SUMMARY
Members of the mammalian phosphoinositide-3-OH kinase (PI3K) family of proteins are critical regulators of various cellular process including cell survival, growth, proliferation and motility. Oncogenic activating mutations in the p110α catalytic subunit of the heterodimeric p110/p85 PI3K enzyme are frequent in human cancers. Here we show the presence of frequent mutations in p85α in colon cancer, a majority of which occurs in the inter-Src homology-2 (iSH2) domain. These mutations uncouple and retain p85α's p110-stabilizing activity, while abrogating its p110-inhibitory activity. The p85α mutants promote cell survival, Akt activation, anchorage independent cell growth, and oncogenesis in a p110-dependent manner.
SIGNIFICANCE
Somatic mutations in the catalytic p110α subunit of PI3K are common in cancers. In this study, we show the occurrence of frequent mutations in the regulatory p85α subunit of PI3K in human cancers. Our data demonstrate an alternate mechanism for PI3K-pathway activation and oncogenesis resulting from the impaired regulation of p110 activity by mutant p85α. Further, p85α mutations are likely to be useful as diagnostic markers for identification of p110-dependent tumors that may not carry an activating p110α mutation, but are candidates for targeted treatment with PI3K pathway inhibitors that are in development.
INTRODUCTION
Phosphoinositide 3-kinase (PI3K) family of lipid kinases are divided into three major classes, based on primary sequence, substrate preference, and regulation (Cantley, 2002; Engelman et al., 2006; Fruman et al., 1998; Hawkins et al., 2006). While class IA PI3Ks are heterodimeric enzymes composed of a catalytic subunit (p110α, p110β or p110δ) complexed with one of five regulatory subunits (p85α, p55α, p50α, p85β or p55γ), the class IB enzyme is a dimer made of p110γ catalytic subunit and p101 or p84 regulatory subunit (Cantley, 2002; Hawkins et al., 2006; Vanhaesebroeck et al., 2005). The class I catalytic subunit polypeptide p110α, p110β, p110δ, and p110γ are encoded by PIK3CA, PIK3CB, PIK3CD, and PIK3CG, respectively (Cantley, 2002; Vanhaesebroeck and Waterfield, 1999). The regulatory subunits are encoded by five genes: PIK3R1 codes p85α, p55α, and p50α; PIK3R2 codes p85β; PIK3R3 codes p55γ; PIK3R5 codes p101; and PIK3R6 codes p84 (Cantley, 2002; Vanhaesebroeck and Waterfield, 1999).
The p110 catalytic subunits of PI3K share a common domain architecture consisting of an N-terminal adapter binding domain (ABD) that binds to p85 regulatory subunits, a Ras binding domain (RBD), a putative membrane binding C2 domain, a helical region that makes a regulatory contact with the p85 nSH2 domain (Miled et al., 2007), and a C-terminal kinase domain. Similarly, the p85 regulatory subunits have in common an N-terminal SH3 domain, a domain homologous to the Rho GTPase-activating protein (GAP) domain of the BCR gene product (BCR domain), and two SH2 domains (nSH2 and cSH2) that flank an intervening antiparallel coiled-coil (iSH2) required for binding to the ABD in p110 (Holt et al., 1994). Besides its role in inhibiting the catalytic activity of p110, in the basal state, the p85 regulatory subunit is required to stabilize the catalytic p110 subunit (Kodaki et al., 1994; Yu et al., 1998). Upon growth factor stimulation, the nSH2 and cSH2 domains of p85 bind to phosphorylated tyrosines (YXXM motif) in activated receptors and adaptors that activate catalytic p110 (Backer et al., 1992; Carpenter et al., 1993; Otsu et al., 1991). Once activated, PI3Kinases phosphorylate phosphoinositide 4,5-bisphosphate (PIP2) leading to the production of phosphoinositide 3,4,5-triphosphate (PIP3) which, serves as an important second messenger that regulates cell survival, growth, proliferation, and motility through a variety of downstream effectors (Cantley, 2002; Carpenter et al., 1993; Engelman et al., 2006; Jimenez et al., 2002; Vanhaesebroeck and Waterfield, 1999; Yu et al., 1998).
Several studies have identified common somatic mutations in PIK3CA in cancers of colon, rectum, breast, ovary, brain, and liver (Bader et al., 2005; Samuels et al., 2004). Several PIK3CA mutants, including hotspot mutations in the helical and kinase domain, show elevated lipid kinase activity in vitro and induce oncogenic transformation in vivo (Gymnopoulos et al., 2007; Ikenoue et al., 2005; Isakoff et al., 2005; Kang et al., 2005; Samuels et al., 2004; Zhao and Vogt, 2008). While oncogenic p110α mutations are common in cancers (Bader et al., 2005; Samuels et al., 2004), such mutations in the regulatory p85α subunit are not as common (Bader et al., 2005; Hennessy et al., 2005). Previously, a truncated form of p85α containing residues 1-571 fused to a fragment of Eph (p65) was identified in an x-ray-irradiated mouse lymphoma model (Borlado et al., 2000; Chan et al., 2002; Jimenez et al., 1998). However, this mutation has so far not been found in human cancers. Although, a p85α truncation mutant was described in a human lymphoma cell line (Jucker et al., 2002) its relevance in oncogenesis is not clear (Horn et al., 2008). A low prevalence of p85α mutation in breast (Wood et al., 2007), colon (Philp et al., 2001), and ovarian (Philp et al., 2001) tumors has been reported, but the functional role of these mutations in tumorigenesis is not known. Recently, frequent occurrence of p85α mutations in glioblastoma was reported (Parsons et al., 2008; TCGA, 2008). However, the ability and role of these mutations in promoting oncogenesis remains to be studied.
In this study we have systematically sequenced a large number of tumors and found mutations in p85α that uncouple its p110-inhbitory effects from the stabilization activity, leading to p110-mediated survival signaling and oncogenesis.
RESULTS
Identification of PI3K regulatory subunit mutations
We sequenced coding exons and a ~50bp region flanking the exons of PIK3R1, PIK3R2, PIK3R3, PIK3R4, and PIK3R5 regulatory subunits of PI3K in primary human cancers. A total of 672 human primary tumor samples consisting of 213 non-small-cell lung (NSCLC), 108 colorectal, 62 breast, 87 renal cell (RCC), 46 ovary, 40 skin (melanomas), 37 gastric, 21 small-cell lung (SCLC), 16 esophageal, 13 bladder, 12 chronic lymphocytic leukemia (CLL), 11 hepatocellular (HCC), and 6 pancreatic cancers were analyzed (Table S1). We found that PIK3R1, which codes for p85α, was mutated in 9 of 108 colorectal (8%), 1 of 62 breast (2%) and 1 of 6 pancreatic (17%) tumor samples (Figure 1, Figure S1, and Tables S1 and S2). All the mutations were confirmed to be somatic by their presence in the original tumor DNA and absence in the matched adjacent normal tissue through additional sequencing or mass spectrometric analysis (Figures S1, S2). A majority of the mutations identified were amino acid substitutions that clustered in the iSH2 domain. Interestingly, one of the mutated residues N564 was within hydrogen bonding distance of p110α C2-domain residue N345 (Huang et al., 2007; Figure 1A, C). Substitutions at residue N345 of p110α are oncogenic (Gymnopoulos et al., 2007), probably due to destabilization of iSH2-C2 inhibitory interactions. Therefore, it is likely that the p85α N564D mutation mimics the effect of N345 substitutions in p110α. Interestingly, both N564 and an adjacent residue D560 of p85α, which is also within hydrogen bonding distance of N345 of p110α, were recently reported to be mutated in glioblastoma (TCGA, 2008). Though the potential effect of these substitutions on relieving the inhibitory contacts have been proposed (TCGA, 2008), the actual functional consequence of the mutation and its role in oncogenesis have not been explored. The regulatory p85α subunit besides regulating p110α also regulates p110β and p110δ (Engelman et al., 2006; Vanhaesebroeck et al., 2005). A similar set of regulatory contacts involving residue N344 in p110β and residue N334 p110δ, analogous to p110α residue N345, and p85α iSH2 residues N564 and D560 has been previously proposed (Amzel et al., 2008). Our modeling of the C2 domain of p110β and p110δ supports these regulatory interactions (Figure 1C) suggesting that the p85α mutations may lead to general activation of all class IA catalytic subunits.
Figure 1. p85α mutations in human cancer.
(A) Cartoon depicting the p85α somatic mutations. Horizontal bar below mutated residues indicates conserved residues across the p85 family members (R1-R3). Black and orange vertical bars indicate previously identified p85α mutations (Jimenez et al., 1998; Jucker et al., 2002; Parsons et al., 2008; Philp et al., 2001; TCGA, 2008; Wood et al., 2007). Orange bars signify hotspots, with the bar height drawn proportional to number of times the residue/region has been altered. (B) p85α somatic mutations modeled on a recent crystal structure (PDB code 2RD0) of p85α iSH2 domain in complex with p110α (Huang et al., 2007). Blue patches on the surface of p110α represent regions that are <3.5Å from p85α iSH2. (C) The p85α residues, N564 and D560 within hydrogen bonding distance of N345 of p110α (Huang et al., 2008; TCGA, 2008) located in the C2 domain is shown. Models of p110β (yellow) and p110δ (purple) C2 domains superimposed on p110α C2 domain (cyan) illustrates the proximity of conserved residue N344 of p110β and N334 of p110δ (corresponding to N345 of p110α) and p85α iSH2 residues D560 and N564. (D) Mutation map of tumors carrying changes in one or more of the genes studied. Each column corresponds to a tumor sample. Pc = pancreatic cancer. 1Nature 455, 1061, 2Science 321, p1807, 3Science 318, p1108 and 4The EMBO journal 17, p743.
In addition to mutations that clustered in iSH2, we identified mutations in the cSH2 domain, specifically a mutation in residue R649, which is part of the conserved FLVERS motif required for phospho-tyrosine engagement (Hoedemaeker et al., 1999). The R649 mutation is likely to alter the activity of the protein, given that analogous conserved Arg residues within SH2 domains of other proteins are mutated in multiple human diseases (Figure S3). Mutations in the nSH2 and BCR domains and in the spacer between those domains were also identified (Figure 1A). It is noteworthy that the majority of the mutated residues identified within nSH2, iSH2 and cSH2 were conserved across the p85 family (Figure 1 and Figure S4). Meta-analysis of mutations in p85α (Jimenez et al., 1998; Jucker et al., 2002; Parsons et al., 2008; Philp et al., 2001; TCGA, 2008; Wood et al., 2007) revealed several recurrent mutation sites that include R348, G376, L380, E439, K459, D560, N564 and R574. Interestingly, these mutated sites are conserved across the three closely related p85 family members (p85α/p55α/p50α, p85β and p55γ) suggesting that these mutations are likely to have a functional role in oncogenesis. We also detected mutations in PIK3R2 (p85β) and two additional regulatory subunits PIK3R4 (p150) and PIK3R5 (p101), albeit at a much lower frequencies (Figure S5; Tables S2 and S3).
In addition to sequencing PI3K regulatory subunit genes, we tested the tumor samples for somatic mutations in PIK3CA, KRAS and PTEN. In colon tumor bearing p85α mutations, we rarely found KRAS or PTEN mutations. However, ~45% of p85α-mutant colon tumor samples had PIK3CA mutations, suggesting that the p110α and p85α mutations are not always mutually exclusive in this tumor type (Figures 1D; Figure S6; Tables S2 and S3). Unlike in colon cancer, published PIK3R1 mutation data shows that it is mutually exclusive with PIK3CA mutations in glioblastomas (Parsons et al., 2008; TCGA, 2008), although the frequency of PIK3CA mutations in glioblastomas is known to be low (Hartmann et al., 2005) compared with colon cancers (Bader et al., 2005; Table S2). PTEN mutations generally are rare in colon cancers (Wang et al., 1998), but co-occurred with PIK3R1 mutations in two of our samples (Table S3). In a similar analysis of the published glioblastoma data, we found only one of the PI3KR1 mutations to co-occur with PTEN mutation, even though PTEN mutations are quite frequent in glioblastomas (Parsons et al., 2008; TCGA, 2008; Yin and Shen, 2008).
p85α mutants interact with and stabilize p110α
To understand the function of p85α mutations, we tested them for their ability to interact with p110α. In co-immunoprecipitation experiments using COS-7 cells, we found that overexpressed HA-tagged p85α wild-type and point mutants that contained an intact p110-binding domain (Dhand et al., 1994; Huang et al., 2007; Miled et al., 2007) were able to interact with p110α (Figure 2A). Similarly, p85α mutants with a short insertion (H669insLKH), deletion (QYL579delL) or a premature stop (R642*) were all able to interact with p110α, as demonstrated by the presence of p110α in the immunoprecipitated p85α (Figure 2B, D). Additionally, the C2-iSH2 interaction region mutants D560Y and D560Y_N564D were able to interact with p110α (Figure 2C). As previously described, a truncated p85 mutant Q572* was also able to interact with p110α (Shekar et al., 2005). However, p85α mutants that lacked the p110α binding region, namely R162*, L380fs, R348* and a dominant negative mutant p85Δ (Dhand et al., 1994), all failed to interact with p110α (Figure 2). As expected, the mock and vector control lanes in the assay did not immunoprecipitate HA-p85α or p110α, confirming the specificity of the p85α-p110α interactions observed.
Figure 2. p85α mutants interact with p110α and stabilize p110α.
(A-D) HA-tagged p85α mutants and wild-type p85α that carry an intact p110α interaction domain immunoprecipitate p110α from Cos7 cells transfected with the constructs indicated in the panels. Expression of all the transfected constructs was confirmed in the lysates as shown below in each of the panels. The dominant negative p85Δ and empty vector transfected cells served as controls. (E) p85α mutants that contain an intact p110α binding domain when transiently expressed in pan-p85 null MEFs stabilize p110α levels.
Given that most of the p85α mutants still could interact with p110α, we next tested their ability to stabilize p110α protein levels in immortalized pan-p85 null MEFs. It was previously shown that the endogenous levels of p110α in the pan-p85 null background are significantly reduced but can be restored by reconstituting these cells with wild-type p85 (Brachmann et al., 2005). We focused primarily on testing p85α iSH2 domain mutants for their ability to stabilize p110α, given that majority of the mutations identified clustered within this domain. As with wild-type p85α, all of the p85α mutants that contained an intact p110α-binding domain stabilized p110α (Figure 2E). Consistent with their inability to bind p110α, L380fs and the dominant negative p85Δ, did not stabilize p110α (Figure 2E).
p85α mutants interact with p110β and p110δ
The p85α regulatory subunit associates with and regulates all class IA PI3K members including p110β and p110δ (Chantry et al., 1997; Engelman et al., 2006; Hu et al., 1993; Vanhaesebroeck et al., 2005). Hence, we tested the ability of myc-tagged p110β or p110δ to interact with a subset of p85α mutants predicted to destabilize the C2 - iSH2 interactions (Figure 1C). We found that overexpressed HA-tagged p85α wild-type, D560Y, N564D, D560Y_N564D, QYL579delL and Q572*, all of which contained an intact p110-binding domain (Dhand et al., 1994; Huang et al., 2007; Miled et al., 2007), were able to interact with both p110β and p110δ (Figure 3A, B). However, as expected dominant negative mutant p85Δ (Dhand et al., 1994), the mock and vector control lanes did not immunoprecipitate p110β or p110δ (Figure 3A, B) confirming the specificity of the p85-p110β/δ interactions observed.
Figure 3. p85α mutants interact with p110β and p110δ.
(A-B) Indicated HA-tagged p85α mutants and wild-type p85α co-transfected with myc-p110β (A) or myc-p110δ (B) immunoprecipitated p110β (A) or p110δ (B). Expression of all the transfected constructs was confirmed in the lysates. The dominant negative p85Δ and empty vector transfected cells served as controls.
Mutant p85α/p110 class IA PI3K holoenzymes are more active
Having established that the p85α mutants can interact with p110α, p110β and p110δ, we tested if the mutations altered the ability of p85α to suppress the activity of the catalytic subunit. In order to study this, we expressed, purified and tested heterodimeric p85α/p110 class IA PI3K wild-type or mutant enzymes for their lipid kinase activity. The mutants tested were selected based on the stabilization activity data (Figure 2E), transient cell-survival analysis (Figure S7), the structural relevance of p85α N564 (Huang et al., 2007) mutation, and the proximity of QYL579delL mutation to the oncogenic murine Q572* mutation (Jimenez et al., 1998).
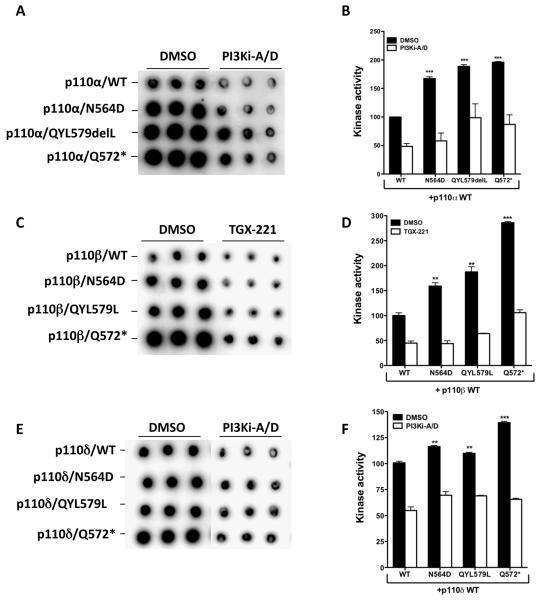
In testing the PI3K activity of purified recombinant iSH2 domain mutant p85α/p110α holoenzyme, we found that relative to the wild-type p85α/p110α, the N564D mutant and QYL579delL iSH2 p85 mutants had ~1.7- to 2.0-fold increase in activity as measured by an in vitro phosphatidylinositol (PI) phosphorylation assay (Knight et al., 2007; Figure 4A and 4B). As expected, the murine Q572* mutant (Chan et al., 2002; Jimenez et al., 1998; Shekar et al., 2005) showed increased kinase activity compared with the wild-type. We further tested the p85α mutants in an alternate assay using nSHi proteins composed a minimal p85α wild-type fragment that spans the nSH2 and iSH2 (amino acids 320-600) domains (Shekar et al., 2005). This minimal nSHi wild-type p85α protein is known to inhibit overexpressed p110α activity in cell lysates (Shekar et al., 2005). Consistent with the holoenzyme activity observed, mutant nSHi proteins showed decreased p110α-activity inhibition (Figure S8). Despite the increased PI3K activity, the mutant p85α/p110α holoenzymes were effectively inhibited by a p110α/δ inhibitor (Folkes et al., 2007).
Figure 4. p85α mutants are impaired in regulating class IA PI3K activity.
(A, C, E) Dot blots show the amount of phosphorylated lipid generated by each of the tested heterodimeric enzymes in triplicates. (B, D, F) Mean ± SEM of the amount of phosphorylated lipid generated in (A, C, E) by each of the enzyme complexes in arbitrary units. Elevated PI3K activity is effectively inhibited by a p110α/δ (B, F) or p110β (TGX-221; D) inhibitor (Folkes et al., 2007; Jackson et al., 2005). Mean ± SEM % control (p85α-WT) activity is shown. (*p<0.05, **p<0.01 and ***p<0.001)
Having established the elevated lipid kinase activity of mutant p85α/p110α holoenzyme, we next measured the PI3K activity of purified recombinant iSH2 domain wildtype or mutant p85α/p110β and p85α/p110δ holoenzymes. We found that relative to the wild-type p85α/p110β, the N564D and QYL579delL iSH2 p85/p110β mutant enzymes had ~1.6- to 1.8-fold increase in activity as measured by an in vitro PI phosphorylation assay (Knight et al., 2007; Figure 4C and 4D). Similarly, relative to wild-type p85α/p110δ holoenzyme, the N564D mutant and QYL579delL iSH2 p85/p110δ mutant enzymes showed elevated lipid kinase activity (~1.2-fold) although the increase in lipid kinase activity was low compared to p85α/p110α or p85α/p110β mutant holoenzymes (Figure 4E and 4F). Consistent with the impaired inhibition by the p85α mutants across all the class IA catalytic subunits, the murine mutant Q572* p85α/p110β and p85α/p110δ enzymes also showed increased kinase activity compared with the wild-type enzyme (Figure 4A-F). Similar to mutant p85α/p110α enzymes, the lipid kinase activity of mutant p85α/p110β or p85α/p110δ was effectively inhibited by a p110β (Jackson et al., 2005) or p110α/δ inhibitor (Folkes et al., 2007), respectively (Figures 4C-F and Figure S9), indicating that pan-p110 class IA inhibitors are likely to be efficacious in patients with p85α mutations.
p85α mutants promote Akt activation and cell survival
In order to further understand the functional relevance of the p85α mutations we investigated their ability to render the IL-3-dependent murine BaF3 cells growth factor-independent by stably expressing the iSH2 mutants either alone or in combination with wild-type p110α̣, p110β or p110δ. We additionally tested a glioblastoma iSH2 domain mutant D560Y and a synthetic-iSH2 double mutant D560Y_N564D for their ability to promote IL-3 independent growth of BaF3 cells. Stable expression of the HA-tagged p85α mutants either alone or in together with myc-tagged p110α, p110β or p110δ in the BaF3 cells was confirmed by Western blotting (Figure 5A-D). Unlike wild-type p85α, all of the p85α mutants tested promoted IL3-independent BaF3 cell survival either on their own or in combination with p110α̣ p110β or p110δ (Figure 5A-H). This survival benefit was blocked by a p110α/δ- or a p110β (TGX-221) inhibitor (Folkes et al., 2007; Jackson et al., 2005), confirming the requirement of p110 activity for the observed p85α mutant-mediated survival effect (Figure 5E-H). Given that elevated class IA PI3K enzyme activity normally results in activation of downstream, pro-survival kinase Akt (Engelman et al., 2006), we tested the stable BaF3 cells expressing the mutant p85α for the phosphoAkt (pAkt) levels following IL3 withdrawal. Consistent with the ability of the mutants to promote cell survival, cells that expressed p85α mutants had increased pAkt when compared to cells expressing wild-type p85α (Figures 5A-D and Figure S10 A-D).
Figure 5. p85α mutants promote p110-dependent cell survival and Akt signaling.
(A-H) Stable expression p85α mutants alone (A, E), together with p110α (B, F) or p110β (C, G) or p110δ (D, H) promote IL3-independent survival of BaF3 cells (E-H) and elevated pAkt (A-D) levels. The cell survival effects (E-H) of p85α mutants are inhibited by a p110α/δ or p110β (TGX-221) inhibitor (Folkes et al., 2007; Jackson et al., 2005). Data shown in (E-H) are mean ± SEM of at least three independent experiments with multiple replicates. (*p<0.05 and **p<0.01)
In addition to cell survival, the p85α mutants D560Y, N564D, N564D_D560Y, and QYL579delL also promoted anchorage-independent growth of BaF3 cells in a class IA PI3K-activity-dependent manner that was blocked by PI3K inhibitors (Figure 6A-F). To further confirm that the p85 iSH2 C2-contact mutants were unable to inhibit p110-activity, we stably reconstituted the expression of the p85α mutants in pan-p85 null MEFs (Brachmann et al., 2005) using a retroviral vector. As observed in our transient reconstitution studies (Figure 2E), all the p85α mutants stabilized the expression of endogenous p110α similar to wild-type p85α (Figure 6G). In addition to stabilizing p110α, wildtype and mutant p85α expression also stabilized p110β and p110δ (Figure 6G). Also, MEFs expressing D560Y, N564D, D560Y_N564D, QYL579delL, and the Q572* murine mutant showed an increase in proliferation when compared to MEFs expressing wild-type p85α or empty vector (Figure 6H). Consistent with this, MEFs expressing mutant p85α showed a higher basal level of pAkt/Akt (Figures 6H and Figure S10E). Taken together, these data indicate that the iSH2 p85α mutations do not affect p85α-mediated p110 stabilization, but impair p85α's ability to negatively regulate class IA PI3K s leading to Akt-mediated cell survival and proliferation.
Figure 6. p85α mutants promote anchorage independent growth, p110 stabilization and proliferation.
(A-F) A representative image showing anchorage-independent growth (A-C) and number of colonies formed in the absence or presence of PI3K inhibitors (D-F) by BaF3 cells stably expressing p85α mutants together with p110α (A), p110β (B) or p110δ (C). (G, H) Pan-p85 null MEFs stably reconstituted with p85α mutants show stabilization of p110α̣ p110β and p110δ (G), elevated pAkt (G) and increased cell proliferation (H). Data shown (D-F, H) are mean ± SEM. (*p<0.05 and **p<0.01).
p85α mutants promote oncogenesis in vivo
BaF3 cells that stably express oncogenic p110α mutants promote leukemia and reduced survival when transplanted into mice (Horn et al., 2008). Since the p85α mutants promote BaF3 cell survival, anchorage independent growth and proliferation in vitro in a p110-dependent manner (Figures 5 and 6), we tested p85α/p110α eGFP/dsRed-tagged BaF3 cells for induction of leukemia-like disease in mice. Mice transplanted with BaF3 cells expressing mutant p85α subunit together with wild-type p110α showed a median survival of 30 to 40 days (Figure 7A). With the exception of one animal that received cells expressing p110α alone, all mice that received BaF3 cells expressing either wild-type p85α or co-expressing wild-type p85α and p110α, or had been transduced with empty vector were still alive at the end of the 55-day study period. Necropsies were conducted at 30 days on an additional cohort of three mice per treatment to follow disease progression. Bone marrow, spleen, and liver samples from these animals reviewed for any pathological abnormalities. As the BaF3 cells were tagged with eGFP and/or dsRed, we examined isolated bone marrow for infiltrating cells by fluorescence-activated cell sorting (FACS). The bone marrow from mice transplanted with cells expressing mutant p85α showed a significant proportion of infiltrating eGFP /dsRed-positive cells compared with bone marrow from mice receiving wild-type p85α or empty-vector control cells (Figure 7B, C). Consistent with the FACS data, in mice receiving cells expressing mutant p85α, histological evaluation of spleen and liver stained with hematoxylin and eosin (H&E) showed significant infiltration of blasts (Figure 7D). However, in mice receiving cells expressing wild-type p85α or empty vector transduced cells, there was little or no evidence of blast infiltration or empty vector transduced cells (Figure 7D). Mice carrying the mutant p85α/p110α-expressing cells also showed increased spleen and liver weight, consistent with an infiltration of cells into the liver and spleen (Table 1). These results demonstrate the in vivo leukemogenic potential conferred by the p85α mutants on hematopoietic cells.
Figure 7. p85α mutants lead to reduced overall survival.
(A) Kaplan-Meier survival curves for cohorts of mice implanted with p85α mutant expressing BaF3 cells compared to vector control cells or p85α wild-type cells or p110α cells alone or p85α /p110α cells showed a significant reduction in overall survival (n = 10 for arms; Log-rank test p<0.0001). (B) Flow cytometric analysis of total bone marrow isolated from mice receiving GFP- and/or dsRed-tagged BaF3 cells expressing the various p85α mutants. (C) Mean number of GFP and/or dsRed positive cells in the bone marrow of mice (n = 3) that received BaF3 cells expressing p85α mutants. Values are mean ± SEM. (D) Representative H&E-stained liver (top) and spleen (bottom) sections from the same mice analyzed in (B). Arrows indicate tumor cells infiltrating the liver. R = red pulp, W = lymphoid follicles of white pulp. In unmarked spleen section, there is a loss of red/white pulp architecture due to disruption by infiltrating tumor cells. Scale bar corresponds to 100μm.
Table 1.
Characterization of BAF3-p85α mutant implanted mice
| Weights |
|||
|---|---|---|---|
| Transplanted cells | Body | Spleen | Liver |
| Empty vector | 20.9 ± 0.7 | 0.44 ± 0.11 | 1.17 ±0.24 |
| p85α-WT | 20.3 ± 0.4 | 0.11 ±0.01 | 1.16 ±0.01 |
| p110α-WT | 22.1 ±0.9 | 0.21 ±0.05 | 1.15 ±0.01 |
| p85α-WT + p110α-WT | 20.3 ± 0.4 | 0.11 ±0.01 | 1.37 ±0.01 |
| p85α-D560Y + p110α-WT | 20.9 ± 0.8 | 1.18 ±0.15** | 2.87 ±0.30** |
| p85α-N564D + p110α-WT | 20.9 ± 0.5 | 0.97 ± 0.14** | 2.88 ± 0.49** |
| p85α D560Y_N564D + p110α-WT | 21.3 ±0.3 | 0.87 ± 0.03** | 2.51 ± 0.32** |
Balb/C mice received 2×106 Ba/F3 cells stably expressing p110α-WT and p85α-WT or mutants (n=3) or the vector (n=3) by tail-vein injection. Allografted mice were sacrificed 5 weeks after transplantations and tissue were collected and analyzed.
p<0.01 vs p85α-WT+p110α-WT
DISCUSSION
Mutations in the p110α catalytic subunit of PI3K are common in colon cancers (Samuels et al., 2004). In this study, we have identified the occurrence of frequent p85α regulatory subunit mutations in colon cancer. The p85α mutants were unable to negatively regulate p110α, p110β and p110δ activity, despite retaining their ability to stabilize them. Of particular interest is a group of mutations in p85α at the interface between p85α-iSH2 and C2 domain of p110α described in a recent crystal structure (Huang et al., 2007; TCGA, 2008). The mutated residues in p85α N564, and D560, are within hydrogen bond distance of residue N345 in p110α, which, when mutated, renders p110α oncogenic (Gymnopoulos et al., 2007). These iSH2 mutations mimic the effects of substitutions at N345 C2 domain of p110α in that they promote elevated p110α activity and therefore cell survival. In vivo, these mutants were capable of promoting oncogenesis, confirming the relevance of these mutants in colon cancer and glioblastoma. In addition, mutation at Q579 within the iSH2 also confers properties similar to mutations at N564 or D560, although the structural implication of this mutation remains to be understood. Taken together with the previously described murine Q572* mutant (Jimenez et al., 1998) these findings suggest that this region of iSH2 is a potential hotspot for the accumulation of p85α mutations that confer a cellular growth advantage.
Though, oncogenic mutations in p110α are common, such mutations in other class IA PI3Ks, p110β and p110δ, are not common (Bader et al., 2005; Samuels et al., 2004). The p85α subunit besides interacting and regulating p110α, can also heterodimerize with p110β and p110δ and regulate their activity (Chantry et al., 1997; Engelman et al., 2006; Hu et al., 1993; Vanhaesebroeck et al., 2005). Consistent with this, as observed with p110α, we found that the p85α mutants were able to interact and stabilize both p110β and p110δ. However, the p85α mutants were impaired in the ability to negatively regulate p110β and p110δ activity. This is supported by our structural modeling and the proposal in a recent review (Amzel et al., 2008) that suggest the presence of a conserved set of regulatory interactions between p110-C2 iSH2-p85α across all the three class IA PI3Ks. Our findings indicated that mutations in p85α could serve as a global mechanism for deregulation of class IA PI3K activity in cancers. This is consistent with the emerging role of p110β and p110δ in cancer (Kang et al., 2006; Zhao and Vogt, 2008).
Besides deregulation of class IA PI3K activity by the p85α mutants, the impact of the p85α mutations on p110-independent p85α functions (Garcia et al., 2006; Taniguchi et al., 2006), the role of the additional p85α mutants, and the activity of co-occurring p85α/p110α double mutant enzymes may all contribute to pathogenesis and remains to be explored. In gliomas, where p110α mutations are not common (Bader et al., 2005), our data on the function of p85α mutations indicates that such mutants may play a major route for PI3K pathway activation, besides PTEN inactivation (Parsons et al., 2008; TCGA, 2008; Yin and Shen, 2008).
Taken together, our data shows that PIK3R1 is a frequently mutated, functionally relevant colon cancer gene. Consistent with our findings, a recent forward genetic screen in mouse for colon cancer genes identified PIK3R1 as a site of transposon insertion in several independent lines (Starr et al., 2009). With the identification of PIK3R1 mutations and the establishment of their functional relevance, it is very likely that the p85 mutations when used as biomarkers should allow the detection of p110-dependent p85-mutant tumors that are very likely to respond to treatment with inhibitors that target PI3Ks or their downstream effectors such as Akt.
EXPERIMENTAL PROCEDURES
Tumor DNA, mutation and genomic amplification
Primary human tumor samples with appropriate IRB approval and patient informed consent were obtained from commercial sources (Table S1). The human tissue samples used in the study were de-identified (double-coded) prior to their use and hence, the study using these samples is not considered human subject research under the US Department of Human and Health Services regulations and related guidance (45 CFR Part 46). Tumor content in all the tumors used was confirmed to be >70% by pathology review. Tumor DNA was extracted using Qiagen Tissue easy kit. (Qiagen, CA). All coding exons of PIK3R1-R5, PIK3CA, KRAS and PTEN were amplified using primers listed in Table S4 and sequenced using 3730xl ABI sequencer (AppliedBiosystems, CA). The PCR products were generated using two pairs of primers, an outer pair and an inner pair to increase the specificity (Table S4), using standard PCR conditions. The sequencing data was analyzed for presence of protein variants not present in the dbSNP database using Mutation Surveyor (Softgenetics, PA) and additional automated sequence alignment programs. The putative variants identified were confirmed for their prescence by DNA sequencing or mass spectrometry analysis (Sequenom, CA) of the original tumor DNA followed by confirmation of its absence in the adjacent matched normal DNA by a similar process applied to the tumor DNA (Figures S1 and S2).
The C2 homology models we generated by ESyPred3D (Lambert et al., 2002) server, using C2 domain sequences and the crystal structure of PI3K alpha (PDB: 2RD0) as a template. Alignment method was PIR target-template alignment with neural net and new screening algorithm. The initial homology model was then improved by manual adjustment and geometric optimization with COOT (Emsley and Cowtan, 2004).
Genomic DNA from tumor samples as annotated in Table S1 was analyzed using an Affymetrix GeneChip mapping 100K arrays (Affymetrix, Santa Clara, CA, USA) as per the manufacturer instructions. Copy number data were generated for each array by using Affymetrix Chromosome Copy Number Tool (CNAT) v.2.0 (Huang et al., 2004) and/or dChipSNP (Lin et al., 2004). The array data from this study was deposited with Gene Expression Omnibus (GEO) database under accession number GSE1852.
Cell lines
The IL-3-dependent mouse pre-B cell line BaF3 and Cos7 cells were purchased from ATCC (American Type Culture Collection, Manassas, VA). BaF3 cells were maintained in RPMI 1640 supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific, Rockford, IL), 2mM L-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin (complete RPMI) and 2ng/mL mouse IL-3. The p85 α, β knockout mouse embryonic fibroblasts (Pan-p85 MEFs) were obtained form Dr Lewis C. Cantley's lab (Harvard Medical School, Boston, MA) and maintained in DMEM supplemented with 15% (v/v) fetal bovine serum, while COS7 cells were maintained in DMEM supplemented with 10% (v/v) fetal bovine serum, 2mM L-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin. Phoneix (Orbigen, San Diego, CA) cells were maintained in DMEM supplemented with 10% FBS. Spodoptera frugiperda (Sf9) insect cells (Vaughn et al., 1977) were grown in ESF 921 serum free medium (Expression systems LLC, Woodland CA).
Plasmids and antibodies
Eukaryotic expression plasmid pRK5E driving the expression of HA- p85α mutants, untagged p110α, N-myc tagged p110α, N-myc tagged p110β, and N-myc tagged p110δ were constructed using standard PCR and cloning strategies. pRetro-IRESdsRed (Clontech, CA) or a version of this retroviral vector with GFP in place of dsRed was used to stably express HA-p85α, N-myc-p110α (Yu et al., 1998), N-mycp110β, and N-myc-p110δ. Recombinant baculovirus expressing N-His6-p110α, N-His6-p110β, N-His6-p110δ and untagged p85α driven by the polyhedrin promoter were prepared according to manufacturer's recommendations using the Bac-to-Bac Expression System (Invitrogen, CA). GST fusion constructs expressing p85 nSHi made of amino acids 320-600 (Shekar et al., 2005) was made in pGEX6p-2 (GE Healthcare, NJ).
Antibodies that recognize pAKT (Ser473) and AKT (Cell Signaling Technology, Beverly, MA), p110β (Cell Signaling Technology, Beverly, MA), p110α and p110δ (Epitomics, South San Francisco, CA), c-Myc (Genentech Inc., South San Francisco), β-actin and Flag M2 (Sigma Life Science, St. Louis, MO) were used in the study. Horseradish peroxidase (HRP)–conjugated HA antibody (Roche Molecular Biochemicals, Indianapolis, IN) and HRP-conjugated secondary antibodies (Pierce Biotechnology, Rockford, IL) were used in western blots. Anti-HA rat monoclonal (clone 3F10) coupled affinity matrix (Roche Diagnostics, Germany) and c-Myc tag IP application kit (Thermo scientific, Waltham, MA) were used for immunoprecipitation studies.
Recombinant proteins
To generate recombinant heterodimeric PI3K enzyme, 1L of 2 × 106 Sf-9 cells/ml were co-infected with either p110α or p110β or p110δ and p85α expressing baculoviruses at an MOI of 0.5 each. Cell pellets were harvested at 72h post infection and lysed in lysis buffer A (50mM Tris HCl pH8.0, 0.3M NaCl, 20mM Imidazole, 1mM TCEP and protease inhibitors). The cell lysate was clarified by spinning at 35000g for 1hr at 4°C. The clarified supernatant was loaded over a Ni-NTA Superflow (Qiagen 30410) column pre-equilibrated with lysis buffer A. The His6P110α/p85α complex was eluted using immidazole gradient (20mM to 250mM) made using buffer B (20mM Tris HCl pH8.0, 0.3M NaCl and 0.25mM TCEP). The purified His6P110/p85α was further purified on a Superdex 200 size exclusion column pre-equilibrated with 25mM Tris pH7.5, 150mM NaCl, 0.25mM TCEP and 10% Glycerol. Purified proteins were serially diluted and resolved on SDS-containing PAGE for concentration assessment (Figure S11).
BL-21 E. coli cells transformed with the appropriate pGEX6p-2 nSHi expression construct were induced to express GST fusion proteins as per the manufacturer recommendations. Cell pellet from one liter culture was lysed in lysis buffer C (PBS, 2mMDTT and protease inhibitors), microfluidized and centrifuged at 35,000g for 1h at 4°C. Clarified lysate was loaded over a GST-Sepharose 4 fast flow (GE Healthcare 17-5132-01) column pre-equilibrated with lysis buffer C. Bound GST-p85 nSHi was eluted using buffer B (50mM Tris pH8.0, 10mM NaCl, 1mM EDTA, 2mM DTT and 10mM GSH). Eluted proteins were dialyzed into buffer D (20mM Tris PH8, 150mM NaCl, 1mM EDTA and 0.25mM TCEP) and incubated with PreScission protease (GE Healthcare 27-0843-01) at 4°C overnight to remove the GST tag. The untagged p85 nSHi in the dialyzed and digested material was recovered by passing it over a GST-Sepharose column. The nSHi protein was further purified by size exclusion on a Superdex 200 column.
Generation of stable cell lines
Retroviral constructs expressing HA-p85-dsRed and/or myc-p110-GFP were transfected into Pheonix amphoteric cells using Lipofectamine (Invitrogen, Carlsbad, CA). The resulting virus was then transduced into BaF3 cells and pan-p85 null MEFs. Top 10% of the infected cells based on the expression of retroviral IRES driven dsRed and/or GFP were sterile sorted by flow cytometry and characterized for expression of proteins by western blot. Pools of these cells were then used in further studies.
Immunoprecipitation and western blot
Cos-7 (2.5 × 105) or pan-p85 null MEFs (10 × 106) cells were transfected or nucleofected with appropriate constructs expressing HA-p85 and/or p110-WT DNA using Fugene 6 (Roche diagnostics) or nucleofection kit 1 (Amaxa Inc., Cologne, Germany) following the manufacturer's protocol. At 48h post transfection/nucleofection cells were washed with PBS and lysed in a lysis buffer I (50mMTrisHCl pH 7.5, 150mM NaCl, 1mM EDTA, 1% Triton X-100). The clarified lysates were incubated for 2h at 4°C with anti-HA antibody coupled beads. The HA beads were then spun down at 500g for 2 min. and washed thrice using the lysis buffer I. The immunoprecipitated proteins remaining on the beads were boiled in SDS-PAGE loading buffer, resolved on a 4-20% SDS-PAGE (Invitrogen, Carlsbad, CA) and transferred onto a nitrocellulose membrane. Immunoprecipitated proteins or proteins from lysates were detected using appropriate primary, HRP-conjugated secondary antibody and chemiluminescences Super signal West Dura chemiluminescence detection substrate (Thermo Fisher Scientific, Rockford, IL).
Survival and proliferation Assay
For the initial characterization of the p85α mutants, 2×106 BaF3 cells were nucleofected with appropriate pRK5E wild-type or mutants constructs expressing HA-p85α and/or myc-p110α DNA using nucleofection kit V (Amaxa Inc. Gaithersburg, MD). After a 3 h recovery period in media without IL-3 following nucleofection cells were plated in 3 × 96-well plates in triplicates and monitored for survival. Similarly, BaF3 cells stably expressing the wild-type and mutant p85α alone or together with mycp110α, myc-p110β or myc-p110δ, were washed twice in PBS and plated in 3 × 96-well plates in replicates of twelve in complete RPMI medium without IL3. 500nM of the p110 small molecule inhibitor PI3Ki-A/D (Folkes et al., 2007) or TGX-221 (Jackson et al., 2005) in 0.1% DMSO was used to test the effects of p110 inhibition, where relevant as depicted in the figures. Viable cells at 0h, 24h, 48h, 96h and 120h were determined using Cell Titer-Glo luminescence cell viability kit (Promega Corp., Madison, WI) and Synergy 2 (Biotek Instrument, CA) luminescence plate reader. All the cell number values were normalized against 0h values. Normalized vector control cell value was used to calculate the relative survival of the various mutant p85α either alone or in combination with class IA PI3Ks. In order to assess proliferation of pan-p85-null MEFs stably expressing p85α-WT or mutants, 5000 cells in triplicates were plated in media with 15% serum and allowed to proliferate for 5 days. Cell numbers were measure at day 0 and day 5 using the luminescence cell viability kit. Data was expressed as Mean ± SEM of fold change over vector or p85-WT. Mean and statistical significance was determined using GraphPad V software (GraphPad, San Diego, CA).
In Vitro PI3K activity assay
Purified PI3K heterodimeric enzyme was quantified on a gel to estimate the concentration of p85α along with either p110α, p110β or p110δ (Figure S11). For each of the p110/p85α purified enzyme combination, 30ng of the purified protein was tested for PI3K activity as described recently (Knight et al., 2007). Briefly, 30 ng of each of the purified enzyme was incubated for 30 min at 25°C with PI and 2 μCi [γ-32P] ATP (Perkin Elmer, Waltham, MA) in 25μl of kinase reaction buffer (10 μM ATP, 1 mg/ml BSA, 25 mM HEPES (pH 7.4) and 10 mM MgCl2) either with or without 500nM PI3KiA/D or TGX-221. At the end of 30 min, 4 μl of each reaction was spotted onto a dry nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA) using a multichannel pipette, dried and washed five times with wash solution (1M NaCl in 1% H3PO4). After the washes the membrane was dried and exposed to a phosphor screen for 1 hr. The exposed phosphor screen was then scanned on a Typhoon phosphorimager (GE Healthcare, Piscataway, NJ). The amount of bound radioactive lipid derived from PI phopsorylation by the different PI3K enzymes studied was quantified using a MATLAB script called Spot (Knight et al., 2007). Alternatively, inhibition of recombinant p110α in baculovirus lysates by E. coli expressed and purified p85α nSHi was directly measured as described previously (Shekar et al., 2005).
BaF3 Colony formation assay
BaF3 cells (2×105), stably expressing p110α, p110β or p110δ along with p85α wild-type or mutants, was mixed with 2 mls of IL3-free Methocult (STEMCELL Technologies, Canada) with or without 500nM PI3Ki-A/D or TGC-221 and plated on to 6 well plates that were then incubated at 37°C for 2 weeks. The presence of colonies was assessed using Gel count imager (Oxford Optronix Ltd, Oxford, UK). The number of colonies in each plate was quantified using Gel count software (Oxford Optronix Ltd, Oxford, UK).
Animal Studies
BaF3 cells (2 × 106) expressing the p85α wild-type or mutants along with p110α were implanted into 8-12 week old Balb/C nude mice by tail vein injection. A total of 13 animals per treatment were injected. Of this 10 mice were followed for survival and 3 were used for necropsy at day 30 to assess disease progression by histological analysis of spleen and liver. Bone marrow single cell suspension obtained from these animals was also analyzed for the presence and proportion of GFP and/or dsRed positive BaF3 cells by FACS analysis. When possible dead or moribund animals in the survival study were dissected to confirm the cause of death. Morphologic and histological analyses of spleen, liver and bone marrow was also done on these animals. Spleen and liver were fixed in 10% neutral buffered formalin, then processed in an automated tissue processor (TissueTek, CA) and embedded in paraffin. Four-micron thick sections were stained with H&E (Sigma), and analyzed histologically for presence of infiltrating tumor cells. Photographs of histology were taken on a Nikon 80i compound microscope with a Nikon DS-R camera. All animal studies were performed under Genentech's Institutional Animal Care and Use Committee (IACUC) approved protocols.
Statistical Analyses
Student's t-test (two tailed) was used for statistical analyses to compare treatment groups using GraphPad Prism 5.00 (GraphPad Software, San Diego, CA). A P-value <0.05 was considered statistically significant (*p<0.05, **p<0.01 and ***p<0.001). For Kaplan-Meier Method of survival analysis, log-rank statistics were used to test for difference in survival.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Genentech DNA Sequencing, Oligo, Microarray, and FACS labs for their help with the project. We thank the Genentech Bioinformatics group for informatics infrastructure support and the Pathology Core Labs for providing histology, immunohistochemistry, and tissue management support. Thanks to Christopher Dant, Roberta Kelly, Kim Newton and Jan Theunissen for their critical reading of the manuscript. Work in JMB's lab was supported by a grant from the Janey Fund and NIH GM55692. Special thanks to Dr. Lewis C. Cantley for the pan-p85 null MEFs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interest statement: Majority of the authors are employees of Genentech Inc., as indicated adjacent to author names.
References
- Amzel LM, Huang CH, Mandelker D, Lengauer C, Gabelli SB, Vogelstein B. Structural comparisons of class I phosphoinositide 3-kinases. Nat Rev Cancer. 2008;8:665–669. doi: 10.1038/nrc2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM, Myers MG, Jr., Shoelson SE, Chin DJ, Sun XJ, Miralpeix M, Hu P, Margolis B, Skolnik EY, Schlessinger J, et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. Embo J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- Borlado LR, Redondo C, Alvarez B, Jimenez C, Criado LM, Flores J, Marcos MA, Martinez AC, Balomenos D, Carrera AC. Increased phosphoinositide 3-kinase activity induces a lymphoproliferative disorder and contributes to tumor generation in vivo. Faseb J. 2000;14:895–903. doi: 10.1096/fasebj.14.7.895. [DOI] [PubMed] [Google Scholar]
- Brachmann SM, Yballe CM, Innocenti M, Deane JA, Fruman DA, Thomas SM, Cantley LC. Role of phosphoinositide 3-kinase regulatory isoforms in development and actin rearrangement. Mol Cell Biol. 2005;25:2593–2606. doi: 10.1128/MCB.25.7.2593-2606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Carpenter CL, Auger KR, Chanudhuri M, Yoakim M, Schaffhausen B, Shoelson S, Cantley LC. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J Biol Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- Chan TO, Rodeck U, Chan AM, Kimmelman AC, Rittenhouse SE, Panayotou G, Tsichlis PN. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide 3-kinase regulatory subunit. Cancer Cell. 2002;1:181–191. doi: 10.1016/s1535-6108(02)00033-8. [DOI] [PubMed] [Google Scholar]
- Chantry D, Vojtek A, Kashishian A, Holtzman DA, Wood C, Gray PW, Cooper JA, Hoekstra MF. p110delta, a novel phosphatidylinositol 3-kinase catalytic subunit that associates with p85 and is expressed predominantly in leukocytes. J Biol Chem. 1997;272:19236–19241. doi: 10.1074/jbc.272.31.19236. [DOI] [PubMed] [Google Scholar]
- Dhand R, Hara K, Hiles I, Bax B, Gout I, Panayotou G, Fry MJ, Yonezawa K, Kasuga M, Waterfield MD. PI 3-kinase: structural and functional analysis of intersubunit interactions. Embo J. 1994;13:511–521. doi: 10.1002/j.1460-2075.1994.tb06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Folkes A, Shuttleworth S, Chuckowree I, Oxenford S, Wan NC, Castanedo G, Goldsmith R, Gunzner J, Heffron T, Mathieu S, et al. Preparation of thienopyrimidines and furopyrimidines as lipid kinase inhibitors for treating cancer and other diseases. PCT Int. Appl. 2007:206. CODEN: PIXXD2 WO 2007127175 A2 20071108. [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Garcia Z, Silio V, Marques M, Cortes I, Kumar A, Hernandez C, Checa AI, Serrano A, Carrera AC. A PI3K activity-independent function of p85 regulatory subunit in control of mammalian cytokinesis. Embo J. 2006;25:4740–4751. doi: 10.1038/sj.emboj.7601324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Bartels G, Gehlhaar C, Holtkamp N, von Deimling A. PIK3CA mutations in glioblastoma multiforme. Acta Neuropathol. 2005;109:639–642. doi: 10.1007/s00401-005-1000-1. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Hoedemaeker FJ, Siegal G, Roe SM, Driscoll PC, Abrahams JP. Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate. J Mol Biol. 1999;292:763–770. doi: 10.1006/jmbi.1999.3111. [DOI] [PubMed] [Google Scholar]
- Holt KH, Olson L, Moye-Rowley WS, Pessin JE. Phosphatidylinositol 3-kinase activation is mediated by high-affinity interactions between distinct domains within the p110 and p85 subunits. Mol Cell Biol. 1994;14:42–49. doi: 10.1128/mcb.14.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Bergholz U, Jucker M, McCubrey JA, Trumper L, Stocking C, Basecke J. Mutations in the catalytic subunit of class IA PI3K confer leukemogenic potential to hematopoietic cells. Oncogene. 2008;27:4096–4106. doi: 10.1038/onc.2008.40. [DOI] [PubMed] [Google Scholar]
- Hu P, Mondino A, Skolnik EY, Schlessinger J. Cloning of a novel, ubiquitously expressed human phosphatidylinositol 3-kinase and identification of its binding site on p85. Mol Cell Biol. 1993;13:7677–7688. doi: 10.1128/mcb.13.12.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Gabelli SB, Amzel LM. Insights into the oncogenic effects of PIK3CA mutations from the structure of p110alpha/p85alpha. Cell Cycle. 2008;7:1151–1156. doi: 10.4161/cc.7.9.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- Huang J, Wei W, Zhang J, Liu G, Bignell GR, Stratton MR, Futreal PA, Wooster R, Jones KW, Shapero MH. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, Ohta M, Jazag A, Guleng B, Tateishi K, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, Cantley LC, Brugge JS. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, Kenche V, Anderson KE, Dopheide SM, Yuan Y, et al. PI 3-kinase p110beta: a new target for antithrombotic therapy. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- Jimenez C, Hernandez C, Pimentel B, Carrera AC. The p85 regulatory subunit controls sequential activation of phosphoinositide 3-kinase by Tyr kinases and Ras. J Biol Chem. 2002;277:41556–41562. doi: 10.1074/jbc.M205893200. [DOI] [PubMed] [Google Scholar]
- Jimenez C, Jones DR, Rodriguez-Viciana P, Gonzalez-Garcia A, Leonardo E, Wennstrom S, von Kobbe C, Toran JL, R. B. L, Calvo V, et al. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. Embo J. 1998;17:743–753. doi: 10.1093/emboj/17.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M, Sudel K, Horn S, Sickel M, Wegner W, Fiedler W, Feldman RA. Expression of a mutated form of the p85alpha regulatory subunit of phosphatidylinositol 3-kinase in a Hodgkin's lymphoma-derived cell line (CO) Leukemia. 2002;16:894–901. doi: 10.1038/sj.leu.2402484. [DOI] [PubMed] [Google Scholar]
- Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Feldman ME, Balla A, Balla T, Shokat KM. A membrane capture assay for lipid kinase activity. Nat Protoc. 2007;2:2459–2466. doi: 10.1038/nprot.2007.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T, Woscholski R, Hallberg B, Rodriguez-Viciana P, Downward J, Parker PJ. The activation of phosphatidylinositol 3-kinase by Ras. Curr Biol. 1994;4:798–806. doi: 10.1016/s0960-9822(00)00177-9. [DOI] [PubMed] [Google Scholar]
- Lambert C, Leonard N, De Bolle X, Depiereux E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- Otsu M, Hiles I, Gout I, Fry MJ, Ruiz-Larrea F, Panayotou G, Thompson A, Dhand R, Hsuan J, Totty N, et al. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, Whitehead RH, Thomas RJ, Phillips WA. The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Shekar SC, Wu H, Fu Z, Yip SC, Nagajyothi, Cahill SM, Girvin ME, Backer JM. Mechanism of constitutive phosphoinositide 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J Biol Chem. 2005;280:27850–27855. doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'Sullivan MG, Matise I, Dupuy AJ, et al. A Transposon-Based Genetic Screen in Mice Identifies Genes Altered in Colorectal Cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Tran TT, Kondo T, Luo J, Ueki K, Cantley LC, Kahn CR. Phosphoinositide 3-kinase regulatory subunit p85alpha suppresses insulin action via positive regulation of PTEN. Proc Natl Acad Sci U S A. 2006;103:12093–12097. doi: 10.1073/pnas.0604628103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Ali K, Bilancio A, Geering B, Foukas LC. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Waterfield MD. Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae) In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- Wang ZJ, Taylor F, Churchman M, Norbury G, Tomlinson I. Genetic pathways of colorectal carcinogenesis rarely involve the PTEN and LKB1 genes outside the inherited hamartoma syndromes. Am J Pathol. 1998;153:363–366. doi: 10.1016/S0002-9440(10)65579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yin Y, Shen WH. PTEN: a new guardian of the genome. Oncogene. 2008;27:5443–5453. doi: 10.1038/onc.2008.241. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.