Abstract
We report here the identification, following expression cloning, of a molecule, STING (STimulator of INterferon Genes) that regulates innate immune signaling processes. STING, comprising 5 putative transmembrane (TM) regions, predominantly resides in the endoplasmic reticulum (ER) and is able to activate both NF-κB and IRF3 transcription pathways to induce type I IFN and exert a potent anti-viral state following expression. In contrast, loss of STING rendered murine embryonic fibroblasts (STING −/−MEFs) extremely susceptible to negative-stranded virus infection, including vesicular stomatitis virus, VSV. Further, STING ablation abrogated the ability of intracellular B-form DNA, as well as members of the herpes virus family, to induce IFNβ, but did not significantly affect the Toll-like receptor (TLR pathway). Yeast-two hybrid and co-immunprecipitation studies indicated that STING interacts with RIG-I and with Ssr2/TRAPβ, a member of the translocon-associated protein (TRAP) complex required for protein translocation across the ER membrane following translation[1, 2]. RNAi ablation of TRAPβ and translocon adaptor Sec61β was subsequently found to inhibit STING’s ability to stimulate IFNβ. Thus, aside from identifying a novel regulator of innate immune signaling, this data implicates for the first time a potential role for the translocon in innate signaling pathways activated by select viruses as well as intracellular DNA.
Cellular host defense responses to pathogen invasion principally involves the detection of pathogen associated molecular patterns (PAMPs) such as viral nucleic acid or bacterial cell wall components including lipopolysaccharide or flagellar proteins that results in the induction of anti-pathogen genes[3–9]. For example, viral RNA can be detected by membrane bound Toll-like receptors (TLR’s) present in the endoplasmic reticulum (ER) and/or endosomes (e.g. TLR 3 and 7/8) or by TLR-independent intracellular DExD/H box RNA helicases referred to as retinoic acid inducible gene 1 (RIG-I) or melanoma differentiation associated antigen 5 (MDA5, also referred to as IFIH1 and helicard)[3–10]. Pathogen DNA can be recognized by TLR 9 present in plasmacytoid dendritic cells, although it is now apparent that important TLR-independent pathways also exist to recognize DNA in alternate tissue, the mechanisms of action of which remain to be determined [3–10]. These events culminate in the activation of downstream signaling events, leading to the transcription of NF-κB and IRF3/7-dependent genes, including type I IFN.
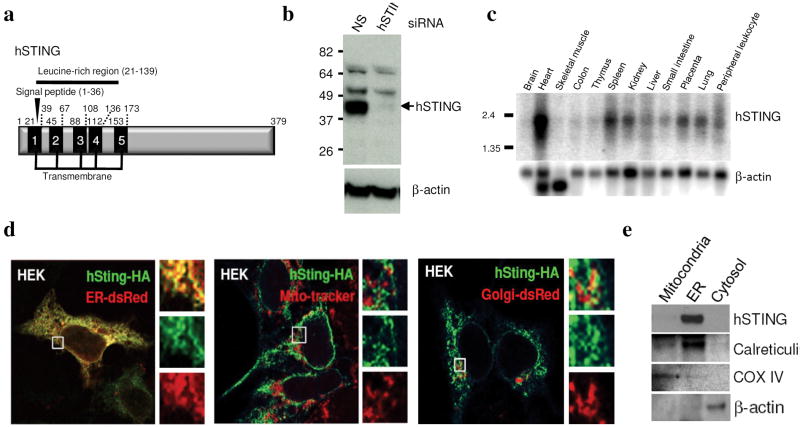
To further determine the mechanisms of innate immune signaling, we employed an expression screening system in which approximately 5,500 human and 9,000 murine full length cDNA’s were individually transfected into 293T cells harboring a luciferase gene under control of the IFNβ promoter (IFNβ-Luc). Five genes whose overexpression lead to the significant induction of IFNβ-Luc was found to be IPS-1 (also referred to as VISA/CARDIF/MAVS) (Supplemental Fig. 1)[11–14]. However, we also isolated a previously uncharacterized molecule (gi:38093659/NP_938023/2610307O08RIK) which we referred to as STING (for STimulator of INterferon Genes) that harbored 5 predicted TM motifs (in humans) and existed as a 379 amino acid protein in human cells and 378 amino acids in murine cells (Fig. 1a and Supplemental Fig 1). A putative signal cleavage motif was found to exist at position 1–36 and a leucine rich region was apparent between amino acids 21–139 (Fig. 1a). The predicted molecular weight of human STING (hSTING) was 42,192 Da, which approximately corresponded to its observed molecular weight in human 293 cells following immunoblot analysis using a rabbit antiserum raised to a STING peptide (Fig. 1b). RNAi studies confirmed that the observed 42kDa band was indeed STING (Fig. 1b). STING was found to be ubiquitously expressed in a variety of tissues as determined by northern analysis and was found to predominantly reside in the ER region of the cell as determined by confocal microscopy and fractionation analysis (Fig. 1c–e).
Fig 1. STING is an ER protein.
a. Schematic of hSTING indicating transmembrane and leucine rich regions. b. Immunoblot analysis of STING in HEK 293 cells treated with RNAi to STING (hSTING) or control RNAi (NS). c. Northern blot analysis of human STING. d. Confocal analysis of HEK 293 cells transfected with hSTING tagged at the carboxyl end with HA. Transfected cells were also analyzed using ER-dsRed, Mitotracker or Golgi-dsRed. e. Fractionation experiments confirm that STING resides in the ER. Control antibodies indicate accuracy of fractionation (Calreticulin- ER, COX IV-mitochondria, β-actin- cytosol).
Overexpression of STING in 293T cells was subsequently confirmed to robustly induce the expression of the IFN promoter (IFNβ-Luc) up to 400-fold, but not a control TK promoter driving luciferase (pRL-TK), interferon regulatory factor 3 (IRF3) responsive promoters (PRD-III-I-Luc) up to 1000-fold, an NF-κB responsive promoter (NF-κB-Luc) 12-fold and interferon-inducible promoters (interferon sensitive response element- ISRE-Luc) up to 800-fold (Fig. 2a–d). STING did not activate control promoters driving luciferase reporters such as those derived from the Rb, p53 or E2F genes (Supplemental Fig. 2). Increased dimerization of IRF3 was also observed in STING expressing 293T cells confirming that STING regulates the induction of type I IFN at upstream of IRF3 activation, dimerization and translocation (Supplemental Fig. 2)[15]. Endogenous IFNβ mRNA and IFNβ protein was induced significantly in MEFs transiently transfected with STING (Fig. 2e,f). DNA microarray analysis of STING expression in 293T cells further emphasized STING’s ability to induce primary innate immune response genes (Fig. 2g). Accordingly, MEFs cells expressing STING or IPS-1 were significantly resistant to VSV infection (Fig. 2h,i). Subsequent analysis indicated that STING function was ablated in the absence of the IκB kinase family member TBK-1, confirming that STING’s activity involved activation of IRF3 and was indeed upstream of this kinase (Fig. 2j)[15]. Finally, we observed that STING did not exert robust activity in the absence of FADD, which has also been shown to be important for efficient innate immune signaling processes (Supplemental Fig. 2)[16]. We also established that while STING, HA tagged at the carboxyl region, retained activity, this activity was lost when STING was tagged at the amino terminus or carboxyl terminus with GFP (data not shown). Subsequent analysis indicated that the amino terminus region of STING containing the 5 putative TM regions (amino acids 1–230) or just the carboxyl region of STING (amino acids 173–379) did not exhibit significant ability, alone, to induce the IFNβ-Luc promoter (Fig. 2k,l). Thus, full length intact STING is required for efficient function. However, we further observed that the carboxyl region of STING exerted a dominant-negative inhibitory effect and could impede the ability of full-length STING to stimulate IFNβ-Luc (Fig. 2m). Collectively, this data indicates that expression of STING activates the innate immune response including type I IFN leading to the induction of an antiviral state.
Fig 2. STING facilitates IFN induction.
a. 293T cells were transfected with an IFNβ-Luc (p110-Luc) plasmid and increasing amounts of human (hSTING), murine (mSTING) or control ΔRIG-I. b. 293T cells transfected as in (a) with either PRD-III-I-Luc (b), NF-κB-Luc (c) or ISRE-Luc (d) reporter plasmids were analyzed similarly. e. MEF’s were transfected as in (a) and IFNβ mRNA was analyzed by qRT-PCR. f. Medium from transfected MEFs was analyzed for IFNβ protein by ELISA. g. Microarray analysis of 293T cells transfected with hSTING. h. MEF’s transfected with STING or ΔRIG-I or IPS-1 are resistant to VSV-GFP infection (MOI 1). i. Plaque assay from (h). j. TBK-1 deficient MEFs do not facilitate STING signaling. k. Schematic of hSTING variants. l. 293T cells were transfected as in (a) with hSTING or variants and luciferase measured. m. 293T cells were transfected with STING and increasing amounts of hSTING-Full, hSTING-N or hSTING-C with luciferase plasmids as in (a). Asterisks indicate significant difference (P < 0.05) as determined by Student’s t-test. Error bars; +/− s.d.
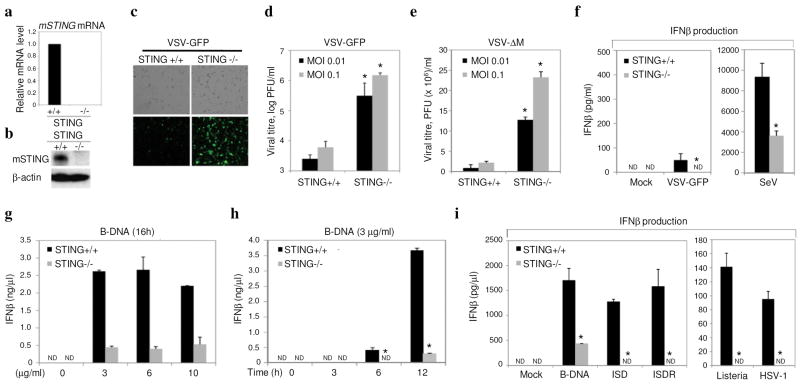
To further analyze STING function, we utilized an RNAi approach to ablate STING in a number of cell-types. Our data indicated that knockdown of STING in HEK 293 cells, modestly reduced the ability of the negative-stranded rhabdovirus VSV-GFP to induce IFNβ, presumably since these viruses are only weak activators of IFNβ (Supplemental Fig. 3). However, such cells were rendered extremely susceptible to virus infection and replication. To confirm a requirement for STING in the regulation of type I IFN induction and in host defense, we generated STING deficient (STING −/−) mice by targeted homologous recombination in ES cells (Supplemental Fig 4). STING −/− animals were born at the Mendelian ratio and developed and bred normally. Accordingly, MEFs from wild type and STING −/− animals were infected with VSV-GFP at varying MOI’s (0.01–1) for up to 36 hours post infection. This study confirmed that more progeny virus was produced in MEFs lacking STING compared to controls (2 logs; 24–36 hours) (Fig. 3a–d). This data was verified using VSV expressing a luciferase reporter gene and VSV-ΔM, which exhibits a defect in the viral matrix protein normally responsible for inhibiting cellular mRNA export from the nucleus[17](Supplemental Fig. 4 and Fig. 3e). Reconstitution of STING to STING −/− MEF’s rescued the susceptibility to VSV infection (Supplemental. Fig. 4). Similar analysis also indicated that loss of STING also reduced the ability of SeV to induce IFNβ (Fig 3f). In contrast, we did not observe a strong requirement for STING to mediate the ability of transfected poly I:C to induce IFNβ induction, which is largely governed by the intracellular RIG-I homologue MDA5[18] (Supplemental Fig. 5). We also observed that the positive-stranded encephalomyocarditis virus (EMCV), a member of the picornavirus family, did not effectively induce IFNβ nor replicate differently regardless of the presence of STING (Supplemental Fig. 5). Thus, we conclude that STING may play a more predominant role in facilitating RIG-I mediated innate signaling rather than MDA5. Interestingly, we did notice a significant defect (>5-fold) in the ability of transfected B-form DNA (poly dA-dT) to induce IFNβ in MEFs lacking STING compared to controls (Fig 3g,h). More strikingly, the non CpG containing interferon stimulatory DNA (ISD) was completely unable to induce IFNβ in STING −/− MEFs, as was the DNA virus herpes simplex virus 1 and bacteria Listeria monocytogenes (Fig. 3i). TLR9 is considered to govern CpG DNA-mediated induction of IFNβ, but is not active in MEFs[19, 20]. Thus, it is plausible that STING may function in TLR9-independent, DNA-mediated induction of type I IFN. This effect was similarly observed in murine STING lacking bone marrow derived macrophages (BMDM) or bone marrow derived dendritic cells (GM-DC) cultured using granulocyte-monocyte colony stimulating factor (GM-CSF) [Supplemental Fig. 6]. However, no significant difference was observed in the ability of exogenous poly I:C or lipopolysaccharide (LPS) to induce IFNβ, when comparing STING −/− BMDM or GM-DC’s to controls, events which depend on TLR 3 and 4 respectively. While, loss of STING rendered MEFs highly susceptible to VSV-GFP, less susceptibility was observed following VSV-GFP infection of STING −/−GM-DC’s or BMDM’s, indicating that STING may be more important in facilitating negative-stranded virus-mediated innate signaling in fibroblasts. Collectively, our data would indicate that loss of STING leads to a defect in RIG-I mediated type I IFN induction but does not affect the TLR pathway. In addition, we report that STING functions in the pathway utilized by intracellular B-form DNA to induce IFNβ.
Fig 3. Loss of STING affects host defense.
a. qRT-PCR analysis of mSTING mRNA in STING −/− or control MEFs. b. Immunoblot of mSTING in −/− cells or controls. c. Fluorescence microscopy (GFP) of mSTING −/− or controls infected with VSV-GFP (MOI 0.1). d. Viral titers from (c). e. Viral titers following VSVΔM infection. f. Endogenous IFNβ levels from STING −/− or controls after infection with VSV-GFP (MOI 1) or Sendai Virus (SeV MOI 1) g. STING −/− MEF’s or controls were treated with transfected B-DNA and IFNβ measured by ELISA. h. Time course analysis of (g). i. STING −/− or controls were exposed to transfected B-form DNA, interferon stimulatory DNA (ISD), or ISD reversed nucleotide sequene (ISDR), Listeria monocytogenes (MOI 50), HSV (MOI 5) for 12 hours, and IFNβ measured by ELISA. Asterisks indicate significant difference (P < 0.05) as determined by Student’s t-test. Error bars; +/− s.d. ND; not detectable.
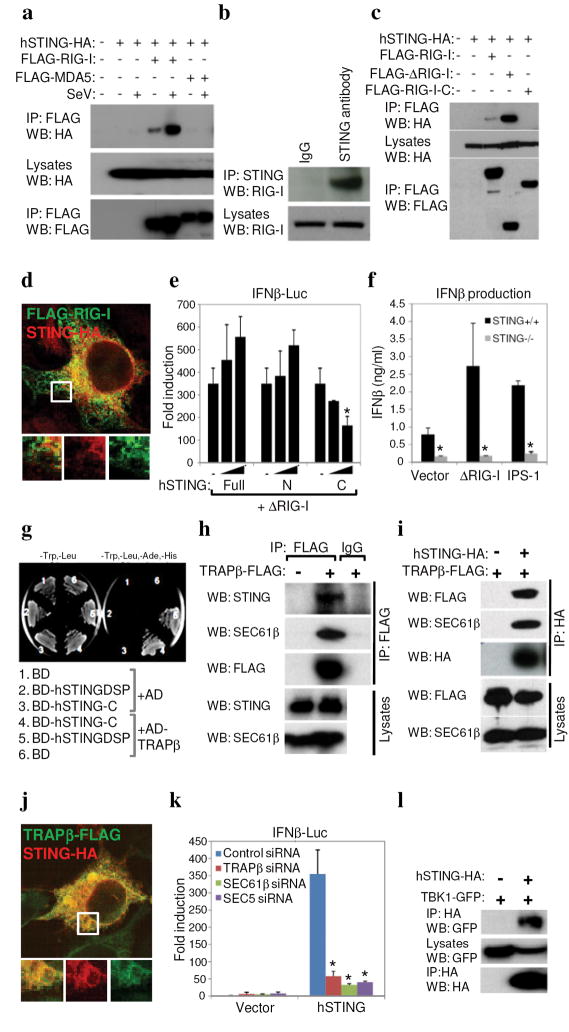
Fig 4. STING associates with the translocon.
a. 293T cells were co-transfected with HA-STING, FLAG-RIG-I or MDA5 and infected with SeV (MOI 1). Lysates were immunoprecipitated (IP) and immunoblotted (IB) using antibodies to HA or FLAG. b. Endogenous hSTING associates with RIG-I in HUVECs. c. ΔRIG-I (aa1-284) and not RIG-I-C (aa218-925) associates with STING in co-transfected 293T cells. d. Confocal image of 293T cells co-transfected with tagged STING and RIG-I. e. 293T cells were co-transfected with control vector (−) or increasing amounts of full-length, amino (aa1-230) or carboxyl (aa173-379) STING and ΔRIG-I, and IFNβ-Luc was measured. f. Control or STING −/− MEFs were transfected with ΔRIG-I (aa1-284) or IPS-1 and IFNβ was measured by ELISA. g. BD-hSTING-C interacts with Ssr-2/TRAPβ (AD-hTRAPβ) in yeast-two hybrid screening (BD-hSTINGΔSP, amino acids 36-369; BD-hSTING-C amino acids 173-379). h. HEK 293 cells were transfected with FLAG-tagged TRAPβ and endogenous STING or Sec61β measured by immunoblot. i. STING and TRAPβ were co-transfected HEK 293 cells and analysis carried out as in (h). j. Co-localization of STING and TRAPβ in 293T cells. k. RNAi to TRAPβ, SEC61β or Sec5 in HEK 293 cells ablates STING signaling. l. HA-STING associates with GFP-TBK-1 in co-transfected HEK 293 cells. Asterisks indicate significant difference (P < 0.05) as determined by Student’s t-test. Error bars; +/− s.d.
To further examine the mechanisms of STINGs function in innate signaling, we attempted to determine if STING interacted with RIG-I and/or MDA5, which are putative innate immune signaling receptors for negative or positive stranded viral RNA respectively[3, 4, 10]. Co-immunoprecipitation experiments in 293T cells principally indicated that FLAG-tagged RIG-I but not MDA5 could associate with HA-tagged STING in co-transfection experiments (Fig 4a). The binding of RIG-I to STING was augmented upon infection of 293T cells with SeV (Fig 4a). We confirmed in normal human umbilical vein endothelial cells (HUVECs) that endogenous RIG-I could associate directly, or indirectly as a complex, with endogenous STING (Fig 4b). This data would be in agreement with previous data indicating that STING seems to preferentially modulate the RIG-I, rather than the MDA5 regulated pathway (Fig. 3d and Supplemental Fig. 5). We also determined that the CARD domains of RIG-I (amino acids 1-284) were preferentially able to associate with HA-STING in transfected 293T cells (Fig. 4c). RIG-I was subsequently shown to colocalize with STING in the co-transfected 293T cells (Fig. 4d). It was similarly observed that the carboxyl region of STING could inhibit the function of RIG-I, again indicating that this region of STING can exhibit a dominant-inhibitory effect (Fig 4e). We also noted association of the RIG-I downstream adaptor IPS-1 with STING although similar to the situation with RIG-I, it is not yet clear whether IPS-1 directly interacts with STING or exists as a complex with RIG-I/STING (Supplemental Fig 7). Accordingly, we observed that STING was able to induce the induction of an IFNβ driven luciferase construct in MEFs lacking RIG-I or IPS-1, likely confirming that that STING functions downstream of these latter molecules (Supplemental Fig 8). The ability of ΔRIG-I or IPS-1 to induce IFNβ appeared diminished in STING −/− MEFs (Fig. 4f). However, there was also a marked reduction in the induction of IFNβ by all transfected plasmids (including vector alone) in the absence of STING compared to control MEFs, since endogenous DNA innate signaling pathways are likely defective (Fig. 3g–i). This data indicates that STING may be an important downstream adaptor molecule that facilitates RIG-I and perhaps IPS-1 function.
To shed further insight into the molecular mechanisms of STING action, we screened an IFN-induced, human fibroblast yeast two-hybrid cDNA library using STING (amino acids 173–379) as a bait and repeatedly isolated Ssr2/TRAPβ, a member of the TRAP complex comprising four subunits (α-Δ) that facilitates translocation of proteins into the ER following translation (Fig. 4g)[1, 2]. The TRAP complex is known to associate with the translocon, comprising three subunits, Sec61 α, β and γ. Given this information, we confirmed that TRAPβ can indeed associate with endogenous STING in HEK 293 cells following co-immunoprecipitation experiments (Fig. 4h). Using this approach, we confirmed that TRAPβ also co-immunoprecipitated with endogenous Sec61β (Fig. 4h). We next verified that STING could also associate not only with TRAPβ but also Sec61β likely as a complex (Fig 4i). STING was also observed to colocalize with TRAPβ in the ER region of the cell (Fig. 4j). Indeed, loss of TRAPβ or SEC61β reduced STING’s, ability to induce an IFNβ promoter driving luciferase (Fig 4k; Supplemental Fig. 9). Taken together, this data would indicate that the STING may be involved in translocon function and that the translocon may be able to influence the induction of type I IFN.
Thus, STING is predominantly an ER resident protein that may link RIG-I and DNA-mediated intracellular innate signaling to the translocon. Speculatively, RIG-I may detect translating viral RNA’s at the intersection of ribosome/ER translocon association and/or with mitochondrial associated ER membranes (MAM’s) and require STING to exert effective function[2]. Alternatively, STING may participate in mediating ER stress response pathways an event that remains to be verified. While it is not clear how signaling from the translocon to IRF-3/NF-kB occurs, it has recently been established that translocon physically associates with the exocyst - the octomeric Sec6-Sec8 complex that also associates with the ER and tethers secretory vesicles to membranes, and facilitates protein synthesis and secretion[21, 22]. Recently, the exocyst complex was found to recruit and activate TBK1 and play a role in type I IFN-β induction[23]. Our preliminary analysis indicates that STING also co-immunprecipitates with TBK1 and that RNAi ablation of Sec5 also rendered cells defective in the STING function (Fig. 4l). Thus, STING may facilitate the detection of intracellular viral RNA species as well as B-form DNA indicating convergence of these intracellular PAMP recognition pathways.
Methods Summary
Plasmid constructs
Human STING (hSTING), murine STING (mSTING), hSTING-N (amino acids 1–230) and hSTING-C (amino acids 173–379) sequences were amplified by PCR and were cloned into pcDNA3 plasmids to generate C-terminally HA-tagged expression constructs. Expression plasmids encoding Flag-tagged RIG-I, ΔRIG-I (amino acids 1–284), IPS-1 and TBK-1 were described previously[24]. GFP-tagged RIG-I and TBK-1 were generated by cloning into pAcGFP1-C1 (Clontech, CA). Other plasmids were obtained as follows: p110-Luc and PRD-III-I-Luc (T. Maniatis), IFNβ-Luc and ISRE-Luc (J. Hiscott), NFκB-Luc (Stratagene, CA), ER-dsRED and Golgi-dsRED (Clontech, CA).
Antibodies
Rabbit polyclonal antibody against a synthetic peptide corresponding to residues 324–340 of human STING was obtained from EvoQuest Custom Antibody Services (Invitrogen, CA). Other antibodies were obtained from following sources: SEC61β (Upstate, NY), β-Actin, HA, FLAG (Sigma, MO), COX IV, Calreticulin, IRF3 and RIG-I (Abcam, MA).
Confocal microscopy
ER-DsRED and Golgi-DsRED (Clontech, CA) were used for ER and Golgi marker, respectively. For mitochondria staining, living cells were incubated with 300 nM of Mito Tracker Red (Invitrogen, CA) for 45 min at 37°C.
RNA interference
Chemically synthesized 21-nucleotide siRNA duplexes were obtained from Dharmacon, Inc (Lafayette, CO). RNA oligonucleotides used for human and murine STING were as follows: hSTING, GCAUCAAGGAUCGGGUUU; mSTING, CCAACAGCGUCUACGAGA.
Generation of the STING knockout mice
The linearized targeting vector was electroporated into E14.1 ES cells originated from 129SvEv strain, followed by the selection in G418. One positive clone was injected into C57BL/6J blastocysts and STING −/− mice generated on a C57BL/6J background.
Yeast two-hybrid analyses
To screen for interacting partners of STING a Yeast two Hybrid approach was utilized, adopting a novel IFN-induced, telomerase immortalized human fibroblast cDNA library, generated by our laboratory. hSTING-C (amino acids 173–379) sequence was amplified by PCR and was cloned into pGBK-T7 (Clontech) to generate a bait plasmid pGBK-T7-hSTING-C. 100μg of the IFN-induced library’s plasmid DNA was used to perform a library-scale transformation of yeast strain Y187.
Statistics
Students t-test was used to analyze data.
Methods
Cells, viruses and reagents
293T and HEK293 cells were obtained from the ATCC and were maintained in DMEM medium supplemented with 10% FBS. Human Umbilical Vein Endothelial Cells (HUVEC) were obtained from Lonza (Basel, Switzerland), and were maintained in endothelial growth medium (EGM-2) (Lonza). FADD+/+, FADD−/−, TBK-1+/+ and TBK-1−/− MEFs were provided by W.-C. Yeh[16]. VSV-GFP was used in infections and titered as described previously[16]. VSV-ΔM was constructed as described in Faria et al[17]. Murine IFN-β ELISA Kit was obtained from PBL (Piscataway, NJ). EMCV was purchased from ATCC. Synthetic ds B-DNA (poly dA-dT) and polyI:C were purchased from GE healthcare. Interferon stimulatory DNA (ISD) and reverse sequence of ISD were described previously[20]. For stimulation of cells, -DNA or polyI:C were mixed with Lipofectamine 2000 (Invitrogen, CA) at a ratio of 1:1 (vol/wt), and then added to cells at a final concentration of 3 μg/ml.
Reporter analysis
293T cells or MEFs seeded on 24-well plates were transiently transfected with 50 ng of the luciferase reporter plasmid together with a total of 250 ng of various expression plasmids or empty control plasmids. As an internal control, 10 ng of pRL-TK was transfected simultaneously. Then, 36 hours later, the luciferase activity in the total cell lysates were measured
Northern blot
Human multiple tissue RNA blots (Clontech, CA) were hybridized with a 32 P-labelled full-length human STING probe.
Real time PCR
Total RNA was isolated from cells using the RNeasy RNA extraction kit (Qiagen, Valencia, CA) and cDNA synthesis was performed using 1μg of total RNA (Roche, Indianapolis, IN). Fluorescence real time PCR analysis was performed using a LightCycler 2.0 instrument (Roche Molecular Biochemicals, Indianapolis, IN) and TaqMan Gene Expression Assays (Applied Biosystems, CA). Relative amounts of mRNA were normalized to the 18S ribosomal RNA levels in each sample.
DNA Microarray analysis
Total RNA was extracted from 293T cells transfected with STING expressing vectors. Preparation of cDNA and microarray analysis was performed at the W.M. Keck Foundation Biotechnology Research Laboratory DNA microarray facility at Yale University (New Haven, CT). The Human Genome U133 Plus 2.0 Array (Affymetrix, CA) was used. Data analysis was performed with GeneSpring software (Silicon Genetics, CA)[16].
Mitochondria and ER fraction isolation
Mitochondria and ER membranes were purified on discontinuous sucrose gradients as previously described[25]. Briefly, HUVEC cells in MTE buffer (0.27M mannitol, 10 mM Tris-HCl, 0.1mM EDTA, pH 7.4) were lysed by sonication. Lysed cells were centrifuged at 700 × g for 10 min to remove Nuclei and cellular debris. Mitochondria were obtained by centrifugation at 15,000 × g for 10 min, and post mitochondrial supernatant was used for purification of ER fractions. Mitochondria pellet was resuspended in MTE buffer, and was layered on discontinuous sucrose gradients consisting of 1.0 M and 1.7M sucrose and banded by centrifugation at 40,000 × g for 22 min. Mitochondria fraction was collected and pelleted by centrifugation at 15,000 × g for 10 min. Purified mitochondria were resuspended in PBS and used for western blot analysis. To isolate ER fractions, postmitochondrial supernatant described above was layered on discontinuous sucrose gradients consisting of 1.3M, 1.5 M and 2.0 M sucrose, and banded by centrifugation at 100,000 × g for 70 min. The ER fraction at the interface between the supernatant and the 1.3 M sucrose was collected, and pelleted by centrifugation at 100,000 × g for 45 min. The ER membranes were resuspended in PBS and were used for western blot analysis.
RNA interference
293T cells were plated on 24-well plates at 5 × 104 cells per well and transfected with 10 pmol of siRNA duplex per well using Lipofectamine RNAiMAX (Invitrogen, CA). MEFs were transfected by using an Amaxa nucleofector apparatus (program A-023) and Amaxa MEF nucleofector kit 1 according to the manufacturer’s recommendations. At 72 hours after transfection, cells were used for further experiments.
Coimmunoprecipitation, native PAGE and immunoblot analysis
Cells were seeded on 100-mm dishes at 1 × 106 cells per dish. Cells were transfected with a total of 10 μg of empty plasmid or various expression plasmids using Lipofectamine 2000. At 36 hours after transfection, cells were lysed in M-PER buffer (PIERCE, IL) or 1% digitonin (Calbiochem, CA) buffer (20 mM Tris-HCl, 150 mM NaCl, 1% digitonin) containing protease inhibitors (Roche, IN). Lysates were incubated with HA affinity matrix (COVANCE, WI) or FLAG affinity beads (SIGMA) overnight. The beads were washed three times by TBS containing 0.05% Tween-20 and immunoprecipitates were eluted with non-reducing sample buffer by boiling for 5 min. For endogenous STING immunoprecipitation, HUVEC were lysed in 1% digitonin lysis buffer. Cleared supernatants were incubated with 10μg of anti-STING antibody, followed by incubation with immobilized protein G (PIERCE). The beads were washed four times by 1% digitonin lysis buffer and immunoprecipitates were eluted with SDS sample buffer by boiling for 5 min. Native PAGE and Immunoblotting was carried out as previously described[24].
Generation of the STING knockout mice
The linearized targeting vector was electroporated into E14.1 ES cells originated from 129SvEv strain, followed by the selection in G418. Targeted clones were screened by PCR. From 90 clones, 1 positive clone was identified. This ES clone was subjected to the generation of chimera mice by injection using C57BL/6J blastocysts as the host. The resulting male chimeras were further mated with C57BL/6J female mice for germline transmission. The heterozygous mice (F1 mice) were interbred to obtain wild-type, heterozygous and homozygous littermates (F2). The genotypes of the mice were determined by PCR. Animals were generated at the University of Miami School of Medicine Transgenic Core Facility. Mice were allowed to freely access to food and water and housed at an ambient temperature of 23°C and at a 12 h light/dark cycle. Animal care and handling was performed as per IACUC guidelines.
IFN induced library construction
To screen for interacting partners of STING a Yeast two Hybrid approach was adopted using a novel IFN-induced, telomerase immortalized human fibroblast cDNA library. To construct this library, cells were treated overnight with 1000 units each of human IFN–α and –β to induce interferon dependent genes. Poly(A)+ RNA was extracted and cDNA synthesis was carried out using ‘BD Powerscript™’ RT (Clontech). The cDNA was then amplified by PCR, digested with SfiI and ligated to the yeast prey vector pGADT7-RecAB. The ligated mixture was transformed into E. coli strain DH10B. The number of independent clones in the unamplified library was estimated to be 3.2 × 106, with an average size of 1.52 kb and inserts ranging in size from 0.8 to 3.0 kb. 15 independent colonies were isolated and DNA extracted to check for size by PCR.
Acknowledgments
We gratefully thank T. Venkataraman, JinHee Hyun, T. Sato, M. Conkright for technical assistance, M Gale for RIG-I and IPS-1 lacking MEFs, Yen Chen Weh for TBK-1 lacking MEFs and S. Nagata, T. Maniatis, J. Hiscott and N. Reich for plasmid constructs.
References
- 1.Hartmann E, et al. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem. 1993;214(2):375–81. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- 2.Menetret JF, et al. Architecture of the ribosome-channel complex derived from native membranes. J Mol Biol. 2005;348(2):445–57. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–24. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B, et al. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7(10):753–66. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 5.Takahasi K, et al. Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses. Mol Cell. 2008;29(4):428–40. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 8.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 9.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–45. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onomoto K, Yoneyama M, Fujita T. Regulation of antiviral innate immune responses by RIG-I family of RNA helicases. Curr Top Microbiol Immunol. 2007;316:193–205. doi: 10.1007/978-3-540-71329-6_10. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 12.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005 doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 13.Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Xu LG, et al. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Mol Cell. 2005;19(6):727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 15.McWhirter SM, et al. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A. 2004;101(1):233–8. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balachandran S, Thomas E, Barber GN. A FADD-dependent innate immune mechanism in mammalian cells. Nature. 2004;432(7015):401–5. doi: 10.1038/nature03124. [DOI] [PubMed] [Google Scholar]
- 17.Faria PA, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17(1):93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 19.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7(1):40–8. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 20.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24(1):93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Guo W, Novick P. The exocyst meets the translocon: a regulatory circuit for secretion and protein synthesis? Trends Cell Biol. 2004;14(2):61–3. doi: 10.1016/j.tcb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Lipschutz JH, V, Lingappa R, Mostov KE. The exocyst affects protein synthesis by acting on the translocation machinery of the endoplasmic reticulum. J Biol Chem. 2003;278(23):20954–60. doi: 10.1074/jbc.M213210200. [DOI] [PubMed] [Google Scholar]
- 23.Chien Y, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127(1):157–70. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 24.Balachandran S, et al. Fas-associated death domain-containing protein-mediated antiviral innate immune signaling involves the regulation of Irf7. J Immunol. 2007;178(4):2429–39. doi: 10.4049/jimmunol.178.4.2429. [DOI] [PubMed] [Google Scholar]
- 25.Mavinakere MS, et al. Processing of human cytomegalovirus UL37 mutant glycoproteins in the endoplasmic reticulum lumen prior to mitochondrial importation. J Virol. 2006;80(14):6771–83. doi: 10.1128/JVI.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]