Summary
Drosophila telomeres are comprised of DNA sequences that differ dramatically from those of other eukaryotes. Telomere functions, however, are similar to those found in telomerase-based telomeres, even though the underlying mechanisms may differ. Drosophila telomeres use arrays of retrotransposons to maintain chromosome length, while nearly all other eukaryotes rely on telomerase-generated short repeats. Regardless of the DNA sequence, several end-binding proteins are evolutionarily conserved. Away from the end, the Drosophila telomeric and subtelomeric DNA sequences are complexed with unique combinations of proteins that also modulate chromatin structure elsewhere in the genome. Maintaining and regulating the transcriptional activity of the telomeric retrotransposons in Drosophila requires specific chromatin structures, and while telomeric silencing spreads from the terminal repeats in yeast, the source of telomeric silencing in Drosophila is the subterminal arrays. However, the subterminal arrays in both species may be involved in telomere-telomere associations and/or communication.
Keywords: Drosophila, telomere, gene silencing, chromatin, capping
Introduction
At the dawn of the genetic age, William Bateson (1861-1926) produced what has become the mantra for formal genetics: “Treasure your exceptions.” By this he meant that a careful examination of a mutant's phenotype would shed light on the normal wild type process. The same can be said of understanding the nature of wild type structures in species that diverge from a consensus structure. Thus, while Drosophila telomeres have been described as unusual (1), they can provide a unique insight into structure function relationships. While it is becoming increasingly obvious that telomere functions are conserved in evolution, the structures that perform these functions are rather flexible (2). Studying Drosophila telomeres may help to understand how certain proteins have evolved to perform essential functions at telomeres with very different DNA organization and answer such questions as, “what is important about telomere structure in terms of specific telomere functions?”
To answer this, we must first ask what are the functions of a telomere. Telomeres were initially described as structures at the ends of linear chromosomes that are necessary for the stability of chromosomes, sometimes referred to as chromosome caps. After irradiation rearranged chromosomes, except ring chromosomes, which have no ends, virtually always carried a previously existing telomere at each end (3). In modern terms, we would say that telomeres are necessary to distinguish a natural chromosome end from a double strand DNA break (DSB). As a result, telomeres are not normally subject to resection necessary for recombination repair; nor are they subject to nonhomologous end joining (NHEJ) or any other mechanism that would tend to remove DSBs by ligation.
Second, as DNA polymerase cannot replicate a linear DNA molecule completely (4,5), and further manipulation at the chromosome end may be necessary to maintain the proper DNA structure at the tip, telomeres are also needed for length homeostasis (6). The so-called end replication problem applies to all linear DNA molecules, but at least three mechanisms to maintain chromosome length, despite the inevitable terminal erosion, have been described. First, a specialized reverse transcriptase, termed telomerase, may add short repeat units to the chromosome end using an RNA contained in the telomerase complex a as template. This mechanism is wide-spread but not exclusively used in eukaryotes. Even in species with telomerase alternative mechanisms may be used in some circumstances. Second, non-reciprocal recombination (gene conversion) between repeated regions on different telomeres may result in the elongation of one participant. Third, targeted transposition of telomere-specific retrotransposons to chromosome ends is used in Drosophila to increase chromosome length.
Other activities have been described for telomeres. In some cell types telomeres tend to associate with each other and with the nuclear envelope. They also inactivate genes in their vicinity, a process known as telomeric silencing or telomeric position effect (TPE). As genes near the nuclear periphery show reduced activity, telomere associations might conceivably play a role in TPE. Telomeres may also be important for homologous chromosome pairing during meiosis (7,8).
Considering function as the defining telomere characteristic, it is convenient to divide the telomere region into three components (Fig. 1). At the extreme end of the chromosome is the cap. This structure comprises several proteins that act in concert. Depending on the species, some of these proteins may recognize specific features of the terminal DNA sequence (9). A second region is defined by the presence of the terminal DNA sequence outside of the cap. As the length of this DNA array may be several kilobases, it may easily be larger than the cap itself. A third region is identified by an irregular array of repeated sequences often termed the subterminal repeat, or the telomere associated sequence (TAS). While TAS has been identified in a wide variety of species (10), their sequence and arrangement varies considerably from one species to another and even from one telomere to another within the same cell.
Figure 1.
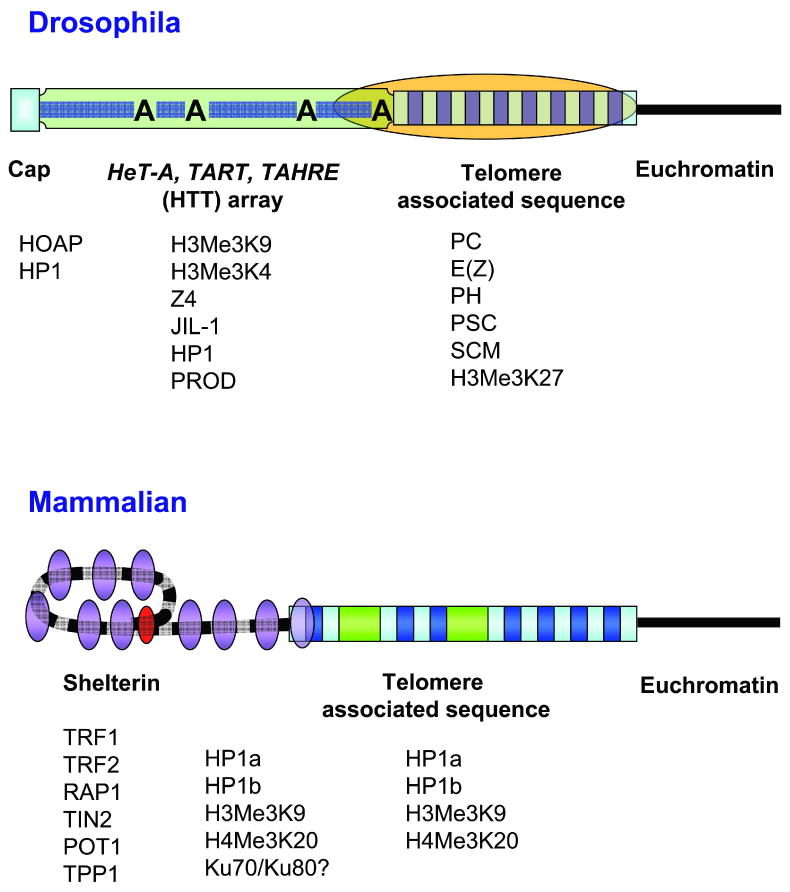
Schematic comparison of the overall structure of the telomere region in Drosophila and in human and their associated proteins. In Drosophila, the terminal capping complex (blue square) consists of proteins that are primarily associated with heterochromatin and bind in a sequence-independent manner to the end of the retrotransposon (HTT) array, which replaces the telomerase-generated repeats found in other eukaryotes. This array consists of tandem non-LTR retrotransposons of irregular length with their oligo(A) tails (indicated by A) facing towards the centromere. It is associated with proteins that are otherwise found in either heterochromatin or euchromatin (green rectangle) and has euchromatic properties. The subtelomeric (TAS) repeats (alternating light and dark blue small rectangles) assemble a distinct silencing complex (orange oval), which spreads into the proximal region of the HTT array. Thus, the capping functions at the chromosome end are clearly separated from the silencing domain of TAS, which has heterochromatic properties. In human cells, the telomerase-generated TTAGGG repeats (alternating black and grey bar) may form a t-loop structure, which is stabilized by specific proteins (red oval). Proteins binding to the telomeric repeats assemble a heterochromatic silencing complex (telosome or shelterin; purple ovals), which may spread into the TAS region. Thus, capping and silencing are centered on the telomeric repeats. Associated proteins are shown below each structure. Not shown: the cap and shelterin are maintained by DNA damage response proteins, ATM and the MRN complex.
While Drosophila terminal repeats consist of arrays of telomere-specific non-long terminal repeat (LTR) retrotransposons, HeT-A, TART and TAHRE (collectively abbreviated here as HTT elements), instead of short telomerase-generated repeats (Fig. 1), Drosophila telomeres appear in other ways to be similar to telomeres in other organisms. The chromosome cap consists of a number of proteins necessary for chromosome stability and to regulate chromosome elongation. Many of these proteins are found at telomeres in other species (11). TPE is also observed at Drosophila telomeres, although it seems to originate in the TAS region rather than the telomeric repeats. Here, we relate the specific structural features of Drosophila telomeres to universal telomere functions.
The chromosome cap
After extensive genetic and cytological analysis of ionizing radiation-induced chromosome rearrangements in Drosophila, Muller (3,12) proposed that a specific structure is required on each chromosome end for viability, which he termed a telomere or cap. These studies were extended by McClintock (13,14), who found that a broken chromosome end in some maize tissues may induce a cycle of chromosome fusion, anaphase bridges and new chromosome breaks. A similar requirement for a structure to distinguish natural ends of linear chromosome from chromosome breaks has been identified in other species with linear chromosomes, from viruses to eukaryotes, although the solution has varied widely (2). Here, the term telomere will refer to the collection of structures and functions that have been ascribed to chromosome ends, while cap will refer to the specific structure that distinguishes a natural chromosome end from a chromosome break.
In eukaryotes that have telomerase, the capping complex specifically recognizes the telomerase-generated terminal repeats, although some components, such as Ku70/80, may bind DNA independently of sequence. Drosophila, however, with an array of retrotransposons, each up to 12 kb in length, does not have a specific terminal sequence. The location of a cap, in fact, may be able to jump short distances, thus allowing the addition of HTT elements to the chromosome end (15), the elongation of chromosomes by gene conversion (16,17), and rapid telomere shortening of up to 50 kb that leaves at least some of the TAS sequence intact (18-20). P transposable element-mediated DSBs near the telomere may facilitate these rapid telomere shortening events, although telomere-linked DSBs may also occur spontaneously (20). However, when chromosome breaks are induced in somatic cells farther from the telomere, they either induce mitotic arrest and apoptosis or in some cases broken chromosomes may be transmitted through mitosis by undergoing a breakage-fusion-bridge cycle (21,22), in which a dicentric chromosome breaks at anaphase, and the resulting chromosome fragment fuses with its sister after replication and forms an new dicentric anaphase bridge, similar to that seen by McClintock (13).
After irradiation of females with a mutation in the mutator gene mu2 it is possible to recover high frequencies of terminal deficiency chromosomes that have lost a telomere (23,24). These deficiencies do not appear to induce cell cycle arrest or end-to-end fusions and are stable over many sexual generations in the absence of a mu2 mutation, even though they may have unique DNA sequence at the chromosome end, rather than the HTT retrotransposons (25). The broken ends may eventually acquire these transposable elements (15,26). Thus, it is clear that the capping function in Drosophila does not require a specific DNA sequence, but instead may act through an epigenetic mechanism similar to that seen for centromeres (27,28).
Stabilization of a broken chromosome end with a new cap, termed healing, appears to be cell type specific. McClintock noted that maize chromosomes broken during meiotic anaphase proceeded through a breakage-fusion-bridge cycle during pollen growth and endosperm development, but healed in the embryo (13,14). Similarly, chromosomes broken in Drosophila oocytes can remain unrepaired for days in a mu2 mutant female, but are healed immediately after fertilization. Further, healing may be nucleus-specific, as chromosome breaks in the oocyte pronucleus after fertilization are healed, while breaks in the sperm pronucleus are not (24).
Proteins associated with the cap
HP1 and HOAP are major components of the Drosophila chromosome cap and are required for chromosome stability (Table 1), independent of the presence of HTT elements (29,30). HP1 is highly conserved, with three family members in both Drosophila and humans. The original HP1 (HP1a) is discussed here. HP1 may bind DNA directly in the formation of the chromosome cap, unlike its binding activity in repressive chromatin, where it interacts with histone H3 methylated at Lys9 (H3Me3K9) (31). HOAP is a DNA binding protein that resembles sequence-specific high mobility group (HMG) proteins (32,33). Loss of either results in telomere fusions, even when the retrotransposon array is still present at the chromosome end. Other proteins necessary for capping function are also major components of the DNA damage response (11), as has been found in mammals (34). In particular, members of the Mre11/Rad50/Nbs (MRN) complex are required to maintain HOAP at the telomere and prevent telomere fusions (35-39), as are the ATM and ATR kinases (39-43). The ATM and ATR DNA damage response pathways are partially redundant. While loss of ATR does not affect chromosome stability, mutations in both pathways simultaneously results in a greater telomere-fusion mutant phenotype than loss of ATM by itself (40). ATM and MRN, however, appear to work in the same damage response pathway (35). In humans and other eukaryotes telomere protection depends on the length of the terminal array, thus it is difficult to determine whether ATM targets enzymes necessary for elongation of the terminal array or proteins that mediate telomere protection. In flies, however, ATM targets protection, not elongation.
Table 1.
Drosophila Genes involved in telomere stability
| Mutant phenotype | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Protein | HOAP, HP1 mislocalization | Telomere fusions | Increased transposition to telomeres | Increased telomeric gene conversion | Human homologue | Reference |
| Su(var)205 | HP1 | Yes | Yes | Yes | Yes | HP1a | (11,29,64) |
| cav | HOAP | Yes | Yes | -- | (39,40,47) | ||
| tefu | ATM | Yes*† | Yes | ATM | (35,39-43) | ||
| eff | UbcD1 | No | Yes | UBE2D2 | (46) | ||
| mre11 | MRE11 | Yes | Yes | Mre11 | (35,36,39,40) | ||
| rad50 | Rad50 | Yes | Yes | Rad50 | (36-38) | ||
| nbs | Nbs | Yes | Yes | NBS1 | (37,39,40) | ||
| woc | WOC | No | Yes | ZMYM4 | (45) | ||
| mei-41 | ATR | Yes* | Yes* | ATR | (39,40) | ||
| mus304 | ATRIP | Yes* | Yes* | ATRIP | (39,40) | ||
| Irbp | Ku70 | Yes | Yes | Ku70 | (53) | ||
| Ku80 | Ku80 | Yes | Yes | Ku86 | (53) | ||
| aub | AUB | Yes | PIWIL1 | (63) | |||
| spn-E | SPN-E | Yes | TDRD9 | (63) | |||
| E(tc) | E(TC) | No | Yes | -- | (66) | ||
The ATM and ATR pathways are partially redundant. The strongest phenotypes are for double mutants with defects in both.
Mislocalization is observed in polytene chromosomes where telomere fusions had not been found. Mislocalizaiton was not observed in diploid chromosomes.
A few other proteins, including the heterochromatin proteins SUUR, HP2 and SU(VAR)3-7 and the transcription factor WOC (Table 1), have been identified at the extreme chromosome ends by immunocytochemistry (44,45), although their functions at the tips are not known. The first three may play a role in heterochromatin formation (Table 2). In addition to telomeres, where it is required to prevent telomere fusions, WOC is found at interband regions along the chromosome arms, where it colocalizes with the initiating forms of RNA polymerase II. woc mutants exhibit normal accumulations of HP1 and HOAP at telomeres. Likewise, mutations in Su(var)205, cav, tefu and rad50 (Table 1) did not affect WOC localization to telomeres, suggesting that WOC and the HP1/HOAP complex act independently to control telomere stability (45).
Table 2.
Other functions of telomere-associated proteins.
| Protein | Full Name | Proposed function outside of telomeres |
|---|---|---|
| The Cap* | ||
| HP1 | Heterochromatin Protein 1 | Forms heterochromatin and silent euchromatin |
| HOAP | HP1-ORC Associated Protein | None known |
| Required to Prevent Telomere Fusions | ||
| ATM | Ataxia Telangiectasia Mutated | kinase; DNA damage response |
| UbcD1 | Ubiquitin conjugating enzyme D1 | Degradation of inhibitor of apoptosis protein 1 |
| MRE11 | Meiotic Recombination 11 | DNA endonuclease; DNA damage response |
| RAD50 | Radiation Sensitive 50 | DNA exonuclease; DNA damage response |
| NBS | Nijmegen Breakage Syndrome | DNA damage response |
| WOC | Without Children | Transcription factor |
| ATR | Ataxia Telangiectasia Related | kinase; DNA damage response |
| ATRIP | ATR Interacting Protein | DNA helicase; DNA damage response |
| Required to Maintain Telomere Length | ||
| Ku70 | Ku subunit 70 | Nonhomologous end joining |
| Ku80 | Ku subunit 80 | Nonhomologous end joining |
| AUB | Aubergine | RNA interference |
| SPN-E | Spindle-E | RNA helicase; RNA interference |
| E(TC) | Enhancer of telomeric gene conversion | None known |
| TEL | Telomere elongation | None known |
| Terminal Binding | ||
| SUUR | Suppressor of under-replication | Formation of interstitial heterochromatin |
| SU(VAR)3-7 | Suppressor of variegation 3-7 | Heterochromatin formation, condensation of male X |
| Retrotransposon Array | ||
| JIL-1 | JIL-1 | Phosphorylates histone H3 at serine 10; maintains open chromatin |
| PROD | Proliferation disrupter | Heterochromatin formation; chromosome segregation |
| Z4 | Z4 | Separates chromatin compaction domains |
| HP1 | Heterochromatin Protein 1 | Forms heterochromatin and silent euchromatin |
| H3Me3K9 | Histone H3 trimethylated at lysine 9 | Heterochromatin formation |
| H3Me3K4 | Histone H3 trimethylated at lysine 4 | Mark for active chromatin |
| Telomere Associated Sequence | ||
| PC | Polycomb | Cellular memory; transcriptional repressor |
| E(Z) | Enhancer of zeste | histone methylation; gene silencing |
| PH | Polyhomeotic | gene silencing |
| PSC | Posterior sex combs | chromatin remodeling; gene silencing |
| SCM | Sex comb on midleg | transcription factor; gene silencing |
| H3Me3K27 | Histone H3 trimethylated at lysine 27 | Mark for active chromatin |
The cap is defined operationally as preferential binding to the chromosome terminus and requirement for the prevention of telomere fusions.
The gene eff (synonym UbcD1) encodes a highly conserved class I ubiquitin (E2) conjugating enzyme. Mutations in this gene cause high frequencies of telomeric fusions independent of the presence of HTT elements (46,47). It is not known whether the UbcD1 protein causes poly-ubiquitination and degradation of a telomere component or mono-ubiquitination resulting in modified allosteric conformation and activity of its substrate, nor is the substrate known. eff mutations, however, do not appear to affect the telomeric localization of either HP1 or HOAP (11,47).
The capping complex appears to cover an extensive region of DNA. First, when a capped broken chromosome end is placed close to the yellow gene, it affects the ability of the tissue specific enhancers to activate yellow transcription. Specifically, when an enhancer upstream of the yellow promoter is located within 4 kb of the chromosome end the enhancer function is inhibited (17). Second, P transposable elements were unable to transpose when they were within 5 kb of the chromosome end (48). Third, transvection, the trans-activation of a promoter by somatic pairing between the gene in question and enhancer elements of the homologous gene, requires at least 6 kb of sequence upstream of the promoter in order to be effective (49); extensive homology, however, was not required. Thus, several chromosomal functions are inhibited when they are too close to a chromosome end.
The retrotransposon array and chromosome elongation
The three retrotransposons at Drosophila telomeres are unusual in a number of respects that may reflect their chromosomal position (50). All three elements have very long 3′ untranslated regions (UTRs) that encompass half the element. The sequence of the these UTRs, as well as the coding sequence, exhibit a strand bias in nucleotide content that resembles that seen in telomerase-generated telomere sequences in other organisms (51) and have the propensity to form G-quadruplex structures (52), suggesting strand-specific constraints on nucleotide content independent of any telomerase RNA template. HeT-A elements also lack a reverse transcriptase gene needed for their transposition.
Drosophilids elongate their telomeres by a combination of targeted transposition of HTT elements and homologous recombination between these elements (1,53,54). A transposition mechanism has been proposed (1,54-57) based on that of other non-LTR retroelements, with the difference that HTT elements would not need a DNA cut for transposition. They instead could use the free 3′ terminus at the chromosome end to prime reverse transcription. Although the details of this process remain unresolved, this transposition mechanism (Fig. 2) predicts that the HTT elements should form a telomeric head-to-tail arrangement, with their oligo(A) tails always facing towards the centromere. The stochastic nature of transposition coupled with the end replication problem predicts that individual elements in the array will be variably 5′ truncated. Both of these predictions are observed (58-60). An important prerequisite in this telomere elongation mechanism is the generation of transcripts from the HTT elements in spite of 5′ truncations. HeT-A elements contain promoter activity within the last 600 bp of their 3′ UTRs, which is directed through their oligo(A) tails (16,61) and has the ability to transcribe downstream elements in the telomeric array (62).
Figure 2.
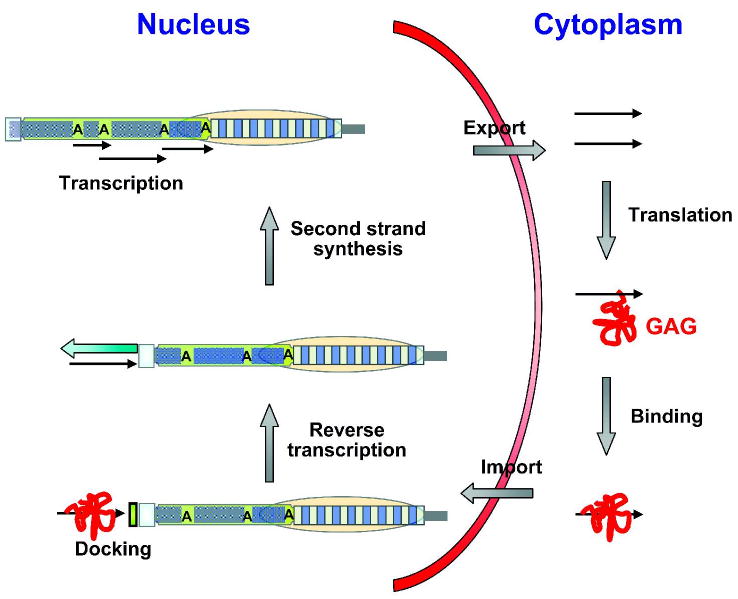
Schematic of the nuclear and cytoplasmic events in Drosophila telomere elongation by retrotransposition. Transcripts are generated from a telomeric retrotransposon using promoter activity located in the 3′ UTR of an upstream HeT-A element (black arrows). The transcripts leave the nucleus to serve as mRNA for the translation of the element-encoded GAG-like protein (red squiggle), which contains three nuclear localization signals. GAG binds the RNA and facilitates re-entry into the nucleus. After docking to a chromosome end, perhaps mediated by a protein-protein interaction between the GAG-protein and the terminal-capping complex, a reverse transcriptase uses the free 3′ hydroxyl group at the chromosome end as primer to copy the RNA intermediate into the first DNA strand. Second strand synthesis occurs by DNA repair and completes the addition of a new HeT-A retrotransposon. It is conceivable that HTT transcripts that have not left the nucleus may also be used a templates for reverse transcription at the chromosome ends. However, sequence analyses of very recently transposed HeT-A elements (15) and of several in native telomeric arrays (58) suggest that there is a selection for the incorporation of elements with a functional GAG open reading frame that have previously been translated in the cytoplasm. Such a positive selection would prevent a gradual inactivation of telomeric transposons due to the accumulation of mutations. Other symbols are as in Fig. 1.
The structure of the Drosophila telomere raises the question of how the transcriptional activity and targeting of the telomeric retrotransposons is controlled. It appears important to regulate the access of transposition intermediates to the chromosome end, which may be an important mechanism for telomere length control, similar to the telomere-telomerase interaction in other eukaryotes (6). HTT transcriptional activity may also play an important role in length maintenance, as both HTT transcript abundance and attachment to chromosome ends may be controlled by an RNAi-based mechanism in the germ line (63). HP1 also plays a critical role in limiting telomere elongation. Heterozygous mutations in the Su(var)205 gene, which encodes HP1, cause an increase in both targeted retrotransposition of HTT elements and telomeric gene conversion (64). Interestingly, the Su(var)20502 mutation, which disrupts the chromodomain (65), does not affect HP1 localization to telomeres or telomere fusions when homozygous (29), although it does affect HeT-A transcript levels (31).
Very few other genes have been tested for their effects on telomere elongation in Drosophila. Of those that have, the Ku70/Ku80 complex, a component of NHEJ, and the E(TC) protein limit chromosome elongation via gene conversion (53,66). PROD binds just upstream of the HeT-A promoter, and prod mutants exhibit increased HeT-A transcript levels, suggesting that PROD acts as a repressor of HeT-A transcriptional activity (67). However, given that prod mutants do not develop long HTT arrays, it appears that increased transcript levels are not sufficient to drive increased transposition of these elements.
Telomere growth does not continue indefinitely in stocks heterozygous for these mutants; rather HTT array length reaches a plateau (68), suggesting a mechanism for limiting telomere length. One candidate, telomere shortening, may occur in a mechanism resembling sporadic shortening described in yeast (69) and seen in some Drosophila stocks, especially those with P elements located in telomere regions within about 50 kb of the chromosome end (18-20). Alternatively, exceptionally long terminal arrays correlate with reduced fertility and fecundity in individual females, which is reflected in abnormal oocyte development (70), suggesting selection against long HTT arrays.
Insertions of other transposons into the HTT array are rare, and only one incident of a roo insertion into a HeT-A at the 3L telomere has been reported (71). The 4R telomere may be an exception in that it lacks TAS and, instead, has a 5.4 kb “transition zone” that consists of a scramble of HTT elements and other non-telomeric transposons between the uninterrupted HTT array and the last gene (58,60). These observations are not surprising given the rapid turnover of telomeric sequences near the chromosome terminus. P elements can also integrate into the HTT array (71). It is possible to directly compare the frequency of insertions into the HTT and TAS regions using the results of the Drosophila Gene Disruption Project in which >30,000 P element insertions were generated. After accounting for the different array lengths and the fact that many of the insertions noted as being in either HTT or TAS were not in fact telomeric, we calculate that there may have been roughly 8 fold more frequent P element insertions into TAS than HTT. It has been noted, however, that TAS is a strong hot spot for P element insertions (72-74). Thus, the HTT array does not appear to be refractory to transposition by other elements.
Proteins associated with the HTT array
To begin to unravel the mechanisms that regulate the transcriptional activity of the telomeric retrotransposons, it is important to understand the structure of the telomeric chromatin. Proteins associated with the HTT array, however, are difficult to identify, because no genetic screen can be performed for HTT-binding proteins owing to the absence of a predictable mutant phenotype. Therefore, immunohistological staining for potential candidate proteins is one of the few options, which so far has identified three proteins at the HTT array: Z4, JIL-1 and PROD (44,67). All three proteins are also associated with a multitude of other sites in the Drosophila genome, suggesting general and widespread functions (Table 2). These studies used extended HTT arrays generated by the Tel mutant that are easily distinguishable from the cap region. It is possible that these long HTT arrays have a chromatin structure that differs from shorter arrays.
The amino-termini of the four core histones can harbor a variety of post-translational modifications, and such modified histones can serve as epigenetic marks in chromatin. For example, histone H3 trimethylated at lysine 4 (H3Me3K4) is associated with transcriptionally active chromatin, while H3Me3K9 and H3Me3K27 are markers for inactive chromatin, and acetylation of several lysine residues of histones H3 and H4 serves to mark actively transcribed regions. Histone modifications in the pericentric heterochromatin of Drosophila and their involvement in gene silencing are well characterized (75), but analyses of the telomeric regions are just beginning. Andreyeva et al. (44) have localized some characteristic histone modifications at the telomeres of polytene chromosomes by immunohistochemistry and found H3Me3K9 at HTT but not at TAS, while H3Me3K27 was absent from HTT but present at TAS and at the cap. The acetylated histone, H4AcK12, which is often associated with heterochromatin, was not found at telomeres. However, H3Me3K4, a euchromatin mark was present in the HTT array. This limited set of modified histones indicates that the HTT array carries both heterochromatic and euchromatic marks similar to the kinetochore region (76). Even though H3Me3K9, which usually binds the heterochromatin protein HP1, is found at the HTT region, HP1 was not seen by immunocytochemistry. Recent chromatin immunoprecipitation experiments, however, show HP1 distributed along the HTT array outside the cap, and disruption of HP1 function increases HeT-A transcription and transcription of reporter genes at several sites in the HTT array (R Capkova Frydrychova, TK Archer and JM Mason, unpublished). Thus, HP1 may play different roles at the cap and along the HTT array outside the cap.
While these studies point to a unique pattern of histone modifications and chromosomal proteins in the chromatin of the HTT array, genetic analysis has clearly shown that white reporter genes inserted into the HTT array exhibit characteristics of euchromatin (71). Many more histone modifications need to be tested and the proteins interacting with these epigenetic imprints need to be identified to gain a clear picture of HTT chromatin. An interesting candidate is the phosphorylated histone H3PhS10, which is generated by the JIL-1 kinase and is influenced by several signaling cascades (77).
Telomere associated repeat sequences
Telomeres in Drosophila and in other organisms have been considered heterochromatic for at least two reasons: they contain repetitive DNA sequences and have the ability to repress the activity of genes inserted into telomeric regions (50). The repressed and variegated expression of telomeric transgenes is phenotypically similar to position effect variegation (PEV), the clonal inactivation of a euchromatic gene positioned close to or within pericentric heterochromatin (78,79).
Detailed genetic analysis of telomeric transgenes revealed that the HTT array is not responsible for telomeric silencing (71). Instead, the source of TPE was localized to TAS, which is a region adjacent to the HTT array (Fig. 1). The TAS region consists of several kilobases of complex satellite sequences, which varies among telomeres, although there are sequence similarities. The repeat unit is 460 bp on the 2L telomere (80), 1 kb on 3R, and two different repeats (400 and 800 bp) are interspersed on XR (72).
Transgenic experiments with P-element constructs bearing a 2L TAS array adjacent to a mini-white reporter gene revealed that TAS silencing is array-length and orientation dependent (81). Similarly, a 1.2 kb fragment from the X-TAS is also able to silence an adjacent transgenic mini-white gene (82). Silencing is inversely related to distance from TAS based on expression of transgenes located in the HTT array (Fig. 3). white reporter genes inserted into the HTT array more than 5-10 kb from TAS did not exhibit repression or variegation, but inserts located closer to TAS were repressed and showed variegated expression (Fig. 3). Spreading of silencing varied, possibly depending on the structure of the transposable element, the telomere affected, or other factors in the genetic background (71). This supports the notion that telomeric silencing is mediated by TAS rather than by the HTT domain and that distance from TAS is a factor that determines the degree of repression.
Figure 3.
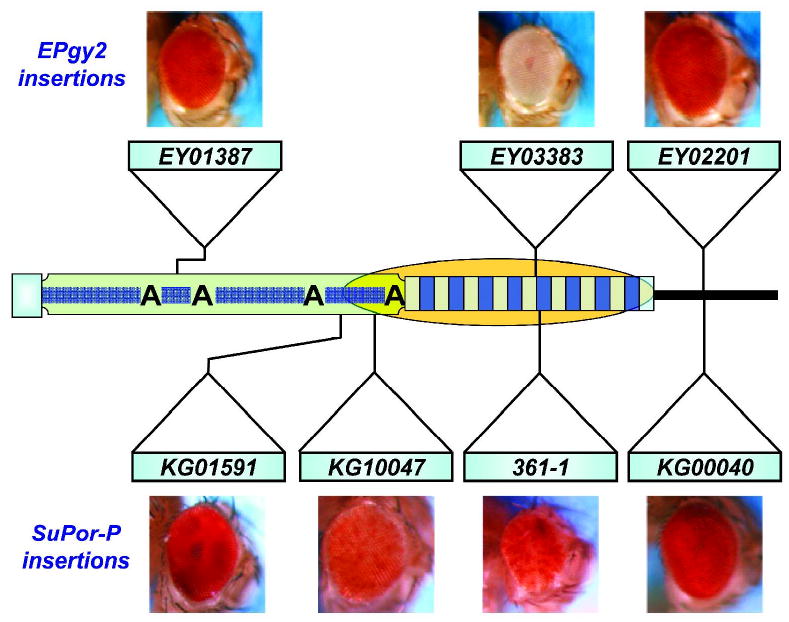
Cis-acting silencing at the telomeres of Drosophila as deduced from expression levels of P element insertions carrying a white (w) reporter gene in various telomeric domains. Silencing effects (orange oval) emerge from TAS and spread a short distance into the HTT array. Two different insertion constructs were used. The EPgy2 element lacks the eye-specific enhancer and expresses w at a relatively low level; the SuPor-P element carries the eye-specific enhancer and expresses w more strongly. Insertions in TAS and in HTT close to TAS are repressed, while those in HTT far from TAS are not. Other symbols are as in Fig. 1.
Screens of eye color phenotypes in experiments with P{wvar}, a white eye color reporter transgene precisely inserted between the terminal retrotransposon array and TAS of the 2L telomere (20,83), revealed TAS-mediated silencing in trans on telomeric transgenes located in both homologous and non-homologous telomeres (20,62,83). Flies with a heterozygous 2L TAS deficiency showed a significant increase in the level of eye pigment in comparison to their siblings bearing a wild type 2L telomere (Fig. 4). PCR analyses showed that transcription of the P{wvar} transgene can be initiated from a promoter of an upstream HeT-A element, and that the level of this HeT-A{P{wvar} read-through transcript is elevated in the presence of a 2L TAS deficiency (62).
Figure 4.
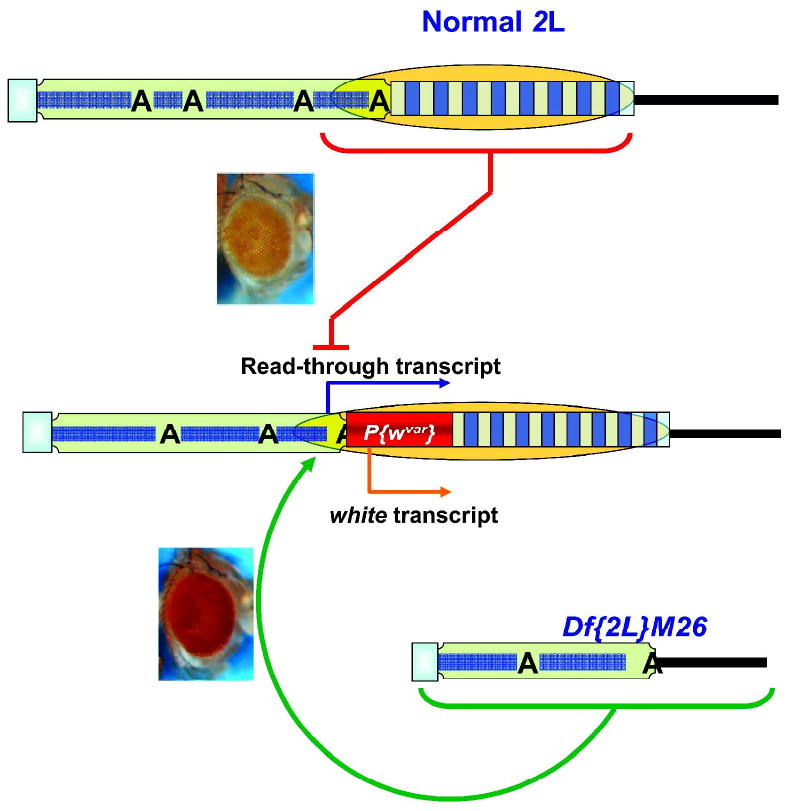
Trans-effect of TAS deletions on the expression of telomeric transgenes. w transgene expression at the 2L telomere occurs primarily from the w promoter (orange bent arrow) with a small contribution from the adjacent HeT-A promoter (blue bent arrow). This expression is repressed by the adjacent TAS in cis (orange oval) in the presence of a full length TAS array (red line). Deletion of 2L TAS in trans (Df(2L)M26) causes higher expression (green round arrow) of the w transgene. Others symbols as in Fig. 1
A large number of dominant mutations that modify PEV have been characterized (78). These mutations are frequently found in genes encoding chromosomal proteins or enzymes that modify histones (84). In spite of phenotypic similarities, however, TPE appears to be qualitatively different from PEV. Suppressors and enhancers of PEV do not modify TPE, which suggests different sets of chromosomal proteins as limiting factors in these two types of silencing (74,82,85). As part of a systematic search for trans-acting TPE modifiers, Mason et al. (85) screened a collection of stocks containing autosomal deficiencies covering 77% of the major autosomes for dominant suppressors of silencing of a telomeric transgene. A large portion of stocks that showed TPE suppression were identified as carrying a 2L TAS deficiency, confirming that 2L TAS plays a significant role in the suppression of telomeric silencing towards both homologous and non-homologous telomeres. Although the screen potentially identified 18 sites of TPE suppressors, these sites did not correspond to the positions of genes known to control chromatin structure, with the exception of one site corresponding to the location of gpp, which was identified as a dominant suppressor of TPE by Schedl's group (86). gpp is the Drosophila homologue of the yeast DOT1 gene, encoding a histone methyltransferase responsible for methylation of Lys 79 of histone H3. There are several possibilities that may explain the low number of potential sites that have been revealed by the deficiency screen, such as the localization of suppressors of TPE primarily on the X or fourth chromosome, the possible recessive character of TPE suppressors (in contrast to dominant PEV modifiers), or the simple chromatin structure in the TAS containing relatively few components (85). In any case, despite intensive efforts in a number of labs, the genetic factors playing a role in telomeric silencing remain unidentified.
PcG proteins and their role in telomeric silencing
Two Polycomb-group (PcG) repressors, PC and E(Z), have been immunolocalized to TAS of all telomeres, except 2R (44), where these proteins were not detected despite the presence of TAS. Consistent with the methyltransferase activity of E(Z) toward histone H3 at Lys 27, H3Me3K27 was also enriched at TAS. While genetic data on silencing of transgenes inserted into TAS support the idea that TAS arrays form heterochromatic domains (71), immunolocalization on polytene chromosomes (44) revealed that TAS regions lack two well-established marks of heterochromatin: HP1 and H3Me3K9. Both proteins are believed to play a crucial role in heterochromatin formation. Their absence in TAS supports the view that TPE and PEV are under different genetic control.
Three similarities, however, exist for transgenes inserted into TAS and near a Polycomb response element (PRE), which is a region that binds PcG proteins. First, both exhibit variegated expression. Second, long-range interactions may occur. Capkova Frydrychova et al. (62) observed genetic interactions between nonhomologous telomeres, while Vazquez et al. (87) reported that the PcG gene mcp can mediate physical interactions between remote chromosomal sites. Transgenes inserted into TAS can silence other genes with the same sequence at many other sites in the genome (88) possibly using an RNAi-based mechanism (89). Third, pairing sensitivity, defined as the lower expression of a flanking reporter gene in a homozygous state than in a hemizygous one, has been observed at telomeres (82) and at PRE-containing transgenes (90). Although these parallels may suggest that PcG proteins are significant players in Drosophila TPE, the deficiency screen for dominant modifiers of TPE (85) did not identify deficiencies of PcG genes as dominant TPE suppressors. Thus, the exact role of these proteins in telomeric silencing remains to be elucidated.
Evolution of Drosophila telomeres
How might telomeres with retrotransposons at the terminus have arisen? One might imagine a simple sequence of events, each with a reasonable probability of occurrence (91). The first departure from a canonical telomere composed of simple repeat sequences generated by telomerase might have been the loss of telomerase. This has happened numerous times during evolution in species from the dipteran insects, including Drosophila, to plants, such as onion (92). There are several examples, from arthropods in general and insects in particular, of independent lost telomerase (93,94).
After the loss of telomerase, the simple telomeric repeat sequence might still be maintained by recombination between these repeats. When human cancer cells become immortalized, for example, the majority of them have reactivated telomerase to maintain chromosome length, but a number of immortalized cancer lines have not. Instead, they use recombination as an alternative mechanism for maintaining chromosome length (95). Some yeast cells that survive after having lost telomerase also show recombination between simple telomeric repeats (96).
Simple telomeric repeats are required at chromosome ends as long as the cap binds to the terminus in a sequence-dependent manner. Loss of sequence specificity would allow the complex repeats of TAS to become positioned at the chromosome ends. Species found to have lost telomerase have also lost simple telomeric repeats. Drosophila also appears to have lost a TRF2-like protein that would stabilize a t-loop. Chironomus telomeres lack the long 3′ overhangs needed for t-loop formation (97). Recombination between these complex repeat sequences has been proposed as a mechanism for chromosome length maintenance in these species. It should be noted that ‘lower’ dipterans, such as the midge Chironomus and the mosquito Anopheles, which may resemble a Drosophila ancestor, fall into this category (55,98,99).
This pattern of chromosome length maintenance by recombination may be augmented by the occasional insertion of transposable elements. In the silkworm, Bombyx mori, two retrotransposons target telomere regions, but insert into the terminal array rather than attach to the chromosome tip (100,101). In addition, mammalian LINE elements have the capability of endonuclease-independent transposition and can attach themselves directly to chromosome ends with disrupted telomeres (102).
Targeting of retrotransposable elements might be achieved by two mechanisms. Loss of the endonuclease gene, such as is seen in Drosophila HeT-A elements, would prevent the transposons from inserting into interstitial regions and allow attachment to the chromosome end, which resembles a permanent DSB. This mechanism would require that the proteins translated from a specific RNA intermediate preferentially bind to that RNA and return with it to the nucleus (Fig. 2). This has been seen for mammalian LINE transposons (103), and there is evidence for this type of mechanism in Drosophila HeT-A transposition (104). A second targeting mechanism may be the homing of the GAG protein from these elements to the chromosome end (105). The HTT arrays found in Drosophila chromosomes could have arisen from canonical telomeres by a series of plausible steps. In fact many of the intermediates proposed here have counterparts in the telomeres of non-Drosophila species. The ‘unusual’ telomeres of Drosophila may represent merely one end of a continuum of possible telomere structures (91), but they provide valuable information on telomere function. In eukaryotes with canonical telomeres, end protection and chromosome elongation both depend on the simple repeats generated by telomerase. Drosophila telomeres show that the two vital functions of telomeres, capping and length maintenance, can be separated.
Conclusions
Drosophila telomeres are functionally equivalent to those from eukaryotes that use telomerase to extend their chromosome ends. Ultimately, it is function that defines the telomere. Natural chromosome ends in Drosophila are required for chromosome stability, as they are in other species, while DSBs induce cell cycle delay and are subject to DNA repair. Drosophila telomeres maintain chromosome length and have several other activities that are less well understood. They form heterochromatin and silence neighboring genes, as do telomeres in yeast and humans. While physical associations among Drosophila telomeres have not been found, other than those that arise from anaphase movements, telomere-telomere communication has been noted in the regulation of TPE.
While at first glance telomeres in Drosophila appear to be physically very different from those in other species, the differences are not as stark as they appear. In yeast, mammals and Drosophila, most of the proteins necessary for telomere stability, including ATM and the MRN complex, perform double duty in the DNA damage response pathways. The terminal DNA array in Drosophila is very different from the canonical telomeric DNA sequence, but numerous other plant and animal species also lack telomerase and the simple telomerase-generated repeats. The complex and variable TAS arrays are not unique to Drosophila. Although the complete chromatin structure of the Drosophila telomere remains to be determined, it appears to be a hybrid of euchromatic and heterochromatic domains. Much needs to be learned, but the small overall size of the Drosophila telomere makes it an excellent region to study complex epigenetic interactions.
Acknowledgments
Thanks are due to Elena Casacuberta, Mike Resnick and Greg Stuart for critical reviews of the manuscript
Funding Agencies: This work was supported by U.S. Public Health Services grant GM-56729 and by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences
Abbreviations
- DSB
double strand DNA break
- HTT
telomere-specific HeT-A, TART and TAHRE retrotransposons
- LTR
long terminal repeat
- MRN
MRE11/RAD50/NBS protein complex
- NHEJ
nonhomologous DNA end joining
- PRE
Polycomb response element
- PcG
Polycomb group
- PEV
position effect variegation
- RNAi
RNA interference
- TAS
telomere associated DNA sequence
- TPE
telomeric position effect
- UTR
untranslated region
References
- 1.Mason JM, Biessmann H. The unusual telomeres of Drosophila. Trends Genet. 1995;11:58–62. doi: 10.1016/s0168-9525(00)88998-2. [DOI] [PubMed] [Google Scholar]
- 2.Louis EJ, Vershinin AV. Chromosome ends: different sequences may provide conserved functions. Bioessays. 2005;27:685–697. doi: 10.1002/bies.20259. [DOI] [PubMed] [Google Scholar]
- 3.Muller HJ. The remaking of chromosomes. The Collecting Net. 1938;8:182–195. [Google Scholar]
- 4.Watson JD. Origin of concatameric T7. DNA Nature New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 5.Olovnikov AM. A theory of marginotomy. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 6.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 7.Bass HW. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol Life Sci. 2003;60:2319–2324. doi: 10.1007/s00018-003-3312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding X, Xu R, Yu J, Xu T, Zhuang Y, et al. SUN1 Is Required for Telomere Attachment to Nuclear Envelope and Gametogenesis in Mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 10.Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Curr Opin Genet Dev. 1997;7:822–828. doi: 10.1016/s0959-437x(97)80046-9. [DOI] [PubMed] [Google Scholar]
- 11.Cenci G, Ciapponi L, Gatti M. The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma. 2005;114:135–145. doi: 10.1007/s00412-005-0005-9. [DOI] [PubMed] [Google Scholar]
- 12.Muller HJ, Herskowitz IH. Concerning the healing of chromosome ends produced by breakage in Drosophila melanogaster. Am Nat. 1954;88:177–208. [Google Scholar]
- 13.McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biessmann H, Champion LE, O'Hair M, Ikenaga K, Kasravi B, et al. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahn T, Savitsky M, Georgiev P. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol Cell Biol. 2000;20:7634–7642. doi: 10.1128/mcb.20.20.7634-7642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikhailovsky S, Belenkaya T, Georgiev P. Broken chromosomal ends can be elongated by conversion in Drosophila melanogaster. Chromosoma. 1999;108:114–120. doi: 10.1007/s004120050358. [DOI] [PubMed] [Google Scholar]
- 18.Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levis RW. Viable deletions of a telomere from a Drosophila chromosome. Cell. 1989;58:791–801. doi: 10.1016/0092-8674(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 20.Mason JM, Konev AY, Golubovsky MD, Biessmann H. Cis- and trans-acting influences on telomeric position effect in Drosophila melanogaster detected with a subterminal transgene. Genetics. 2003;163:917–930. doi: 10.1093/genetics/163.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad K, Golic KG. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics. 1998;148:775–792. doi: 10.1093/genetics/148.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad K, Golic KG. Telomere loss in somatic cells of Drosophila causes cell cycle arrest and apoptosis. Genetics. 1999;151:1041–1051. doi: 10.1093/genetics/151.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason JM, Strobel E, Green MM. mu-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc Natl Acad Sci USA. 1984;81:6090–6094. doi: 10.1073/pnas.81.19.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason JM, Champion LE, Hook G. Germ-line effects of a mutator, mu2, in Drosophila melanogaster. Genetics. 1997;146:1381–1397. doi: 10.1093/genetics/146.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biessmann H, Mason JM. Progressive loss of DNA sequences from terminal chromosome deficiencies in Drosophila melanogaster. EMBO J. 1988;7:1081–1086. doi: 10.1002/j.1460-2075.1988.tb02916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traverse KL, Pardue ML. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres. Proc Natl Acad Sci USA. 1988;85:8116–8120. doi: 10.1073/pnas.85.21.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biessmann H, Carter SB, Mason JM. Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci USA. 1990;87:1758–1761. doi: 10.1073/pnas.87.5.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 29.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The Heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 30.Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol. 2003;5:82–84. doi: 10.1038/ncb902. [DOI] [PubMed] [Google Scholar]
- 31.Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, et al. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 32.Badugu R, Shareef MM, Kellum R. Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J Biol Chem. 2003;278:34491–34498. doi: 10.1074/jbc.M305262200. [DOI] [PubMed] [Google Scholar]
- 33.Shareef MM, King C, Damaj M, Badagu R, Huang DW, et al. Drosophila heterochromatin protein 1 (HPI)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12:1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slijepcevic P. The role of DNA damage response proteins at telomeres- an “integrative” model. DNA Repair. 2006;5:1299–1306. doi: 10.1016/j.dnarep.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 35.Bi XL, Wei SCD, Rong YKS. Telomere protection without a telomerase: The role of ATM and Mre11 in Drosophila telomere maintenance. Curr Biol. 2004;14:1348–1353. doi: 10.1016/j.cub.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 36.Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, et al. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol. 2004;14:1360–1366. doi: 10.1016/j.cub.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Ciapponi L, Cenci G, Gatti M. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics. 2006;173:1447–1454. doi: 10.1534/genetics.106.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorski MM, Romeijn RJ, Eeken JCJ, de Jong AWM, van Veen BL, et al. Disruption of Drosophila Rad50 causes pupal lethality, the accumulation of DNA double-strand breaks and the induction of apoptosis in third instar larvae. DNA Repair. 2004;3:603–615. doi: 10.1016/j.dnarep.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Oikemus SR, Queiroz-Machado J, Lai K, McGinnis N, Sunkel C, et al. Epigenetic telomere protection by Drosophila DNA damage response pathways. Plos Genetics. 2006;2:693–706. doi: 10.1371/journal.pgen.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi XL, Srikanta D, Fanti L, Pimpinelli S, Badugu R, et al. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2005;102:15167–15172. doi: 10.1073/pnas.0504981102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oikemus SR, McGinnis N, Queiroz-Machado J, Tukachinsky H, Takada S, et al. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Gene Dev. 2004;18:1850–1861. doi: 10.1101/gad.1202504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva E, Tiong S, Pedersen M, Homola E, Royou A, et al. ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr Biol. 2004;14:1341–1347. doi: 10.1016/j.cub.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 43.Song YH, Mirey G, Betson M, Haber DA, Settleman J. The Drosophila ATM ortholog, dATM, mediates the response to ionizing radiation and to spontaneous DNA damage during development. Curr Biol. 2004;14:1354–1359. doi: 10.1016/j.cub.2004.06.064. [DOI] [PubMed] [Google Scholar]
- 44.Andreyeva EN, Belyaeva ES, Semeshin VF, Polkholkova GV, Zhimulev IF. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J Cell Sci. 2005;118:5465–5477. doi: 10.1242/jcs.02654. [DOI] [PubMed] [Google Scholar]
- 45.Raffa GD, Cenci G, Siriaco G, Goldberg ML, Gatti M. The putative Drosophila transcription factor Woc is required to prevent telomeric fusions. Mol Cell. 2005;20:821–831. doi: 10.1016/j.molcel.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, et al. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Gene Dev. 1997;11:863–875. doi: 10.1101/gad.11.7.863. [DOI] [PubMed] [Google Scholar]
- 47.Cenci G, Siriaco G, Gatti M. The role of HeT-A and TART retrotransposons in Drosophila telomere capping. Genetica. 2003;117:311–318. doi: 10.1023/a:1022972902263. [DOI] [PubMed] [Google Scholar]
- 48.Melnikova L, Biessmann H, Georgiev P. The vicinity of a broken chromosome end affects P element mobilization in Drosophila melanogaster. Mol Genet Genomics. 2004;272:512–518. doi: 10.1007/s00438-004-1072-y. [DOI] [PubMed] [Google Scholar]
- 49.Savitsky M, Kahn T, Pomerantseva E, Georgiev P. Transvection at the end of the truncated chromosome in Drosophila melanogaster. Genetics. 2003;163:1375–1387. doi: 10.1093/genetics/163.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biessmann H, Prasad S, Walter MF, Mason JM. Euchromatic and heterochromatic domains at Drosophila telomeres. Biochem Cell Biol. 2005;83:477–485. doi: 10.1139/o05-053. [DOI] [PubMed] [Google Scholar]
- 51.Danilevskaya ON, Lowenhaupt K, Pardue ML. Conserved subfamilies of the Drosophila HeT-A telomere- specific retrotransposon. Genetics. 1998;148:233–242. doi: 10.1093/genetics/148.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abad JP, Villasante A. The 3′ non-coding region of the Drosophila melanogaster HeT-A telomeric retrotransposon contains sequences with propensity to form G-quadruplex. DNA FEBS Lett. 1999;453:59–62. doi: 10.1016/s0014-5793(99)00695-x. [DOI] [PubMed] [Google Scholar]
- 53.Melnikova L, Biessmann H, Georgiev P. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics. 2005;170:221–235. doi: 10.1534/genetics.104.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Ann Rev Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 55.Biessmann H, Mason JM. Telomerase-independent mechanisms of telomere elongation. Cell Mol Life Sci. 2003;60:2325–2333. doi: 10.1007/s00018-003-3247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 57.Pardue ML, Rashkova S, Casacuberta E, DeBaryshe PG, George JA, et al. Two retrotransposons maintain telomeres. in Drosophila Chromosome Res. 2005;13:443–453. doi: 10.1007/s10577-005-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abad JP, de Pablos B, Osoegawa K, de Jong PJ, Martin-Gallardo A, et al. Genomic analysis of Drosophila melanogaster telomeres: Full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol. 2004;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
- 59.Biessmann H, Kasravi B, Jakes K, Bui T, Ikenaga K, et al. The genomic organization of HeT-A retroposons in Drosophila melanogaster. Chromosoma. 1993;102:297–305. doi: 10.1007/BF00661272. [DOI] [PubMed] [Google Scholar]
- 60.George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 62.Capkova Frydrychova R, Biessmann H, Konev AY, Golubovsky MD, Johnson J, et al. Transcriptional activity of the telomeric retrotransposon HeT-A in Drosophila melanogaster is stimulated as a consequence of subterminal deficiencies at homologous and nonhomologous telomeres. Mol Cell Biol. 2007;27:4991–5001. doi: 10.1128/MCB.00515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Gene Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Platero JS, Hartnett T, Eissenberg JC. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melnikova L, Georgiev P. Enhancer of terminal gene conversion, a new mutation in Drosophila melanogaster that induces telomere elongation by gene conversion. Genetics. 2002;162:1301–1312. doi: 10.1093/genetics/162.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Török T, Benitez C, Takacs S, Biessmann H. The protein encoded by the gene proliferation disrupter (prod) is associated with the telomeric retrotransposon array in Drosophila melanogaster. Chromosoma. 2007;116:185–195. doi: 10.1007/s00412-006-0090-4. [DOI] [PubMed] [Google Scholar]
- 68.Siriaco GM, Cenci G, Haoudi A, Champion LE, Zhou C, et al. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics. 2002;160:235–245. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li BB, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Gene Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 70.Walter MF, Biessmann MR, Benitez C, Torok T, Mason JM, et al. Effects of telomere length in Drosophila melanogaster on life span, fecundity, and fertility. Chromosoma. 2007;116:41–51. doi: 10.1007/s00412-006-0081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Biessmann H, Prasad S, Semeshin VE, Andreyeva EN, Nguyen Q, et al. Two distinct domains in Drosophila melanogaster telomeres. Genetics. 2005;171:1767–1777. doi: 10.1534/genetics.105.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by Single-P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roseman RR, Johnson EA, Rodesch CK, Bjerke M, Nagoshi RN, et al. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics. 1995;141:1061–1074. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallrath LL, Elgin SCR. Position effect variegation in Drosophila is associated with an altered chromatin structure. Gene Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 75.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 76.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 78.Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 79.Schulze SR, Wallrath LL. Gene regulation by chromatin structure: Paradigms established in Drosophila melanogaster. Annu Rev Entomol. 2007;52:171–192. doi: 10.1146/annurev.ento.51.110104.151007. [DOI] [PubMed] [Google Scholar]
- 80.Walter MF, Jang C, Kasravi B, Donath J, Mechler BM, et al. DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma. 1995;104:229–241. doi: 10.1007/BF00352254. [DOI] [PubMed] [Google Scholar]
- 81.Kurenova E, Champion L, Biessmann H, Mason JM. Directional gene silencing induced by a complex subtelomeric satellite from Drosophila. Chromosoma. 1998;107:311–320. doi: 10.1007/s004120050313. [DOI] [PubMed] [Google Scholar]
- 82.Boivin A, Gally C, Netter S, Anxolabehere D, Ronsseray S. Telomeric associated sequences of Drosophila recruit Polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics. 2003;164:195–208. doi: 10.1093/genetics/164.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Golubovsky MD, Konev AY, Walter MF, Biessmann H, Mason JM. Terminal retrotransposons activate a subtelomeric white transgene at the 2L telomere in Drosophila. Genetics. 2001;158:1111–1123. doi: 10.1093/genetics/158.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wallrath LL. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 85.Mason JM, Ransom J, Konev AY. A deficiency screen for dominant suppressors of telomeric silencing in Drosophila. Genetics. 2004;168:1353–1370. doi: 10.1534/genetics.104.030676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, et al. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics. 2005;169:173–184. doi: 10.1534/genetics.104.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vazquez J, Muller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marin L, Lehmann M, Nouaud D, Hassan I, Anxolabéhère D, et al. P-element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics. 2000;155:1841–1854. doi: 10.1093/genetics/155.4.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reiss D, Josse T, Anxolabehere D, Ronsseray S. aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol Genet Genomics. 2004;272:336–343. doi: 10.1007/s00438-004-1061-1. [DOI] [PubMed] [Google Scholar]
- 90.Kassis JA. Pairing-sensitive silencing, Polycomb group response elements, and transposon homing in Drosophila. Homology Effects. 2002;46:421–438. doi: 10.1016/s0065-2660(02)46015-4. [DOI] [PubMed] [Google Scholar]
- 91.Louis E. Are Drosophila telomeres an exception or the rule? Genome Biol. 2002;3:reviews0007.1–0007.6. doi: 10.1186/gb-2002-3-10-reviews0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pich U, Schubert I. Terminal heterochromatin and alternative telomeric sequences in Allium cepa. Chromosome Res. 1998;6:315–321. doi: 10.1023/a:1009227009121. [DOI] [PubMed] [Google Scholar]
- 93.Sahara K, Marec F, Traut W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999;7:449–460. doi: 10.1023/a:1009297729547. [DOI] [PubMed] [Google Scholar]
- 94.Frydrychova R, Grossmann P, Trubac P, Vitkova M, Marec FE. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome. 2004;47:163–178. doi: 10.1139/g03-100. [DOI] [PubMed] [Google Scholar]
- 95.Stewart SA, Weinberg RA. Telomeres: Cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–557. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 96.Lundblad V. Telomere maintenance without telomerase. Oncogene. 2002;21:522–531. doi: 10.1038/sj.onc.1205079. [DOI] [PubMed] [Google Scholar]
- 97.Rosen M, Edstrom JE. Chromosome ends in Chironomus tentans do not have long single-stranded overhangs characterizing canonical telomeres. Chromosome Res. 2002;10:21–31. doi: 10.1023/a:1014257808705. [DOI] [PubMed] [Google Scholar]
- 98.Lopez CC, Kamnert I, Scherbik SV, Edstrom JE. Interspersed DNA element restricted to a specific group of telomeres in the dipteran Chironomus pallidivittatus. Gene. 1999;233:249–259. doi: 10.1016/s0378-1119(99)00129-8. [DOI] [PubMed] [Google Scholar]
- 99.Biessmann H, Donath J, Walter MF. Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol Biol. 1996;5:11–20. doi: 10.1111/j.1365-2583.1996.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 100.Takahashi H, Okazaki S, Fujiwara H. A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori. Nucleic Acids Res. 1997;25:1578–1584. doi: 10.1093/nar/25.8.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fujiwara H, Osanai M, Matsumoto T, Kojima KK. Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori. Chromosome Res. 2005;13:455–467. doi: 10.1007/s10577-005-0990-9. [DOI] [PubMed] [Google Scholar]
- 102.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 103.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–27. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 104.Biessmann H, Kasravi B, Bui T, Fujiwara G, Champion LE, et al. Comparison of two active HeT-A retroposons of Drosophila melanogaster. Chromosoma. 1994;103:90–98. doi: 10.1007/BF00352317. [DOI] [PubMed] [Google Scholar]
- 105.Rashkova S, Karam SE, Kellum R, Pardue ML. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J Cell Biol. 2002;159:397–402. doi: 10.1083/jcb.200205039. [DOI] [PMC free article] [PubMed] [Google Scholar]


