Abstract
Rationale and Objectives
The authors present their initial experience using a 3-T whole-body scanner equipped with a 128-channel coil applied to lung motion assessment. Recent improvements in fast magnetic resonance imaging (MRI) technology have enabled several trials of free-breathing three-dimensional (3D) imaging of the lung. A large number of image frames necessarily increases the difficulty of image analysis and therefore warrants automatic image processing. However, the intensity homogeneities of images of prior dynamic 3D lung MRI studies have been insufficient to use such methods. In this study, initial data were obtained at 3 T with a 128-channel coil that demonstrate the feasibility of acquiring multiple sets of 3D pulmonary scans during free breathing and that have sufficient quality to be amenable to automatic segmentation.
Materials and Methods
Dynamic 3D images of the lungs of two volunteers were acquired with acquisition times of 0.62 to 0.76 frames/s and an image matrix of 128 × 128, with 24 to 30 slice encodings. The volunteers were instructed to take shallow and deep breaths during the scans. The variation of lung volume was measured from the segmented images.
Results
Dynamic 3D images were successfully acquired for both respiratory conditions for each subject. The images showed whole-lung motion, including lifting of the chest wall and the displacement of the diaphragm, with sufficient contrast to distinguish these structures from adjacent tissues. The average time to complete segmentation for one 3D image was 4.8 seconds. The tidal volume measured was consistent with known tidal volumes for healthy subjects performing deep-breathing maneuvers. The temporal resolution was insufficient to measure tidal volumes for shallow breathing.
Conclusion
This initial experience with a 3-T whole-body scanner and a 128-channel coil showed that the scanner and imaging protocol provided dynamic 3D images with spatial and temporal resolution sufficient to delineate the diaphragmatic domes and chest wall during active breathing. In addition, the intensity homogeneities and signal-to-noise ratio were adequate to perform automatic segmentation.
Keywords: Lung, magnetic resonance imaging, image segmentation, lung volume measurement
Magnetic resonance imaging (MRI) is a suitable option to analyze lung motion (1– 8). MRI has an advantage over other imaging modalities in motion analysis because it does not expose subjects to ionizing radiation, allowing baseline studies to be obtained in healthy subjects and compared to disease studies. As reported elsewhere, three-dimensional (3D) MRI is especially useful in studying the movement of the diaphragm and the rib cage (1,9 –12). One limitation of current 3D MRI of the lung is that it requires subjects to hold their breath at multiple respiratory phases (1,10,11). It is well known from lung motion analysis using biplanar fluoroscopy that the motion of the lung during normal breathing has hysteresis that does not appear in breath-hold imaging (13). Furthermore, subjects with chronic obstructive pulmonary disease typically have difficulty holding their breath, limiting the feasibility of MRI motion analysis in these subjects. Other investigators have tried to overcome these problems by performing dynamic 3D image studies by recording two-dimensional multislice imaging taken in free-breathing subjects (12,14).
Recent improvements in fast imaging have enabled an advanced form of dynamic 3D imaging in free-breathing subjects (15–17). Blackall et al (17) proposed the use of a fast field echo and echo-planar imaging (EPI) sequence and achieved an imaging speed of 330 ms/frame, with acquisition matrix size of 128 × 256 for 25 to 27 slices. This imaging method was sufficiently fast to investigate the intracycle and intercycle reproducibility of respiratory motion, including hysteresis analysis. Parallel imaging is another possible solution to enable lung MRI in free-breathing subjects, but without the serious image distortion likely found in EPI-based sequences. Plathow et al (18) reported motion analysis of the lung using a 3D fast low-angle shot (FLASH) sequence combined with a parallel imaging technique and view sharing on a 1.5-T whole-body magnetic resonance scanner with a 6-channel coil. This imaging speed was also sufficiently fast to monitor lung motion and to correlate pulmonary function and intrathoracic tumor mobility.
As the image speed increases, image quality is compromised, making it difficult to meet the image quality necessary for motion analysis. Blackall et al (17) stated that the EPI-based sequence showed considerably less detail within the lung. Images acquired with the view-sharing technique were also not applicable for fully automatic segmentation because of intensity inhomogeneities and motion artifacts (16).
The objective of this study, on the basis of this background, was to assess whether dynamic 3D MRI of the lung could be performed using 128-channel, 3-T MRI to produce images of sufficiently good quality to apply automatic image segmentation. We propose an imaging method using a 3D FLASH sequence combined with a generalized auto-calibrating partially parallel acquisition (GRAPPA) technique. We also report volunteer studies (n = 2) to assess whether the combination of the proposed 3-T parallel imaging method and an image segmentation method allows comprehensive lung motion analysis in free-breathing subjects.
MATERIALS AND METHODS
Subjects
The two subjects were a healthy male (age, 35 years) and female (age, 22 years) volunteers. The study protocol was approved by the institutional review board prior to the experiment. Informed consent for dynamic MRI of the lung was obtained from the subjects after the nature of the procedures and potential hazards had been fully explained.
MRI Protocol
Imaging was carried out on a 3.0-T prototype whole-body scanner (Tim TRIO; Siemens Medical Systems, Erlangen, Germany) extended to accommodate 128 independent receiver channels at Massachusetts General Hospital. A 128-channel receive-only coil developed by Schmitt et al (19) was connected to the scanner to perform dynamic parallel MRI. The coil elements of the 128-channel coil are mounted on a fiberglass mold adapted to a male subject (Fig 1). The scanner system allows the signal from each individual coil element to be detected with a single radiofrequency receiver channel. Dynamic MRI was performed using a 3D FLASH sequence combined with the GRAPPA technique. Frame rates of 0.62 to 0.76 frames/s (3D) were achieved, with the following imaging parameters: repetition time, 1.66 ms; echo time, 0.72 ms; flip angle, 9°; receiver bandwidth, 1560 Hz/pixel; GRAPPA acceleration factor, 4; field of view, 400 × 400 mm; acquisition matrix, 128 × 128; slab thickness, 150 mm; 24 to 30 slice encodings; and voxel size, 3.125 × 3.125 × 5 mm. The images were acquired with the subject in the supine position. During the scan, the subjects were instructed to keep breathing, and 20 frames of 3D images were obtained for each examination. Two different breathing maneuvers were examined for each subject: shallow breathing in the range from the functional residual capacity (FRC) to the FRC plus tidal volume and deep breathing from the residual volume (RV) to the total lung capacity. Two examinations were performed for each breathing maneuver, yielding a total of four series of 3D images from the two subjects.
Figure 1.
A 128-channel receiver coil with 60 coil elements on the front side (left) and 68 on the back side (right) was used for imaging. All the coil elements have a diameter of 75 mm and are arranged in a continuous array of hexagonal symmetry to minimize next-neighbor coupling.
Postprocessing and Visualization of Magnetic Resonance Images
The magnetic resonance images were processed on a Linux-based workstation (Dell Precision 470, Dell, Inc., Round Rock, TX; central processing unit, Intel Xeon [Dual Core, 64 bit] at 2.8 GHz, Intel Corporation, Santa Clara, CA; random-access memory, 4 GB; operating system, Fedora Core 7, Red Hat, Inc., Raleigh, NC). The lung area in each frame was segmented on the basis of a combination of confidence-connectedness and fuzzy-connectedness algorithms, which were implemented in the Insight Segmentation and Registration Toolkit (Kitware, Inc., Clifton Park, NY) (20). In this combination, a rough segmentation of the lung is performed using the confidence-connectedness algorithm to estimate the mean and variance of gray values in the lung area; these values are then used for the fuzzy-connectedness algorithm to compute an affinity map, which represents degrees of adjacency and the similarity of pairs of nearby voxels. The basic idea of the confidence-connectedness algorithm is that the region grows from predefined seed points toward the boundaries of the desired area, which is determined on the basis of simple statistics of current regions, through iterations. The range of intensities included to the region is controlled by the multiplier parameter. The final region is extracted by thresholding the affinity map. The multiplier and threshold parameters were tuned so that the lung area was correctly delineated from the surrounding tissue but not from the blood vessels in the lung to calculate the lung volume. The parameters were constant for every frame except several frames in the deep-breathing protocol near maximum inhalation. For those frames, the parameters were manually adjusted to obtain a segmented area. Note that in all cases, judgment about whether the segmentation algorithm “worked” was based on a comparison of the segmented lung area to that determined by visual observation of the original images. The time required for the segmentation was measured using the internal clock of the workstation. After the segmentation, a marching-cubes algorithm (21) was applied to the segmented data to extract the surface of the lung area and visualized by surface rendering using the Visualization Toolkit (Kitware, Inc.) (22). The number of voxels in the segmented lung area from each frame was summed to measure the lung volume.
RESULTS
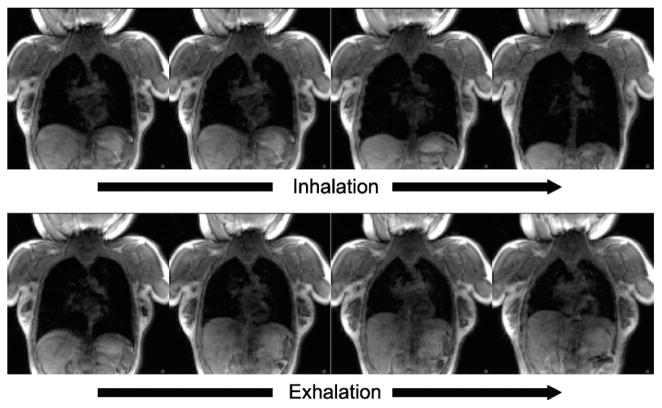
Dynamic 3D images were successfully acquired for both volunteers in two respiratory conditions. The whole-lung motions, including lifting of the chest wall and displacement of the diaphragm, were visualized. Figure 2 shows consecutive coronal sections of the dynamic 3D images from the female subject representing the whole breathing cycle during the deep-breathing protocol. Note the sharp delineation of the diaphragmatic domes and chest wall during active breathing. The lung area in each frame of the dynamic 3D images was successfully segmented (Fig 3). The average time to complete segmentation for one 3D frame was 4.8 seconds. For the shallow-breathing protocol (from FRC to FRC plus tidal volume), lung volume varied from approximately 2000 to 2500 cm3 for the subject and from 1300 to 1800 cm3 for the female subject (Fig 4). In deep breathing (from RV to total lung capacity), lung volume ranged from 2300 to 3200 cm3 and from 1200 to 3500 cm3, respectively. Although minimum lung volume during deep breathing (RV) is supposed to be less than minimum volume during shallow breathing (FRC), the results indicate that the minimum lung volume of the male volunteer was approximately the same as FRC.
Figure 2.
The selected coronal images from the consecutive frames of free-breathing dynamic three-dimensional images of the lung of a 28-year-old female volunteer acquired using a 3-T magnetic resonance imaging scanner equipped with a 128-channel receiver coil are shown. The images were acquired every 1.6 seconds in exhalation and inhalation during deep breathing.
Figure 3.
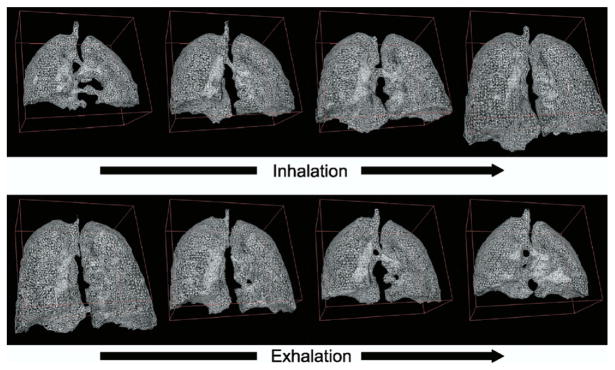
Triangular representation of the lung surface during deep breathing was generated using a marching-cubes algorithm (21) applied to the segmented lung images. After segmenting the lung in each frame, the marching-cubes algorithm was applied.
Figure 4.
The temporal lung volume changes during deep (a) and shallow (b) breaths are plotted. The lung volumes were calculated by counting the numbers of voxels belonging to the lung area on the segmented three-dimensional images.
DISCUSSION
We report our initial experience of lung motion analysis on a 3-T MRI scanner equipped with a 128-channel receiver coil. We achieved a temporal resolution for dynamic 3D imaging of 0.62 to 0.76 frames/s, which allowed us to observe free-breathing lung motion in three dimensions. At the same time, the image quality was sufficient to see the boundary of the lung area as well as to apply an automatic segmentation algorithm to extract the lung volume. This then allowed us to determine the time-dependent lung volume.
The applicability of automatic postprocessing is critical for motion analysis using dynamic MRI, in which tens of 3D image frames consisting of tens of slices must be processed. The lung area was segmented automatically with the constant multiplier and threshold parameters throughout the series of frames except several frames near maximum inhalation of the deep-breathing protocol. Examination of Figure 4 shows that the time spent at maximal inhalation for the deep-breathing protocol was very short, with subsequent large changes in lung volume per unit time. This resulted in motion artifacts around the lung boundary. The blurred boundary impeded the growth of the region during the segmentation by the confidence-connectedness algorithm. The multiplier and threshold parameters were adjusted so that the region grew toward the true boundaries even if the region had larger statistical variation of the pixel intensities.
Surface rendering of the segmented image enables the detailed qualitative assessment of free-breathing 3D lung motion, which could never have been achieved with either free-breathing dynamic two-dimensional images or breath-hold 3D images acquired at multiple respiratory phases. Figure 3 shows the nonuniformity of motion of the diaphragm during deep respiration. In the initial stage of inhalation, when the diaphragm starts contracting to enlarge the thoracic cavity in an inferior direction, major displacement of the diaphragm along the superior-inferior direction is observed around the lumbar spine, and afterward, the displacement propagates to the costal area of the diaphragm. During the earlier exhalation state, the costal area of the diaphragm was lower (more on the inferior side) than the other area. These actions of the diaphragm can be explained by a contraction of the muscle connected to the lumbar spine and the friction between the pleurae under the ribcage. Intracycle hysteresis of lung motion has been discussed in previous publications (13,17).
The segmented lung image depicts a variation of the lung volume during deep breathing that is consistent with known tidal volumes of healthy adults. The merit of a lung volume measurement based on MRI is that it provides absolute volume data, including RV, which cannot be measured by spirometry. It would be interesting to perform further validation by comparing tidal volumes measured by spirometry. Although several studies comparing spirometry-based measurement with four-dimensional computed tomography (23–25) and dynamic MRI (15,16) have been published, to the best of our knowledge, no MRI studies have compared MRI-based volume measurement with spirometry in an MRI scanner, partly because MRI-compatible spirometers are not widely available.
Although the proposed imaging protocol provides temporal resolution to capture lung motion, it was not sufficient to measure the volume variation in shallow breathing, which is the most natural way for a subject to breathe. In addition, the limited temporal resolution also meant that there was substantial heart motion during the acquisition of each slide, thereby causing motion artifacts around the heart. It is clear that one can trade spatial resolution for higher temporal resolution. And for the task demonstrated here (ie, to measure the lung volume as a function of time), lower spatial resolution would suffice. For example, the number of phase encoding steps could be reduced by a factor of two and the temporal resolution increased by the same factor. For future diagnostic use of the 3D free-breathing protocols, however, high spatial resolution will be required to observe the anatomic structures of the lung. This study, with a voxel size of 3.125 × 3.125 × 5 mm, achieved spatial resolution close to that obtained under normal breath-hold MRI pulmonary examinations. Thus, one of the purposes of this study was to demonstrate automatic segmentation in dynamic 3D free breathing with this type of spatial resolution. Temporal resolution is also improved by increasing the acceleration factor for the parallel imaging, which was limited to 4 in this study. Schmitt et al (19) demonstrated an acceleration factor as high as 8 with this 128-channel receiver coil, with a maximum geometry factor of 2.3 for coronal imaging with a one-dimensional parallel acquisition technique. Moreover, the design of the coil allows the use of a two-dimensional parallel acquisition technique with an acceleration factor of 20, which can dramatically improve temporal resolution.
In conclusion, the initial experience of the 3-T whole-body scanner with a 128-channel coil showed that the scanner and imaging protocol provided dynamic 3D images with spatial and temporal resolution sufficient to delineate the diaphragmatic domes and chest wall during active breathing and intensity homogeneities and a signal-to-noise ratio to perform automatic segmentation. There is potential to acquire dynamic 3D images with higher temporal resolution, and its application to lung motion analysis and diagnosis is promising.
Footnotes
This work was supported in part by grant R21CA116271-02 from the National Institutes of Health, Bethesda, MD.
References
- 1.Gierada DS, Curtin JJ, Erickson SJ, Prost RW, Strandt JA, Goodman LR. Diaphragmatic motion: fast gradient-recalled-echo MR imaging in healthy subjects. Radiology. 1995;194:879–884. doi: 10.1148/radiology.194.3.7862995. [DOI] [PubMed] [Google Scholar]
- 2.Gierada DS, Curtin JJ, Erickson SJ, Prost RW, Strandt JA, Goodman LR. Fast gradient echo magnetic resonance imaging of the normal diaphragm. J Thorac Imaging. 1997;12:70–74. doi: 10.1097/00005382-199701000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Gierada DS, Hakimian S, Slone RM, Yusen RD. MR analysis of lung volume and thoracic dimensions in patients with emphysema before and after lung volume reduction surgery. AJR Am J Roentgenol. 1998;170:707–714. doi: 10.2214/ajr.170.3.9490958. [DOI] [PubMed] [Google Scholar]
- 4.Suga K, Tsukuda T, Awaya H, et al. Impaired respiratory mechanics in pulmonary emphysema: evaluation with dynamic breathing MRI. J Magn Reson Imaging. 1999;10:510–520. doi: 10.1002/(sici)1522-2586(199910)10:4<510::aid-jmri3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 5.Iwasawa T, Yoshiike Y, Saito K, Kagei S, Gotoh T, Matsubara S. Paradoxical motion of the hemidiaphragm in patients with emphysema. J Thorac Imaging. 2000;15:191–195. doi: 10.1097/00005382-200007000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu S, Shirato H, Aoyama H, et al. High-speed magnetic resonance imaging for four-dimensional treatment planning of conformal radiotherapy of moving body tumors. Int J Radiat Oncol Biol Phys. 2000;48:471–474. doi: 10.1016/s0360-3016(00)00624-6. [DOI] [PubMed] [Google Scholar]
- 7.Gee J, Sundaram T, Hasegawa I, Uematsu H, Hatabu H. Characterization of regional pulmonary mechanics from serial magnetic resonance imaging data. Acad Radiol. 2003;10:1147–1152. doi: 10.1016/s1076-6332(03)00329-5. [DOI] [PubMed] [Google Scholar]
- 8.Kiryu S, Loring SH, Mori Y, Rofsky NM, Hatabu H, Takahashi M. Quantitative analysis of the velocity and synchronicity of diaphragmatic motion: dynamic MRI in different postures. Magn Reson Imaging. 2006;24:1325–1332. doi: 10.1016/j.mri.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Paiva M, Verbanck S, Estenne M, Poncelet B, Segebarth C, Macklem PT. Mechanical implications of in vivo human diaphragm shape. J Appl Physiol. 1992;72:1407–1412. doi: 10.1152/jappl.1992.72.4.1407. [DOI] [PubMed] [Google Scholar]
- 10.Gauthier AP, Verbanck S, Estenne M, Segebarth C, Macklem PT, Paiva M. 3-dimensional reconstruction of the in-vivo human diaphragm shape at different lung-volumes. J Appl Physiol. 1994;76:495–506. doi: 10.1152/jappl.1994.76.2.495. [DOI] [PubMed] [Google Scholar]
- 11.Cluzel P, Similowski T, Chartrand-Lefebvre C, Zelter M, Derenne JP, Grenier PA. Diaphragm and chest wall: assessment of the inspiratory pump with MR imaging-preliminary observations. Radiology. 2000;215:574–583. doi: 10.1148/radiology.215.2.r00ma28574. [DOI] [PubMed] [Google Scholar]
- 12.Craighero S, Promayon E, Baconnier P, Lebas JF, Coulomb M. Dynamic echo-planar MR imaging of the diaphragm for a 3D dynamic analysis. Eur Radiol. 2005;15:742–748. doi: 10.1007/s00330-004-2482-2. [DOI] [PubMed] [Google Scholar]
- 13.Seppenwoolde Y, Shirato H, Kitamura K, et al. Precise and real-time measurement of 3D tumor motion in lung due to breathing and heartbeat, measured during radiotherapy. Int J Radiat Oncol Biol Phys. 2002;53:822–834. doi: 10.1016/s0360-3016(02)02803-1. [DOI] [PubMed] [Google Scholar]
- 14.von Siebenthal M, Szekely G, Gamper U, Boesiger P, Lomax A, Cattin P. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol. 2007;52:1547–1564. doi: 10.1088/0031-9155/52/6/001. [DOI] [PubMed] [Google Scholar]
- 15.Plathow C, Fink C, Sandner A, et al. Comparison of relative forced expiratory volume of one second with dynamic magnetic resonance imaging parameters in healthy subjects and patients with lung cancer. J Magn Reson Imaging. 2005;21:212–218. doi: 10.1002/jmri.20255. [DOI] [PubMed] [Google Scholar]
- 16.Plathow C, Schoebinger M, Fink C, et al. Evaluation of lung volumetry using dynamic three-dimensional magnetic resonance imaging. Invest Radiol. 2005;40:173–179. doi: 10.1097/00004424-200503000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Blackall JM, Ahmad S, Miquel ME, McClelland JR, Landau DB, Hawkes DJ. MRI-based measurements of respiratory motion variability and assessment of imaging strategies for radiotherapy planning. Phys Med Biol. 2006;51:4147–4169. doi: 10.1088/0031-9155/51/17/003. [DOI] [PubMed] [Google Scholar]
- 18.Plathow C, Fink C, Ley S, et al. Measurement of tumor diameter-dependent mobility of lung tumors by dynamic MRI. Radiother Oncol. 2004;73:349–354. doi: 10.1016/j.radonc.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt M, Potthast A, Sosnovik DE, et al. A 128 channel receive-only cardiac coil for highly accelerated cardiac MRI at 3 tesla. Magn Reson Imaging. 2008;59:1431–1439. doi: 10.1002/mrm.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo T. Insight into images principles and practice for segmentation, registration and image analysis. Wellesley, MA: A.K. Peters; 2004. [Google Scholar]
- 21.Lorensen WE, Cline HE. Marching cubes: a high resolution 3D surface construction algorithm. Comput Graph. 1987;21:163–169. [Google Scholar]
- 22.Schroeder W, Martin K, Lorensen B. The Visualization Toolkit: an object-oriented approach to 3-d graphics. 4. Clifton Park, NY: Kitware, Inc; 2006. [Google Scholar]
- 23.Low DA, Parikh PJ, Lu W, et al. Novel breathing motion model for radiotherapy. Int J Radiat Oncol Biol Phys. 2005;63:921–929. doi: 10.1016/j.ijrobp.2005.03.070. [DOI] [PubMed] [Google Scholar]
- 24.Lu W, Parikh PJ, El Naqa IM, et al. Quantitation of the reconstruction quality of a four-dimensional computed tomography process for lung cancer patients. Med Phys. 2005;32:890–901. doi: 10.1118/1.1870152. [DOI] [PubMed] [Google Scholar]
- 25.Simon L, Giraud P, Servois V, Rosenwald JC. Lung volume assessment for a cross-comparison of two breathing-adapted techniques in radiotherapy. Int J Radiat Oncol Biol Phys. 2005;63:602–609. doi: 10.1016/j.ijrobp.2005.05.020. [DOI] [PubMed] [Google Scholar]