Abstract
We reported here a novel, ready-to-use bioarray platform and methodology for construction of sensitive carbohydrate cluster microarrays. This technology utilizes a 3-dimensional (3-D) poly(amidoamine) starburst dendrimer monolayer assembled on glass surface, which is functionalized with terminal aminooxy and hydrazide groups for site-specific coupling of carbohydrates. A wide range of saccharides, including monosaccharides, oligosaccharides and polysaccharides of diverse structures, are applicable for the 3-D bioarray platform without prior chemical derivatization. The process of carbohydrate coupling is effectively accelerated by microwave radiation energy. The carbohydrate concentration required for microarray fabrication is substantially reduced using this technology. Importantly, this bioarray platform presents sugar chains in defined orientation and cluster configurations. It is, thus, uniquely useful for exploration of the structural and conformational diversities of glyco-epitope and their functional properties.
Keywords: Carbohydrates, Carbohydrate Microarray, Glycan, Glycan arrays, Glycomics Oligosaccharides, Microwave assisted reaction, Polysaccharides, Postgenomics
Introduction
Carbohydrates are prominently displayed on the surface of cell membranes and on the exposed regions of macromolecules in bodily fluids. Their structural diversity and abundant presence in surface-display make them suitable for storing biological signals in the forms that are recognizable by other biological systems. Carbohydrate-mediated molecular recognition underlies many aspects of biological processes.1–6 These include important cellular events, such as fertilization,6a, 7 embryonic development,8 cell differentiation and cell–cell communication,9–11 and many molecular processes that are mediated or regulated by carbohydrate biosignals. The latter may include but not limited to blood coagulation,12, 13 inflammation,3, 14 chemotaxis,15 as well as host recognition and immune responses to microbial pathogens,16–18 Exploration of sugar chain diversities and their roles in molecular recognition and bio-signaling is one of the current challenges to functional Glycomics investigation.
Carbohydrate-based microarray technologies have recently emerged as promising tools for glycomics research.19–49 These bioarrays present tens or hundreds of different sugars as microarrays on solid surfaces in a spatially discrete pattern. Technically, they are well-suited for monitoring multiplex molecular interactions involving carbohydrates, such as DNA– sugar, protein – sugar, sugar – sugar, and sugar –metabolite interactions. In addition, the miniaturized methodology of bioarrays is particularly important for Glycomics research, because no biological amplification strategy, such as the Polymerase Chain Reaction (PCR) or cloning, exists to produce usable quantities of complex oligosaccharides. Practically, carbohydrate microarrays may have advantage in rapid determination of the binding profile of carbohydrate-binding proteins or anti-glycan antibodies, detection of disease-associated or pathogen-specific antibodies for the diagnostic and/or prognostic applications, and characterization of carbohydrate-cell or carbohydrate-microbe recognition events and the high-throughput screening of inhibitors for blocking carbohydrate-mediated molecular recognition in a specific biomedical application.
A number of experimental approaches have been explored for construction of carbohydrate microarrays19–34 Methods of non-covalent attachment (i.e., massive adsorption), such as nitrocellulose-coated glass slides35, 36 and hydrophobic black polystyrene substrate,37 are practical approaches to high-throughput production of polysa ccharide and glyconjugate microarrays. These technologies are, however, not suitable for direct immobilization of mono- and oligosaccharide microarrays. This is owing to the fact that carbohydrate binding on these surfaces becomes less stable as the molecular weight decreases.35 Mono- and oligosaccharides may be chemically modified with reactive tags, such as amine6b, 38–41, thiol,42 cyclopentadiene,43 lipid,44, 45 fluorous,46–48 azide,49 photoactive,50, 51 biotin,52, 53 etc. These chemically “activated” carbohydrates can be immobilized onto substrates that were functionalized with compatible chemical groups. Efforts have been also made to devise methods that allow for covalent immobilization of carbohydrates on substrates without prior chemical derivatization. Carroll et al reported a method for the covalent patterning of unmodified oligosaccharides onto substrates functionalized with photoactive phthalimide chromophores.54, 55 Upon exposure to UV light, the photoactive aromatic carbonyls react with C-H groups of the sugars to form a covalent bond. The photocoupling procedure may target more than one CH- group on the sugar rings. This strategy may improve sugar coupling efficiency by grapping saccharides with multiple attachment sites. However, it is unlikely suitable for production of the single epitope glycan arrays since the coupled saccharides are presented in more than one configurations in a given microspot.
It is desirable to develop bioarray platforms that display structurally well-defined sugar epitopes in given micro-spots in order to facilitate the fine-specificity analysis of the carbohydrate-mediated molecular recognition. This goal can be achieved by site-specific coupling of saccharides in a bioarray substrate. Shin’s group reported hydrazide-coated glass slides for immobilization of unmodified mono-, di- and oligosaccharides under long reaction times (~12 hours) and high temperatures (~50 °C), but high sugar concentration (~30 mM was required for the immobilization).56 Zhi et al also reported hydrazide-derivatized monolayer on a gold surface for fabrication of carbohydrate microarrays with unmodified oligosaccharides.57 Zhou and his collaborators reported the development of self-assembled monolayers containing aminooxy-functionalized glass slides as a platform for immobilizing unmodified oligosaccharides through the formation of oxime bonding with the carbonyl group at the reducing end of the suitable carbohydrates.58 A technical issue in exploring the application of the aminooxy- and hydrazide- functionalized surfaces is to improve their sugar coupling efficiency. We have noticed that the reactions of native carbohydrates with surface-bearing aminooxy or hydrazide groups are significantly slower than in solution, especially when the carbohydrate molecular weight increases because of the reduced activity of the reducing group of the carbohydrates.
In this paper, we report development of novel bioaray substrates for the site-directed immobilization of carbohydrates and presentation of saccharides in defined cluster configuration. This technology utilizes a 3-dimensional (3-D) poly(amidoamine) starburst dendrimer monolayer assembled on glass surface, which is functionalized with terminal aminooxy and hydrazide groups for site-specific coupling of carbohydrates. We have demonstrated that glass slide surfaces modified with these functionalized 3-D dendrimeric monolayer support effective immobilization of mono-, oligo- and polysaccharide without prior chemical modification. Its efficacy of carbohydrate coupling is accelerated by conventional microwave radiation energy.
Experimental Section
Materials and Reagents
All chemicals unless stated were purchased from Sigma-Aldrich-Fluka Chemical Corp. (Milwaukee, WI) and were used as received: (3-Glycidyloxypropyl) trimethoxysilane (GPTS) (98%), Boc-aminooxy acetic acid, adipic acid dihydrazide (ADHZ), 1-ethyl-3-(3-dimethylaminopropylcarbodimide) (EDC), N-hydroxy- succinimide (NHS), and PAMAM starburst dendrimers of generation 1 to Generation 5. Lectins and biotin-conjugated lectins were purchased from Sigma or EY Laboratories (San Mmateo, CA). Mouse Anti-Sialyl Lewis-× monoclonal antibody was purchased from GenWay Biotech, Inc. Carbohydrate probes were collected from Aldrich, Sigma, Calbiochem, Dextra lab, Acros and Senn Chemicals. Cy3-labeled streptavidin, bovine serum albumin, and BiotinTag™ Micro Biotinylation Kit are also purchased from Sigma. Biotinylation of lectins and antibody were conducted under manufactor’s protocol. Multi-well (1×16) microarray hybridization Cassettes was obtained from ArrayIt Inc.( Sunnyvale, CA).
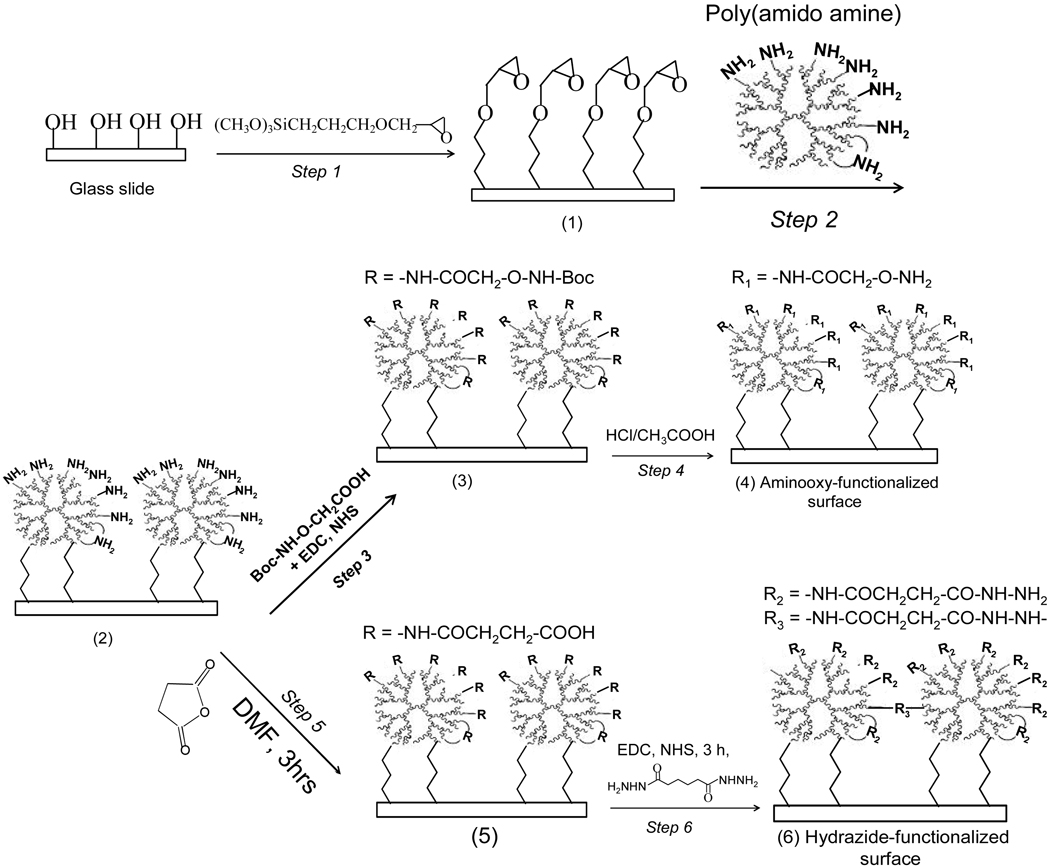
Preparation of Aminooxy and Hydrazine Functionalized Dendrimeric Substrates (Scheme 1)
Scheme 1.
Chemical process for preparation of 3-dimensioanl aminooxy- and hydrazide functionalized glass slides.
Generating dendrimer monolayer coating on GPTS silanized slide (Scheme 1, surface (2))
Microscope glass slide is cleaned with hot Piranha solution (30% H2O2: H2SO4/1:3), and then thoroughly rinsed with distilled water and HPLC purified ethanol. The cleaned slide substrate is immersed into a 1mM of GPTS/toluene solution for 30 minutes to form a monolayer on the glass surface with Epoxy functional groups towards the outside(step 1, Scheme 1). The GPTS functionalized slide are then incubated overnight in solution containing dendritic PAMAM starburst monomer (1% (w/v) Sigma-Aldrich) and 5 wt% KOH in menthol at room temperature under gentle shaking(step 2, Scheme 1). Excessive monomer is removed by washing with methanol and DI water and the dendrimer-coated slides are dried in a nitrogen stream.
Functionalization of dendrimer film with aminooxy groups (Scheme 1, surface (4))
The PAMAM dendrimer coated glass slides (Scheme 1, surface (2)) were placed into a 50 mM phosphate buffer solution (pH =6.0) containing 1mM of Boc-aminooxyacetic acid (Sigma), 1 mM EDC and 1.5 mM NHS and allowed to react for 2.5 h (step 3, Scheme 1). The glass slides were then washed with water and immersed into 2 M HCl/acetic acid solution for 2 hrs to remove the Boc- group (step 4, Scheme 1). The aminooxy-functionalized dendrimeric slides were washed with ethanol and water and dried under N2 blow.
Functionalization of dendrimer film with hydrazide groups (Scheme 1, surface (6))
The PAMAM dendrimer coated glass slides (Scheme 1, surface (2)) were placed into a saturation solution of succinic anhydride (GA) in N,N-dimethylformaide (DMF) for 3 hrs. The slides were then carefully washed with pure DMF several times. Subsequently, the carboxyl groups on the substrate surface were activated with 1 mol L−1 N-hydroxysuccinimide (NHS)/ 1 mol L−1 1-ethyl-3-(3-dimethylaminopropylcarbodimide) (EDC) in DMF for 2 h and washed with pure DMF. The activated slides (Scheme 1, surface (5)) were then immersed into a solution of 10 mg/mL aqueous solution of adipic acid dihydrazide (ADHZ, Sigma-Aldrich) for 2.5 h. Finally, the slides were washed with water and dried under N2 blow.
Fabrication of Carbohydrate Microarrays
Microspotting
The carbohydrate probes were dissolved in five types of printing buffers of: (1), Formamide, (2), 50%DMSO, 50%H2O, (3), 1%NaCl aqueous solution, (4), 0.1 mM sodium phosphate buffer (pH 7.4) 20%glycerol; and (5), 0.1 mM sodium phosphate buffer (pH 5). Polysaccharide probes were usually spotted at the initial concentration of 0.5 mg /ml, and mono- and oligosaccharides were spotted in the molar concentrations in 100 µM. They were further series diluted at ten fold. A given concentration of each preparation is repeated at least three times to support statistic analysis. 1 nL of the carbohydrates solution from a 384-well plate was printed onto aminooxy or hydrazide functionalized dendrimeric substrates by using a MicroGrid Array Printer (Ann Arbor, MI) in 60% relative humidity.
Immobilization of spotted carbohydrates on slide under microwave wave radiation energy
A GE domestic microwave oven was used for providing microwave radiation energy for immobilization of spotted carbohydrates on the aminooxy or hydrazide functionalized dendrimer-coated slides. For immobilization, the slides with spotted carbohydrate microarrays were placed on the plates of microwave oven. Energy power level was set at 50%. The microwave radiation dosage was controlled by the microwave time. After treatment of the microarray with microwave radiation, the slide was immersed into PBS (pH 7.4) containing 0.1% Tween 20 with gentle for 2 mins shaking to remove unbound carbohydrates.
Detection of Carbohydrate-Protein Interactions with Carbohydrate Microarrays
Blocking of slides surface
To optimize blocking protocols, four blocking buffers were tested on the microarrays fabricated on the hydrazide-functionalized dendrimer substrate. The blocking buffers were as follows: (1) Superblocking buffer from Pierce; (2) 0.1× PBS buffer containing 1% BSA, 0.1% Tween 20; (3) 0.1 × PBS buffer containing 3% casein, 0.1% Tween 20; and (4) 0.1 × PBS buffer containing 3% nonfat milk. Spotted substrates were immersed into one of the above blocking buffer for 60 mins, and then rinsed with 0.1×PBST buffer before immunoassay.
Immunoassays on carbohydrate microarrays
Both direct immunoassay and sandwich immunoassay were tested on the fabricated carbohydrate microarray for detection of the lectin-carbohydrate interaction. For direct immunoassay, the carbohydrate microarrays were incubated with individual or mixed Cy3-labeled lectins in 0.1 × PBST (pH 7.4) containing 0.1% Tween 20, 1 mM MnCl2 and CaCl2 for 1 h at room temperature. For sandwich immunoassays, the carbohydrate microarrays were incubated with cocktail solution of individual or mixed biotinalyted lectins and Cy3- streptavidin in 0.1 × PBST (pH 7.4) containing 1 mM MnCl2 and CaCl2 for 1 h. Following incubation, slides were washed twice in 0.1 × PBST buffer for 10 min, and then rinsed briefly (30 sec) in deionized water to remove PBST buffer salts prior to spin dried.
Microarray Imaging and Data Analysis
Microarrays were scanned at 10 µm resolution with the scanning laser confocal fluorescence microscope of a ScanArray 5000 System (PerkinElmer Life Science, Boston, MA) or AlphaScan™ (Alpha Innotech Inc, San Leandro, CA). The emitted fluorescent signal was detected by a photomultilier tube (PMT) at 570 nm for Cy3. For all microarray experiments, the laser power was 75% and the PMT gain was 65%. The fluorescent signals were analyzed by quantifying the pixel density (intensity) of each spot using ImaGene 3.0 (Biodiscovery, Inc., Los Angeles, CA). Fluorescence intensity values for each array spot and its background were calculated using ScanArray Express software. The local background signal was automatically subtracted from the immunoassay signal of each separate spot and the mean signal intensity of each spot was used for data analysis. Statistical analyses were performed using SigmaPlot 5.0 (Jandel Scientific, San Rafael, CA) or by Microsoft Excel®.
RESULTS AND DISCUSSION
Preparation of Aminooxy and Hydrazide Functionalized 3-D Dendrimeric substrates
We have developed chemical processes for preparation of the aminooxy and hydrazine functionalized dendrimer coatings on microscope slides. Scheme 1 briefly outlines the processes used to prepare aminooxy- and hydrazide- functionalized dendrimeric substrates. The concept for the using of dendrimeric surface as platform for carbohydrate microarray fabrication was based on the assumption that the higher functionalized surface of PAMAM starburst dendrimers attached to a solid support should help to maximize the immobilization capacity for carbohydrate probes and in turn to provide the polyvalency(i.e., the cluster effect) required for probing carbohydrate-protein interaction. The dendrimers are a class of synthetic polymers with an unusual three-dimensional highly branched structure composed of a core, repeating units, and end groups. Dendrimers such as Poly(amido amine) (PAMAM) are an ideal material for the construction of 3D films.59a, 59b Furthermore, dendrimers have the feasibility to be further functionalized with active groups due to their well-defined composition and constitution, as well as narrow molecular distribution.59c–59e To prepare the dendrimer coated slides, the surface amine-riched PAMAM were first covalently bound to an epoxy-modified surface that was formed by 3-glycidyloxypropyl)trimethoxysilane (GPTS) self-assembled on glass slide surface (Step1 and 2, Scheme 1). This procedure to yield dendrimer-activated glass slides is simple, fast, and highly effective. The PAMAM dendrimers tested in this study include generation 1 to 5 (G1–G5). They differ in the sizes of dendrimers and in the number of available sites in each dendrimer for further activation with either aminooxy- or hydrazine-groups.
To generate the aminooxy functional groups on the dendrimer coated slides, the amine groups on the dendrimer coated slides were coupled to the carboxyl groups of the N-Boc-Aoa-OH that was activated with hydroxylsuccimide group (Step3, Scheme 1). Free aminooxy groups were then generated upon treatment of the glass slide with HCl/acetic acid for the purpose to remove the Boc- group (Step 4, Scheme 1).
To generate the hydrazide functional groups on the dendrimer coated slides, the terminal amino groups of the dendrimers attached to the slides were converted into carboxyl groups by treatment with succinic anhydride in N,N-dimethylformaide (DMF) overnight, followed by the final activation step with EDC/NHS allowing for covalent link with adipic acid dihydrazide (ADHZ) (Step 5, and 6, Scheme 1). Lee et al. reported the use of succinic anhydride and hydrazine in DMF solution to convert the surface bounded NH2 groups to hydrazide groups.56 However, this process involved the use of high toxic hydrazine (NH2NH2), which raises the concern safety control in manufacturing process, especially in large scale manufacturing process. To reduce the environmental pollution, we used low toxic material of adipic acid dihydrazide (ADHZ) to prepare the hydrazide-functionalized 3-dimesional substrates.
Another benefit of using our proposed process is that the PAMAM dendrimeric monomers were inter-molecularly cross-linked by treatment with the homobifuctional reagents ADHZ, after coupling of the dendritic monomers to slide surfaces (as shown in Scheme 1, by the -R3- linking). We expect that the cross-linked polymer film will improve the stability of the surface against harsh washing conditions applied during immunoassay process.
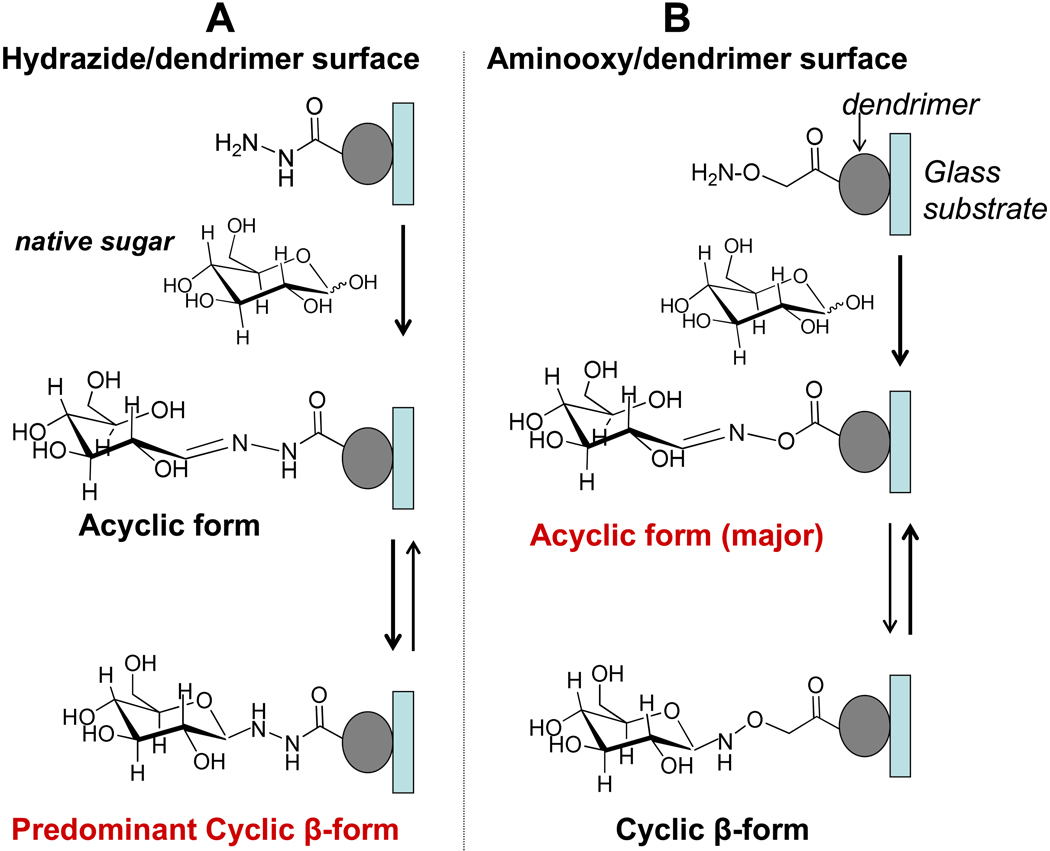
Carbohydrates Immobilization Mechanism
The immobilization of carbohydrates onto aminooxy and hydrazide surfaces are based on the hydrazone formations between the highly reactive nucleophilic aminooxy/hydrazide and the carbonyl group at the reducing end of suitable carbohydrates via irreversible condensation(Figure 1). Thus, the monosaccharides and oligosaccharides immobilized on these two types of substrates are in defined orientation. Reactions of aminooxy or hydrazide groups with free carbohydrates are known to be chemoselective and have been used for the synthesis of various glycoconjugates60, 61 and carbopeptides.62 The efficiency of the method relies on the ease of formation and on the good stability of the oxime linkage up to pH 9. It is known that cyclic adducts with β-configuration are produced predominantly in reactions of carbohydrates with hydrazide-containing compounds (Figure 1A).56, 60, 63 In contrast, it has been reported that reactions between carbohydrates and substances containing aminooxy groups generate acyclic products preferentially (Figure 1B).56, 61, 64 This carbohydrate immobilization method requires only a few derivatization steps on slide surface treatments but without the requirement of prior chemical modification of carbohydrate probes, which make it very attractive for preparing carbohydrate microarray in individual laboratories.
Figure 1.
The immobilization mechanism of oligosaccharides onto Aminooxy-dendrimer and Hydrazide-dendrimer coated slides.
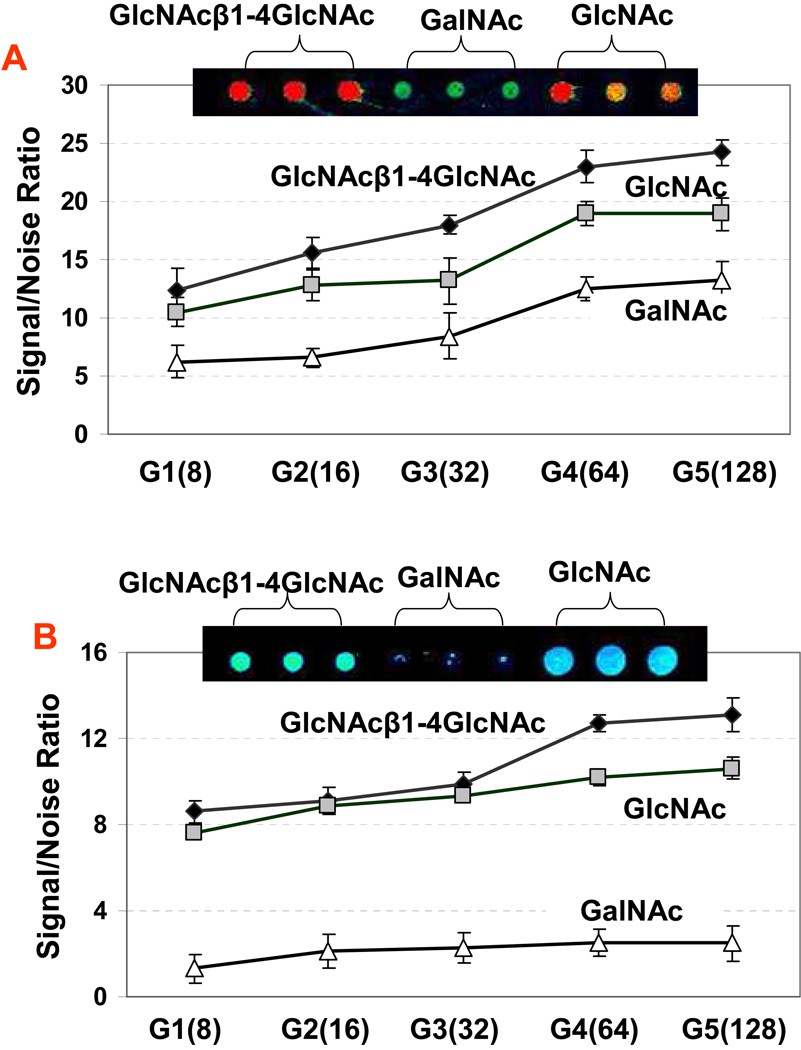
The Cluster Effect of Carbohydrates Immobilized on 3D Dendrimeric Film for Probing Carbohydrate-Protein Interactions
The affinities of most oligosaccharide-protein interactions are usually lower than the affinities from DNA-DNA or protein-protein interactions. This fact has pointed the requirement of forming multivalent interaction between the immobilized carbohydrates and proteins for detecting their interactions.65 The high density of surface functional groups on PAMAM starburst dendrimers should help to maximize the immobilization capacity of carbohydrates and thus provide the multivalency (i.e., the cluster effect) required for detectable carbohydrate-protein interaction. We have tested the contribution of cluster effect from dendrimer films by comparing the lectin-binding reactivities with the glycan arrays that were constructed using different generations of PAMAM dendrimer substrates. An example of this study is shown in Figure 2A and 2B. The binding reactivities of lectin WGA with spotted saccharides (GlcNAcβ1-4GlcNAc, GalNAc and GlcNAc) are clearly increased with the increasing generation number of PAMAM dendrimer. Both hydrazide-dendrimer coated substrate (Figure 2A) and the aminooxy-dendrimer coated substrate (Figure 2B showed consistently that PAMAM dendrimer generation 4 (containing 64 surface groups) showed almost twice the signal of the carbohydrates PAMAM dendrimer generation 1 (containing 8 surface groups). The cluster effect of dendrimer films especially benefits the detection of protein interactions with monosaccharides that were immobilized onto the 3-D dendrimer surfaces. This was evidenced by the successful detection of lectin-monosaccharide interactions on monosaccharides such as the GlcNAc and GalNAc that were immobilized on the two types of dendrimer-coated substrates. These protein-monosaccharide interactions were previously reported to be undetectable with the monosaccharides immobilized on a 2-dimensional surface.56 The slides coated with PAMAM dendrimer generation 4 (G4, containing 64 surface groups) showed almost the same signal intensity compared to the carbohydrates immobilized on the slides coated with PAMAM dendrimer G5 (containing 128 surface groups). Since the PAMAM dendrimer G4 is cheaper than the G5, the G4 was selected for preparing the aminooxy-dendrimer and hydrazide-dendrimer-functionalized substrates. Figure 2 also reveals that the hydrazide-dendrimer coated slides provided higher fluorescence signal/noise ratios for the mono-, disaccharides(GlcNAc, GalNAc, GlcNAcβ1-4GlcNAc) than that on the aminooxy-dendrimer coated slides, which indicated that the mono-, disaccharides immobilized on the hydrazide-dendrimer coated slides showed stronger binding of proteins than these carbohydrates immobilized on aminooxy-dendrimer coated slides. This can be attributed to the fact that cyclic adducts with β-configuration are produced predominantly in reactions of carbohydrates with hydrazide-dendrimer surface, while the acyclic adducts are produced in reactions of carbohydrates with aminooxy-dendrimer surface. This result is consistent with the carbohydrate in the immobilization mechanism discussed above. Therefore, it is desirable that the carbohydrates are immobilized in a site-specific format and preserve the monosaccharide’s structure for elucidation of the structurally specific protein interaction.
Figure 2.
Cluster effect of the oligosaccharides immobilized on the 3-D dendrimeric substrates for protein affinities. Oligosaccharides of GlcNAcβ1-4GlcNAc, GalNAc and GlcNAc were spotted on the glass slides coated with hydrazide-derivatized PAMAM dendrimer Generation 1 to 5 (A), and aminooxy-derivatized PAMAM dendrimer Generation 1 to 5 (B). The arrayed slides were microwave treated for 10 mins in 800W oven. The microarrays were then probed with 0.1 µg/mL of Cy3-labeled WGA lectin at 30 °C for 60 min, followed by washing with PBST buffer for 5 mins twice and 0.01 × PBS for 1 min. The micraorrays were measured with confocal scanner under identical layer power and PMT. The number showed in the bracket of axis is the surface functional groups of dendrimer generation. The noise (i.e., background) was determined as the fluorescent intensity around the spots. Each data shown represent means ± SD of 12 replicates from four slides. Insert images were obtained from slides coated with hydrazide- and aminooxy- functionalized PAMAM dendrimer Generation 4, respectively.
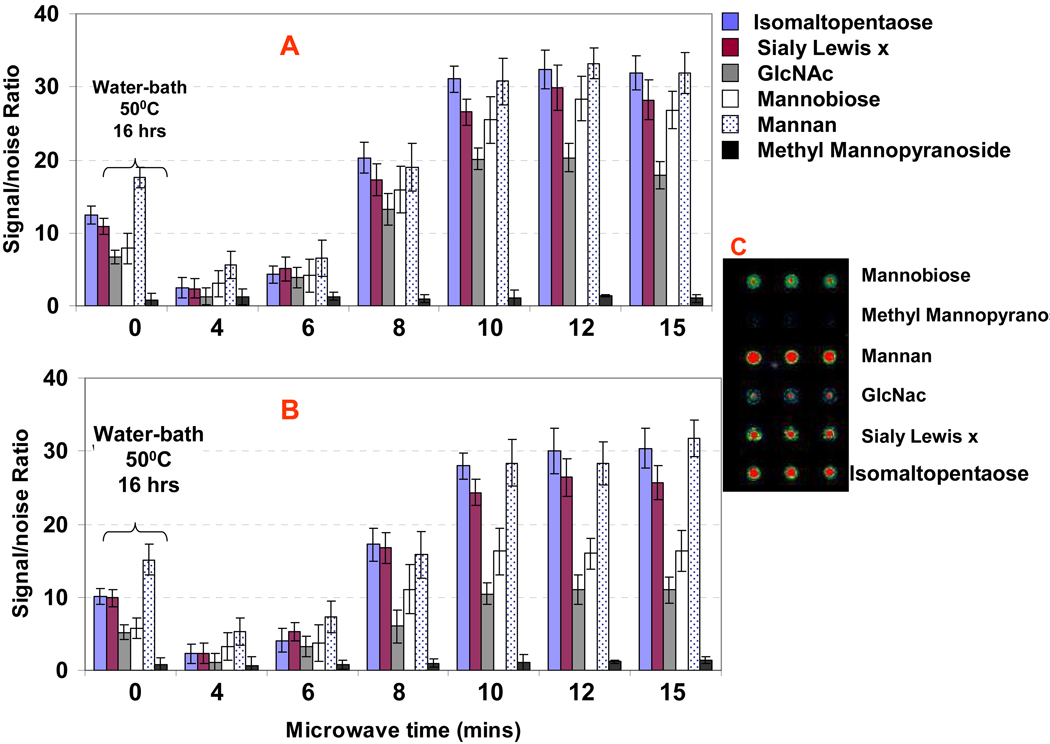
Microwave Radiation Energy for Rapid and Effective Immobilization of Unmodified Carbohydrates onto Aminooxy and Hydrazide Functionalized 3-D Substrates
Microwaves (~0.3–300 GHz) lie between the infrared and radio frequency (RF) electromagnetic spectrum. In the past two decades, the use of microwave radiation has greatly increased in its application to accelerating reactions in synthetic organic chemistry,66, 67 nanomaterial synthesis,68, 69 and biochemistry.70, 71 The “Technology Vision 2020” of the U.S. chemical industry believes that microwave heating will soon replace traditional heating techniques in chemical and biochemical synthesis. It is widely thought that microwaves accelerate chemical reactions by increased rate of mutarotation (the process of equilibration between unreactive closed ring structures that proceeds via the reactive open chain intermediate72) above that achieved by equivalent thermal heating, and by accelerated solvent heating which increases the rate of mutarotation.
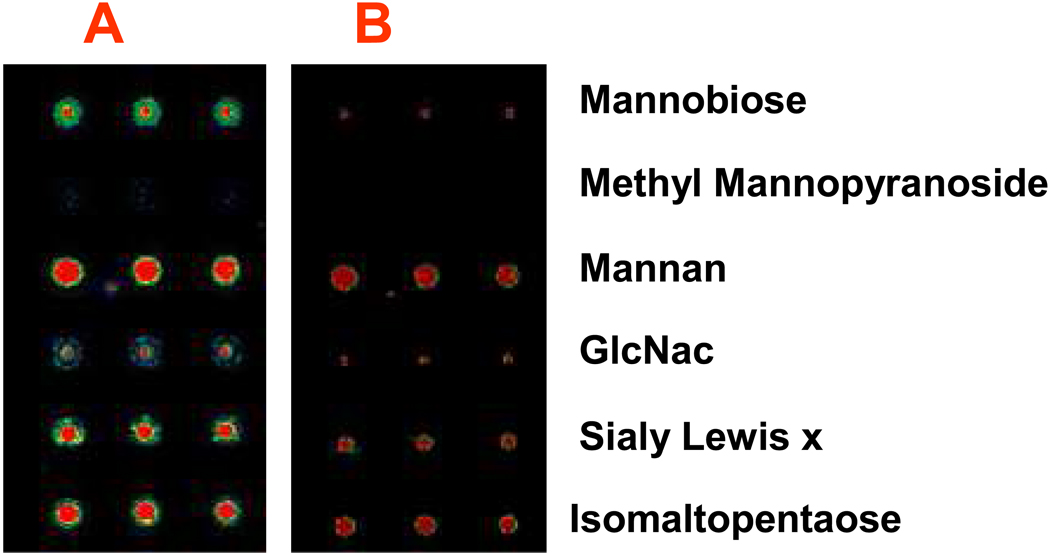
Microwave is clean, convenient, and effective energy for chemical reaction. The use of microwave energy for immobilization of carbohydrates has not been tested before. To find the microwave radiation energy for rapid immobilization of carbohydrates spotted onto hydrazide/dendrimer and aminooxy/dendrimer surface, representative carbohydrate probes include Mannose(monosaccharides), Mannobiose(disaccharides), Sialyl Lex(trisaccharide), isomaltopentaose(oligossachride), and Mannan (polysaccharide) were printed onto the two types of 3-D substrates. The printed slides were placed into GE domestic microwave oven and treated with microwave at different time course. The microwave radiation dosage was controlled by the microwave time and power level. In the experiment, the power level was set at 50% of the GE domestic microwave oven having a maximum power of about 800 watts, and the exposure time varied from 4 to 15 minutes. After treating the microarray with microwave radiation, the microarray slides were washed with 0.1 × PBS buffer to remove unbound carbohydrates, and blocked with 1% BSA PBST blocking buffer for 1 hr. The microarrays were then probed with cocktail solution of 1 µg/mL of biotin-ConA, biotin-WGA (wheat germ agglutinin), and Cy3-streptavidin for 1 h, respectively. After washing the microarray, the fluorescent intensities were measured with fluorescent scanner. Figure 3 (A) and (B) illustrated the effect of microwave energy for immobilization of carbohydrate probes on the two types 3-D substrates. It clearly showed that the efficiency of immobilizing carbohydrate probes on both of the hydrazide-dendrimer and aminooxy-dendrimer surface increased with the increase of microwave energy (i.e., the microwave time). Maximum fluorescent intensities were obtained for 10 mins of microwave treatment on both the aminooxy and hydrazide dendrimer slides, which indicated the effective immobilization of carbohydrates probe on the slide surfaces. We also tested the immobilization of carbohydrates on these slides by traditional heating through incubation of the printed slides (the slides were sealed into casket) in water-bath for 16 hrs under 50°C. These slides showed much lower signals in lectin detection as compared to the slides under microwave for 10 mins, which indicated fewer carbohydrates were immobilized on the slide by traditional heating. These results showed that the microwave radiation energy is not only more convenient but also more effective than traditional water-bath heating for accelerating the reaction of carbohydrates onto aminooxy and hydrazide-functionalized surface. The spots arrayed with methyl mannopyranoside where the reducing end is blocked by methyl group, showed no detectable signal for lectin binding, which indicated methyl mannopyranoside was not immobilized on the slides. This verified that the immobilization of carbohydrate probes on the hydrazide-dendrimer and aminooxy-dendrimer slide surface was through the formation of covalent bonding between the carbohydrates and slide surface, which resulted in site-specific immobilization of mono- and oligosaccharides. For the polysaccharides that the activities of reducing end are known very weak, it may involved the formation of covalent binding between the hydroxyl and aminooxy or hydrazide groups on the slide surfaces. However, this will not affect the epitope display of polysaccharide as it was known that a polysaccharide may present multiple glyco-epitopes, including terminal non-reducing epitopes and internal chain epitopes.18 Interestingly, higher fluorescence signals after lectin probing were observed for the mono-, disaccharides (such as GlcNAc and mannobiose) immobilized on hydrazide-coated glass slides than those immobilized on aminooxy-coated slides. But for polysaccharides (Mannan) and oligosaccharides, fluorescence signals after lectin probing were similar between the two types of slides.
Figure 3.
Microwave radiation energy for effective immobilization of carbohydrates on (A)hydrazide-dendrimer coated slides, and (B) aminooxy-dendrimer coated slides. The slides spotted with 100 µM of mono-, di-, oligosaccharides and polysaccharides in Formamide were placed into domestic microwave oven, and under microwave treatment for 4–15 mins. (C) is the representative image obtained after the microarray on hydrazide-dendrimer coated slide was subjected 10 min microwave treatment and then probed with cocktail solution of 0.1 µg/mL of biotin–ConA, biotin-WGA lectins, and Cy3-Streptavidin for 1 h at 30 °C. Fluorescence intensity values of the arrays after washing (with PBST buffer for 5 mins twice and 0.01 × PBS for 1 min) were measured with confocal scanner under identical layer power and PMT. The noise (i.e., background) was determined as the fluorescent intensity around the spots. Each data shown represent means ± SD of 12 replicates from four slides.
To verify that the carbohydrates were covalently immobilized on the hydrazide-dendrimer and aminooxy-dendrimer slide surface after microwave radiation, we compared the stability of the carbohydrates immobilized on hydrazide-dendrimer coated slide by microwave energy after extensive washing of the slides with strong ionic solution (i.e., 2 M NaCl aqueous solution for 10 mins) and after gentle washing (i.e., 0.1 × PBS buffer for 2 mins). The carbohydrate microarrays were then probed with same concentrations of lectins under identical condition. Figure 4(A) and (B) showed the microarray images. The signal intensities of carbohydrate microarrays after extensive washing of the slides with strong ionic solution is ~80% of the signal intensities of carbohydrate microarrays after gentle washing. This result showed that that extensive washing of the slides with strong ionic solution (i.e., 2 M NaCl aqueous solution) does not remove the carbohydrate probes from the microarray substrates. Although we cannot rule out possible physical adsorption of the polysaccharides on the slides surface, this experiment has demonstrated that the immobilization of carbohydrates on the hydrazide-dendrimer slide by microwave energy are stable against harsh washing conditions and supports the conclusion that the spotted mono- and oligosaccharides are covalently bound on the substrate surface. The ~20% reduction of intensities after extensive washing is likely owing to removal of remaining non-covalently bound saccharides, especially those with larger molecular weights.
Figure 4.
Stability of the carbohydrates immobilized on hydrazide-dendrimer coated slide facilitated by microwave energy. (A) Microarray image of microwave treated array slide after washing with 0.1 × PBS buffer for 2 mins; (B) microarray image of microwave treated array slide after washing with 2M NaCl solution for 10 mins. The assay protocol is as same as described in Figure 3.
Printing Buffer Selection
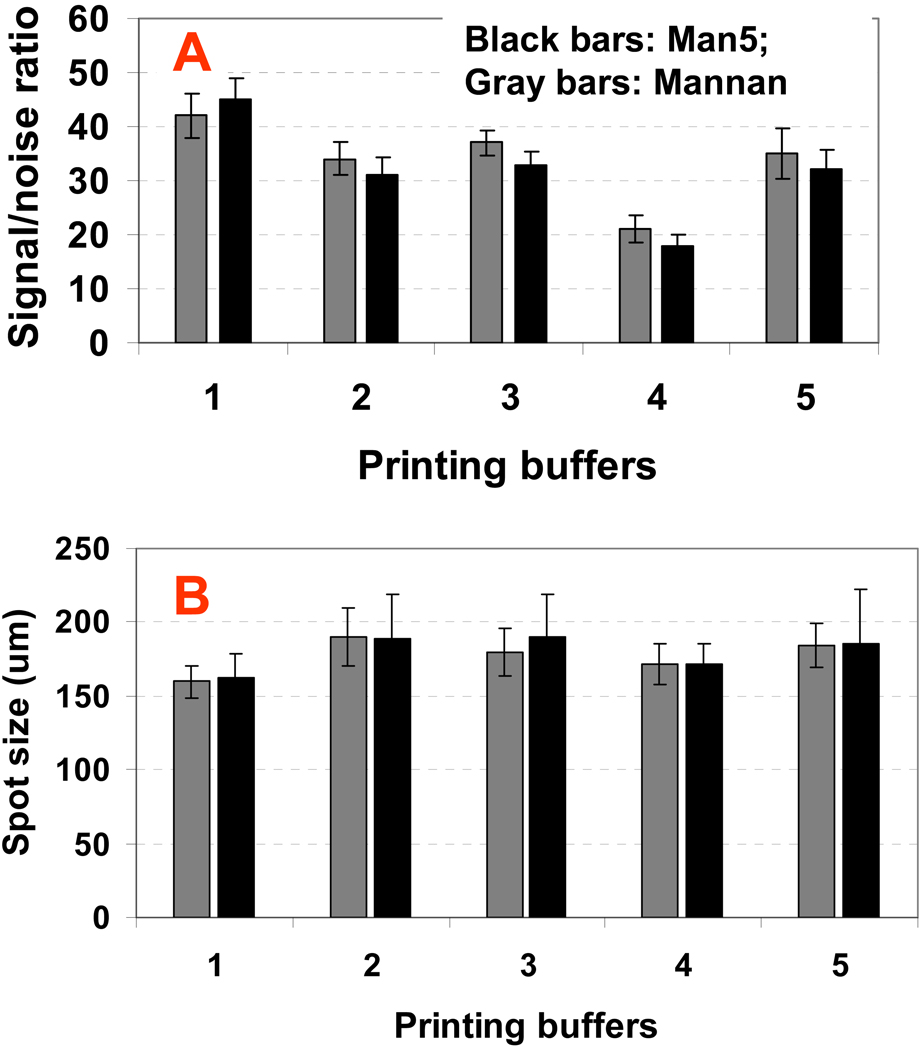
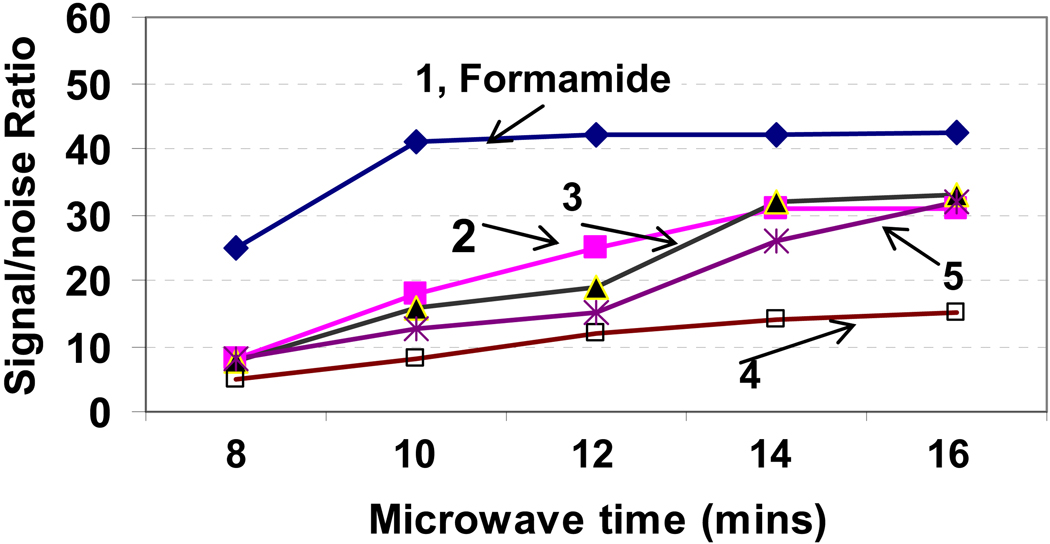
The characteristic of the printing buffer may affect the immobilization efficiency of carbohydrates on slide surface and quality of the spots produced. Printing buffers need to be optimized for carbohydrate solubilization, immobilization, spot uniformity, and stablity under microwave treatment. We compared five types of printing buffers for carbohydrate microarray printing with representative oligosaccharide (mannopentaose) and polysaccharides (Mannan) in. These are: 1, Formamide, 2, 50%DMSO, 50%H2O, 3, 1%NaCL aqueous solution, 4, 0.1 mM sodium phosphate buffer (pH 7.4) 20%glycerol; and 5, 0.1 mM sodium phosphate buffer (pH 5). The printing solutions were prepared by dissolving 100 µM of carbohydrate probes into the printing buffer. The printed microarrays were subjected to microwave treatment for 10 mins, and then probed with 1 µg/mL Cy3-ConA. The signal intensity and spot size were analyzed and compared as shown in Figure 5. The formamide printing buffer gave the best signal-to-noise ratios for both the oligosaccharide and polysaccharide (Figure 5 A). The spot sizes using formamide as printing buffer were ~160 um (diameter) and are more uniform (i.e., lower standard derivation) than the spot sizes produced using other three types of printing buffer (Figure 5, B). The printing buffer of 0.1 mM sodium phosphate (pH 7.4) containing 20% glycerol gave good spot size uniformity, but gave the lowest signal-to-noise ratios. Glycerol was widely used as a printing buffer component for protein microarrays to avoid the evaporation of protein printing solution. However, our results have showed that glycerol is not a good component in printing buffer for fabrication of carbohydrate microarrays, probably due to the adherence of glycerol on the slide surface after microwave radiation, which might block the carbohydrate’s binding sites. By contrast, Formamide as printing buffer shows improved reaction rate of the carbohydrate with hydrazide groups (Figure 6). Under same microwave radiation time, the sugar (mannopentaose) spotted in formamide is more efficiently immobilized as was evidenced by the higher lectin-binding signals compared to sugar spotted in other four buffers. The result of formamide being the best printing buffer is not surprising, because formamide has good solubility to carbohydrates, a high volatile point, and is a good microwave adsorbing solvent due to its high dielectric constant.
Figure 5.
Effect of printing buffers for fabrication of carbohydrate microarrays on hydrazide-dendrimer coated slides regarding to the buffer component to probe immobilization efficiency (A), and to microarray spot size uniformity (B). Black bar presents oligosaccharide Man-5; while gray bar represent polysaccharides(Manna). Printing buffers are: 1, Formamide, 2, 50%DMSO, 50%H2O, 3, 1%NaCL aqueous solution, 4, 0.1 mM sodium phosphate buffer (pH 7.4) 20%glycerol; and 5, 0.1 mM sodium phosphate buffer (pH 5). The slides spotted with 100 µM of mono-, did-oligosaccharides and polysaccharides in Formamide were placed into domestic microwave oven, and under microwave treatment for 4–15 mins. Carbohydrate probes dissolved into any of the five printing buffer at 100 µM were spotted on hydrazide-dendrimer coated slides and were subjected 10 min microwave treatment and then probed with mixture solution of 0.1 µg/mL of Cy3–labeled ConA for 1 h. Fluorescence intensity values of the arrays after washing (with PBST buffer for 5 mins twice and 0.01 × PBS for 1 min) were measured with confocal scanner under identical layer power and PMT. The noise (i.e., background) was determined as the fluorescent intensity around the spots. Each data shown represents means ± SD of 36 replicates from four slides.
Figure 6.
Comparison of the effect of printing buffers to carbohydrate immobilization rates on the hydrazide-dendrimer coated slide. Printing buffers components and assay protocols are same as described in Figure 5. Formamide as printing buffer was found to improve the immobilization rate and efficiency. Each data shown represents means of 36 replicates from four slides.
Carbohydrate Concentration Required for Immobilization on 3-D Substrates Under Microwave Energy
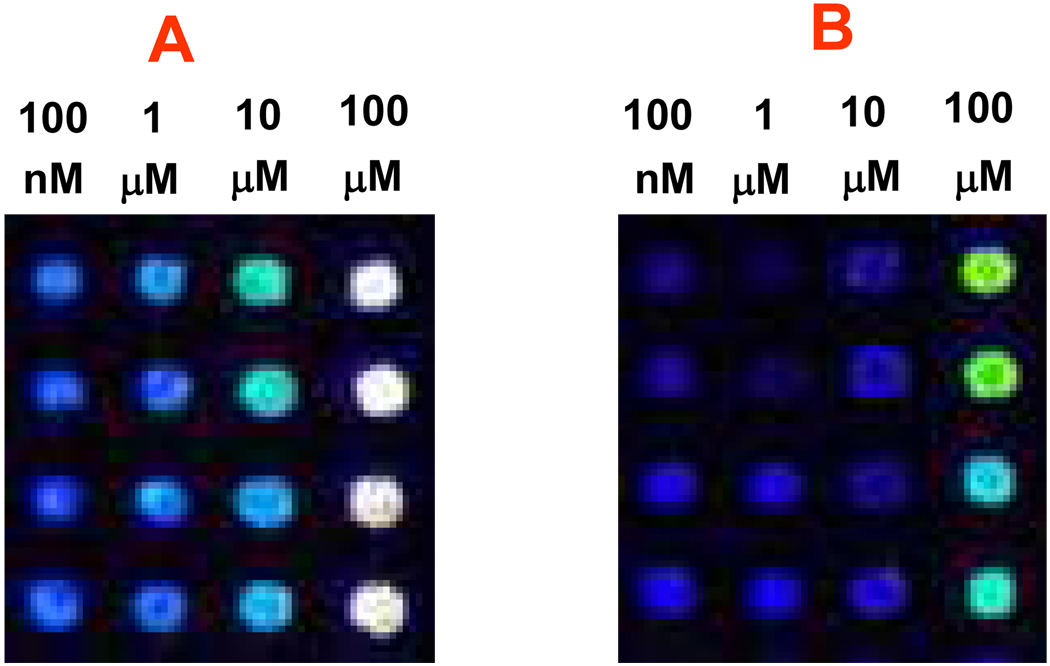
We compared the carbohydrate probe concentration required to effectively be immobilized on the hydrazide-dendrimer substrate under the treatments of microwave and water-bath heating. A series of diluted carbohydrate probe (Sialyl Lewis X) in formamide printing buffer were spotted in a 4 × 4 array-format with four replicates for each concentration onto the hydrazide-dendrimer coated slides. The arrayed slides were then either under microwave treatment for 10 mins or incubated in 50°C water-bath for 16 hrs. After blocking the slides, both of the arrays treated with microwave and water-bath heating were probed with cocktail solution having same concentration (1 µg/mL) of biotin-labeled anti-Sialyl Lex antibody and Cy3-Streptavidin, respectively. The fluorescent intensities were analyzed after scanning the arrays. The signal intensity of the bound antibody correlates with the concentration of carbohydrates immobilized on the hydrazide/dendrimer coated slides. Figure 7 showed the array images obtained from slides treated with microwave for 10 mins and water-bath heating at 50°C for 16hrs. It illustrated that good signals were obtained using pickup solution concentrations of sugar in the range 1–10 µM (or 0.82– 8.2 µg/mL) for array treated with microwave energy for only 10 mins, while detectable signal were obtained using pickup solution concentrations of sugar in 100 µM (82 µg/mL) for array subjected to heating at 50°C for 16 hrs. Since 1 nL of this solution was spotted by the arraying robot, this equates to 1–10 fmol of spotted Sialyl-Lewis-X and indicates the very high level of detection sensitivity achieved in the microarray that was fabricated by immobilizing sugar probes on the hydrazide-dendrimer substrate with microwave energy assistance. The concentrations of spotting solutions (0.82– 8.2 µg/mL or 1–10 µM) required for providing detectable signals on this platform are much lower than that of many other previously described 2-dimesional substrates, such as hydrazide-functionalized glass slides for unmodified oligosaccharides (30 mM),56 hydrazide-functionalized gold substrate for unmodified heparin saccharides (3–33 µM),57 and aminooxy-functionalized glass slide for unmodified oligosaccharides (20–500 µM),58 as well as derivatized carbohydrates on various functionalized substrate surface via different coupling chemistry, e.g., Houseman et al. (2 mM)42a, 43, Xia et al. (0.1–10 mM).38 Overall, the carbohydrate concentration (~1–10 µM) required for microarray fabrication is substantially reduced by using the microwave radiation energy to immobilize the sugars on the 3-D substrate.
Figure 7.
Comparison of concentration of carbohydrate probes required for effective immobilization on hydrazide-dendrimer coated slide under 10 mins microwave treatment and normal water-bath heating. ~1 nL of Sialyl Lex from concentration of 100 no to 100 µM in Formamide was arrayed on the slides and subjected to (A) 10 mins microwave treatment, and (B) by water-bath heating (50 °C, 16 hrs). The treated slides were incubated with 35 µL of 1 Ug/mL mixture of Biotin-labeled anti-Sialyl Lex antibody and Cy5-Streptavidin solution at 30 °C for 60 min. The microarrays were washed with PBST buffer for 5 mins twice and 0.01 × PBS for 1 min. Images were scanned with confocal scanner under same laser power and PMT, and were recorded in rainbow color display for better vision with naked eyes.
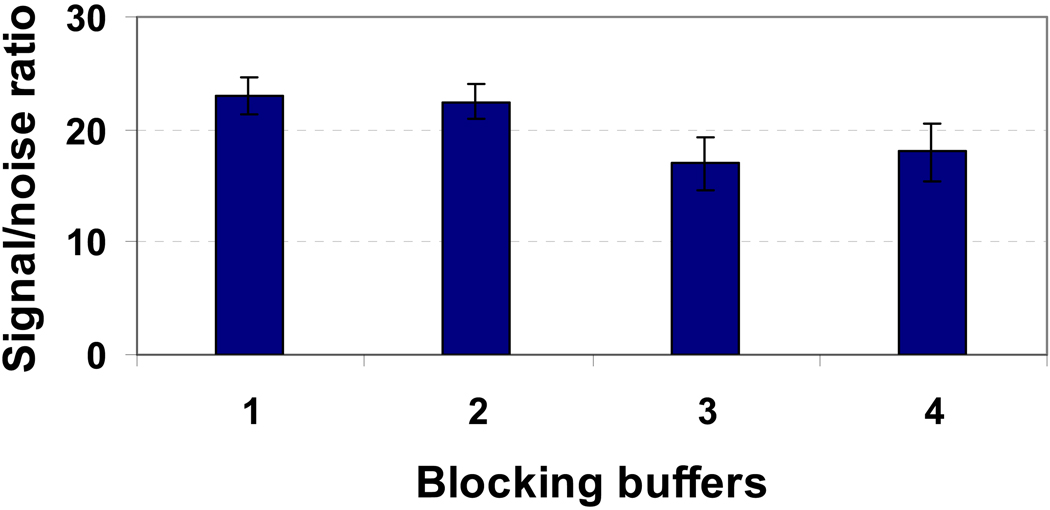
Blocking Buffer Selection
A non-specific background signal caused by the binding of cognate proteins on the slide surface is one of the problems encountered in microarray technology. Effective blocking of the microarray on the 3D slides is needed to avoid nonspecific adsorption of target samples caused by the positively charged aminooxyhydrazide groups of the 3D slides. We therefore tested the following blocking buffers: (1) Superblocking buffer from Pierce; (2) 0.1 × PBS buffer containing 1% BSA, 0.1% Tween 20; (3) 0.1 × PBS buffer containing 3% casein, 0.1% Tween 20; and (4) 0.1 × PBS buffer containing 3% nonfat milk. Among them, nonfat milk and BSA are classical blocking reagents that have been found effective in blocking nonspecific adsorption for many immunoassays on protein microarrays. Typical results are shown in Figure 8. We found that 0.1 × PBS buffer containing 1% BSA, 0.1% Tween 20 (called PBST buffer) provided a simple and effective method for blocking the hydrazide-dendrimer 3D slide, which also provided a higher signal-to-noise ratio and did not affect specificity. However, the blocking buffers containing casein and non-fat milk showed relative low signal-to-noise ratio (blocking buffer 3 and 4). This result showed that the blocking buffer should be carefully selected for blocking carbohydrate microarrays because some blocking proteins traditionally used in ELISA assays on protein microarrays contain glycans that may interfere with the immunoassays. BSA is known as glycan-free protein, thus would not affect the specificity of bioassays on carbohydrate microarrays.
Figure 8.
Selection of blocking reagents for the carbohydrate microarray on the hydrazide-dendrimer coated slide. GlcNAcβ1-4GlcNAc arrays were spotted on hydrazide-dendrimer coated slides (12 replicates per slide), and subjected to 10 min microwave treatment and the slides were blocked with one of the blocking reagents before immunoassay: (1) Superblocking buffer from Pierce; (2) 0.1 × PBS buffer containing 1% BSA, 0.1% Tween 20; (3) 0.1 × PBS buffer containing 3% casein, 0.1% Tween 20; and (4) 0.1 × PBS buffer containing 3% nonfat milk. The immunoassay fluorescent signals were obtained by incubation of the microarray with 35 µL of cocktail solution containing 1 µg/mL of Biotin-WGA and Cy3-Streptavidin solution in a 0.1× PBST solution at 30 °C for 60 min. The microarrays were washed with PBST buffer for 5 mins twice and 0.01 ×PBS for 1 min before they scanned with confocal scanner under identical layer power and PMT. The noise signal was determined to be the fluorescent intensity around the spots. Each data shown represent means ± SD of 48 replicates from four slides.
Specificity of Carbohydrate-Protein Interactions on Model Carbohydrate Microarrays Containing Mono-, Oligo-, and Poly-saccharides
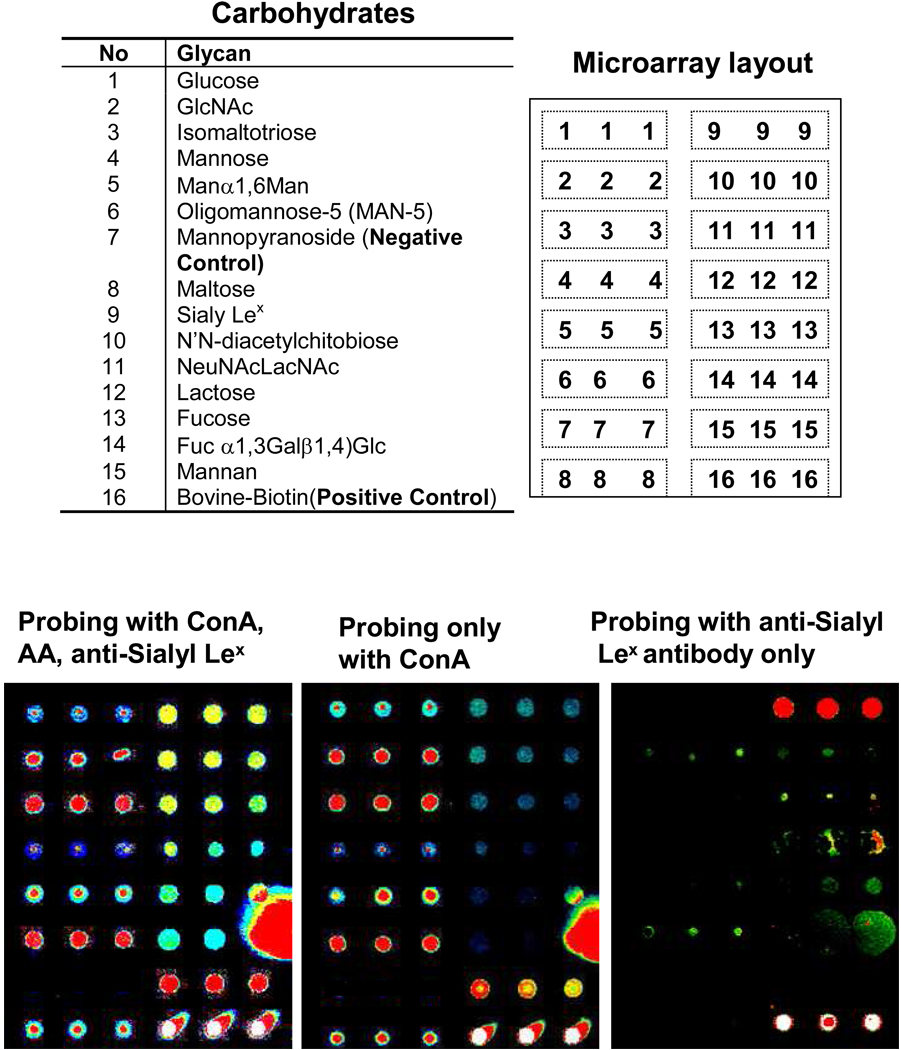
To demonstrate that the new slide surface is applicable for high-throughput production of carbohydrate microarrays, we examined the carbohydrate microarrays containing a panel of mono-, oligo-, and polysaccharides. Fifteen mono-, oligo-, and polysaccharides (see glycan list in Figure 9) in formamide buffer at a concentration of 100 µM were microspotted on the hydrazide/dendrimer slide surface. After being covalently bound under microwave radiation, the arrayed slides were blocked by treating with 0.1 × PBST, 1% BSA buffer for one hour. To examine the glyco-epitopes preserved by these carbohydrate microarrays, we probed them with either a mixed solution of biotin-labeled lectins and anti-Sialyl Lex antibody or probed with individual lectin/antibody separately, and finally stained with Cy3-Streptavidin. The binding profiles, as shown in Figure 9, showed that all these expected specific interactions are detected. For example, those probed with the mixed solution of Concanavalin A (ConA), A. aurantia (AA), Wheat Germ Agglutinin (WGA), and anti-Sialyl Lex showed detectable signals for all of the carbohydrates(Right image), except the Methyl Mannoside that could not be immobilized on the substrate due to the blocked reducing end and served as negative control in these arrays. As expected, microarrays probed with ConA only showed strong lectin binding to Isomaltotriose, Mannan, Man5, and less tight binding to maltose, Manα1,6Man, and Sialyl Lex, and very weak binding to mannose and Lactose (middle image). ConA (Concanavalin A) is Man-and/ or G1c-specific lectin requires the C-3, C-4, and C-5 hydroxyl groups of the Man or G1c ring for binding. Importantly, we observed significant ConA reactivities with monosaccharides, such as mannose and glucose, which was not detectable on 2-dimensional hydrazide/surfaces.45 The improved saccharide binding reactivities can be attributed to the multivalency (i.e., cluster effect) of the monosaccharides immobilized on the hydrazide/dendrimer surface. Lectin AA exhibited only strong binding to Fucose arrays, while lectin WGA exhibited strong binding to the arrays of oligosaccharides containing terminal N-acetylglucosamine or chitobiose, such as NeuNAcLacNAc, GlcNAc, and diacetlychitobiose (images not showed here). When probing with anti-Sialyl Lex antibody only, it showed that only the Sialyl Lex spots were recognized by the antibody (Left image). These results clearly verified that the immobilized carbohydrate probes on the hydrazide-dendrimer substrate surface preserved the specific glyco-epitopes of carbohydrates.
Figure 9.
Specificity of carbohydrate-lectin/antibody interactions on the carbohydrate microarrays fabricated on the hydrazide-dendrimer coated slide by microwave assisted immobilization. Spotted carbohydrate microarrays were blocked with 0.1× PBS buffer containing 1% BSA, 0.1% Tween 20 after treatment for 10 mins microwave radiation. The microarrays were then incubated with either a cocktail of 1 µg/mL of biotin-labeled lectins of ConA, AA. WGA and anti-Sialyl Lex antibody or individual lectins/antibody solution at 30 °C for 60 min, respectively, followed by stained with 0.1 µg/mL of Cy3-streptavidin at 30 °C for 60 min. The microarrays were washed with PBST buffer for 5 mins twice and 0.01 × PBS for 1 min before they scanned with confocal scanner under identical layer power and PMT. The binding profiles showed that all these interactions have been detected successfully, indicating the retention of specific binding activity of the immobilized carbohydrates.
Conclusion and Perspectives
We presented here development of 3-dimensional hydrazide-dendrimer and aminooxy-dendrimer substrates for construction of carbohydrate cluster microarrays. These technologies provide site-specific and covalent attachments of unmodified carbohydrates and support presentation of carbohydrates in a unique orientation and defined cluster configurations. Microwave radiation energy was introduced to facilitate saccharide immobilization. Optimum printing buffer (Formamide) and blocking buffer (0.1 × PBS buffer containing 1% BSA, 0.1% Tween 20) were selected for these novel carbohydrate microarray platforms. We further demonstrated (1) a wide range of unmodified carbohydrates of diverse structures are applicable for these 3-D substrates; (2) immobilization of the spotted carbohydrates are achieved by subjecting the arrayed slide to conventional microwave radiation for less than ten minutes; (3) the carbohydrate concentration required for microarray fabrication in these substrates is substantially reduced (~1–10 µM); (4) the carbohydrate cluster microarrays produced by this technology present glyco-epitopes for highly sensitive detection by specific carbohydrate-binding proteins; and (5) the 3-D hydrazide-dendrimer substrate gave better detection sensitivity than the aminooxy-dendrimer functionalized substrate for the panel of oligosaccharides tested in this study. Although the initial approach for preparing hydrazide-dendrimer and aminooxy-dendrimer functionalized substrates involved a three-step platform surface modification prior to robotic spotting of the sugars, the process can be simplified in future by using synthesized hydrazide-dendrimer or aminooxy-dendrimer as starting materials. Overall, this work has established novel bioarray substrates and methodology for construction of carbohydrate cluster microarray, which may serve as new analytical tools for high throughput functional glycomics exploration.
Acknowledgement
This work was supported by Grant Number 1R43 RR023763-01 and 2R44 RR023763-02 (X.C. Zhou) from the National Center for Research Resources Services (NCRR), and U01CA128416 (D. Wang) from National Cancer Institute (NCI), components of the National Institutes of Health (NIH),. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCRR or NCI or the National Institutes of Health.
References
- 1.Bertozzi CR, Kiessling LL. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 2.Lis H, Sharon N. Chem. Rev. 1998;98:77–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 3.Varki A, Cummings R, Esko JD, Freeze H, Hart G, Marth J. Essentials in Glycobiology. Plainview, NY: Cold Spring Harbor Laboratory Press; 2002. [PubMed] [Google Scholar]
- 4.Roseman S. J. Biol. Chem. 2001;276:41527–41542. doi: 10.1074/jbc.R100053200. [DOI] [PubMed] [Google Scholar]
- 5.(a) Seeberger PH, Werz DB. Nat. Rev. Drug Discovery. 2005;4:751–763. doi: 10.1038/nrd1823. [DOI] [PubMed] [Google Scholar]; (b) Werz DB, Seeberger PH. Chemistry-A European J. 2005;11:3194–3206. doi: 10.1002/chem.200500025. [DOI] [PubMed] [Google Scholar]; (c) Seeberger PH, Werz DB. Nature. 2007;446:1046–1051. doi: 10.1038/nature05819. [DOI] [PubMed] [Google Scholar]
- 6.(a) Stevens J, Blixt O, Paulson JC, Wilson IA. Nat. Rev. Microbiol. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rosati F, Capone A, Della Giovampaola C, Brettoni C, Focarelli R. Int. J. Dev. Biol. 2000;44(6):609–618. [PubMed] [Google Scholar]
- 7.Focarelli R, La Sala GB, Balasini M, Rosati F. Cells Tissues Organs. 2001;168(1–2):76–81. doi: 10.1159/000016808. [DOI] [PubMed] [Google Scholar]
- 8.Crocker PR, Feizi T. Curr. Opin. Struct. Biol. 1996;6(5):679–691. doi: 10.1016/s0959-440x(96)80036-4. [DOI] [PubMed] [Google Scholar]
- 9.Ziska SE, Henderson EJ. Proc. Natl. Acad. Sci. U.S.A. 1988;85(3):817–821. doi: 10.1073/pnas.85.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geijtenbeek TBH, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GCF, Middel J, Cornelissen ILMHA, Nottet HSLM, KewalRamani VN, Littman DR, Figdor CG, van Kooyk Y. Cell. 2000;100(5):587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 11.Geijtenbeek TBH, Torensma R, van Vliet SJ, van Duijnhoven GCF, Adema GJ, van Kooyk Y, Figdor CG. Cell. 2000;100(5):575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl U, Thunberg L, Backstrom G, Riesenfeld J, Nordling K, Bjork I. J. Biol. Chem. 1984;259(20):12368–12376. [PubMed] [Google Scholar]
- 13.Capila I, Linhardt RJ. Angew. Chem. Int. Ed. Engl. 2002;41(3):391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 14.Kansas GS. Blood. 1996;88(9):3259–3287. [PubMed] [Google Scholar]
- 15.Gardinali M, Padalino P, Vesconi S, Calcagno A, Ciappellano S, Conciato L, Chiara O, Agostoni A, Nespoli A. Arch. Surg. 1992;127(10):1219–1224. doi: 10.1001/archsurg.1992.01420100077014. [DOI] [PubMed] [Google Scholar]
- 16.Karlsson KA, Angstrom J, Bergstrom J, Lanne B. APMIS. 1992;100 Suppl.27:71–83. [PubMed] [Google Scholar]
- 17.Feizi T, Loveless RW. "Carbohydrate recognition by Mycoplasma pneumoniae and pathologic consequences.". Am. J. Respir. Crit. Care. Med. 1996;154(4 Pt 2):S133–S136. doi: 10.1164/ajrccm/154.4_Pt_2.S133. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, Kabat EA. Carbohydrate antigens (polysaccharides) In: Regenmortal MHVV, editor. Structure of Antigens. Vol. 3. Boca Raton. New York. London. Tokyo: CRC Press; 1996. pp. 247–276. [Google Scholar]
- 19.Paulson JC, Blixt O, Collins BE. Nat. Chem. Biol. 2006;2(5):238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 20.Wang D. Proteomics. 2003;3:2167–2175. doi: 10.1002/pmic.200300601. [DOI] [PubMed] [Google Scholar]
- 21.Ratner DM, Adams EW, Disney MD, Seeberger PH. ChemBioChem. 2004;5:1375–1383. doi: 10.1002/cbic.200400106. [DOI] [PubMed] [Google Scholar]
- 22.de Paz JL, Horlacher T, Seeberger PH. Methods Enzymol. 2006;415:269–292. doi: 10.1016/S0076-6879(06)15017-X. [DOI] [PubMed] [Google Scholar]
- 23.Feizi T, Chai W. Nat. Rev. Mol. Cell Biol. 2004;5(7):582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 24.Ratner DM, Seeberger PH. Curr. Pharm. Des. 2007;13(2):173–183. doi: 10.2174/138161207779313650. [DOI] [PubMed] [Google Scholar]
- 25.Horlacher T, Seeberger PH. OMICS. 2006;10(4):490–498. doi: 10.1089/omi.2006.10.490. [DOI] [PubMed] [Google Scholar]
- 26.Branderhorst HM, Ruijtenbeek R, Liskamp RMJ, Pieters RJ. ChemBioChem. 2008;9(11):1836–1844. doi: 10.1002/cbic.200800195. [DOI] [PubMed] [Google Scholar]
- 27.Zhou XC, Carroll GT, Turchi C, Wang D. “arbohydrate Microarrays as Essential Tools of Postgenomic Medicine”. In: Garg Hari G, Cowman Mary K, Hales Charles A., editors. Carbohydrate Chemistry and Biology for Physicians. New Jersey, USA: The Humana Press Inc; 2007. [Google Scholar]
- 28.de Paz JL, Seeberger PH. QSAR Comb. Sci. 2006;25:1027–1032. [Google Scholar]
- 29.Monzo A, Guttman A. QSAR Comb. Sci. 2006;25:1033–1038. [Google Scholar]
- 30.Culf AS, Cuperlovic-Culf M, Ouellette RJ. OMICS. 2006;10(3):289–310. doi: 10.1089/omi.2006.10.289. [DOI] [PubMed] [Google Scholar]
- 31.Park S, Lee MR, Shin I. Chem. Commun. 2008;37:4389–4399. doi: 10.1039/b806699j. [DOI] [PubMed] [Google Scholar]
- 32.(a) Park S, Lee MR, Shin I. Chem. Soc. Rev. 2008;37:1579–1591. doi: 10.1039/b713011m. [DOI] [PubMed] [Google Scholar]; (b) Park S, Lee MR, Shin I. Nature Protocols. 2007;2(11):2747–2758. doi: 10.1038/nprot.2007.373. [DOI] [PubMed] [Google Scholar]
- 33.Laurent N, Voglmeir J, Flitsch SL. Chem. Commun. 2008;37:4400–4412. doi: 10.1039/b806983m. [DOI] [PubMed] [Google Scholar]
- 34.(a) Horlacher T, Seeberger PH. Chem. Soc. Rev. 2008;37:1414–1422. doi: 10.1039/b708016f. [DOI] [PubMed] [Google Scholar]; (b) Vegas AJ, Fuller JH, Koehler AN. Chem. Soc. Rev. 2008;37:1385–1394. doi: 10.1039/b703568n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Nat. Biotechnol. 2002;20(3):275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 36.Wang D, Lu J. Physiol. Genomics. 2004;18(2):245–248. doi: 10.1152/physiolgenomics.00102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willats WGT, Rasmussen SE, Kristensen T, Mikkelsen JD, Knox JP. Proteomics. 2002;2:1666–1671. doi: 10.1002/1615-9861(200212)2:12<1666::AID-PROT1666>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 38.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Nat. Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 39.Song XZ, Xia BY, Lasanajak Y, Smith DF, Cummings RD. Glycoconjugate J. 2008;25:15–25. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- 40.Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, Cummings RD. J. Biol. Chem. 2008;283(15):10109–10123. doi: 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings RD, Bovin N, Wong CH, Paulson JC. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.(a) Houseman B, Gawalt E, Mrksich M. Langmuir. 2003;19:1522–1531. [Google Scholar]; (b) Ratner DM, Adams EW, Su J, O’Keefe BR, Mrksich M, Seeberger PH. ChemBioChem. 2004;5:379–382. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 43.Houseman BT, Mrksich M. Chem. Biol. 2002;9:443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 44.(a) Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Nat. Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]; (b) Liu Y, Feizi T, Carnpanero-Rhodes MA, Childs RA, Zhang YN, Muiioy B, Evans PG, Osborn HMI, Otto D, Crocker PR, Chai WC. Chem. Biol. 2007;14:847–859. doi: 10.1016/j.chembiol.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Palma AS, Feizi T, Zhang Y, Stoll MS, Lawson AM, Diaz-Rodriguez E, Campanero-Rhodes MA, Costa J, Gordon S, Brown GD, Chai W. J. Biol. Chem. 2006;281:5771–5779. doi: 10.1074/jbc.M511461200. [DOI] [PubMed] [Google Scholar]
- 46.Pohl NL. Angew. Chem., Int. Ed. 2008;47:3868–3870. doi: 10.1002/anie.200704801. [DOI] [PubMed] [Google Scholar]
- 47.Chen GS, Pohl NL. Org. Lett. 2008;10:785–788. doi: 10.1021/ol702915e. [DOI] [PubMed] [Google Scholar]
- 48.(a) Mamidyala SK, Ko KS, Jaipuri FA, Park G, Pohl NL. J. Fluorine Chem. 2006;127:571–579. [Google Scholar]; (b) Nicholson RL, Ladlow ML, Spring DR. Chem. Comm. 2007;38:3906–3908. doi: 10.1039/b712906h. [DOI] [PubMed] [Google Scholar]
- 49.Sun XL, Stabler CL, Cazalis CS, Chaikof EL. Bioconjugate Chem. 2006;17:52–57. doi: 10.1021/bc0502311. [DOI] [PubMed] [Google Scholar]
- 50.Kohn M, Wacker R, Peters C, Schroder H, Soulere L, Breinbauer R, Niemeyer C, Waldmann H. Angew. Chem., Int. Ed. 2003;42:5830–5834. doi: 10.1002/anie.200352877. [DOI] [PubMed] [Google Scholar]
- 51.(a) Pei Z, Yu H, Theurer M, Walden A, Nilsson P, Yan M, Ramstrom O. ChemBioChem. 2007;8:166–168. doi: 10.1002/cbic.200600447. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pei Y, Yu H, Pei Z, Theurer M, Ammer C, Andre S, Gabius H-J, Yan M, Ramstrom O. Anal. Chem. 2007;79(18):6897–6902. doi: 10.1021/ac070740r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leteux C, Stoll MS, Childs RA, Chai W, Vorozhaikina M, Feizi TJ. Immunol. Methods. 1999;227:109–119. doi: 10.1016/s0022-1759(99)00077-0. [DOI] [PubMed] [Google Scholar]
- 53.Leteux C, Childs RA, Chai WG, Stoll MS, Kogelberg H, Feizi T. Glycobiology. 1998;8:227–236. doi: 10.1093/glycob/8.3.227. [DOI] [PubMed] [Google Scholar]
- 54.Carroll GT, Wang D, Turro NJ, Koberstein JT. Langmuir. 2006;22(6):2899–2905. doi: 10.1021/la0531042. [DOI] [PubMed] [Google Scholar]
- 55.Wang D, Carroll GT, Turro NJ, Koberstein JT, Kovac P, Saksena R, Adamo R, Herzenberg LA, Herzenberg LA, Steinman L. Proteomics. 2007;7(2):180–184. doi: 10.1002/pmic.200600478. [DOI] [PubMed] [Google Scholar]
- 56.Lee M-r, Shin I. Org. Lett. 2005;7:4269–4272. doi: 10.1021/ol051753z. [DOI] [PubMed] [Google Scholar]
- 57.Zhi ZL, Powell AK, Turnbull JE. Anal. Chem. 2006;78(14):4786–4793. doi: 10.1021/ac060084f. [DOI] [PubMed] [Google Scholar]
- 58.Zhou XC, Zhou JZ. Biosens. Bioelectron. 2006;21:1451–1458. doi: 10.1016/j.bios.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 59.(a) Zhou XC, Zhou JZ. Anal. Chem. 2004;76(18):5302–5312. doi: 10.1021/ac049472c. [DOI] [PubMed] [Google Scholar]; (b) vanHest JCM, Delnoye DAP, Baars MWPL, van Genderen MHP, Meijer EW. Science. 1995;268:1592–1595. doi: 10.1126/science.268.5217.1592. [DOI] [PubMed] [Google Scholar]; (c) Le Berre V, Trevisiol E, Dagkessamanskaia A, et al. Nucleic Acids Res. 2003;31(16):e88. doi: 10.1093/nar/gng088. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Fréchet JMJ. Science. 1994;263:1710–1715. doi: 10.1126/science.8134834. [DOI] [PubMed] [Google Scholar]; (e) Zhou XC, Wu LY, Zhou JZ. Langmuir. 2004;20(20):8877–8885. doi: 10.1021/la048950b. [DOI] [PubMed] [Google Scholar]
- 60.(a) Leteux C, Childs RA, Chai W, Stoll MS, Kogelberg H, Feizi T. Glycobiology. 1998;8:227–236. doi: 10.1093/glycob/8.3.227. [DOI] [PubMed] [Google Scholar]; (b) Shinohara Y, Sota H, Gotoh M, Hasebe M, Tosu M, Nakao J, Hasegawa Y. Anal. Chem. 1996;68:2573–2579. doi: 10.1021/ac960004f. [DOI] [PubMed] [Google Scholar]
- 61.(a) Hatanaka Y, Kempin U, Park J-J. J. Org. Chem. 2000;65:5639–5643. doi: 10.1021/jo000414a. [DOI] [PubMed] [Google Scholar]; (b) Zhao Y, Kent SB, Chait BT. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1629–1633. doi: 10.1073/pnas.94.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brask J, Jensen KJ. J. Pept. Sci. 2000;6:290–299. doi: 10.1002/1099-1387(200006)6:6<290::AID-PSC257>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 63.(a) Bendiak B. Carbohydr. Res. 1997;304:85–90. doi: 10.1016/s0008-6215(97)00213-9. [DOI] [PubMed] [Google Scholar]; (b) Ernholt BV, Thomsen IB, Lohse A, Plesner IW, Jensen KB, Hazell RG, Liang X, Jakobsen A, Bols M. Chem. Eur. J. 2000;6(2):278–287. doi: 10.1002/(SICI)1521-3765(20000117)6:2<278::AID-CHEM278>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]; (c) Ojala CR, Ostman JM, Ojala WH. Carbohydr. Res. 2002;337:21–29. doi: 10.1016/s0008-6215(01)00278-6. [DOI] [PubMed] [Google Scholar]
- 64.(a) Carrasco MR, Nguyen MJ, Burnell DR, MacLaren MD, Hengel SM. Tetrahedron Lett. 2002;43(33):5727–5729. [Google Scholar]; (b) Carrasco MR, Brown RT, Serafimova IM, Silva O. J. Org. Chem. 2003;68:195–197. doi: 10.1021/jo026641p. [DOI] [PubMed] [Google Scholar]; (c) Peri F, Dumy P, Mutter M. Tetrahedron. 1998;54:12269–12278. [Google Scholar]
- 65.(a) Branderhorst HM, Ruijtenbeek R, Liskamp RMJ, Pieters RJ. ChemBioChem. 2008;9:1836–1844. doi: 10.1002/cbic.200800195. [DOI] [PubMed] [Google Scholar]; (b) Kikkeri R, Hossain LH, Seeberger PH. ChemComm. 2008:2127–2129. doi: 10.1039/b802177e. [DOI] [PubMed] [Google Scholar]
- 66.Caddick S. Tetrahedron. 1995;51:10403–10432. [Google Scholar]
- 67.Varma RS. Advances in Green Chemistry: Chemical Synthesis Using Microwave Irradiation. Banglore, India: Astrazeneca Research Foundation; 2002. [Google Scholar]
- 68.Gerbec JA, Magana D, Washington A, Strouse GF. J. Am. Chem. Soc. 2005;127:15791–15800. doi: 10.1021/ja052463g. [DOI] [PubMed] [Google Scholar]
- 69.Tsuji M, Hashimoto M, Nishizawa Y, Tsuji T. Mater. Lett. 2004;58:2326–2330. [Google Scholar]
- 70.Roy I, Gupta MN. Curr. Sci. 2003;85(12):1685–1693. [Google Scholar]
- 71.Bismuto E, Mancinelli F, d'Ambrosio G, Massa R. Eur. Biophy. J. 2003;32:628–634. doi: 10.1007/s00249-003-0310-2. [DOI] [PubMed] [Google Scholar]
- 72.(a) Pagnotta M, Pooley CLF, Gurland B, Choi M. J. Phys. Org. Chem. 1993;6:407–411. [Google Scholar]; (b) Yates EA, Jones MO, Caroline E, Clarke CE, Powell AK, Johnson SR, Porch A, Edwards PP, Turnbull JE. J. Mater. Chem. 2003;13:2061–2063. [Google Scholar]