast were considered to be GMC clones, and these GMC clones generated at most just two cells (100%, n=9; Figure 1B). Thus, type I lineages are identical to those reported for Drosophila embryonic neuroblasts (Goodman and Doe, 1993; Pearson and Doe, 2003), larval mushroom body neuroblasts (Lee and Luo, 1999), and grasshopper neuroblasts (Goodman and Spitzer, 1979).
Type II neuroblast lineages
Type II neuroblast clones always contained one large (>8µm diameter) Dpn+ neuroblast near the surface of the brain, but also contained a distinctive group of small (<5µm diameter) Dpn+ cells that lack nuclear Pros (100%; n=17; Figure 1C; Supplemental Table 1). There are also usually 1–2 small cells in direct contact with the neuroblast that lack both Dpn and nuclear Pros (Figure 1C, arrows). These two types of small cells are never observed in type I clones and are a defining feature of type II clones. Type II neuroblast clones are found in several brain regions, including a cluster within the DPM region (Figure 2, yellow shading). One type II neuroblast appears to be the previously identified DPMpm1 neuroblast (Pereanu and Hartenstein, 2006) based on its distinctive axon projection that bifurcates over the medial lobe of the mushroom body before crossing the midline (Figure 2C', inset; Movie 1).
Type II GMC clones were identified by the lack of a large Dpn+ neuroblast. All brain regions that contained type II neuroblast lineages produced GMC clones of greater than two cells (range, 3–10 neurons; average 4.8 ± 0.4; n= 25; Figure 1D,E; Supplemental Table 1); all brain regions that lacked type II neuroblast lineages never generated >2 cell GMC clones (see above). Type II GMC clones often contained Dpn+ Proscyto small cells that are unique to type II neuroblast lineages (Figure 1D; arrowhead), confirming that these clones are sublineages of a type II neuroblast lineage. We conclude that type II neuroblasts generate GMCs that produce more than two neurons. Because type II GMC clones could generate several fold more neurons than a type I GMC, we call them "transit amplifying GMCs" or TA-GMCs.
TA-GMC clones also contained small cells with nuclear Pros (Figure 1D,E); we suggest that these cells are equivalent to type I GMCs based on their cell division profile (see next section), and because we observed two cell clones in regions of the brain that contained type II neuroblast lineages. However, we can't rule out the possibility that some of these nuclear Pros cells are post-mitotic immature neurons (see Discussion).
If type II lineages generate TA-GMCs that make an average of twice as many neurons as a type I lineage, we would expect type II lineages to generate approximately twice as many neurons over the same timespan compared to type I lineages. Indeed, we find that when type I or type II clones are grown for the same length of time (between clone induction and analysis), type II clones generate approximately twice as many neurons. Type I clones in the DAL generate 40.4 ± 3.1 cells (n=16; clone developing 24–72 h after larval hatching [ALH]), whereas type II lineages in the DPM generate 71.2 ± 6.3 cells (n=5; clone developing 24–72h ALH). In all cases the final type I and type II neuroblast clones contained a single large >8µm diameter Dpn+ neuroblast, ensuring that only single neuroblast clones were counted. We conclude that type II TA-GMCs generate more neurons than type I GMCs, and that type II lineages generate more neurons than type I lineages.
Asymmetric cell division within type I and type II lineages
Here we characterize the cell division patterns within type I and type II lineages to help understand the relationship between different cell types in a lineage. We first ask, what cell type is directly produced by type I and type II neuroblasts? We found that type I neuroblasts in the DAL region always segregate Pros protein into the newborn GMC (100%, n=9; Figure 3A) resulting in easily detectable levels of Pros in neuroblast progeny (see Figure 1A). Thus, type I neuroblasts in the DAL generate nuclear Pros+ GMCs, as previously reported (Spana and Doe, 1995; Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006c). In contrast, type II neuroblasts of the DPM region often fail to segregate Pros protein (50%; n=14; Figure 3C), despite proper localization of other apical/basal proteins (100%; n=14; Figure 3C), which would account for reduced Pros levels in newborn progeny (Figure 1C, arrows). The variation in Pros localization among DPM neuroblasts could be due to the presence of some type I neuroblasts in the region, or actual variation among type II neuroblasts. We conclude that type II neuroblasts divide asymmetrically, but can fail to segregate Pros protein into their newborn progeny (see Discussion).
Figure 3. Asymmetric cell division within type I and type II neuroblast lineages.
Mitotic larval neuroblasts and GMCs stained for the apical protein Partner of Inscuteable (Pins) and the basal proteins Miranda (Mira) and Prospero (Pros), in some cases the mitotic marker phosphohistone H3 (PH3) is shown. Pros/Mira or Pros/PH3 panels always show the same neuroblast.
(A) Type I mitotic neuroblast in the DAL region shows apical Pins and basal Mira/Pros. Pros/Mira panels show the same NB.
(B) Type I mitotic GMC in the DAL region shows diffuse cytoplasmic Pros (bright punctate staining in the Pros panel is DNA-associated Pros protein).
(C) Type II mitotic neuroblast in the DPM region identified by lack of asymmetric Pros localization, despite completely normal localization of Pins and Mira. Pros/Mira panels show the same NB.
(D) Type II mitotic TA-GMCs in the DPM identified by their small size and asymmetric localization of Pros and Mira to the cortex opposite the Pins.
Scale bar = 6.24 µm.
We next investigated the relationship between the type II small cells that have high Dpn, low Pros (Dpn+ Proscyto) and those that contain high Pros, but no Dpn (Dpn− Prosnucl). We found that mitotic Dpn+ small cells always form Mira/Pros cortical crescents (100%, n=50; Figure 3D), with Pins protein localized to the opposite cortical domain (100%, n=18; Figure 3D), and the spindle aligned along this cortical polarity axis (data not shown). This type of division is unique to type II lineages, as all type I GMCs always showed diffuse cytoplasmic Pros during mitosis (100%, n=6 in DAL; Figure 3B). We conclude that type II Dpn+ small cells undergo molecularly asymmetric cell divisions to generate a Pros+ sibling and a Pros- sibling. We propose that the sibling with little or no Pros remains a Dpn+ TA-GMC, whereas the Pros+ sibling generates one or two post-mitotic neurons, similar to Pros+ GMCs in type I lineages (see Discussion).
Type II neuroblast progeny are proliferative but can generate differentiated neurons
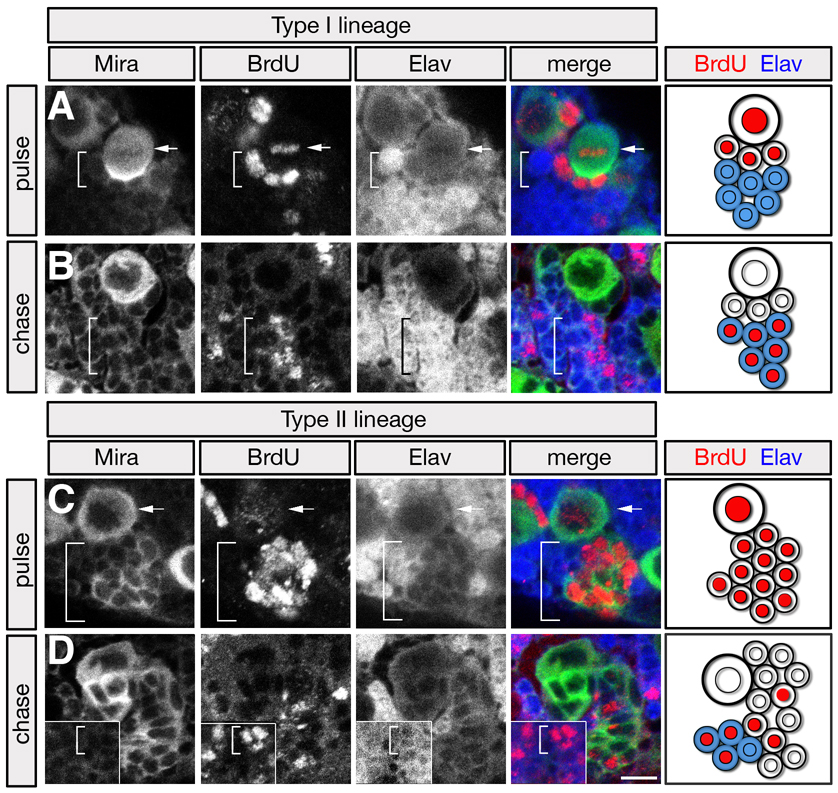
To characterize the cell cycle kinetics of type I GMCs and type II TA-GMCs, we performed BrdU labeling experiments. We exposed larvae to a 4.5 h BrdU pulse and then immediately fixed and assayed for BrdU incorporation. As expected, both type I and type II neuroblasts always incorporated BrdU (Figure 4A,C; arrow). Type I neuroblasts showed only a few closely-associated GMCs labeled (Figure 4A; bracket), whereas type II neuroblasts had a much larger number of labeled progeny (Figure 4C; bracket). It is unlikely that the type II neuroblasts generate all of these progeny during the 4.5 h labeling window, because the shortest neuroblast cell cycle time we have observed in any brain region is ~50 min (C. Cabernard and C.Q.D., unpublished results), and thus we conclude that type II neuroblast progeny undergo more rounds of cell division that type I GMCs.
Figure 4. BrdU pulse/chase analysis of type I and type II neuroblast lineages.
Larvae were pulsed with BrdU for 4.5h and then either fixed immediately ("pulse"; A, C) or grown without BrdU for 18h before fixing ("chase"; B, D). Larvae were stained for Miranda (Mira, green), BrdU (red), and the neuronal marker Elav (blue); neuroblasts (NBs), arrows; neuroblast progeny, brackets; schematics are shown to the right. Type I lineage data were collected in the DAL brain region; type II lineage data were collected in the DPM brain region.
(A–B) Type I neuroblasts always incorporate BrdU during the pulse and dilute it out during the chase, whereas only a few type I GMCs contacting the neuroblast incorporate BrdU during the pulse (A, brackets); following the chase, BrdU is maintained in Elav+ post-mitotic neurons (B, brackets).
(C–D) Type II neuroblasts always incorporate BrdU during the pulse and dilute it out during the chase; many type II progeny incorporate BrdU during the pulse (C, brackets); following the chase, BrdU is maintained in Elav+ post-mitotic neurons (D, brackets; shown in an inset, because the neurons are at the bottom of this confocal image stack).
Scale bar = 6.24 µm.
To determine if the proliferative type II neuroblast progeny are competent to differentiate into neurons, we performed a BrdU pulse/chase experiment. Larvae were fed BrdU for 4.5 h as described above, but then allowed to develop for 18 h without BrdU. Type I neuroblasts lacked BrdU incorporation, as expected due to label dilution during the chase interval, but BrdU was maintained in the Elav+ post-mitotic neurons born during the pulse window (Figure 4B; bracket). Type II neuroblasts and most of their progeny also diluted out BrdU, confirming their status as proliferative cells (see above), and some Elav+ post-mitotic neurons were born during the pulse interval and maintained BrdU labeling (Figure 4D; bracket). We conclude that type II neuroblast progeny are proliferative but can still give rise to differentiated neurons.
DISCUSSION
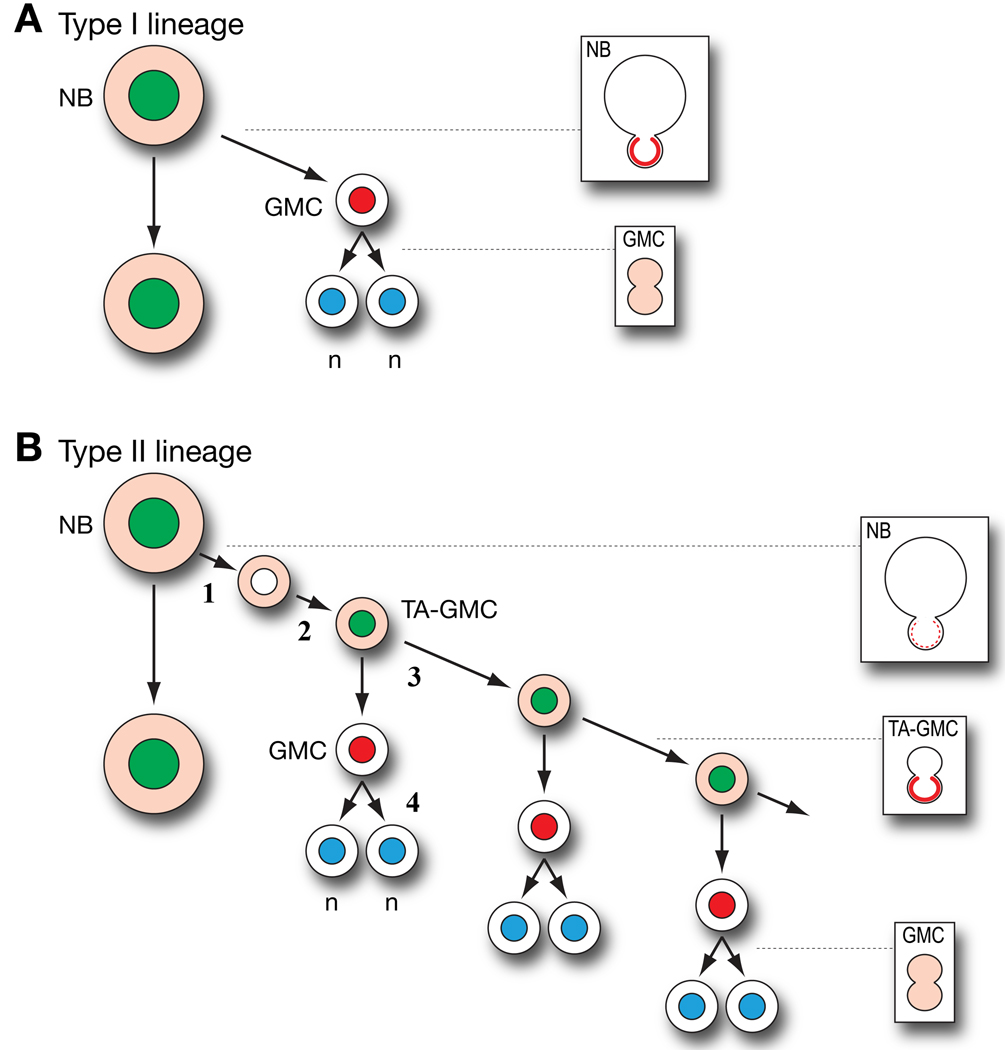
We have identified a novel "type II" neuroblast lineage within the Drosophila larval brain (Figure 5). Although we have not documented this lineage by timelapse imaging, we have the following evidence for each step in the lineage (steps marked by numbers in Figure 5):
Type II neuroblast → Dpn− Proscyto TA-GMC. We place the Dpn− Proscyto TA-GMC as the newborn progeny because this is the only cell type always observed in direct contact with the neuroblast, and because the neuroblast can divide without segregating Pros protein into the newborn GMC (consistent with the low levels of Pros in the Dpn− Proscyto TA-GMC).
Dpn− Proscyto TA-GMC → Dpn+ Proscyto TA-GMC. We propose that Dpn is rapidly upregulated in the newborn TA-GMC because (a) Dpn+ small cells are often located close to the neuroblast; (b) pros mutant type I GMCs will upregulate Dpn levels (Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006c); and (c) in other regions of the CNS Dpn protein is detected in mitotic progenitors and not post-mitotic neurons (Bier et al., 1992).
All Dpn+ Proscyto small cells divide asymmetrically to generate one Pros+ cell and one Pros− cell. We propose that the Pros- cell remains a TA-GMC.
Dpn− Prosnucl GMC divides to form two post-mitotic neurons. This part of the lineage is based on analogy with type I GMCs, which have nuclear Pros and divide symmetrically to generate two neurons (Spana and Doe, 1995). Consistent with this model, we can observe small Dpn− Pros+ cells dividing symmetrically with cytoplasmic Pros closely associated with the pool of Dpn+ TA-GMCs in the DPM. Nevertheless, it remains possible that some or all Dpn− Prosnucl cells directly differentiate into neurons.
Figure 5. Summary of type I and type II larval neuroblast lineages.
(A) Proposed type I neuroblast lineage. Nuclear Dpn (green), nuclear Pros (red), cytoplasmic or undetectable Pros (light red), cortical Prospero (red crescent), neuronal marker Elav (blue). Mitotic profiles are shown in boxes at right. See text for details.
(B) Proposed type II neuroblast lineage. Nuclear Dpn (green), nuclear Pros (red), cytoplasmic or undetectable Pros (light red), cortical Prospero (red crescent), weak or undetectable cortical Pros (dashed red crescent), neuronal marker Elav (blue). Mitotic profiles are shown in boxes at right. See Discussion for details of each numbered step in the lineage.
The most striking feature of the type II lineages is that they contain TA-GMCs that have features of both neuroblasts and GMCs. TA-GMCs resemble neuroblasts in containing nuclear Dpn, low levels of cytoplasmic Pros, their ability to asymmetrically localize Pros during mitosis, and their ability to divide multiple times; yet they resemble GMCs in their small size, physically symmetric cell division, and relatively limited mitotic potential.
There are currently no molecular markers that can be used to unambiguously identify type II neuroblasts. The inability to form Pros crescents may be shared by all type II neuroblasts, but even so, it would only be a useful marker for mitotic neuroblasts. In the DPM brain region (enriched for type II lineages) we find about 50% of the mitotic neuroblasts have little or no Pros crescent, and based on the distinctive lack of Pros in some type II neuroblast progeny, we conclude that these are type II neuroblasts. (The 50% of the DPM neuroblasts that form Pros crescents may be type I neuroblasts within the region, a special subset of type II neuroblasts, or there may be stochastic variability in Pros crescent-forming ability among type II neuroblasts.) In any case, our findings may explain why some labs report seeing Pros crescents (Bello et al., 2006; Betschinger et al., 2006; Choksi et al., 2006; Lee et al., 2006c) whereas others report that neuroblasts do not form Pros crescents (Ceron et al., 2001); both are correct because there are two types of larval neuroblast lineages.
It is unknown whether neuroblasts can switch back and forth between type I and type II modes of cell lineage. If the level of Pros in the neuroblast is the key factor distinguishing these modes of division, then experimentally raising Pros levels in type II lineages may switch them to type I lineages; conversely, reducing Pros levels in type I lineages may switch them to type II lineages. As more brain neuroblasts become uniquely identifiable it will be interesting to address this question. It will also be interesting to search for type II neuroblast lineages in other insects or crustaceans where type I neuroblast lineages have been documented (Goodman and Spitzer, 1979; Goodman and Doe, 1993; Ungerer and Scholtz, 2007).
What terminates the TA-GMC lineage? The TA-GMC may fall below a size threshold for continued proliferation. Alternatively, TA-GMCs may lose contact with a niche-derived signal that maintains their proliferation; Hedgehog, Fibroblast growth factor (Park et al., 2003), and Activin (Zhu et al., 2008) are all required for larval brain neuroblast proliferation, but none have been tested for a role in TA-GMC proliferation. Lastly, there may be lineage-specific factors segregated into the TA-GMCs that limit their mitotic potential. TA-GMCs may die at the end of their lineage, as do some neuroblasts (Bello et al., 2003), or they may differentiate.
It has been shown that loss of Pros and Brat together can generate a more severe neuroblast tumor phenotype than either alone (Betschinger et al., 2006). This suggests that the type II lineages may be especially sensitive to further loss of differentiation promoting factors due to their low levels of endogenous Pros. Indeed, we have observed a dramatic neuroblast tumor phenotype in type II lineages in lethal giant discs mutants (J.Q.B. and C.Q.D., in preparation). This raises the question of how type II lineages benefit the fly. They have the ability to generate more neurons in a faster period of time, due to the presence of TA-GMCs, and may be an evolutionary adaptation to the rapid life cycle of Drosophila. Slower developing insects may not require such rapid modes of neurogenesis.
Supplementary Material
Confocal image stack of the brain shown in Figure 2C' inset to illustrate the axon projections of the DPMpm1 neuroblast clone. The medial half of the brain lobe is shown; DPMpm1 clone (white; right-most clone in the brain), Dpn+ neuroblasts and TA-GMCs, red (labeled in the Figure 2C' inset); Fasciculin II+ neuropile, blue. Movie steps from dorsal surface to ventral surface.
Rotation of the brain shown in Figure 2A',B'. Large Dpn+ neuroblasts (>8µm, green); small Dpn+ TA-GMCs (<5µm, silver); Fasciculin II+ mushroom body (red). The first frame of the movie is the same orientation as shown in Figure 2B'.
ACKNOWLEDGEMENTS
We thank Clemens Cabernard, Chiswili Chabu, and Sen-Lin Lai for comments on the manuscript and helpful discussions. We thank the Bloomington stock center for flies and the Developmental Hybridoma Center (Iowa) for antibodies. J.Q.B. was supported by a NSF IGERT pre-doctoral training grant. C.Q.D. is an Investigator of Howard Hughes Medical Institute.
REFERENCE
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Younger-Shepherd S, Jan LY, Jan YN. deadpan, an essential pan-neural gene in Drosophila, encodes a helix-loop-helix protein similar to the hairy gene product. Genes Dev. 1992;6:2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila. Dev Cell. 2008 doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E, Hirth F. Asymmetric stem cell division in development and cancer. Prog Mol Subcell Biol. 2007;45:205–225. doi: 10.1007/978-3-540-69161-7_9. [DOI] [PubMed] [Google Scholar]
- Ceron J, Gonzalez C, Tejedor FJ. Patterns of cell division and expression of asymmetric cell fate determinants in postembryonic neuroblast lineages of Drosophila. Dev Biol. 2001;230:125–138. doi: 10.1006/dbio.2000.0110. [DOI] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: Balancing self-renewal with differentiation. Development. 2008 doi: 10.1242/dev.014977. in press. [DOI] [PubMed] [Google Scholar]
- Goodman CS, Doe CQ. Embryonic development of the Drosophila central nervous system. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 1131–1207. [Google Scholar]
- Goodman CS, Spitzer NC. Embryonic development of identified neurones: differentiation from neuroblast to neurone. Nature. 1979;280:208–214. doi: 10.1038/280208a0. [DOI] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006a;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006b;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006c;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li L, Vaessin H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 2000;14:147–151. [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Park Y, Rangel C, Reynolds MM, Caldwell MC, Johns M, Nayak M, Welsh CJ, McDermott S, Datta S. Drosophila perlecan modulates FGF and hedgehog signals to activate neural stem cell division. Dev Biol. 2003;253:247–257. doi: 10.1016/s0012-1606(02)00019-2. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Ungerer P, Scholtz G. Filling the gap between identified neuroblasts and neurons in crustaceans adds new support for Tetraconata. Proc Biol Sci. 2007 doi: 10.1098/rspb.2007.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K, Liu TH, Vaessin H. The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis. 2000;26:77–85. doi: 10.1002/(sici)1526-968x(200001)26:1<77::aid-gene10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, Boone JQ, Jensen PA, Hanna S, Podemski L, Locke J, Doe CQ, O'Connor MB. Drosophila Activin and the Activin-like product Dawdle function redundantly to regulate proliferation in the Drosophila larval brain. Development. 2008;135 doi: 10.1242/dev.010876. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confocal image stack of the brain shown in Figure 2C' inset to illustrate the axon projections of the DPMpm1 neuroblast clone. The medial half of the brain lobe is shown; DPMpm1 clone (white; right-most clone in the brain), Dpn+ neuroblasts and TA-GMCs, red (labeled in the Figure 2C' inset); Fasciculin II+ neuropile, blue. Movie steps from dorsal surface to ventral surface.
Rotation of the brain shown in Figure 2A',B'. Large Dpn+ neuroblasts (>8µm, green); small Dpn+ TA-GMCs (<5µm, silver); Fasciculin II+ mushroom body (red). The first frame of the movie is the same orientation as shown in Figure 2B'.