Abstract
The cerebral cortex has diverse types of inhibitory neurons. In rat cortex, past research has shown that parvalbumin (PV), somatostatin (SOM), calretinin (CR), and cholecystokinin (CCK) label four distinct chemical classes of GABAergic interneurons. However, in contrast to rat cortex, previous studies indicate that there is significant co-localization of SOM and CR in mouse cortical inhibitory neurons. In the present study, we further characterized immunochemical distinctions among mouse inhibitory cortical neurons by double immunochemical labeling with chemical markers. We found that, PV, SOM and vasointenstinal peptide (VIP) reliably identify three non-overlapping distinct subpopulations, as there was no overlap of immunoreactivity between PV and all the other chemical markers tested, and SOM and VIP did not show any overlap in labeled neurons in all the cortical areas. In comparison, there was significant overlap in combinations of other chemical markers. With some laminar and regional variations, the average overlap of SOM/CR (percentage of SOM+ cells expressing CR) and SOM/neuropeptide tyrosine (NPY) across all examined layers and cortical regions was 21.6% and 7.1%, respectively. The average overlap of VIP/CR, VIP/NPY, and CR/NPY was 34.2%, 9.5% and 10%, respectively. We quantified and assessed the percentages of marker-positive GABAergic cells, and showed that the non-overlapping subpopulations (i.e., PV+, SOM+ and VIP+ cells) accounted for about 60% of the GABAergic cell population. Taken together, our data reveal important chemical distinctions between mouse inhibitory cortical neurons and indicate that PV, SOM, and VIP can differentially label a majority of mouse inhibitory cortical neurons.
Keywords: interneurons, immunoreactivity, frontal cortex, barrel cortex, visual cortex
Introduction
Inhibitory neurons comprise about 20% of all cortical neurons and are crucial to cortical function. They can be further subdivided into numerous classes or subtypes based on their immunochemical, morphological, and physiological properties (Somogyi et al., 1998; Kawaguchi and Kondo, 2002; Markram et al., 2004; Ascoli et al., 2008; Burkhalter, 2008). Immunostaining for chemical markers is an important tool for inhibitory cell type classification and identification; because these markers are often closely correlated with morphology and/or physiology, they can serve as a proxy for complete characterization when other measures are not practical. The most informative markers are those that have minimal overlap with other markers and that correlate closely with other features. These include parvalbumin (PV), calretinin (CR), somatostatin (SOM), vasointenstinal peptide (VIP) and cholecystokinin (CCK), among others (DeFelipe, 1993; Kubota et al., 1994; Kawaguchi and Kubota, 1996; Gonchar and Burkhalter, 1997; Kawaguchi and Kubota, 1997; Gonchar et al., 2007). For example, most PV-immunopositive neurons are fast-spiking (FS) basket cells or chandelier cells; most SOM-immunopositive neurons are Martinotti cells; and CR-immunopositive neurons have been characterized as bipolar or double-bouquet cells; large CCK-immunopositive neurons have been characterized as non-FS basket cells including irregular spiking basket cells expressing type 1 G-protein-coupled cannabinoid receptors (CB1) (Kawaguchi and Kubota, 1996, 1997; Oliva et al., 2000; Kawaguchi and Kondo, 2002; Chattopadhyaya et al., 2004; Galarreta et al., 2004; Xu et al., 2006; Miyoshi et al., 2007). As a result, these markers have emerged as extremely valuable tools for the classification and identification inhibitory neuron subtypes.
Despite the value of these markers, it has been clear for many years that in some cases cell types which appear morphologically or physiologically similar in different species may not stain for the same chemical markers in one species as they do in another (e.g., cats, monkeys and rats) (Hendry et al., 1989; Hendry and Jones, 1991; Kubota et al., 1994; Meskenaite, 1997). Nevertheless, considerable progress has been made in correlating cell types with chemical markers, particularly in the cortex of the rat, where PV, SOM, CR and CCK label four distinct and non-overlapping classes of GABAergic interneurons, which account for a great majority of the whole inhibitory neuronal population (Kubota et al., 1994; Gonchar and Burkhalter, 1997; Kawaguchi and Kubota, 1997; Xu et al., 2006). Furthermore, VIP expression in the rat cortex overlaps extensively with both CCK and CR, but not with PV or SOM (Kawaguchi and Kubota, 1997). Thus an alternative parcellation in rat cortex would create 3 non-overlapping groups: PV+, SOM+, and VIP+ inhibitory neurons.
As the mouse has emerged as an increasingly important model for the study of cortical circuits, it has been revealed that, even amongst relatively closely related rodent species, rat and mouse, there is variability in the expression of chemical markers in cortex (Xu et al., 2006; Gonchar et al., 2007; Miyoshi et al., 2007). The most striking difference is that, in contrast to the rat, in mouse cortex there is significant co-localization of SOM and CR (Xu et al., 2006; Xu and Callaway, 2009). This suggests that CR is not a particularly useful marker in the mouse because it does not distinguish a population that is distinct from SOM+ cells. But this left open the possibility that VIP might be a useful alternative which could identify a population of PV-negative and SOM-negative cells, as in the rat. Recently this possibility was questioned by a study of mouse visual cortex, where considerable overlap between SOM and several other markers, most prominently VIP, was observed (Gonchar et al., 2007).
To further understand these relationships, we embarked on a systematic and quantitative analysis of the expression of chemical markers across cortical areas and layers in the mouse. Our data reveal important chemical distinctions among mouse cortical inhibitory neurons, and indicate that immunoreactivity for PV, SOM and VIP distinguishes three major non-overlapping classes of mouse inhibitory cortical neurons.
Materials and Methods
Tissue preparation
All animals were handled and experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee at the Salk Institute. Eight adult C57BL/6 mice and 2 GAD67-GFP knock-in (Δneo) transgenic mice (Tamamaki et al., 2003) were use for this study. In the GAD67-GFP knock-in mouse, it is believed that all the GABAergic cells express GFP, and all the GFP-positive cells are positive for GAD67 and GABA (Tamamaki et al., 2003).
Following anesthetization with over-dosage of Nembutal (sodium pentobarbital, >100mg/kg, i.p.), mice were transcardially perfused with (1) 0.1M phosphate buffered saline (PBS, pH 7.3–7.4, 10–20 ml) followed by 100–150ml of a fixative containing 4% paraformaldehyde (PFA) in 0.1M PBS, or (2) 0.1M PBS followed by 100–150ml of a fixative containing 4% PFA and 0.2% glutaraldehyde in 0.1M PBS. The latter perfusion better preserved neurotransmitters in brain tissues and were specifically designated for GABA immunohistochemistry. The brain was removed from the skull, post-fixed overnight in the same fixative (4°C) and then transferred to 30% sucrose in PBS for at least two days. Parasaggital sections were cut on a freezing microtome at 25 μm in thickness for immunostaining.
Antibody information
In order to study the colocalization of two neurochemical markers in wild type mouse cortex, we immunochemically double labeled sections using different combinations of parvalbumin (PV), somatostatin (SOM), calretinin (CR), vasoactive intestinal peptide (VIP), and neuropeptide tyrosine (NPY) antibodies originated in different host species. As shown in Table 1, the primary antibodies used in this study include: rat α-somatostatin antibody(Chemicon, Temecula, CA), rabbit α-parvalbumin antibody(Swant, Bellinzona, Switzerland), goat α-parvalbumin antibody (Swant), rabbit α-calretinin antibody(Swant), goat α-calretinin antibody(Swant), rabbit α-neuropeptide tyrosine (NPY) antibody(Chemicon), sheep α-neuropeptide tyrosine antibody(Chemicon), and rabbit α-vasoactive intestinal peptide (VIP) antibody(ImmunoStar, Hudson, WI). Additionally, in order to determine the chemical content of GABA immunoreactive cells, we double labeled sections either with rabbit α-γ aminobutyric acid (GABA) polyclonal antibody (Sigma, St. Louis, MO) in wild type mouse cortex, or with chicken anti-GFP antibody (for GFP expressing GABAergic cells)(Aves Labs, Tigard, OR) in the transgenic mouse cortex and one of the primary antibodies of the chemical markers. Then the secondary antibodies for fluorescent visualization included Cy2-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA; 711-225-152,1:200), Cy3-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch, 711-165-152, 1:200), Cy2-conjugated donkey anti-goat IgG (Jackson ImmunoResearch, 805-225-180, 1:200), Cy3-conjugated donkey anti-goat IgG (Jackson ImmunoResearch, 805-165-180, 1:200), Cy2-conjugated donkey anti-rat IgG (Jackson ImmunoResearch, 712-225-150, 1:200), Cy3-conjugated donkey anti-rat IgG (Jackson ImmunoResearch, 712-165-150, 1:200), Cy2-conjugated donkey anti-sheep IgG (Jackson ImmunoResearch, 713-225-003, 1:200), and donkey anti-sheep Alexa Fluor 488 IgG (Invitrogen, Carlsbad, CA; A-11015, 1:200). None of the observed labeling was due to non-specific binding of secondary antibodies or auto-fluroscence in the fixed tissue because sections labeled with secondary antibodies alone showed no detectable labeling. Please also note that secondary antibody specificity for double label experiments was tested by the manufacturer (Jackson ImmunoResearch) by ELISA and/or solid-phase adsorbed to ensure minimal cross reaction with the other antibody hosts we used, which prevents the secondary antibodies from cross-reacting with IgG raised in different species.
Table 1.
Primary antibodies used for immunostaining
| Anitgen | Immunogen | Manufacturing details | Working dilution |
|---|---|---|---|
| CCK | Synthetic human gastrin/CCK 2–17 conjugated with carbodiimide to keyhole limpet hemocyanin | UCLA Cure (Los Angeles, CA), mouse monoclonal, 9303 | 1:1000 |
| CR | Human recombinant calretinin | Swant (Bellinzona, Switzerland), goat polyclonal, CG1 | 1:500 |
| CR | Human recombinant calretinin | Swant, rabbit polyclonal, 7699/4 | 1:1000 |
| GABA | (γ-Aminobutyric Acid)-BSA | Sigma-Aldrich (St. Louis, MO), rabbit polyclonal, A2052 | 1:5000 |
| GFP | Purified recombinant green fluorescent protein | Aves Labs (Tigard, OR), chicken polyclonal, GFP-1020 | 1:500 |
| NOS | Synthetic peptide corresponding to amino acid residues 1414–1429 of the rat NOS-I protein | Millipore/Chemicon (Temecula, CA), rabbit polyclonal, AB1552 | 1:250 |
| NPY | Synthetic porcine neuropeptide tyrosine (NPY) | Millipore/Chemicon, rabbit polyclonal, AB1915 | 1:1000 |
| NPY | Synthetic NPY | Millipore/Chemicon, sheep polyclonal, AB1583 | 1:1000 |
| PV | Rat muscle parvalbumin | Swant, goat polyclonal, PVG-214 | 1:1000 |
| PV | Rat muscle parvalbumin | Swant, rabbit polyclonal, PV-28 | 1:1000 |
| SOM | Synthetic 1–14 cyclic somatostatin | Millipore/Chemicon, rat monoclonal, MAB354 | 1:200 |
| VIP | Porcine VIP conjugated to bovine thyroglobulin with carbodiimide | ImmunoStar, Inc. (Hudson, WI), rabbit polyclonal, 20077 | 1:500 |
Antibody characterization and specificity
All primary antibodies used in our staining experiments are commercially available from major companies. According to the technical datasheet from Sigma, the GABA antibody is isolated from antiserum by immunospecific methods of purification; and the antibody shows positive binding with GABA in a dot blot assay, and negative binding with bovine serum albumin (BSA). Based upon technical information from Swant, the specificity of parvalbumin antibody is determined by immunoblots of brain and muscle extracts; the antibody stains a subpopulation of neurons in the normal brain with high efficiency, but does not stain the brain tissue of parvalbumin knock out mice. The calretinin antibody reacts specifically with calretinin in tissue originating from primates, the rat and the mouse. This antibody does not cross-react with calbindin D-28k or other known calcium binding-proteins, as determined by immunoblots and by its distribution in the brain, with the antibody specifically recognizing a band of 29–30k Da in the Western blot. We also carried out immunoabsorption tests to confirm specificity of the parvalbumin and calretinin antibodies. The preincubation of the calretinin antibody with purified proteins, recombinant human calretinin (Swant, 5 μg/mL) and the preincubation of the parvalbumin antibody with recombinant rat parvalbumin (Swant, 5 μg/mL) completely blocked the immunoactivity of the parvalbumin or calretinin antibody, respectively, by showing no detectable labeling in frontal cortex, somatosensory and visual cortical regions on the mouse sections.
According to technical information from Millipore/Chemicon, the somatostatin antibody recognizes somatostatin, and does not show cross-reactivity to enkephalins, other endorphins, substance P. We preincubated the somatostatin antibody with 10 μg/mL synthetic somatostatin peptide (Sigma, S9129), which entirely blocked the antibody’s immunoactivity in the cortical regions examined on the mouse sections.
The specificity of the NPY antibodies supplied by Millipore/Chemicon was determined by radio immuno assays (Millipore/Chemicon data sheet). The antibodies showed less than 0.005% cross reactivity with synthetic peptides somatostatin-14, VIP, angiotensin II, and oxytocin and less than 0.02% cross reactivity with substance P. Synthetic NPY completely abolishes radio labeled NPY binding to NPY antibody in RIA and antibody binding to tissue antigen. Specificity of the VIP antibody is determined by pre absorption with synthetic VIP peptides at a concentration of 10−5 M; staining of VIP is completely blocked (ImmunoStar data sheet). We independently confirmed the specificity of the VIP antibody and the NPY antibody by immunoabsorption tests, using 5–10 μg/mL of synthetic peptides (VIP: Sigma, V6130; NPY: Sigma, N3266).
In addition, we compared our antibody staining results with published examples using the same antibodies. For example, the staining of SOM cells in mouse hippocampus was consistent with that reported by Oliva et al. (2000). Some SOM-stained cells in mouse frontal cortex and S1 were morphologically similar to the SOM-positive cells reported in rat cortex, which are bitufted or multipolar and have a large soma (Kubota et al., 1994; Kawaguchi and Kubota, 1996; Miyoshi et al., 2007). The CR-stained cells in mouse visual cortex tended to be uni- or bipolar and slender, which is similar to that reported in rat visual cortex by Gonchar and Burkhalter (1997). The PV-stained cells resembled basket cells reported in both mouse and rat cortex (Kubota et al., 1994; Miyoshi et al., 2007; Uematsu et al., 2008). None of the observed labeling was due to nonspecific binding of secondary antibodies or autofluorescence in the fixed tissue because sections labeled with secondary antibodies alone showed no detectable labeling.
Antibodies against cholecystokinin (CCK) and nitric oxide synthase (NOS) have also been used in examining chemical properties of rodent cortical inhibitory neurons (Kawaguchi and Kubota, 1997; Gonchar et al., 2007). We examined immunostaining against CCK using the mouse monoclonal CCK antibody (The Center for Ulcer Research and Education (CURE), University of California (Los Angeles, CA), 9303, 1:1000) and the NOS antibody (Chemicon, AB1552, 1:250) in mouse cortical sections, but found that the CCK+ or NOS+ cells were quite scarce in mouse cortex. We did not further quantify their distribution or examine their distinction/overlap with other markers.
Immunochemical staining procedures
To stain tissue with antibodies, conventional immunofluorescent staining was performed. Free-floating sections placed in clean 24-well plates were initially rinsed 3–5 times with PBS. Sections were then rinsed 3–5 times with PBS with 0.1% Triton X-100 and then incubated in a blocking solution for 2 hours at room temperature. The blocking solution contained 5% normal donkey serum (Jackson ImmunoResearch, 017-000-121), 0.2% cold water fish skin gelatin (Sigma, G7765), and 0.25% Triton X in PBS. Following incubation in blocking solution, sections were then transferred to a blocking solution containing the primary antibodies for 36 hours at 4°C. After incubation in primary antisera, sections were set out at room temperature and allowed to equilibrate to the temperature for an hour. The sections were then rinsed 5 times in PBS and then 5 times in a working buffer containing 10% blocking solution in PBS. Following a thorough rinsing, sections were then incubated with appropriate secondary antibodies in blocking solution for 3–5 hours. After incubation with secondary antibodies and another rinse with PBS, sections were counterstained with a 10μM 4′,6-diamidino-2-phenylindole (DAPI, Sigma, D-9542) solution for 10 minutes in order to help distinguish laminar boundaries of cortex. Sections were then rinsed 3 times with PBS and wet mounted onto subbed slides. Following air-drying overnight, sections were dehydrated, defatted, and cover-slipped with Krystalon mounting medium (EMD Chemicals Harleco, 64969-95).
For immunoabsorption control tests, the primary antibodies were first preincubated with synthetic peptides or purified proteins overnight in blocker solution, and this solution was then applied to the sections for 24 hours and the remaining staining procedures were completed as described above. We did not see any specific immunolabeling in the sections used for these control tests.
Quantification and image acquisition
For cell counting and quantification of immunolabeled neurons, sections were viewed with a Nikon Optiphot-2 upright epifluorescent microscope equipped with a digital camera (Optronics, Goleta, CA), using filter sets from Omega Optical Inc. (Brattleboro, VT), and Neurolucida software (MicroBrightField, Colchester, VT) was used to acquire data images and plot the distributions of immunopositive cells. Stained cells were counted across all the layers in the cortical regions. The data were exported, and custom programs in Matlab (MathWorks, Inc., Natick MA) were used to do further quantitative analysis such as laminar distributions and percent co-localization of chemical markers. The final cell counts were corrected using the Abercrombie method, i.e., the ratio of the “real” number to the observed number is T/(T+h), where T = section thickness and h = the mean diameter of the nuclei of cells along the axis perpendicular to the cutting plane of the section (Guillery, 2002). The T in our study was 25 μm, and the average h used in the study was 8 μm.
We determined cortical layers in sections at a low magnification (4x or 10x objective) based upon DAPI staining for fluorescent sections. We examined the anterior medial portion of frontal cortex, which lacks a granular layer 4, but DAPI staining does reveal a homologous region which we identified as layer 4 using the same criteria as for other areas.
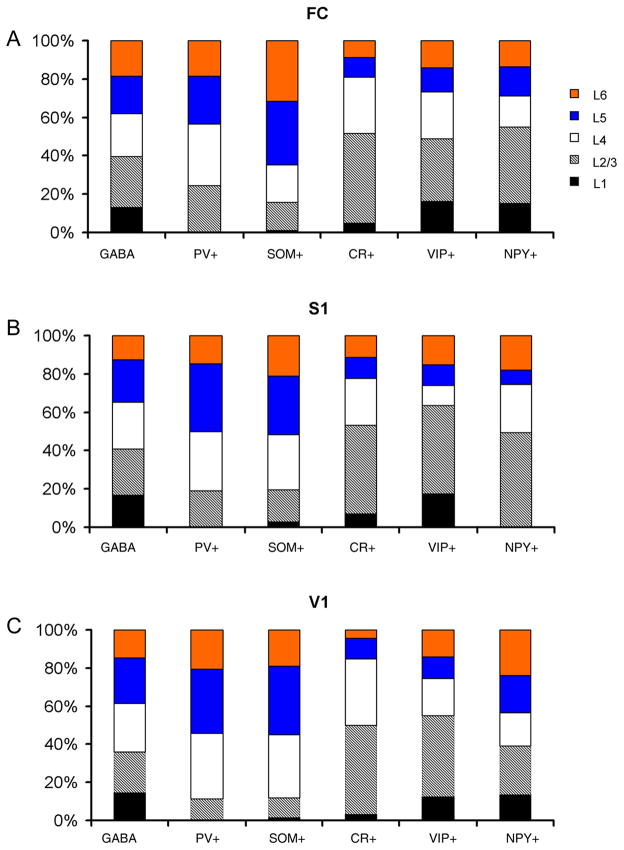
Analysis of co-localization of chemical markers was performed at higher magnification (20x objective) by examining well-stained cells which had visible cell bodies and dendritic processes. The cells were recorded using Neurolucida’s cell counting utility. In order to determine if a cell was indeed positive both for one marker and the other of the selected neurochemical markers, images from the same region of the section were acquired at the same focal plane using separate filter sets to identify Cy2 or Alexa Fluor 488(green) and Cy3 (red) labeled cells, and then the two images were overlaid in Adobe Photoshop (Adobe, San Jose, CA) to determine co-localization. This allowed for accurate identification of double labeled cells. We measured the overlap percentages of marker expressions, and marker-positive cell densities (Tables 2 and 3). At least 4 sections from different regions for each marker were quantified; and data was reported as the values of mean ± SD (Table 2). We compared the density of marker-positive cells with the density of GABAergic cells to estimate the individual percentages of marker specific cells (Table 4). The laminar percentages of marker-positive cells were calculated as [individual laminar densities/all summed densities of all the layers] × 100% (Figure 5).
Table 2.
GABAergic and marker-positive cell densities in different cortical layers of mouse frontal cortex (FC), primary somatosensory cortex (S1) and visual cortex (V1)
| FC | ||||||
|---|---|---|---|---|---|---|
| GABAergic | PV | SOM | CR | NPY | VIP | |
| across layers | 434.4 (49.9) | 171.6 (26.2) | 85 (12.1) | 78.4 (3.8) | 45.9 (3.2) | 32.5 (1.8) |
| layer 1 | 283.2 (41.2) | 0 (0.0) | 4.9 (4.6) | 16.7 (1.8) | 42.4 (7.2) | 22.4 (6.9) |
| layer 2/3 | 581 (82.6) | 215 (45.2) | 58.2 (15.0) | 178.1 (2.5) | 90.1 (7.3) | 62.1 (11.2) |
| layer 4 | 486.2 (74.4) | 242.6 (1.9) | 76.9 (22.8) | 124.9 (18.5) | 40.3 (2.3) | 25.5 (20.7) |
| layer 5 | 407.2 (43.1) | 226.6 (54.4) | 130 (23.3) | 28.6 (14.2) | 27.1 (13.7) | 23.4 (3.7) |
| layer 6 | 387.5 (56.5) | 154.8 (20.9) | 124.6 (16.2) | 30.7 (3.1) | 33.7 (1.5) | 29.8 (8.6) |
| S1 | ||||||
|---|---|---|---|---|---|---|
| GABAergic | PV | SOM | CR | NPY | VIP | |
| across layers | 541.2 (88.1) | 204.7 (33.7) | 88.8 (5.7) | 86 (2.1) | 53 (9.6) | 37.9 (9.2) |
| layer 1 | 468.8 (61.4) | 0 (0.0) | 10.7 (18.6) | 30.2 (2.7) | 0 (0.0) | 33.4 (3.8) |
| layer 2/3 | 674.3 (111.9) | 179.5 (51.1) | 67.1 (7.0) | 195.3 (18.4) | 117.4 (33.5) | 87.6 (26.3) |
| layer 4 | 680.8 (143.5) | 295.8 (69.6) | 115.2 (29.3) | 104.4 (3.4) | 60.3 (6.0) | 19.7 (9.4) |
| layer 5 | 607.9 (109.6) | 342.1 (14.1) | 120.8 (38.7) | 46.1 (14.7) | 17.5 (6.9) | 21.4 (12.0) |
| layer 6 | 355.1 (76.8) | 138.1 (31.5) | 85.1 (14.6) | 48.9 (10.2) | 43.3 (1.6) | 28.5 (9.4) |
| V1 | ||||||
|---|---|---|---|---|---|---|
| GABAergic | PV | SOM | CR | NPY | VIP | |
| across layers | 646 (23.5) | 262.1 (3.3) | 109.5 (21.9) | 136.4 (15.3) | 45.2 (2.1) | 54.2 (5.8) |
| layer 1 | 486.3 (46.7) | 0 (0.0) | 8.7 (9.9) | 50.6 (41.2) | 20.9 (14.1) | 25.2 (7.3) |
| layer 2/3 | 677.9 (78.1) | 140.4 (11.3) | 53.4 (13.6) | 296.2 (15.0) | 70.9 (18.2) | 113.9 (14.7) |
| layer 4 | 805.1 (63.1) | 405.5 (6.9) | 169.1 (32.4) | 221.9 (8.8) | 38.9 (1.8) | 52.1 (7.7) |
| layer 5 | 735 (41.8) | 399.3 (1.4) | 182.9 (48.2) | 76.7 (15.0) | 24.9 (26.4) | 30.4 (3.1) |
| layer 6 | 475 (60.0) | 243.8 (0.0) | 97.1 (32.9) | 33.1 (9.0) | 55.7 (1.3) | 38.8 (6.5) |
Note that the numbers in the tables above are formatted as mean ± (SD). The densities were obtained from measurement of 4–6 cortical sections. The GABAergic cell density refers to the density of GFP-expressing GABAergic cells measured from the GAD-GFP transgenic mouse cortex.
Table 3.
The average percentage overlap of chemical markers across all cortical layers (1–6) in mouse frontal cortex (FC), primary somatosensory cortex (S1) and visual cortex (V1)
| I. FC | ||||||
|---|---|---|---|---|---|---|
| SOM | CR | VIP | NPY | PV | ||
| All Layers | SOM | 26.8(294/1098) | 0(0/960) | 9.3(49/526) | 0.1(1/781) | |
| CR | 41.2(294/713) | 29.9(155/519) | 8.5(56/662) | 0(0/432) | ||
| VIP | 0(0/391) | 30.3(155/512) | 10.4(41/394) | 0(0/370) | ||
| NPY | 25.4(49/193) | 12.4(56/452) | 8.4(41/489) | 0.5(2/389) | ||
| PV | 0.1(1/1048) | 0(0/1337) | 0(0/1463) | 0.2(2/1266) | ||
| Layer 1 | SOM | 57.1(4/7) | 0(0/10) | 16.7(1/6) | 0(0/5) | |
| CR | 17.4(4/23) | 5.6(1/18) | 14.3(2/14) | 0(0/24) | ||
| VIP | 0(0/37) | 2.3(1/44) | 2.5(1/40) | 0(0/19) | ||
| NPY | 5.9(1/17) | 5.7(2/35) | 3.1(1/32) | 0(0/31) | ||
| PV | 0(0/3) | 0(0/4) | 0(0/1) | 0(0/0) | ||
| Layer 2/3 | SOM | 81.4(149/183) | 0(0/124) | 11.5(9/78) | 0(0/138) | |
| CR | 41.2(149/362) | 28.6(71/248) | 5.1(16/314) | 0(0/234) | ||
| VIP | 0(0/207) | 26.3(71/270) | 12.9(26/201) | 0(0/208) | ||
| NPY | 12.9(9/70) | 10.2(16/157) | 14.6(26/178) | 0(0/159) | ||
| PV | 0(0/250) | 0(0/326) | 0(0/280) | 0(0/280) | ||
| Layer 4 | SOM | 44(73/166) | 0(0/158) | 2.8(3/107) | 0(0/132) | |
| CR | 41(73/178) | 27.2(37/136) | 4.4(7/159) | 0(0/98) | ||
| VIP | 0(0/41) | 53.6(37/69) | 17.5(10/57) | 0(0/45) | ||
| NPY | 9.7(3/31) | 9.5(7/74) | 14.5(10/69) | 1.7(1/60) | ||
| PV | 0(0/285) | 0(0/334) | 0(0/364) | 0.3(1/329) | ||
| Layer 5 | SOM | 11.5(51/443) | 0(0/148) | 6.5(13/200) | 0.4(1/259) | |
| CR | 51.5(51/99) | 39.5(30/76) | 14.5(16/110) | 0(0/57) | ||
| VIP | 0(0/62) | 42.9(30/70) | 3.8(2/52) | 0(0/49) | ||
| NPY | 33.3(13/39) | 18.4(16/87) | 1.8(2/109) | 1.5(1/66) | ||
| PV | 0.3(1/344) | 0(0/448) | 0(0/563) | 0.2(1/439) | ||
| Layer 6 | SOM | 5.7(17/299) | 0(0/265) | 17(23/135) | 0(0/247) | |
| CR | 33.3(17/51) | 39(16/41) | 23.1(15/65) | 0(0/19) | ||
| VIP | 0(0/44) | 27.1(16/59) | 4.5(2/44) | 0(0/49) | ||
| NPY | 63.9(23/36) | 15.2(15/99) | 2(2/101) | 0(0/73) | ||
| PV | 0(0/166) | 0(0/225) | 0(0/255) | 0(0/218) | ||
| II. S1 | ||||||
|---|---|---|---|---|---|---|
| SOM | CR | VIP | NPY | PV | ||
| All Layers | SOM | 15.1(240/1587) | 0(0/1482) | 6.5(50/770) | 0(0/1220) | |
| CR | 33(240/728) | 32.4(149/460) | 12.3(72/585) | 0(0/290) | ||
| VIP | 0(0/510) | 26.5(149/562) | 11.8(52/441) | 0(0/332) | ||
| NPY | 19.1(50/262) | 11.8(72/609) | 7.7(52/672) | 0.5(3/657) | ||
| PV | 0(0/1975) | 0(0/1883) | 0(0/1264) | 0.1(3/2512) | ||
| Layer 1 | SOM | 90.9(10/11) | 0(0/11) | 0(0/3) | 0(0/3) | |
| CR | 40(10/25) | 0(0/18) | 0(0/13) | 0(0/7) | ||
| VIP | 0(0/16) | 0(0/17) | 21.4(3/14) | 0(0/7) | ||
| NPY | 0(0/4) | 0(0/26) | 13.6(3/22) | 0(0/26) | ||
| PV | 0(0/3) | 0(0/2) | 0(0/0) | 0(0/1) | ||
| Layer 2/3 | SOM | 57.8(129/223) | 0(0/199) | 9.6(11/115) | 0(0/186) | |
| CR | 37.1(129/348) | 28(66/236) | 9.5(27/285) | 0(0/131) | ||
| VIP | 0(0/284) | 21.5(66/307) | 12.2(30/245) | 0(0/152) | ||
| NPY | 10.6(11/104) | 9.9(27/272) | 10.5(30/285) | 1(3/290) | ||
| PV | 0(0/302) | 0(0/255) | 0(0/164) | 0.8(3/385) | ||
| Layer 4 | SOM | 7.1(20/283) | 0(0/206) | 0.7(1/142) | 0(0/216) | |
| CR | 13.7(20/146) | 35.1(27/77) | 7.8(9/115) | 0(0/72) | ||
| VIP | 0(0/79) | 35.5(27/76) | 15.6(12/77) | 0(0/70) | ||
| NPY | 2.2(1/46) | 10(9/90) | 13.5(12/89) | 0(0/70) | ||
| PV | 0(0/587) | 0(0/612) | 0(0/338) | 0(0/797) | ||
| Layer 5 | SOM | 8(47/588) | 0(0/545) | 2.1(6/283) | 0(0/430) | |
| CR | 37.9(47/124) | 38.8(31/80) | 12.6(12/95) | 0(0/59) | ||
| VIP | 0(0/58) | 41.3(31/75) | 9.1(5/55) | 0(0/51) | ||
| NPY | 12.8(6/47) | 14.5(12/83) | 4.9(5/102) | 0(0/102) | ||
| PV | 0(0/634) | 0(0/586) | 0(0/412) | 0(0/821) | ||
| Layer 6 | SOM | 7.1(34/482) | 0(0/521) | 14.1(32/227) | 0(0/385) | |
| CR | 40(34/85) | 51(25/49) | 31.2(24/77) | 0(0/21) | ||
| VIP | 0(0/73) | 28.7(25/87) | 4(2/50) | 0(0/52) | ||
| NPY | 52.5(32/61) | 17.4(24/138) | 1.1(2/174) | 0(0/169) | ||
| PV | 0(0/449) | 0(0/428) | 0(0/350) | 0(0/508) | ||
| III. S1 | ||||||
|---|---|---|---|---|---|---|
| SOM | CR | VIP | NPY | PV | ||
| All Layers | SOM | 27.1(221/814) | 0(0/785) | 5.8(33/569) | 0(0/666) | |
| CR | 34.3(221/645) | 39.7(229/577) | 9.5(66/697) | 0(0/388) | ||
| VIP | 0(0/416) | 47.4(229/483) | 6.2(27/433) | 0(0/307) | ||
| NPY | 21.6(33/153) | 21.9(66/302) | 8.4(27/323) | 0(0/218) | ||
| PV | 0(0/979) | 0(0/973) | 0(0/1072) | 0(0/925) | ||
| Layer 1 | SOM | 75(3/4) | 0(0/8) | 0(0/6) | 0(0/7) | |
| CR | 18.8(3/16) | 18.5(5/27) | 9.5(2/21) | 0(0/10) | ||
| VIP | 0(0/22) | 14.3(5/35) | 0(0/24) | 0(0/16) | ||
| NPY | 0(0/6) | 25(2/8) | 0(0/11) | 0(0/10) | ||
| PV | 0(0/0) | 0(0/0) | 0(0/2) | 0(0/3) | ||
| Layer 2/3 | SOM | 63.2(67/106) | 0(0/104) | 4(2/50) | 0(0/85) | |
| CR | 26.2(67/256) | 45.1(114/253) | 9.4(30/318) | 0(0/163) | ||
| VIP | 0(0/232) | 43.7(114/261) | 8.7(21/241) | 0(0/173) | ||
| NPY | 2.9(2/70) | 20.7(30/145) | 13.2(21/159) | 0(0/108) | ||
| PV | 0(0/106) | 0(0/119) | 0(0/133) | 0(0/119) | ||
| Layer 4 | SOM | 39.5(100/253) | 0(0/252) | 2.1(4/192) | 0(0/240) | |
| CR | 39.2(100/255) | 27.6(58/210) | 6.6(16/244) | 0(0/154) | ||
| VIP | 0(0/78) | 59.8(58/97) | 4.7(4/86) | 0(0/54) | ||
| NPY | 14.3(4/28) | 30.2(16/53) | 7.4(4/54) | 0(0/37) | ||
| PV | 0(0/368) | 0(0/349) | 0(0/333) | 0(0/358) | ||
| Layer 5 | SOM | 13.6(44/324) | 0(0/275) | 5.2(11/213) | 0(0/214) | |
| CR | 54.3(44/81) | 50(26/52) | 9.7(7/72) | 0(0/41) | ||
| VIP | 0(0/51) | 56.5(26/46) | 2.8(1/36) | 0(0/39) | ||
| NPY | 52.4(11/21) | 17.5(7/40) | 1.9(1/54) | 0(0/30) | ||
| PV | 0(0/277) | 0(0/340) | 0(0/375) | 0(0/286) | ||
| Layer 6 | SOM | 5.5(7/127) | 0(0/146) | 14.8(16/108) | 0(0/120) | |
| CR | 18.9(7/37) | 74.3(26/35) | 26.2(11/42) | 0(0/20) | ||
| VIP | 0(0/33) | 59.1(26/44) | 2.2(1/46) | 0(0/25) | ||
| NPY | 57.1(16/28) | 19.6(11/56) | 2.2(1/45) | 0(0/33) | ||
| PV | 0(0/228) | 0(0/165) | 0(0/229) | 0(0/159) | ||
Note that the overlap percentage indicates % of the marker populations in rows positive for the markers in columns. The numbers inside the parenthesis () indicate the numbers of cells measured to calculate the percentage.
Table 4.
The estimated percentage compositions of marker-positive GABAergic cells across cortical layers in the cortical regions of FC, S1 and V1
| FC | ||||||
|---|---|---|---|---|---|---|
| PV (%) | SOM (%) | CR (%) | NPY (%) | VIP (%) | PV, SOM, VIP (%) | |
| across layers | 39.5 | 19.6 | 18 | 10.6 | 7.5 | 66.6 |
| layer 1 | 0 | 1.7 | 5.9 | 15 | 7.9 | 9.6 |
| layer 2/3 | 37 | 10 | 30.7 | 15.5 | 10.7 | 57.7 |
| layer 4 | 49.9 | 15.8 | 25.7 | 8.3 | 5.2 | 71 |
| layer 5 | 55.6 | 31.9 | 7 | 6.7 | 5.7 | 93.3 |
| layer 6 | 39.9 | 32.2 | 7.9 | 8.7 | 7.7 | 79.8 |
| S1 | ||||||
|---|---|---|---|---|---|---|
| PV (%) | SOM (%) | CR (%) | NPY (%) | VIP (%) | PV, SOM, VIP (%) | |
| across layers | 37.8 | 16.4 | 15.9 | 9.8 | 7 | 61.2 |
| layer 1 | 0 | 2.3 | 6.5 | 0 | 7.1 | 9.4 |
| layer 2/3 | 26.6 | 10 | 29 | 17.4 | 13 | 49.6 |
| layer 4 | 43.5 | 16.9 | 15.3 | 8.9 | 2.9 | 63.3 |
| layer 5 | 56.3 | 19.9 | 7.6 | 2.9 | 3.5 | 79.7 |
| layer 6 | 38.9 | 24 | 13.8 | 12.2 | 8 | 70.9 |
| V1 | ||||||
|---|---|---|---|---|---|---|
| PV (%) | SOM (%) | CR (%) | NPY (%) | VIP (%) | PV, SOM, VIP (%) | |
| across layers | 40.6 | 17 | 21.1 | 7 | 8.4 | 65.9 |
| layer 1 | 0 | 1.8 | 10.4 | 4.3 | 5.2 | 7 |
| layer 2/3 | 20.7 | 7.9 | 43.7 | 10.5 | 16.8 | 45.4 |
| layer 4 | 50.4 | 21 | 27.6 | 4.8 | 6.5 | 77.8 |
| layer 5 | 54.3 | 24.9 | 10.4 | 3.4 | 4.1 | 83.3 |
| layer 6 | 51.3 | 20.4 | 7 | 11.7 | 8.2 | 79.9 |
Note that the estimated percentages are based upon comparing the densities of marker-positive cells with the densities of GABAergic cells described in Table 1. The last columns of the tables, [PV, SOM, VIP(%)], indicate the summed percentages of GABAergic cells positive for PV, SOM or VIP.
Fig. 5. The cumulative histograms showing percentage laminar distributions of marker-specific GABAergic cells in mouse FC, S1 and V1.
The labels on the horizontal axis indicate different markers. The histograms indicate the percentages of marker-positive cells with different colors and patterns coding different cortical layers.
We also used a Leica TCS SP2 AOBS spectral confocal microscope (Leica Microsystems Inc., Wetzlar, Germany) to examine the fluorescent sections and acquire data images. We avoided signal carryover across channels by using sequential scanning for different channels and adjusting detection window widths and gains. Narrow focal planes and projection images from the scanning system helped to clearly identify co-localization of chemical markers in mouse sections.
Photomicrographs were taken with the Leica confocal microscope. Digital images were compiled in Adobe Photoshop or Illustrator (Adobe, San Jose, CA).
Results
In the present study, we examined immunochemical properties of subtypes of GABAergic inhibitory neurons in mouse cortex using double immunolabeling with different combinations of the markers of GABA, PV, SOM, VIP, CR, and NPY. First we confirm the validity of these chemical markers to identify GABAergic inhibitory neurons in mouse cortex. Then we describe immunochemical distinction and overlap revealed by double staining of the various chemical markers for mouse inhibitory cortical neurons. Across all layers in different regions, we found that PV was an exclusive marker of inhibitory neurons, as it had no overlap with all the other markers tested, including SOM, VIP, CR and NPY. We also found that SOM and VIP were differential markers as neither of them overlapped with PV, and SOM did not overlap with VIP, although both SOM and VIP overlapped with CR and NPY. In addition, we quantify densities of marker-specific cells and assess the percentages of marker-specific GABAergic cells. Our data indicate that PV, SOM and VIP labeled 3 different chemical classes of inhibitory cortical neurons, which account for a majority of mouse inhibitory cortical neurons.
Co-localization of chemical markers with GABAergic cells
Previous studies of mammalian cortex have shown that many neurochemical markers are co-localized in GABAergic cells; however, there are few detailed studies examining the extent of their co-localization and confirming the validity of the chemical markers to identify GABAergic inhibitory neurons (Kubota et al., 1994; Gonchar and Burkhalter, 1997; Gonchar et al., 2007). Therefore, in mouse cortex, we started to examine how effective these chemical markers were to identify inhibitory neurons, and examine to what extent these chemical markers were co-localized with GABAergic cells.
We performed two different sets of immunochemical staining experiments. We first examined the co-localization of GABA with each of the chemical markers. As shown in Figure 1, most of the cells positive for PV, SOM, CR, or NPY were also positive for GABA. The arrows point to some clear examples of co-localization. Quantitatively, through cell counting and measurements under conventional wide-field fluorescent microscopy, across frontal cortex (FC), primary somatosensory cortex (S1) and primary visual cortex (V1), 97% (1839 of 1900) of PV+ neurons were positive for GABA. However, we found that NPY+, SOM+ and CR+ cells tended to have less robust GABA immunoreactivity, although a great majority of them, i.e., 88% (322 of 377) of NPY+ cells, 61% (358 of 588) SOM+ cells and 73% (305 of 418) CR+ cells, still could be clearly identified as immunopositive for GABA. We often noted that neurons with strong SOM+ immunoreactivity tended to be weak in GABA immunoreactivity, which is consistent with previous reports that GABAergic neurons co-localized with neuropeptides have lower levels of GABA content (Kubota et al., 1994; Gonchar and Burkhalter, 1997).
Fig. 1. Sub-populations of GABA immunopositive neurons in mouse cortex.
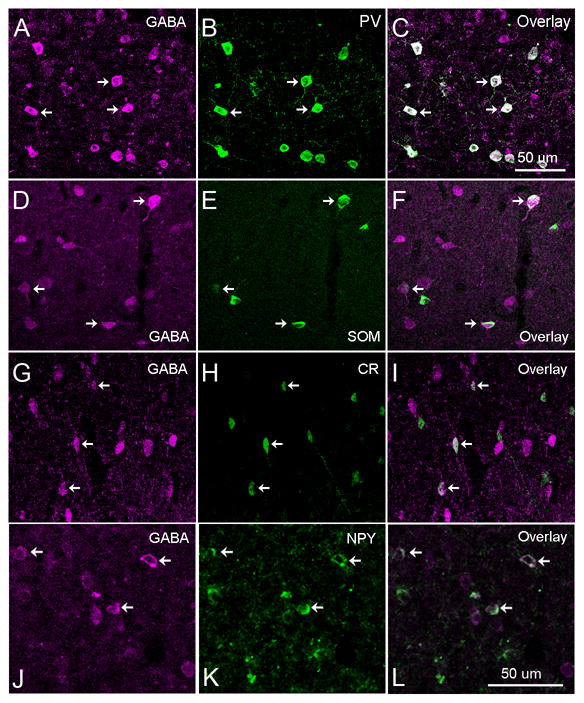
As shown in A–C, almost all the cells immunopositive for parvalbumin (PV) were also positive for GABA. D–F, G–I and J–L show that a great majority of neurons expressing somatostatin (SOM), calretinin (CR), and neuropeptide tyrosine (NPY), respectively, were also immunopositive for GABA. The scale bar in C applied to A–I; and the scale bar in L applies to J–L.
Considering the relatively low sensitivity of GABA immunoreactivity, we also utilized a line of GAD-GFP transgenic mouse in which all the GABAergic inhibitory neurons express GFP (Tamamaki et al., 2003), and stained the transgenic mouse cortical sections with all the 5 different chemical markers (PV, SOM, VIP, CR, or NPY). As demonstrated in Figure 2, the advantage of using the GAD67-GFP mouse was that GFP expression of GABAergic cells was strong and robust compared to GABA immuno-fluorescence seen in wild type mouse cortex, and this facilitated our examination of co-localization of the markers with GABAergic cells throughout cortical regions and layers. Across different cortical areas, immunoreactivity of PV, SOM, CR, NPY or VIP was co-localized with GFP expression of GABAergic neurons in GAD-GFP transgenic mouse cortex. Specifically, 93.4% (1286 out of 1377) of PV+ cells, 99.7% (309 of 310) VIP+ cells, 92.1% (385 of 418) CR+ cells, 93.1% (216 of 232) NPY+ cells, and 96.7% (355 of 367) SOM+ cells were GFP expressing. Therefore, nearly all the cells immunopositive for PV, SOM, CR, NPY or VIP expressed GFP, confirming that these markers are effective for identification of GABAergic cells.
Fig. 2. Co-localization of PV, SOM, CR, NPY and VIP with GFP-expressing GABAergic neurons in GAD-GFP transgenic mouse cortex.
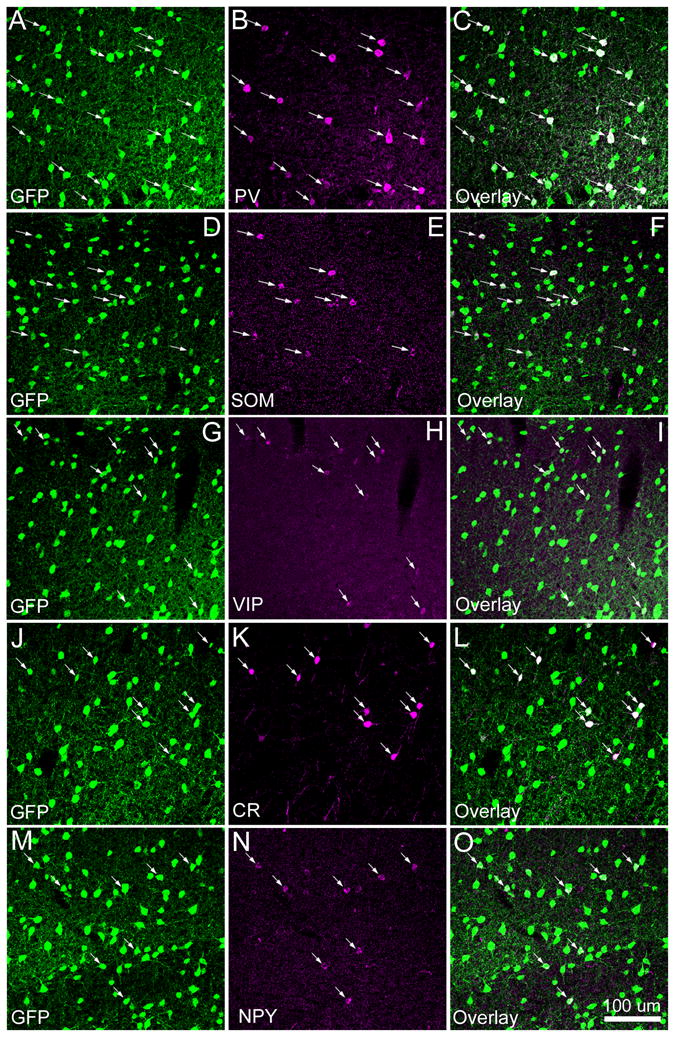
As shown in A–C, essentially all the cells immunopositive for parvalbumin (PV) were co-localized with GFP expressing GABAergic cells in the transgenic mouse in which basically all the GABAergic cells express GFP. The arrows in A–C point to the PV-immunopositive cells overlapping with GFP expression. Similarly, D–F, G–I, J–L and M–O show that nearly all the cells immunopositive for somatostatin (SOM), calretinin (CR), neuropeptide tyrosine (NPY) and vasoactive intestinal peptides (VIP), respectively, were also co-localized with GFP expressing GABAergic cells. The scale bar in O applies to all panels.
Immunochemical distinctions/overlap among inhibitory cortical neurons
To examine if the chemical markers are expressed in mutually exclusive or overlapping populations of inhibitory cortical neurons, we double labeled mouse cortical sections with different combinations of the chemical markers. We found that PV+ cells were the most abundant inhibitory cells in all the cortical regions, and on average, their densities were 171.6 ± 26.2/mm2, 204.7 ± 33.7/mm2 and 262.1 ± 3.3/mm2 (examined in 25 μm sections) for FC, S1 and V1, respectively (see Table 2). As for the laminar distribution of PV+ cells, there were no PV+ cells in layer 1, and layers 4, 5 had much higher densities than layers 2/3 and 6 (Table 2, Figure 5). For example, in S1, the densities of PV+ cells were 295.8 ± 69.6/mm2, 342.1 ± 14.1/mm2 for layers 4 and 5, respectively, and 179.5 ± 51.1/mm2, and 138.1 ± 31.5/mm2 for layer 2/3 and 6, respectively. The percentages of PV+ cells for layer 2/3, 4, 5 and 6 in S1 were 18.8%, 31%, 35.8% and 14.5%, respectively (Figure 5B). A similar trend was also seen in FC and V1 (Figure 5A, C).
PV was an exclusive marker for identifying a subpopulation of GABAergic inhibitory neurons, as double staining revealed that PV immunopositive cells lacked immunoreactivity for SOM, CR, NPY or VIP. As shown in Figure 3 and Table 3, we found that there was essentially no co-localization between PV immunopositive cells and other cells immunopositive for SOM (1/2661), CR (0/4193), NPY (5/4703), or VIP (0/3799) across cortical layers in all three brain regions. Note that PV immunoreactivity has been reported in a small population of pyramidal neurons in some cortical areas in different species (Stichel et al.,1987; Spatz et al., 1994; Johnson and Casagrande, 1995; Van Brederode et al., 1991; Wouterlood et al., 1995; Jinno and Kosaka, 2004). The most relevant study to ours is Jinno and Kosaka (2004), in which they reported a small number of layer 5 corticostriatal projection neurons in mouse somatosensory cortex (S1 and S2) are PV immunopositive. Although the possibility of PV labeling of a small subset of pyramidal cells is not a significant concern in our present study, it should be acknowledged that PV may label pyramidal cells in deep cortical layers.
Fig. 3. No overlap between expressions of parvalbumin (PV) and the other chemical markers tested in mouse cortex.
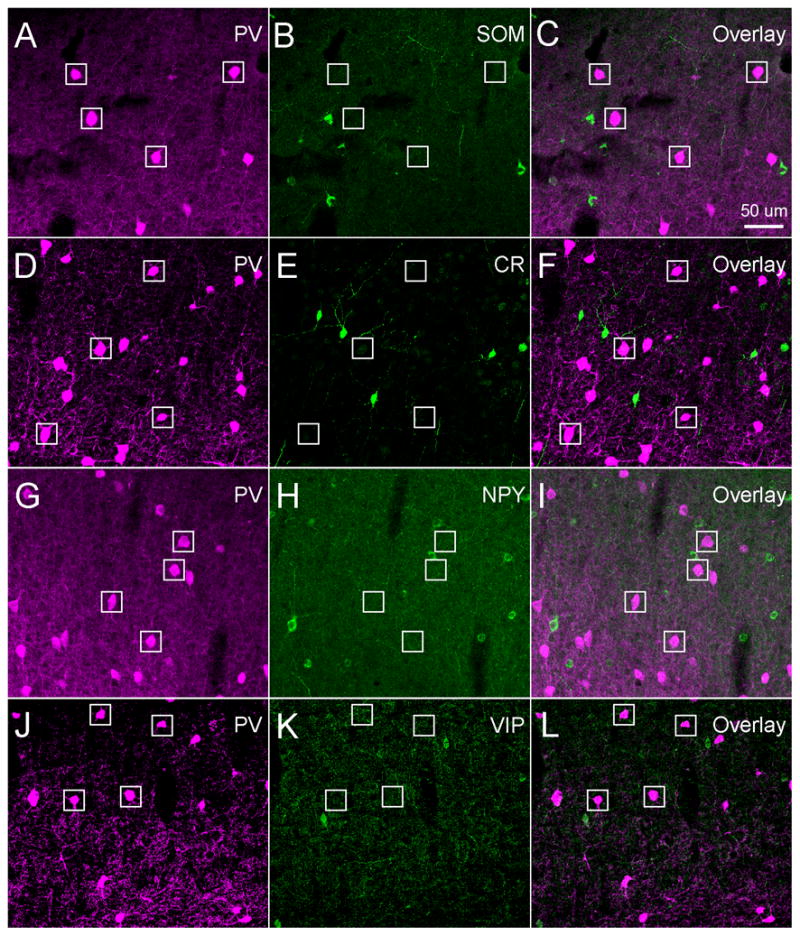
A–C, D–F, G–I and J–L show that PV immunopositive cells were not co-localized with somatostatin (SOM), calretinin (CR), neuropeptide tyrosine (NPY), or vasoactive intestinal peptide (VIP), respectively. White boxes denote the locations of PV+ cells. The sections were from mouse S1 cortex. Scale bar in C applies to all panels.
SOM+ cells had a lower density than PV+ cells across all the cortical regions, and their average values was 85 ± 12.1/mm2, 88.8 ± 5.7/mm2 and 109.5 ± 21.9/mm2, for FC, S1 and V1, respectively (see Table 2). SOM+ cells were distributed much less in upper layers (layers 1 and 2/3) than deeper layers (4, 5 and 6) (Table 3, Figure 5). As seen in other cortical regions, SOM+ cell densities in S1 were 10.7 ± 18.6/mm2, 67.1 ± 7.0/mm2, 115.2 ± 29.3/mm2, 120.8 ± 38.7/mm2, and 85.1 ± 14.6/mm2 for layers 1, 2/3, 4, 5 and 6, respectively. The percentages of SOM+ cells in each layer were 2.7%, 16.8%, 28.9%, 30.3% and 21.3% for layers 1, 2/3, 4, 5 and 6, respectively.
SOM was a differential marker for inhibitory neurons in mouse cortex, as SOM+ cells lacked PV immunoreactivity (see above), and they also lacked VIP immunoreactivity (Figure 4G–I, Table 3). However, SOM was not exclusive, as some SOM+ cells were positive for CR or NPY (Figure 4A–F, Table 3). The average overlap of SOM/CR (percentage of SOM+ cells expressing CR) was 26.8% (294/1098), 15.1% (240/1587) and 27.1% (221/1814) across layers for FC, S1 and V1, respectively. As noted in our previous study (Xu et al., 2006), there was the largest degree of SOM and CR overlap in cortical layer 2/3, as the overlap rate of SOM/CR was 81.4%, 57.8%, and 63.2% for FC, S1 and V1, respectively (Table 3); the overlap rate of SOM/CR was dramatically lower in deeper layers, with average overlap of 20.4%, 7.3% and 19.5% for layers 4, 5 and 6, respectively. Compared to CR, the overlap of SOM/NPY (percentage of SOM+ cells expressing NPY) was much less, with the average overlap being 9.3%, 6.5% and 5.8% across layers for FC, S1 and V1, respectively (Table 3).
Fig. 4. Double immunochemical staining of mouse cortical sections with different combinations of somatostatin (SOM), calretinin (CR), neuropeptide tyrosine (NPY) and vasoactive intestinal peptide (VIP).
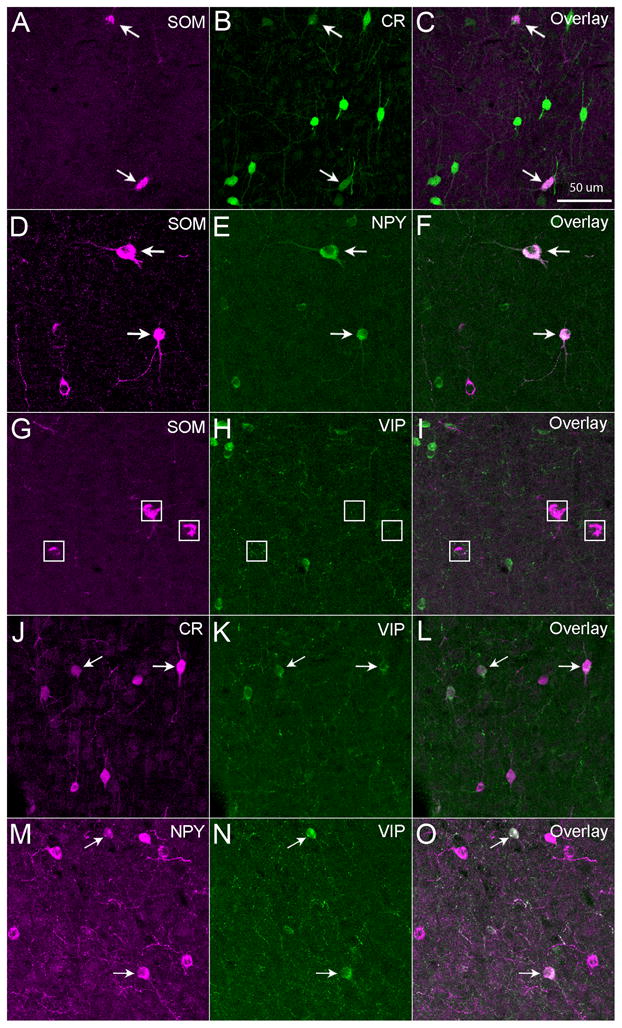
Some SOM+ neurons were immunopositive for CR (A–C) or NPY (D–F), but SOM+ neurons were not immunopositive for VIP (G–I). Some CR+ cells were immunopositive for VIP (J–L), and some NPY+ cells were also immunopositive for VIP (M–O). Arrows point to double-labeled cells. White boxes in panels G–I denote locations of SOM+ cells. The sections were from mouse S1 cortex. The scale bar in C applies to all panels.
The densities of CR+ cells were comparable to SOM+ cells. On average, the densities of CR+ cells were 78.4 ± 3.8/mm2, 86 ± 2.1/mm2 and 136.4 ± 15.3/mm2 for FC, S1 and V1, respectively (Table 2). The average densities of NPY+ cells were 45.9 ± 3.2/mm2, 53 ± 9.6/mm2 and 45.2 ± 2.1/mm2 across these regions, respectively (Table 2). The average densities of VIP+ cells were 32.5 ± 1.8/mm2, 37.9 ± 9.2/mm2 and 54.2 ± 5.8/mm2 for the three cortical regions, respectively (Table 2).
In contrast to SOM+ and PV+ cells, CR+, VIP+ and NPY+ cells had different laminar distributions; they were preferentially distributed in upper layers (Tables 2 and 4; Figure 5). There were many VIP+, CR+ and NPY+ cells distributed in layers 1 and 2/3 in all cortical regions; a notable exception is the absence of NPY+ cells in layer 1 of S1. The CR+, VIP+ and NPY+ cells had much higher densities in layer 2/3 than other layers (4, 5 and 6) across the cortical regions (Tables 2 and 4; Figure 5). For example, in FC, the density of VIP+ cells in layer 2/3 was 62.1 ± 11.2/mm2, while the densities of layers 1, 4, 5 and 6 were 22.4 ± 6.9/mm2, 25.5 ± 20.7/mm2, 23.4 ± 3.7/mm2, and 29.8 ± 8.6/mm2, respectively. The percentages of VIP+ cells for layers 1, 2/3 through 6 in FC were 16.4%, 32.6%, 24.1%, 12.5% and 14.3%, respectively. In FC, layer 2/3 density of CR+ cells was 178.1 ± 2.5/mm2, with the lower densities of layers 1, 4, 5 and 6 being 16.7 ± 1.8/mm2, 124.9 ± 18.5/mm2, 28.6 ± 14.2/mm2, and 30.7 ± 3.1/mm2, respectively. Their percentages for layers 1, 2/3 through 6 were 4.8%, 46.75, 29.65, 10.2% and 12.5%, respectively. Similarly, in FC, the NPY+ cell density in layer 2/3 was 90.1 ± 7.3/mm2, while the densities for layers 1, 4, 5 and 6 were 42.4 ± 7.2/mm2, 40.3 ± 2.3/mm2, 27.1 ± 13.7/mm2, and 33.7 ± 1.5/mm2, respectively. The percentages of NPY+ cells for layers 1, 2/3 through 6 were 15.5%, 39.6%, 16.1%, 15.3% and 13.6%, respectively.
VIP, like SOM, was another differential chemical marker for GABAergic inhibitory neurons in mouse cortex, as VIP+ cells were not co-localized with PV+ or SOM+ cells. Some of VIP+ cells overlapped with CR+ cells or NPY+ cells (Figure 4J–O, Table 3). There was considerable overlap between VIP/CR (percentage of VIP+ cells expressing CR) with the average being 30.3%, 26.5%, and 47.4% for FC, S1 and V1, respectively (Table 3). The overlap was particularly high in layers 4 and 5, as, for example, in V1, the average overlap of VIP/CR in layers 4 and 5 was about 51.8%. In addition, there was some overlap between VIP/NPY (percentage of VIP+ cells expressing NPY), with the average overlap being 10.4%, 11.8% and 6.2% for FC, S1 and V1, respectively (Table 3). Finally, some CR+ cells were co-localized with NPY+ cells, with the average overlap of CR/NPY (percentage of CR+ cells expressing NPY) being 8.5%, 12.3% and 9.5% for FC, S1 and V1, respectively (see Table 3).
Percentages of marker-specific GABAergic cells
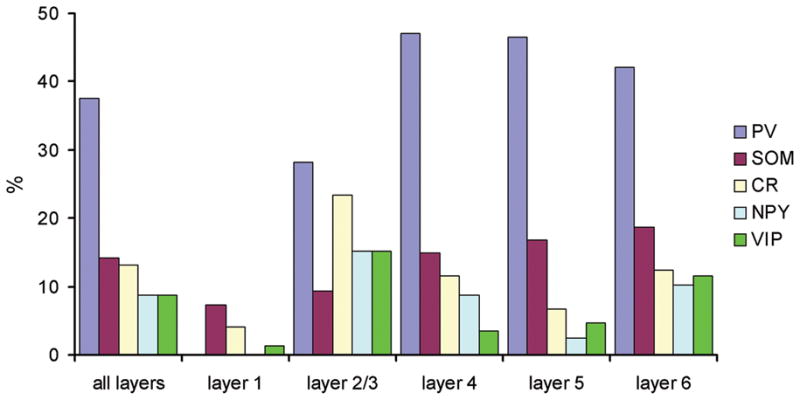
Having established that the PV, SOM, and VIP antibodies that we used identify non-overlapping inhibitory neuron populations, we quantified the percentage of inhibitory neurons that are accounted for by these markers, as well as by CR and NPY. These measures were made separately for each cortical area and layer (Figure 6 and Table 4). We first compared the density of marker-specific cells with the density of GABAergic cells (GFP expressing cells in the GAD-GFP mice) to estimate the individual percentages of marker specific cells. Comparing densities across regions when all layers are combined (Table 4), the estimated percentages of PV+ inhibitory cells were 39.5%, 37.8% and 40.5% for FC, S1, and V1 regions, respectively. The estimated percentages of SOM+ cells were 19.6%, 16.4%, and 17.0%, and the percentages for VIP+ cells were 7.5%, 7.0%, and 8.4% across these regions, respectively. The estimated percentages of CR+ cells were 18.0%, 15.9%, and 21.1%, respectively and the estimated percentages of NPY+ cells were 10.6%, 9.8%, and 7.0% for these regions, respectively.
Fig. 6.
The histogram showing percentage compositions of marker-positive GABAergic cells across S1 cortical layers.
We also directly measured the percentages of marker-specific GABAergic cells by quantifying marker-positive cells in sections double labeled with GFP in the GAD-GFP mice, and one of the chemical markers. These direct measurements matched well with the estimated values (shown above). As seen in Figure 6, in S1, across all the layers, the percentages of PV+, SOM+, VIP+, CR+, and NPY+ GABAergic cells were 37.6%, 14.1%, 8.8%, 13.2%, and 8.8%, respectively. In FC, PV+, VIP+, CR+ and NPY+ cells accounted for 39.3%, 8.1%, 14.6% and 10.9% of the GABAergic cells, respectively. And in V1, PV+, VIP+, CR+, and NPY+ cells accounted for 40.1%, 8.4%, 20.6% and 6.6% of the GABAergic cells, respectively. Due to relatively strong staining artifacts in FC and V1, we did not directly measure the percentages of SOM+ GABAergic cells in those areas. Overall, based upon the estimated and directly measured percentages of marker specific cells, the PV+, SOM+ and VIP+ cells account for about 61–67% of the GABAergic cell population (Table 4), which consisted of three non-overlapping classes across FC, S1 and V1.
The compositions of GABAergic cells positive for the markers varied across cortical layers. Layer 1 was particularly interesting. As shown in Table 4 and Figure 6, we found that most layer 1 GABAergic cells were not labeled by these markers. For example, this is indicated in Figure 6, as the percentages of PV+, VIP+, CR+, NPY+ and SOM+ GABAergic cells (based upon direct measurements from S1 layer 1) were 0%, 7.3%, 4.0%, 0% and 1.3%, respectively. The density of GABAergic cells in S1 layer 1 was 468.8/mm2, but the densities of marker-positive cells were quite low (PV+, 0/mm2, VIP, 33.4/mm2, CR+, 30.2/mm2, NPY+, 0/mm2, SOM+, 10.7/mm2). Thus, a great majority of layer 1 GABAergic cells did not express detectable levels of any of these markers. But note that in rat frontal cortex, actinin-2 is a useful chemical marker, as it labels about 60% of layer 1 inhibitory neurons (Uematsu et al., 2008).
The sum of the percentages for the three markers (PV, SOM and VIP) was 57.7%, 49.65 and 45.4% for layer 2/3 of FC, S1 and V1, respectively. And the summed percentages in layers 4, 5 and 6 were larger, ranging from 63.3% (S1 layer 4) to 93.3% (FC layer 5) (see Table 4).
Discussion
We used double immunolabeling of chemical markers to study immunochemical distinction and overlap among mouse inhibitory cortical neurons, by using antibodies against PV, SOM, CR, NPY or VIP. Our major findings were that three distinct chemical classes of inhibitory cortical neurons can be separately identified by PV, SOM or VIP across cortical regions of FC, S1 and V1, and that overall, these three classes constituted about 60% of the whole GABAergic inhibitory neuronal population. Based upon immunolabeling results of combinations of PV, SOM, CR, NPY and VIP, the PV+ cells were an exclusive subpopulation; the SOM+ cells could be divided into SOM+ only, SOM+/CR+, and SOM+/NPY+ groups; and the VIP cells could be divided into VIP+ only, VIP+/CR+, VIP+/NPY+ groups.
In rodents, previous studies of the immunochemical properties of inhibitory neurons have been focused on rat cortex. GABAergic neurons in rat cortex can be divided into at least 4 non-overlapping (PV+, SOM+, CR+, and CCK+) subpopulations. In rat visual cortex, PV+ cells are the most numerous, making up about 51 % of the GABAergic population; SOM+ cells account for about 17%, and CR+ cells constitute about 17% of the population (Gonchar and Burkhalter, 1997). In rat frontal cortex, CCK+ cells make up a small additional percentage (about 5%) of the GABAergic population (Kubota et al., 1994; Kawaguchi and Kubota, 1997). In addition, in rat frontal cortex, most CR+ cells show VIP immunoreactivity; there is a substantial overlap between SOM+ and NPY+ cells; and some CCK+ cells show VIP immunoreactivity (Kubota et al., 1994; Kawaguchi and Kubota, 1997).
Similar to our observations in mouse cortex, VIP expression in rat cortex does not overlap with SOM or PV (Kubota et al., 1994; Kawaguchi and Kubota, 1997). Thus, previous observations in the rat indicate that PV, SOM, and VIP can identify three non-overlapping populations. Although PV, SOM and VIP identify three separate populations in both rat and mouse cortex, these species appear to differ in the proportion of all GABAergic cells accounted for by these populations. We find that in mouse cortex, across FC, S1 and V1, these 3 non-overlapping GABAergic neuronal populations constitute about 60% of the GABAergic population. On average, the PV+, SOM+ and VIP+ cells accounted for about 40%, 18%, and 8% of the GABAergic population, respectively. In addition, the percentages of CR+ and NPY+ GABAergic cells were about 18.0% and 9%, respectively. This contrasts with observations in rat, where PV, SOM, CR, and CCK are reported to account for more than 90% of all inhibitory neurons in rat frontal cortex (Kawaguchi and Kubota, 1997). In a recent study examining cortical GABAergic neurons in the transgenic rat expressing venus under the control of the vesicular GABA transporter promoter, Uematsu et al. (2008) found that the 6 chemical markers (PV, SOM, CCK, VIP, CR and actinin-2) cover more than 90% of the total GABAergic populations in layers I, II/III, and V, and nearly 90% of cells in VI. It is unlikely that CCK cells in the mouse would be sufficient to account for this 30% difference, as we found that in mouse cortical sections, CCK immunopositive cells were quite sparse (data not shown), which suggests that identification of other markers or marker combinations would be helpful. It would be of interest to quantify actinin-2 expressing neurons in mouse cortex to determine what proportion of interneurons they represent.
Our observation that PV, SOM and VIP are non-overlapping markers across mouse cortex is supported by distinct clusters of PV, SOM and VIP mRNA expression patterns of interneurons (Bernard et al., 2007, Society for Neuroscience Abstract 134.8), and by immunostaining profiles of fate-mapped cortical interneurons (Miyoshi et al., 2007). However, using triple labeling protocols, Gonchar et al. (2007) found extensive overlap of SOM and VIP expression in mouse visual cortex. The neurons expressing SOM accounted for 13.6 % of all inhibitory neurons, and SOM/VIP double-labeled cells accounted for 8.4%. Thus, more than 60% of all SOM+ cells were also identified as VIP positive (double-labeled). This striking discrepancy between our results cannot be explained by sample sizes or cortical areas studied because we also studied mouse visual cortex (as well as FC and S1) and found that out of more than 1200 V1 neurons that were positive for VIP or SOM, none were double-labeled. Given our sample sizes, we would have expected to see more than 700 SOM/VIP double-labeled cells, if the proportions were the same as reported by Gonchar et al.
It therefore appears that the most likely differences between these studies were not minor technical differences, but instead were more fundamental. The most clear difference is that we used different anti-VIP antibodies. For the antibodies we used, we independently tested their specificity using immunoabsorption assays (see the Methods). Thus we are confident in our results. Gonchar et al. (2007) did not perform independent tests of the specificity of antibodies, including anti-VIP antibodies. Such tests would be helpful for identifying whether this difference might be responsible.
In the present study, we focused on identification and characterization of chemical properties of inhibitory cortical neurons. The study was based on double immunostaining and therefore cannot independently identify correlations between immunostaining and cell types. However, when combined with previous studies correlating immunostains with morphological and electrophysiological analyses in mice (Oliva et al., 2000; Chattopadhyaya et al., 2004; Galarreta et al., 2004; Xu et al., 2006; Miyoshi et al., 2007; Xu and Callaway, 2009), they provide a greater quantitative insight and can serve as a guide for the choice of informative chemical markers in future studies of the mouse cortex.
Acknowledgments
This work was supported by grants from the National Institutes of Health [MH063912 (EMC), EY010742 (EMC), and DA023700 (XX)].
References
- Ascoli GA, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nature reviews. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhalter A. Many specialists for suppressing cortical excitation. Front Neurosci. 2008;2:155–167. doi: 10.3389/neuro.01.026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, Huang ZJ. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci. 2004;24:9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J. Neocortical neuronal diversity: chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell surface molecules. Cereb Cortex. 1993;3:273–289. doi: 10.1093/cercor/3.4.273. [DOI] [PubMed] [Google Scholar]
- Galarreta M, Erdelyi F, Szabo G, Hestrin S. Electrical coupling among irregular-spiking GABAergic interneurons expressing cannabinoid receptors. J Neurosci. 2004;24:9770–9778. doi: 10.1523/JNEUROSCI.3027-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y, Burkhalter A. Three distinct families of GABAergic neurons in rat visual cortex. Cereb Cortex. 1997;7:347–358. doi: 10.1093/cercor/7.4.347. [DOI] [PubMed] [Google Scholar]
- Gonchar Y, Wang Q, Burkhalter A. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat. 2007;1:3. doi: 10.3389/neuro.05.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery RW. On counting and counting errors. The Journal of comparative neurology. 2002;447:1–7. doi: 10.1002/cne.10221. [DOI] [PubMed] [Google Scholar]
- Hendry SH, Jones EG. GABA neuronal subpopulations in cat primary auditory cortex: co-localization with calcium binding proteins. Brain research. 1991;543:45–55. doi: 10.1016/0006-8993(91)91046-4. [DOI] [PubMed] [Google Scholar]
- Hendry SH, Jones EG, Emson PC, Lawson DE, Heizmann CW, Streit P. Two classes of cortical GABA neurons defined by differential calcium binding protein immunoreactivities. Exp Brain Res. 1989;76:467–472. doi: 10.1007/BF00247904. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. Physiological and morphological identification of somatostatin- or vasoactive intestinal polypeptide-containing cells among GABAergic cell subtypes in rat frontal cortex. J Neurosci. 1996;16:2701–2715. doi: 10.1523/JNEUROSCI.16-08-02701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kondo S. Parvalbumin, somatostatin and cholecystokinin as chemical markers for specific GABAergic interneuron types in the rat frontal cortex. J Neurocytol. 2002;31:277–287. doi: 10.1023/a:1024126110356. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Hattori R, Yui Y. Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex. Brain research. 1994;649:159–173. doi: 10.1016/0006-8993(94)91060-x. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nature reviews. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Meskenaite V. Calretinin-immunoreactive local circuit neurons in area 17 of the cynomolgus monkey, Macaca fascicularis. The Journal of comparative neurology. 1997;379:113–132. [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Tamas G, Lujan R, Buhl EH. Salient features of synaptic organisation in the cerebral cortex. Brain Res Brain Res Rev. 1998;26:113–135. doi: 10.1016/s0165-0173(97)00061-1. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. The Journal of comparative neurology. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Uematsu M, Hirai Y, Karube F, Ebihara S, Kato M, Abe K, Obata K, Yoshida S, Hirabayashi M, Yanagawa Y, Kawaguchi Y. Quantitative chemical composition of cortical GABAergic neurons revealed in transgenic venus-expressing rats. Cereb Cortex. 2008;18:315–330. doi: 10.1093/cercor/bhm056. [DOI] [PubMed] [Google Scholar]
- Xu X, Callaway EM. Laminar specificity of functional input to distinct types of inhibitory cortical neurons. J Neurosci. 2009;29:70–85. doi: 10.1523/JNEUROSCI.4104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. The Journal of comparative neurology. 2006;499:144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]